Abstract
Quiescent Escherichia coli cells are generated by overexpressing the Rcd transcript in an hns-205 mutant host. The resulting nongrowing, metabolically active cells were used here to express a single-chain antibody fragment (scFv) in shake flask and fermentor cultures. The expression system is based on two plasmids; one carries the product gene expressed from λPL under the control of the cI857 temperature-sensitive repressor, while the second expresses Rcd from λPR. Shifting the culture from 30 to 42°C induces Rcd expression and product expression simultaneously. Our scFv carried a PelB leader, and 90% of the protein was secreted into the culture supernatant. In a batch culture, the supernatant concentration of scFv in the quiescent-cell culture (optical density at 600 nm [OD600] of 3.5) was 37 mg liter−1, compared to a maximum of 13 mg liter−1 in the control culture (final OD600 of 20). In a fed-batch fermentor culture, quiescent cells were held at an OD600 of 20 for 24 h and the extracellular scFv concentration reached a maximum of 150 mg liter−1. A control culture with a similar feed reached an OD600 of 80, but despite the higher density, the extracellular scFv concentration did not exceed 35 mg liter−1. Quiescent cells at an OD600 of 50 exhibited a small decline in the specific product formation rate, but nevertheless, an extracellular scFv concentration of 160 mg liter−1 was achieved in 8 h. The rate of extracellular accumulation was 10-fold greater in the quiescent culture than in the control culture. This study demonstrates that it is possible to establish high-density quiescent E. coli cultures that are capable of efficient synthesis, folding, and export of proteins.
Escherichia coli continues to occupy center stage as a host for large-scale recombinant protein production (21). This is especially true for simple proteins, where posttranslational modifications are not essential for biological activity. Recent developments in our understanding of protein folding have allowed the expression of many proteins in their biologically active soluble form rather than as inclusion bodies (2, 3, 19). Coupled with more efficient secretion mechanisms, it is now possible to obtain a recombinant protein as a periplasmic or even extracellular fraction, which significantly eases the problems of downstream processing (4, 14). While sophisticated vectors with strong and tightly regulated promoters have been developed to maximize product expression, comparatively little attention has been paid to the bacterial host. An improved host could potentially offer gains in product quantity and quality and could be used in conjunction with a wide range of existing vectors.
We have been exploring the conflict between the biotechnologist's desire for more and better product and the inclination of the bacterium to produce biomass. In an attempt to shift the balance in favor of the biotechnologist, we developed the quiescent-cell (Q-cell) expression system, in which a plasmid-encoded protein is expressed in nongrowing but metabolically active cells. By redirecting metabolic flux from biomass production to recombinant protein synthesis, Q-cells offer the prospect of more efficient resource utilization than a conventional E. coli cell factory. The quiescent state is established by overexpressing Rcd, a regulatory transcript encoded by E. coli plasmid ColE1, in an hns-205 mutant host (16). This leads to severe nucleoid condensation and global repression of chromosomal genes, although plasmid gene expression continues (20). In experiments to investigate protein synthesis in Q-cells, we studied the production of plasmid-encoded chloramphenicol acetyltransferase. Rcd was expressed from the λPR promoter under the control of a plasmid-encoded cI857 temperature-sensitive repressor and was induced by a temperature shift from 30 to 42°C. The culture density increased approximately fourfold for 2 to 3 h after Rcd induction and then remained constant as the culture became quiescent. Despite the down-regulation of chromosomal genes, synthesis of the plasmid-encoded chloramphenicol acetyltransferase protein continued for several hours, eventually reaching 30 to 40% of the total cell protein (16).
These initial investigations left unanswered many questions about the utility of Q-cells. They were restricted to low-density shake flask cultures, and the product was a prokaryotic protein of no commercial significance. In the investigation reported here, we have explored the potential of Q-cells to express a commercially significant product in high-density shake flask and fermentor cultures. We have also investigated the capacity of Q-cells to export protein to the periplasm and to form appropriate disulfide bonds. As our model product, we chose a single-chain antibody fragment (scFv), a class of proteins required in large quantity for medical and biotechnological applications. In cancer therapy, for example, repeated doses of 1 g per patient are commonplace (12). Our scFv of choice, clone 3PF12 (24), is being developed as a diagnostic reagent for HLA typing and as a therapeutic reagent. It has been used to model the interaction of anti-HLA-A2 with the HLA molecule, and this information is being used to produce antibodies that specifically recognize HLA-plus peptide. For both of these applications, large quantities of protein with a high specific activity are required (N. A. Watkins, unpublished data). High-level expression of scFv in E. coli often leads to the formation of inclusion bodies, which then require in vitro refolding to recover biological activity. In such cases, the final yields of active protein may be fairly low, around 25 to 30 mg liter−1 (13, 18). Chaperone-assisted refolding and secretion to the periplasm to obtain a better oxidative environment have been used successfully to improve biological activity in vivo (9, 11). Large-scale fermentation of scFvs with different vectors and optimized fermentation strategies has also been reported (1, 8). In this work, we first monitored scFv 3PF12 expression in a high-density shake flask culture and showed that the protein is exported efficiently into the culture supernatant. The system was then scaled up in a fermentor, where fed-batch techniques allowed us to use Q-cell cultures at an optical density at 600 nm (OD600) of up to 50. Under these conditions, the rate of synthesis of biologically active scFv in a fed-batch Q-cell culture was more than 10-fold greater than that of a culture with conventional growth kinetics.
MATERIALS AND METHODS
Strains and plasmids.
W3110 is an E. coli K-12 strain (6). The hns-205::Tn10 derivative of W3110 was constructed by P1 transduction from strain GM230 (5), selecting for mucoid, tetracycline-resistant colonies.
Plasmid pRcd1 expresses Rcd from the λPR promoter in a derivative of plasmid pUC18ΔlacZ (in which most of the lacZ gene and upstream DNA containing the lac promoter between two PvuII restriction sites of plasmid pUC18 have been deleted by PvuII digestion and religation; D. C. D. Rowe, unpublished data). The PCR was used to place rcd under the control of the λPR promoter with oligonucleotide primers 5′-ATGCATATGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTGATAATGGTTGCAGGCGCGATCGCGGCAG-3′ and 5′-ATGCATATGATTTACCATAATCCC-3′ and ColE1 as template DNA. PCR was carried out with Herculase polymerase mix (Stratagene, La Jolla, Calif.) under the following cycling conditions: initial denaturing at 94°C for 5 min; 10 cycles of denaturing at 94°C for 30 s, annealing at 44°C for 1 min, and extension at 72°C for 1 min; followed by 20 cycles of denaturing at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min; and a final extension at 72°C for 5 min. A 194-bp PCR product was generated and cloned by blunt-end ligation into the PvuII site of pUC18ΔlacZ to make pRcd1 (2,558 bp). Plasmids were transformed into E. coli strain GI724 (Invitrogen, Paisley, United Kingdom), which contains a chromosomal copy of the cI gene expressed from the trp promoter, and transformants were selected on M9 minimal medium agar plates containing ampicillin (75 μg ml−1) without tryptophan supplementation. Transformants were then screened at 30°C on M9 minimal medium agar plates containing ampicillin, with and without tryptophan supplementation. The structure of plasmids isolated from transformants unable to grow on medium with tryptophan supplementation was checked by MluI restriction digestion and DNA sequencing.
Plasmid pCMT2b-scFv was constructed by cloning the single-chain antibody 3PF12 plus a pelB leader sequence and 3′ myc and his tags (24) under control of the λPL promoter in expression vector pCMT2b. 3PF12 is a human scFv specific for HLA-A2/A28, isolated by variable gene phage display. The vector pCMT2b (4,453 bp) is a derivative of pCMT2 (4,426 bp) in which the cI857 gene under control of the Plac promoter was replaced with the cI857 gene expressed from its native promoter, PRM. Plasmid pCMT2 was constructed as follows. Plasmid pLEX (Invitrogen) was cut with NdeI and SalI to remove most of the polylinker and religated with an NdeI/SnaBI/SalI DNA linker made by annealing the oligomers 5′-TACGTAGCG-3′ and 5′-TCGACGCTACG-3′ to make pLEXSnaBI. Oligonucleotide primers 5′-CGCGGGATCCGGTGATAAATTATC-3′ and 5′-CGCGGAATCCAGTTGGGTAACGCC-3′ were used to generate a 326-bp PCR product containing the λPL promoter, the λcII ribosome-binding site, and the aspA terminator with pLEXSnaBI as template DNA. PCR was performed with Herculase polymerase mix and the following cycling conditions: initial denaturing at 94°C for 5 min; 25 cycles of denaturing at 94°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min; and a final extension at 72°C for 3 min. The PCR product was cut with EcoRI and BamHI and ligated to the 4,070-bp EcoRI-BamHI fragment of pGP1-2 (22). pCMT2 was then cut with FspI and EcoRI to remove the Plac cI857 gene. PCR was used to amplify the λPRM-cI857 fragment from lambda (cIts857) DNA (Promega, Southampton, England) with oligonucleotide primers 5′-GCGAATTCAGGTGATGATTATCAGCC-3′ and 5′-GATCTTTAGCTGTCTTGGTTTGC-3′. PCR was performed with Herculase polymerase mix and the following cycling conditions: initial denaturing at 94°C for 5 min; 25 cycles of denaturing at 94°C for 30 s, annealing at 54°C for 1 min, extension at 72°C for 1 min; and a final extension at 72°C for 3 min. A 1,098-bp PCR product was cut with EcoRI to generate a blunt sticky-end fragment for ligation to pCMT2 cut with FspI and EcoRI to generate vector pCMT2b. The 3PF12 gene cassette was amplified by PCR with oligonucleotide primers 5′-ATGAAATACCTATTGCCTACG-3′ and 5′-GCGAATTCGTCGACTTATTAATGGTGATGATGGTG-3′ and 3PF12 in pUC119myc-his as template DNA. PCR was carried out with Herculase polymerase mix and the following cycling conditions: initial denaturing at 94°C for 5 min; 25 cycles of denaturing at 94°C for 30 s, annealing at 56°C for 1 min, and extension at 72°C for 1 min; and a final extension at 72°C for 3 min. An 879-bp PCR product was generated and cut with SalI to give a blunt sticky-end fragment for directional cloning into pCMT2b cut with SnaBI and SalI. Plasmids were selected in E. coli strain DS941 on L agar containing kanamycin (30 μg ml−1) at 30°C.
For expression experiments, plasmids were introduced consecutively into W3110 hns-205::Tn10 by electroporation with a Gene Pulser electroporator (Bio-Rad Laboratories Ltd., Hemel Hempstead, England) in accordance with the manufacturer's instructions. pCMT2b or pCMT2b-scFv was introduced first, followed by pRcd1. Transformants were selected on LA plates (17) at 30°C so that Rcd expression and scFv expression were both repressed by the cI857 protein, allowing colonies to form. When appropriate, media were supplemented with kanamycin (25 μg ml−1), ampicillin (75 μg ml−1), or chloramphenicol (30 μg ml−1).
Cell culture.
Luria broth (LB) (10) was used for small-scale shake flask cultures, and Terrific broth (TB) (23) supplemented with 10 mM MgSO4, glycerol, glucose, and yeast extract was used to achieve higher cell densities in shake flask and fermentor cultures. For fed-batch studies, the feed consisted of glucose or glycerol and yeast extract, which were autoclaved separately and mixed in appropriate proportions. When appropriate, media were supplemented with kanamycin (35 μg ml−1) and ampicillin (100 μg ml−1). Small-scale growth and expression studies were conducted initially with 10-ml cultures in 50-ml conical flasks in a shaking water bath. For Rcd and product gene induction, these were transferred from a 30°C water bath to a 42°C water bath. Later, to achieve better aeration, 50-ml cultures were grown in 1-liter flasks in a shaking incubator (250 rpm) at 30°C. To achieve the temperature upshift, these were transferred to a 42°C water bath for 10 min and then placed in a 42°C shaking incubator.
Large-scale batch and fed-batch studies were carried out with an NBS BIOFLO 3000 5-liter fermentor with a working volume of 4 liters. The NBS BIOFLO 3000 had proportional integral derivative-based control loops for temperature, pH, antifoam, and dissolved-oxygen (DO) regulation. The airflow rate was kept at 1 volume of air/volume of medium/min. The DO concentration was set at 40% of saturation and was initially controlled by stirrer speed until the stirrer speed reached a maximum at 1,000 rpm, after which a two-gas mixer was used to pulse pure oxygen for aeration. The metabolic activity of the cultures was estimated indirectly by observing the oxygen uptake rate (OUR), which was a function of the stirrer speed and, later, the frequency of the pure oxygen that was pulsed into the system. The medium pH was set at 7.2 and controlled by automatic addition of 1 N HCl or NaOH. Sigma Antifoam 289 (catalog no. A-5551; Sigma-Aldrich, Poole, England) was added when required. The residual glucose concentration in the culture was measured with a Pocket Scan handheld blood glucose monitor using strips containing immobilized glucose oxidase (LifeScan, High Wycombe, England). Glucose-6-phosphate dehydrogenase was assayed with a detection kit (Sigma) in accordance with the manufacturer's instructions. A peristaltic pump (Watson-Marlow Bredel Pumps Ltd., Falmouth, England) was used for feeding concentrated substrate at the desired flow rate for fed-batch cultures.
Protein analysis by SDS-PAGE and Western blotting.
Cell density was measured spectrophotometrically with a Beckman DU 530 Life Science UV/Vis Spectrophotometer (Beckman Coulter Ltd., High Wycombe, England). Cultures were diluted as necessary in 0.95% (wt/vol) saline, and the OD600 was measured. Simultaneously, the dry cell weight (DCW) was measured by harvesting the cells in a preweighed centrifuge tube, washing them with deionized water, and drying them at 100°C for 24 h. A cell culture OD600 of 1.0 corresponded to 0.38 g of DCW liter−1. This value was used in calculations of glucose uptake and specific rates of product formation. To estimate 3PF12 scFv expression, cells were extracted from culture samples by centrifugation at 8,500 × g for 5 min. The pellet and supernatant were stored at −20°C. The pellet was resuspended in Bug Buster HT protein extraction reagent (Novagen, CN Biosciences Ltd., Beeston, England). Cells were lysed with 200 μl of Bug Buster reagent for a quantity of cells equivalent to 1 ml of a culture with an OD600 of 1.0. The suspension was shaken at room temperature for 20 min, and lysozyme was added to a final concentration of 1 mg ml−1. The suspension was incubated for a further 5 min and then centrifuged at 8,500 × g for 5 min. The cytoplasmic fraction and the extracellular supernatant were run on duplicate 18% Criterion sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels (Bio-Rad). For high cell density batch and fed-batch fermentations, the supernatants were diluted 10-fold prior to loading onto an SDS-PAGE gel with 2× Laemmli sample buffer (Bio-Rad) and the Tris-glycine-SDS buffering system (Bio-Rad). Cytoplasmic fractions were also diluted 10-fold when the culture OD600 was above 50, so that the pellet from a 20-μl culture (equivalent to 1 ml of cells at an OD600 of 1.0) was resuspended in 200 μl of lysis buffer. One gel was used for Bio-Safe G250 Coomassie blue staining or silver staining (Bio-Rad), and the other gel was used for transfer of the proteins onto Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore Ltd., Watford, England) with a Transblot SD blotter (Bio-Rad) and a semidry transfer technique as recommended by the manufacturer.
Western analysis was carried out by binding with a primary antibody consisting of cell culture supernatant containing anti-c-myc (clone 9E10) (24) and a horseradish peroxidase-labeled goat anti-mouse immunoglobulin G antibody (Sigma-Aldrich). A Pierce 3,3′-diaminobenzidine tetrahydrochloride substrate kit (Perbio Science Ltd., Tattenhall, England) was used to develop Western blots.
Measurement of scFv biological activity.
The supernatant and soluble cytoplasmic protein fractions of cultures expressing 3PF12 were tested for biological activity with a platelet immunofluorescence test (24). scFv binding to HLA-A2-positive blood platelet cells was detected with mouse monoclonal antibody 9E10 and fluorescein isothiocyanate-labeled goat-anti-mouse antibody in a three-step process. Processed samples were then analyzed with a Coulter XL flow cytometer running system II software (Beckman-Coulter, Luton, United Kingdom). For all samples, 10,000 events were analyzed and median fluorescence intensity was determined. The amount of active protein in the samples was determined by using a standard curve obtained by titrating purified 3PF12 in a platelet immunofluorescence test (24).
RESULTS
Host-vector combinations for Q-cell culture.
In this study, the Q-cell expression system comprises the host strain E. coli W3110 hns-205 containing two compatible plasmids. The first, pCMT2b-scFv, contains the scFv product gene (with an N-terminal pelB export signal) expressed from the λPL promoter. Transcription of the product gene is controlled by the cI857 temperature-sensitive repressor, which is on the same plasmid. Rcd is expressed from λPR on the second plasmid (pRcd1) and controlled by cI857 acting in trans from pCMT2b-scFv. Thus, a temperature shift from 30 to 42°C simultaneously induces transcription of rcd and that of the product gene. Plasmid pRcd1 is omitted in control cultures in which establishment of quiescence is not required.
Shake flask studies.
We first confirmed that the quiescent state could be established and maintained with our chosen host-vector combination. W3110 hns-205/pRcd1/pCMT2b-scFv was grown overnight in LB supplemented with ampicillin and kanamycin. This culture was used to inoculate 50 ml of the same medium in a 1-liter shake flask, and the culture was grown to mid-log phase at 30°C. Rcd expression and product expression were induced by transferring the flask to a shaking water bath at 42°C when the OD600 of the culture was approximately 0.5. A control culture (lacking pRcd1) was treated similarly, except that the broth was supplemented with kanamycin alone. Following the temperature upshift, the control culture eventually reached stationary phase at an OD600 of 2, while the pRcd1-containing cells entered quiescence at an OD600 of 1. To ensure that the quiescent state could also be achieved in a richer growth medium, the experiment was repeated with TB plus glycerol (0.4%) in place of LB. In this case, the cultures were shifted to 42°C at an OD600 of 1.0 and the pRcd1-containing cells entered quiescence at an OD600 of 5 approximately 3 to 4 h after the temperature upshift. The control culture reached stationary phase at an OD600 of 10.
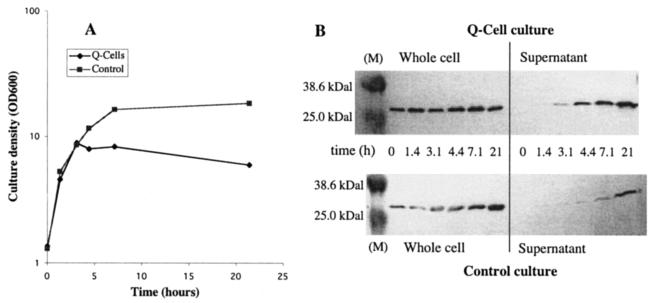
To establish whether quiescence could be achieved at higher cell densities, cultures were grown in TB plus glycerol (0.4%) and the temperature shift was delayed until an OD600 of 2 was reached (Fig. 1A). A pulse of glycerol (final concentration, 0.8%) was added 3 h later, when the OD had reached 8. The control culture entered stationary phase at an OD600 of 18, while the pRcd1-containing culture became quiescent at an OD600 of 8. The quiescent state was stable for at least 21 h after the temperature shift. To confirm that both cultures were expressing scFv, samples were taken soon after the temperature shift and at intervals over the next 21 h. The samples were centrifuged to separate cells from the culture medium, and proteins from whole-cell lysates and culture supernatants were separated by SDS-PAGE and probed for scFv by Western blotting (Fig. 1B). Whole-cell lysates from both cultures showed a strong band for scFv. There appeared to be significantly more product in the Q-cell supernatant, although the culture density was less then half that of the control culture.
FIG. 1.
scFv expression in a shake flask culture. (A) Growth of Q-cell and control cultures. W3110 hns-205/pCMT2b-scFv (control culture; squares) and W3110 hns-205/pCMT2b-scFv/pRcd1 (Q-cell culture; diamonds) cells were grown initially at 30°C and shifted to 42°C at t = 0. Culture OD600 was monitored over the subsequent 24 h. (B) Culture samples were taken at 0, 1.4, 3.1, 4.4, 7.1, and 21.4 h and separated into whole-cell and supernatant fractions. Samples were separated by SDS-PAGE and transferred to PVDF membranes for Western blotting. M, markers.
Fermentor studies of Q-cells.
An important objective of this work was to explore whether the Q-cell system could be scaled up to higher volumes and culture densities. The two strains from the shake flask study were therefore grown in a batch culture in a 5-liter fermentor. The growth medium was TB supplemented with glucose (0.5%) and MgSO4 (10 mM). The culture was further supplemented with 0.5% glucose at an OD600 of 10 (approximately), when the glucose concentration (measured with a handheld blood glucose monitor) fell below 0.05%. In our initial experiments, we noticed that when the temperature was shifted from 30 to 42°C, the stirrer speed increased transiently and then fell sharply, showing that the OUR (a measure of the metabolic activity of the cells) declined rapidly after heat shock. Although the metabolic activity of the control culture recovered subsequently, that of the pRcd1-containing culture did not. One possible cause for the loss of metabolic activity in the Q-cell culture was that the rise in temperature was too rapid. We therefore repeated the experiment, raising the temperature in three stages of 20 min each: 30 to 38°C, 38 to 40°C, and 40 to 42°C. Under these phased heat shock conditions, we no longer observed a fall in the metabolic activity of the Q-cells, presumably because the cells were more able to adapt to the higher temperature (data not shown).
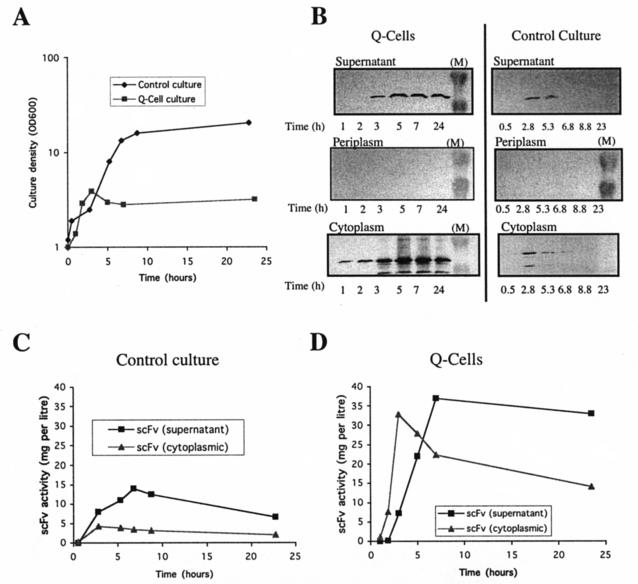
When the phased temperature upshift was initiated at an OD600 of 1 to 2, the density of the Q-cell culture reached a plateau at an OD600 of 3.5 (approximately) while the control culture reached stationary phase at an OD600 of 20 (Fig. 2A). Samples were taken at intervals during the 24 h after the temperature upshift, and supernatant, periplasmic, and cytoplasmic protein fractions were prepared. Western blotting (Fig. 2B) of samples from both Q-cell and control cultures revealed scFv in the supernatant and the cytoplasmic fractions but no detectable amount in the periplasmic fraction. Antibody fragment detected by Western blotting may be in an unfolded and inactive state. We therefore used a platelet-binding assay for biologically active scFv in the supernatant and cytoplasmic fractions (Fig. 2C and D). In the Q-cell cytoplasm, activity rose to 33 mg liter−1 after 3 h and declined thereafter. In the control culture cytoplasm, activity remained within the range of 3 to 5 mg liter−1 throughout. Activity in both supernatants peaked at approximately 7 h, when the Q-cell supernatant contained 37 mg of scFv liter−1, compared to 12 mg liter−1 in the control. Subsequently, the activity declined in both cultures, presumably reflecting degradation of scFv in the supernatant. These calculations of supernatant activity take no account of the sixfold greater density of the control culture, and the productivity of supernatant protein per OD unit was almost 20 times greater for Q-cells than for the control culture.
FIG. 2.
scFv expression in a fermentor batch culture. (A) W3110 hns-205/pCMT2b-scFv (control culture; diamonds) and W3110 hns-205/pCMT2b-scFv/pRcd1 (Q-cell culture; squares) cells were grown initially at 30°C, and a phased temperature shift to 42°C (see text) was initiated at t = 0. Culture OD600 was monitored over the subsequent 24 h. (B) Supernatant, periplasmic, and cytoplasmic protein samples from Q-cell and control cultures were taken at intervals after initiation of the temperature shift. Samples were separated by SDS-PAGE and transferred to PVDF membranes for Western blotting. For the supernatant samples, equal volumes of culture supernatant were loaded onto the gel. For periplasmic and cytoplasmic samples, protein was prepared from equal cell biomasses (estimated from OD600 measurements). The bands in the marker (M) tracks are 38.6 (upper) and 25.0 (lower) kDa. (C) Accumulation of biologically active scFv in the supernatant (squares) and cytoplasmic (triangles) fractions of the control culture (W3110 hns-205/pCMT2b-scFv). Data represent protein activity per liter of culture. (D) Accumulation of biologically active scFv in the supernatant (squares) and cytoplasmic (triangles) fractions of a Q-cell culture (W3110 hns-205/pCMT2b-scFv/pRcd1). Data represent protein activity per liter of culture.
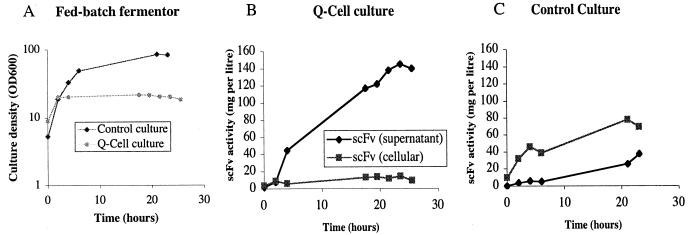
We noted that in batch culture, product accumulated for only the first 5 to 7 h after the temperature shift (Fig. 2D) and wondered whether nutrient depletion might be responsible. The glucose uptake rate of the Q-cell culture was measured by adding pulses of glucose to the medium and monitoring the subsequent decline in its concentration. The initial rate was 0.3 g of glucose per g of DCW per h, which fell slowly over 24 h to less than 10% of this value. In an attempt to prevent substrate starvation during quiescence, we used fed-batch fermentation with a continuous feed (Fig. 3). The cells were grown at 30°C in a 3-liter batch culture to an OD600 of 7, when a feed of concentrated substrate (20% glucose, 20% yeast extract) was started at 40 ml h−1. This matched our estimate of the initial glucose uptake rate of the Q-cell culture with an OD600 of 20. A phased temperature shift was initiated at an OD600 of 10, and the Q-cell culture reached a maximum OD600 of 20 (Fig. 3A). The residual glucose in the medium during the fed-batch phase remained close to 0.1% throughout, demonstrating efficient uptake by the Q-cells. The platelet-binding assay revealed that most of the biologically active scFv was extracellular, rising continuously throughout the experiment to a final concentration of 150 mg liter−1 (Fig. 3B). This corresponds to a rate of accumulation of approximately 6 mg liter−1 h−1 or a specific productivity of 0.3 mg liter−1 h−1 OD unit−1. Intracellular activity (there was no separation of the cytoplasmic and periplasmic fractions in this experiment) remained at a low level (<20 mg liter−1) throughout. When the control culture was subjected to the same feed and temperature shift regimen, it reached a final density more than fourfold higher than that of the Q-cell culture (OD600 of 84; Fig. 3A). In the control culture supernatant, scFv activity rose to approximately 40 mg liter−1; a rate of accumulation of 1.7 mg liter−1 h−1 or a specific productivity of 0.02 mg liter−1 h−1 OD unit−1. Higher scFv activity was detected inside the cell, peaking at 80 mg liter−1 after 20 h (Fig. 3C). Although the intracellular activity of the control culture increased over time, this mainly reflected the increase in culture density. Indeed, the intracellular specific activity increased only during the first 4 h after the temperature shift and thereafter fell from 4.8 mg g of DCW−1 to 2.2 mg g of DCW−1 over the next 20 h.
FIG. 3.
scFv expression in a fed-batch fermentor culture. (A) W3110 hns-205/pCMT2b-scFv (control culture; diamonds) and W3110 hns-205/pCMT2b-scFv/pRcd1 (Q-cell culture; circles) cells were grown initially at 30°C with a feed of 20% glucose-20% yeast extract (see text for details). A phased temperature shift to 42°C was initiated at t = 0, and culture OD600 was monitored over the next 24 h. Culture samples were taken at intervals and separated into supernatant and whole-cell fractions for further analysis. (B) scFv activity of Q-cell supernatant (diamond) and whole-cell (square) fractions. (C) scFv activity of control culture supernatant (diamond) and whole-cell (square) fractions. Supernatant data represent protein activity per liter of culture. For whole-cell assays, protein samples were prepared from equal cell biomasses (estimated from OD600 measurements).
Expression from a high-density Q-cell culture.
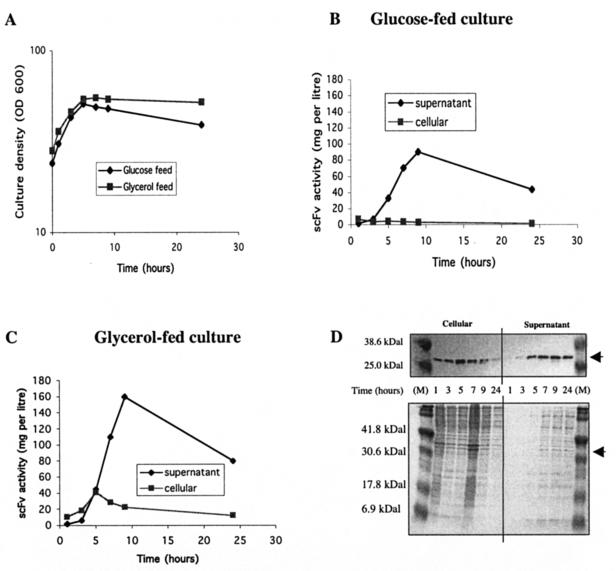
As long as individual cell productivity is maintained, the simplest way to improve the Q-cell product yield would be to increase the density of the Q-cell culture. We therefore modified our fed-batch experiment so that the final OD600 of the culture was 50 rather than 20 (Fig. 4). Initially, cells were grown in a batch culture at 30°C as described for the previous experiment, reaching an OD of 25 in 14 h (early stationary phase). At this point, the feed (40 ml h−1) was started and the culture reentered log phase. Since there is no increase in biomass in quiescence, it was decided to increase the glucose and lower the proportion of yeast extract in the feed (the new composition was 50% glucose and 10% yeast extract). Stoichiometric calculations suggested that the reduced concentration of yeast extract should be sufficient to satisfy the nitrogen requirements of the culture. The feed rate was matched to the initial glucose uptake rate of a culture with an OD600 of 50 so that, compared to that in the low-density fed-batch experiment, the yeast extract feed was reduced fivefold per unit of biomass. After 1 h of feeding, the culture OD600 had risen to 35 and a phased temperature increase from 30 to 42°C was initiated. After a further 3 h, the cells had reached quiescence at an OD600 of 50. The slow decline in culture density observed thereafter reflected the dilution of the culture by the feed (Fig. 4A). The platelet-binding assay revealed a rapid accumulation of active product in the supernatant, i.e., approximately 11 mg liter−1 h−1, over the first 8 h (Fig. 4B). This corresponds to a specific productivity of 0.22 mg liter−1 h−1 OD unit−1. Comparison with the result of the low-density fed-batch experiment suggests that individual cell productivity declined by approximately 30% as the culture OD increased from 20 to 50. Unlike that of the culture with an OD600 of 20, the product concentration did not increase throughout the experiment but, having reached 100 mg liter−1 in 8 h, it declined to less than half of this value over the next 16 h. This presumably reflects product instability in the supernatant. The metabolic activity of the culture (measured by the OUR) declined to 25% of its initial value in 8 h, and a slow buildup of residual glucose was observed (reaching 2% at the end of the fermentor run).
FIG. 4.
scFv expression in a high-density Q-cell culture. (A) W3110 hns-205/pCMT2b-scFv/pRcd1 (Q-cell culture) was grown initially at 30°C, and when the culture reached an OD600 of 25, a feed of yeast extract (10%) and either glucose (50%; diamonds) or glycerol (50%; squares) was initiated. A phased temperature shift (see text) was started at t = 0 (OD600 of 35), and culture density was monitored over the next 24 h. Culture samples were taken at intervals and separated into supernatant and whole-cell fractions for further analysis. (B) scFv activity of supernatant (diamonds) and whole-cell (squares) fractions of the glucose-fed Q-cell culture. (C) scFv activity of supernatant (diamonds) and whole-cell (squares) fractions of the glycerol-fed Q-cell culture. (D) Whole-cell and supernatant fractions from the glycerol-fed culture were separated by SDS-PAGE, stained with Coomassie blue (bottom), and then transferred to PVDF membranes for Western blotting (top). Arrowheads indicate the positions of the scFv band. M, molecular weight markers.
A possible reason for the decline in specific productivity and metabolic activity in the culture with an OD600 of 50 is the reduced yeast extract concentration in the feed. To test this, the experiment was repeated with a feed in which the glucose-to-yeast extract ratio was restored to 1:1. Since it is difficult to prepare a highly concentrated mixture of glucose and yeast extract, the necessary high feed level was achieved by doubling the flow rate to 80 ml h−1. For this reason, and because the fed-batch run was terminated after 8 h, the result must be seen as preliminary. Nevertheless, under these conditions, the rate of product accumulation in the supernatant increased to 15 mg liter−1 h−1 and the specific productivity was restored to the value obtained with the culture with an OD600 of 20 (0.3 mg liter−1 h−1 OD unit−1).
A final high-density fed-batch experiment was performed with glycerol substituted for glucose in the feed (50% glycerol, 10% yeast extract). Once again, the culture entered quiescence at an OD600 of approximately 50 (Fig. 4A). The glycerol feed resulted in an improved rate of product accumulation in the supernatant (20 mg liter−1 h−1) and a higher absolute yield (160 mg liter−1 after 8 h). However, as with the glucose-fed culture with an OD600 of 50, approximately 50% of the biological activity in the supernatant was lost over the next 16 h (Fig. 4C). SDS-PAGE and Western blot analysis mirrored the rapid accumulation and subsequent decline of product in the supernatant (Fig. 4D). However, the Western blot revealed that over the first 5 h, product accumulated more rapidly in the cytoplasm than in the supernatant. Comparison of the bioassay and Western blot results for the cytoplasmic and supernatant fractions suggests that, as expected, the specific activity of the scFv was much lower in the reducing environment of the cytoplasm.
DISCUSSION
In this report, we have shown that productive, stable quiescence can also be established in a high-density shake flask culture at an OD600 of 8 and in a fermentor culture at an OD600 of up to 50. The ease with which the system can be scaled up suggests that Q-cells are suitable for a range of applications, from small-scale analytical work to large-scale fermentation. Behind our development of the Q-cell system was the hope that nongrowing cells would channel a greater proportion of resources into recombinant protein expression than a conventional bacterial cell factory. We use the term resources in a broad sense to encompass not only carbon and energy but also the availability of cellular machinery required for the export and folding of proteins. Our calculation that the specific productivity of Q-cells (approximately 1 mg of product liter−1 h−1 g of DCW−1 in a batch culture) is 10- to 20-fold greater than that of the control culture suggests that this may indeed be the case. Moreover, the specific productivity of Q-cells was relatively constant across a wide range of culture densities (OD600 of 0.5 to 50), and preliminary data suggest that the 30% falloff observed when the density of a fed-batch glucose cultures was increased from an OD600 of 20 to an OD600 of 50 can be reversed by increasing the yeast extract concentration in the feed. The behavior of Q-cells at high density contrasts strongly with that of conventional cultures, in which protein expression falls off as the cells approach and enter stationary phase.
The availability of a simple, quantitative bioassay for our scFv allowed us to assess the abilities of Q-cells to export protein from the cytoplasm and fold it into a biologically active form. In these experiments, the scFv gene was tagged with a pelB leader, but recent work in this laboratory has shown that the ompA and phoA leaders also function efficiently in Q-cells (D. C. D. Rowe and D. K. Summers, unpublished data). We did not detect a significant accumulation of pelB-tagged product in the periplasm of Q-cells or in the control culture. It appears that in both cases, pelB-mediated transport across the inner membrane is followed by folding and movement into the supernatant. In fed-batch fermentor cultures, Q-cells accumulated active protein in the supernatant more than 10 times as fast as the control did. Faster accumulation of product in the supernatant could simply reflect increased lysis of Q-cells, but two pieces of evidence oppose this. First, assays for glucose-6-phosphate dehydrogenase (an intracellular enzyme) in the supernatants of Q-cell and conventional cultures gave similar results. Second, alkaline phosphatase (expressed with its native leader) accumulates in the Q-cell periplasm but virtually no activity is detectable in the supernatant (K. J. Mukherjee, D. C. D. Rowe, and D. K. Summers, unpublished data). It seems likely that Q-cells are capable of faster export because the product is not in competition with other host proteins for access to the export machinery.
The choice of which bacterial strain to use for these studies was guided by the widespread use of W3110 in fermentor work and the requirement for the hns-205 mutation for cells to enter quiescence in response to Rcd induction. To the best of our knowledge, the suitability of hns mutant strains for protein overexpression has not been addressed, but this study shows that they are capable of producing and secreting a high level of biologically active scFv. Hommais et al. (7), by using two-dimensional gel analysis and macroarrays, have demonstrated the wide-ranging changes in protein and transcript levels that occur in hns mutant cells. Their analysis showed changes in the levels of at least 60 proteins and the expression profiles of approximately 250 genes. With such large changes in gene expression, the physiology of hns mutant cells is clearly different from that of wild-type cells. For example, hns mutant cells are able to tolerate higher osmolarities and survive at lower pHs, both indicative of the ability to adapt to environmental stress, which may be advantageous in fermentation systems. hns mutant cells also show a large number of changes in the expression of genes encoding proteins implicated in cell wall composition and found in the periplasm. For example, expression of gspG, which codes for a protein involved in the general secretory response, is increased 17-fold in hns mutant cells. Strains carrying the hns-205 mutation are mucoid on agar plates, and it may be that changes in membrane composition associated with this mutation may assist the secretion of overexpressed proteins. Our data suggest that hns mutant cells may be well suited for protein overexpression.
In earlier shake flask studies (16), it was found that Q-cells were most active in protein synthesis for 3 to 4 h after the induction of Rcd and that the rate declined significantly thereafter. In the present study, we sought to extend the useful period of protein synthesis. It is clear that altering culture conditions can have a significant effect. Fed-batch cultures sustained protein expression over a longer period than batch cultures, and substitution of glycerol for glucose in the fed-batch culture gave a significant increase in the rate of product accumulation. If we compare the carbon flux needed for recombinant protein synthesis with the rate of glucose consumption during quiescence, it is clearly a very small fraction. Even allowing for other proteins that may be synthesized during quiescence (the recombinant protein forms 5 to 10% of the extracellular protein fraction), we can account for less than 10% of the observed carbon flux. Thus, even taking into account the normal maintenance requirements of the cell (around 0.02 g of glucose g of DCW−1 h−1 for E. coli) (15), Q-cells have a metabolic rate that could potentially support still higher rates of product synthesis.
In conclusion, this study has demonstrated that quiescent E. coli can be generated in a variety of media and at high density in both shake flask and fermentor cultures. The cultures are capable of expressing and exporting a biologically active antibody fragment with a specific productivity 10-fold higher than that of a control culture with a conventional growth profile. The higher productivity of Q-cells seems to confirm our expectation that in Q-cells, resources that would otherwise have been used in biomass production are instead available for product formation. We are optimistic that the system may prove beneficial for the expression of a wide range of recombinant proteins.
Acknowledgments
This work was supported by a Biotechnology and Biosciences Research Council grant to D.K.S and D.C.D.R. and financial support to K.J.M. from the Association of Commonwealth Universities.
We thank Anton Middleberg and Nigel Slater, Department of Chemical Engineering, University of Cambridge, for providing facilities for carrying out fermentor work. The expression system described here is protected by international patent PCT/GB97/00731 (16).
REFERENCES
- 1.Bayly, A. M., A. A. Kortt, P. J. Hudson, and B. E. Power. 2002. Large-scale bacterial fermentation and isolation of scFv multimers using a heat-inducible bacterial expression vector. J. Immunol. Methods 262:217-227. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Zvi, A. P., and P. Goloubinoff. 2001. Mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol. 135:84-93. [DOI] [PubMed] [Google Scholar]
- 3.Bothmann, H., and A. Pluckthun. 2000. The periplasmic Escherichia coli peptidylpropyl cis, trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J. Biol. Chem. 275:17100-17105. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, P. 2000. Expressing genes in different Escherichia coli compartments. Curr. Opin. Biotechnol. 11:450-454. [DOI] [PubMed] [Google Scholar]
- 5.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed] [Google Scholar]
- 6.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 8.Horn, U., W. Strittmatter, A. Krebber, U. Knupfer, M. Kujau, R. Wenderoth, K. Muller, S. Matzku, A. Pluckthun, and D. Riesenberg. 1996. High volumetric yields of functional dimeric miniantibodies in Escherichia coli, using an optimised expression vector and high-cell-density fermentation under non-limited growth conditions. Appl. Microbiol. Biotechnol. 46:524-532. [DOI] [PubMed] [Google Scholar]
- 9.Jurado, P., D. Ritz, J. Beckwith, V. de Lorenzo, and L. A. Fernandez. 2002. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J. Mol. Biol. 320:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy, C. K. 1971. Induction of colicin production by high temperature or inhibition of protein synthesis. J. Bacteriol. 108:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knappik, A., and A. Pluckthun. 1995. Engineered turns of a recombinant antibody improves its in vivo folding. Protein Eng. 8:81-89. [DOI] [PubMed] [Google Scholar]
- 12.Kuzel, T. M., and S. T. Rosen. 1994. Antibodies in the treatment of human cancer. Curr. Opin. Oncol. 6:622-626. [DOI] [PubMed] [Google Scholar]
- 13.Lee, M. H., T. I. Park, Y. B. Park, and J. W. Kwak. 2002. Bacterial expression and in vitro refolding of a single-chain Fv antibody specific for human plasma apolipoprotein B-100. Protein Expr. Purif. 25:166-173. [DOI] [PubMed] [Google Scholar]
- 14.Pines, O., and M. Inouye. 1999. Expression and secretion of proteins in E. coli. Mol. Biotechnol. 12:25-34. [DOI] [PubMed] [Google Scholar]
- 15.Roels, J. A. 1983. The linear equation for substrate consumption. In Energetics and kinetics in biotechnology. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 16.Rowe, D. C. D., and D. K. Summers. 1999. The quiescent-cell expression system for protein synthesis in Escherichia coli. Appl. Environ. Microbiol. 65:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sanchez, L., M. Ayala, F. Freyre, I. Pedroso, H. Bell, V. Falcon, and J. V. Gavilondo. 1999. High cytoplasmic expression in E. coli, purification, and in vitro refolding of a single chain Fv antibody fragment against the hepatitis B surface antigen. J. Biotechnol. 72:13-20. [DOI] [PubMed] [Google Scholar]
- 19.Schaffner, J., J. Winter, R. Rudolph, and E. Schwarz. 2001. Cosecretion of chaperones and low-molecular-size medium additives increases the yield of recombinant disulfide-bridged proteins. Appl. Environ. Microbiol. 67:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summers, D. K., and D. C. D. Rowe. 15 March 1997. International patent application, PCT/GB97/00731. Methods and means relating to quiescent cells and uses thereof. Cambridge University Technical Services, Ltd., Cambridge, United Kingdom.
- 21.Swartz, J. R. 2001. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 12:195-201. [DOI] [PubMed] [Google Scholar]
- 22.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmids and cosmid clones. Focus Life Technol 9:12. [Google Scholar]
- 24.Watkins, N. A., C. Brown, C. Hurd, C. Navarrete, and W. H. Ouwehand. 2000. The isolation and characterisation of human monoclonal HLA-A2 antibodies from an immune V gene phage display library. Tissue Antigens 55:219-228. [DOI] [PubMed] [Google Scholar]