Abstract
Phenazine antibiotic production in the biological control bacterium Pseudomonas aureofaciens 30-84 is regulated in part via the PhzR/PhzI N-acyl homoserine lactone (AHL) system. Previous work showed that a subpopulation of the wheat rhizosphere community positively affected phenazine gene expression in strain 30-84 via AHL signals (E. A. Pierson, D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson III, Mol. Plant-Microbe Interact. 11:1078-1084, 1998). In the present work, a second subpopulation, one that negatively affected phenazine gene expression, was identified from this rhizosphere community. Strain 30-84 grown in conditioned medium (CM) from several strains produced lower levels of phenazines (1.5- to 9.3-fold) than control when grown in CM from the strain 30-84I1/I2. Growth of the phzB::lacZ reporter strain 30-84Z in this CM resulted in decreased lacZ expression (4.3- to 9.2-fold) compared to growth of the control strain in CM, indicating that inhibition of phzB occurred at the level of gene expression. Preliminary chemical and biological characterizations suggested that these signals, unlike other identified negative signals, were not extractable in ethyl acetate. Introduction of extra copies of phzR and phzI, but not phzI alone, in trans into strain 30-84Z reduced the negative effect on phzB::lacZ expression. The presence of negative-signal-producing strains in a mixture with strain 30-84 reduced strain 30-84's ability to inhibit the take-all disease pathogen in vitro. Together, the results from the previous work on the positive-signal subpopulation and the present work on the negative-signal subpopulation suggest that cross-communication among members of the rhizosphere community and strain 30-84 may control secondary metabolite production and pathogen inhibition.
Quorum sensing, the regulation of gene expression in response to the intracellular concentration of N-acyl homoserine lactones (AHLs), is a highly conserved mechanism utilized by a diverse range of gram-negative bacteria (4, 13, 20, 34). Examples of plant-associated bacteria that utilize AHL signals to regulate the expression of traits important in plant-microbe interactions include Agrobacterium tumefaciens (14), Erwinia carotovora (29), Erwinia chrysanthemi (2), Pantoea stewartii (5), Pseudomonas aureofaciens (37), Pseudomonas syringae (9), Ralstonia solanacearum (11), and Rhizobium spp. (36).
These regulatory systems share several conserved features. They are comprised of two genes, one that encodes a transcriptional regulatory protein and a second that encodes an AHL synthase (4, 12, 13, 20, 34). The AHL synthase converts cellular precursors into specific AHL signals that vary in different bacteria by length, degree of saturation, and specific side chain substitutions. Upon sufficient AHL signal accumulation within the cell, synthesis of the cognate regulatory protein is stabilized and becomes activated (40). In turn, the activated regulatory protein binds to specific palindromic DNA sequences located within the promoters of genes, and this binding serves to activate or repress target gene expression.
Several examples of compounds that can interfere with AHL-mediated gene regulation, including AHLs, halogenated furanones, and cyclic dipeptides, have been described (3, 15). Other bacteria produce AHL-degrading enzymes, including AiiA by a Bacillus sp. (8) and AiiD by a Ralstonia sp. (19). In addition, some plants produce AHL signal mimics that can negatively affect AHL-mediated gene regulation (32). The identification of molecules that interfere with quorum-sensing systems has received great attention recently, as AHL-mediated gene regulation has been shown to be involved in pathogenesis, colonization, and biofilm formation (13, 16).
The rhizosphere-colonizing biological control bacterium P. aureofaciens strain 30-84 inhibits Gaeumannomyces graminis var. tritici, the causative agent of take-all disease of wheat and barley. Strain 30-84 produces broad-spectrum phenazine antibiotics (27). Phenazine production is important for the survival of strain 30-84 on wheat roots in natural soil (22), and it has the ability to inhibit G. graminis var. tritici (27). The AHL regulatory system PhzR/PhzI encoded by phzR and phzI is required for expression of the phenazine biosynthesis genes in P. aureofaciens (27, 37, 38). Inactivation of phzI resulted in a 1,000-fold decrease in phzB expression in situ in sterile soil (38). It was shown previously (26) that a subpopulation from a random collection of bacteria isolated from a wheat rhizosphere community produced AHL signals that restored phenazine gene expression in vitro and in situ to strain 30-84Ice/I, a phzI, phzB::inaZ reporter derivative of strain 30-84 unable to produce its cognate AHL signal (i.e., positive cross-communication).
In this report, we rescreened this collection of rhizosphere bacteria and identified a number of isolates that produced extracellular molecules that inhibited antibiotic production in wild-type P. aureofaciens strain 30-84 by interfering with phenazine gene expression (i.e., negative cross-communication). We demonstrated that these strains have the potential to reduce the ability of strain 30-84 to inhibit the take-all disease pathogen in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aureofaciens strain 30-84, its derivatives, and the negative-signal-producing strains were grown at 28°C on agar plates or in broth with shaking at 200 rpm in Luria-Bertani (LB) medium (30) or pigment production medium D (PPMD) (38). Strains were also grown on skim milk or King's medium B (27). Escherichia coli was grown at 37°C in LB medium. Antibiotics were used where appropriate at the following concentrations: for E. coli in all media, 50 μg of kanamycin sulfate/ml, 25 μg of tetracycline/ml, and 100 μg of ampicillin/ml; for P. aureofaciens, 50 μg of kanamycin sulfate/ml and 50 μg of tetracycline/ml in LB medium and PPMD or 30 μg of tetracycline/ml in M9 minimal medium. Rifampin was used at 100 μg/ml to counterselect against donor and helper E. coli organisms in triparental matings. β-Galactosidase activity was visualized qualitatively on agar plates supplemented with 40 mg of 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal)/ml. Pathogen inhibition assays were performed using Kanner minimal/potato extract (KMPE) agar plates (27).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(argF-lacZYA) UI169 φ80lacZ ΔM15 λ− | GIBCO-BRL |
| HB101 | F−hsdS20 (rB− mB−) supE44 recA1 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-5 λ− | GIBCO-BRL |
| MC1061 | F−araD139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi | GIBCO-BRL |
| P. aureofaciens | ||
| 30-84 | Phz+ Rifr; wild type | W. W. Bockus |
| 30-84Z | Phz− Rifr; phzB::lacZ genomic fusion | 27 |
| 30-84I1/I2 | Phz− Rifr; phzI::Kmr and csaI::Gmr genomic fusion | 39 |
| 30-84Z/I | Phz− Rifr; phzB::lacZ, phzI::Kmr genomic fusions | 38 |
| 30-84R | Phz− Rifr Kmr; phzR::Tn5lacZ genomic fusion | 27 |
| Plasmids | ||
| pLAFR3 | IncP1 cos+rlx+ Tcr | 31 |
| pRK2013 | IncP1 tra oriE1 Kmr | 10 |
| pDW7311uidA | pLAFR3 with phzI::uidA fusion | 6 |
| pLAF-2.7RVΔ3 | pLAFR3 containing 2.2-kb genomic fragment containing phzI | 37 |
| pLSP10-21 | pLAFR3 containing the 11.2-kb EcoRI fragment of pLSP259 with the phzI and phzR genes | 27 |
Rifr, rifampicin resistant, Kmr, kanamycin resistant; Tcr, tetracycline resistant; Apr, ampicillin resistant; Gmr, gentamicin resistant.
Negative-signal spot plate bioassay.
Five microliters of overnight LB broth cultures of wheat rhizosphere isolates were spotted onto plates containing PPMD or PPMD plus X-Gal. After 24 h, the plates were sprayed using thin-layer chromatography sprayers with strain 30-84 or strain 30-84Z LB broth cultures. The plates were screened at 24 and 48 h. Negative cross-communication was indicated by the presence of a white halo of strain 30-84 or 30-84Z surrounding the spotted bacteria. Those isolates that created a kill zone in addition to a white halo were not studied further.
CM preparation.
Conditioned medium (CM) was prepared as follows. Twenty-five-milliliter volumes of LB or PPMD broth cultures of negative communicators and controls were grown separately with shaking for 48 h. Cultures were centrifuged at 11,200 × g for 10 min at 22°C. Cell-free supernatants were collected, filtered (0.45-μm pore size), and stored at 4°C. CM was made by mixing equal volumes of sterile cell-free supernatants and fresh medium.
Quantification of phenazine production in CM.
Four milliliters of CM prepared from negative-signal-producing strains and controls was inoculated separately with 20 μl of a strain 30-84 LB broth culture and grown with shaking for 24 h. Before inoculation, 20 μl of a 2-ml overnight culture of strain 30-84 cells was washed, centrifuged (15,700 × g) for 1 min, and resuspended in an equal volume (20 μl) of fresh LB broth. The optical density at 620 nm (OD620) was measured to ensure that cell densities were within 0.1 of each other for all cultures. Phenazine extractions were carried out as described previously (27), with the following modifications. Ten-milliliter overnight cultures were centrifuged (13,800 × g) for 10 min. Three milliliters of each supernatant was collected. Two drops of 12 N HCl was added, and the tubes were vortexed briefly. Three milliliters of benzene was added, and the tubes were shaken at 50 rpm for 2 h. The tubes were centrifuged (11,200 × g) for 15 min, and the benzene phase was transferred to new tubes and evaporated under air. Phenazines were resuspended in 1 ml of 0.1 N NaOH, and total phenazine was quantified at OD367.
Quantification of phzB, phzR, or phzI expression in CM.
CM prepared from negative-signal-producing strains grown in LB broth was inoculated with 20 μl of an overnight culture of either strain 30-84Z, strain 30-84R, or strain 30-84(pDW7311uidA), washed in LB broth as described above, and grown with shaking. The OD620 was measured to ensure that cell densities were within 0.1 of each other for all cultures, as described above. β-Galactosidase activity or β-glucuronidase activity was quantified after 24 h as described by Miller (24) and Wilson et al. (35). Treatments were replicated three times, and the experiments were repeated at least twice.
Characterization of isolates.
Five-microliter volumes of overnight LB broth cultures of negative communicators were spotted onto plates containing skim milk, King's medium B, LB broth plus X-Gal, M9 medium, or LB broth plus ampicillin. All plates were screened at 24 and 48 h for zones of clearing, fluorescence, color, or growth. For 16S rRNA gene sequence determinations, the following ingredients were combined in 200-μl PCR tubes: 25.5 μl of sterile deionized water, 10 μl of Eppendorf 5× Taq buffer, 5 μl of Eppendorf 10× Taq buffer, 4 μl of a 10 mM deoxynucleoside triphosphate mixture, 2 μl of primer 1, 2 μl of primer 2, 1 μl of the template, and 0.5 μl of Eppendorf DNA Taq polymerase. The template used was a 1:10 dilution (in deionized water) of an LB broth overnight culture. Identical PCRs were run in the cited and present studies using the following primers: SDBact051aA19 (5′-GTGCCSGCMGCCGCGGTAA-3′) (18), SDBact137aS20 (5′-AGGCCCGGGAACGTATTCAC-3′) (18); fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) (32), rP1 (5′-TACGGTTACCTTGTTACGACTT-3′) (32), 645f (5′-GTAGCGGTGAAATGCGTAG-3′) (this study), or 687r (5′-GCCACTGGTGTTCCTTCC-3′) (this study). Strain PU-15 was subjected to all primers listed; the other strains were subjected to fD1 and rP1 only. All PCRs were run using an Eppendorf Mastercycler gradient thermocycler under the following conditions: one cycle of 95°C for 3 min; 36 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 2 min; and one final cycle of 72°C for 10 min. PCR products were run on a 0.7% agarose gel. Bands were excised and purified by using QIAGEN′s QIAQuick gel extraction kit (according to the manufacturer's instructions). Purified PCR product and primers were sent to the University of Arizona DNA sequencing facility (http://uofadna.arl.arizona.edu/) and sequenced with an Applied Biosystems 377 automated DNA sequencer. Sequences from primers rP1, SDBact1371aS20, and 687r were reverse complemented by using ReadSeq version 1.2 (http://www.nih.go.jp/∼jun/cgi-bin/readseq.pl). Sequences from each primer pair were combined and submitted for database comparison to the NCBI nucleotide-nucleotide BLAST sequences (1). The sequence entry with the lowest expectation (E) value and the highest percentage of nucleotides identical to the query sequence was recorded.
Preliminary characterization of negative signals.
Cultures of several negative-signal-producing strains were grown for 24 h, the cells were removed via centrifugation and filtration (0.45-μm pore size), and the filtrates were extracted with acidified ethyl acetate to isolate any AHLs, furanones, and cyclic dipeptides. The ethyl acetate extracts were dried under a stream of nitrogen. The dried extracts were resuspended in fresh LB medium and inoculated with strain 30-84Z. To further characterize the negative signal produced by one strain, PU-15, 25-ml LB broth cultures were centrifuged, and supernatants were collected as described in “CM preparation.” Two milliliters of supernatant was boiled for 10 min or inoculated with 5 μl of pronase (Boehringer Mannheim; 5 mg/ml) or proteinase K (Roche; 20 mg/ml) and incubated at 37°C for 1 h. Two milliliters of supernatant was also extracted three times with 2 ml of ethyl acetate, chloroform, or hexanes by shaking the mixture at 50 rpm for 1 h and centrifuging it (5,000 × g) for 5 min. The phases were separated, and the solvent phases were pooled and evaporated under N2 gas. Two milliliters of LB broth was added to all samples and inoculated with 20 μl of washed strain 30-84Z. Controls of untreated CM and 2 ml of LB broth were included. After 24 h, β-galactosidase activity was measured as described above.
In vitro G. graminis var. tritici inhibition assays.
Overnight cultures of strain 30-84 and specific negative-signal-producing strains were grown individually and mixed immediately prior to plating at ratios of 25:75 (1:3), 50:50 (1:1), and 75:25 (3:1). Prior to being mixed, each culture's cell density (OD620) was measured, and the optical densities of all cultures were adjusted to within 0.1 of each other. Five microliters of each mixture was spotted twice onto opposite sides of a KMPE plate. As a control, ratios of strain 30-84 and 30-84Z (Phz− PhzI+) or 30-84Z/I (Phz− PhzI−) were spotted. Strains 30-84, 30-84Z, 30-84Z/I and each negative-signal-producing strain alone were also spotted as controls. After 24 h of incubation, a 5-mm-thick plug of G. graminis var. tritici, taken from the leading edge of a fresh potato dextrose agar culture plate, was placed in the center of each KMPE plate. A KMPE plate with G. graminis var. tritici only was included as a control. The plates were incubated at 28°C, and fungal mycelial growth and the distances (in millimeters) between the margin of growth and the bacteria were recorded after 5 days. Samples of the cells were collected, diluted, and counted to ensure that the populations recovered were close to the original ratios (data not shown). The experiment was replicated at least twice.
Statistical analysis.
Treatment effects were determined by analysis of variance, using SAS (version 6.12 for UNIX, 1993; SAS Institute Inc., Cary, N.C.). Means were compared by analysis of variance after multiple comparisons of least significant differences.
RESULTS
Identification of negative-signal-producing strains.
A collection of bacteria isolated from a wheat rhizosphere community (26) was screened for the ability of isolates to inhibit phenazine production by the negative-signal spot plate bioassay. Of 629 strains tested, 43 (7%) negatively affected phenazine production, as judged by the presence of a white halo within the 30-84 lawn surrounding the spotted rhizosphere strains (Fig. 1). Strains that caused a zone of growth inhibition of strain 30-84 were excluded from further analysis. Several strains (PU-5, PU-15, PU-43, PU-295, and PU-761) were chosen for further characterization. For all strains examined, the inhibition of phenazine production persisted for at least 28 days (when the experiment was terminated). Restreaking strain 30-84 from the white zone and from orange regions of the plate resulted in qualitatively similar rates of phenazine production on fresh LB medium, indicating that inhibition was not permanent and only occurred while the strains were in close proximity (data not shown).
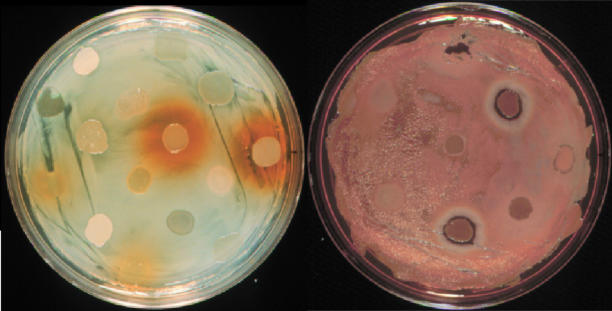
FIG. 1.
Positive and negative signaling by wheat rhizobacteria. Random strains from a collection of rhizosphere bacteria isolated from wheat roots were spotted onto a white lawn of strain 30-84I (PhzI−) (left) or an orange lawn of wild-type strain 30-84 (right). (Left) Positive cross-communication is indicated by the orange halos surrounding the spots, indicating rescue of phenazine production. (Right) Negative cross-communication is indicated by the white halos surrounding the spots, indicating inhibition of phenazine production.
Effect of CM on phenazine production.
In order to collect potential diffusible signals, CM was prepared from several of the negative cross-communicating strains and was inoculated with an overnight culture of strain 30-84. Strain 30-84I1/I2 (which produces no AHL or phenazines) and strain 30-84Z (which produces AHL but no phenazines) were included as negative and positive controls, respectively. The total amount of phenazines produced by strain 30-84 was determined after 24 h (Table 2). Compared to the phenazine production in CM by the control strain 30-84I1/I2, the negative control strains PU-15, PU-43, PU-295, and PU-761 reduced phenazine production 1.5- to 9.3-fold. The positive control strain 30-84Z that produces AHLs resulted in a 1.4-fold increase.
TABLE 2.
Phenazine production by strain 30-84 in CM
| CM sourcea | OD367b | Fold changec |
|---|---|---|
| 30-84I1/I2 (control)d | 1.76 ± 0.12 | |
| PU-5 | 0.73 ± 0.06 | −2.4 |
| PU-15 | 0.77 ± 0.12 | −2.3 |
| PU-43 | 0.19 ± 0.02 | −9.3 |
| PU-295 | 0.24 ± 0.06 | −7.3 |
| PU-761 | 1.2 ± 0.06 | −1.5 |
| 30-84Z (positive control)e | 2.4 ± 0.12 | +1.4 |
Strain used to prepare CM.
OD367 indicates relative phenazine antibiotic production. Values shown are the averages of results for three replicates ± standard errors.
Fold change indicates phenazine production relative to that of the control.
Strain 30-84I1/I2 is a derivative of P. aureofaciens that is defective in phzI and csaI. It does not produce detectable phenazines or AHL signals but does deplete the CM of nutrients.
Strain 30-84Z is a phzB::lacZ reporter that does not produce phenazines but does produce both P. aureofaciens AHL signals.
CM reduces phenazine gene expression.
To determine whether the negative-signal-producing strains were affecting phenazine gene expression, phenazine synthesis, or phenazine stability, β-galactosidase activity in strain 30-84Z, a phzB::lacZ genomic fusion, was assayed in CM (Table 3). CM prepared from the negatively cross-communicating bacteria reduced lacZ expression 4.3- to 9.2-fold more than the CM of the control strain 30-84I1/I2. In contrast, E. coli DH5α had no effect on phzB expression, while the positive control strain 30-84Z resulted in a 2.4-fold increase in β-galactosidase activity. These data suggest that the negatively cross-communicating strains produced a diffusible signal that interfered with phenazine gene expression in strain 30-84.
TABLE 3.
Effect of conditioned medium on phzB expression in strain 30-84Z
| CM sourcea | β-Galactosidase activity (Miller units)b | Fold changec |
|---|---|---|
| 30-84I1/I2 (control)d | 193 ± 23 | |
| DH5α | 211 ± 18 | +1.1 |
| PU-5 | 44 ± 1 | −4.3 |
| PU-15 | 33 ± 5 | −5.8 |
| PU-43 | 34 ± 4 | −5.7 |
| PU-177 | 33 ± 2 | −5.8 |
| PU-295 | 21 ± 2 | −9.2 |
| PU-761 | 32 ± 5 | −6.0 |
| 30-84Ze | 466 ± 46 | +2.4 |
Strain used to prepare CM.
β-Galactosidase activity was measured after 24 h. Values shown are the average of results for three replicates ± standard errors.
Fold change indicates the level of β-galactosidase activity relative to that of the 30-84I1/I2 control.
Strain 30-84I1/I2 is a derivative of P. aureofaciens defective in phzI and csaI. It does not produce phenazines or AHL signals but does deplete the CM of nutrients.
Strain 30-84Z is a phzB::lacZ reporter that does not produce phenazines but does produce both P. aureofaciens AHL signals.
Identification of negatively cross-communicating strains.
Strains PU-5, PU-15, PU-43, PU-177, PU-295, and PU-761 were grown, and a region of the 16s rRNA gene was amplified using the universal 16S sequencing primers fD1 and rP1 (33). The amplified PCR products were cloned, and their DNA sequences were determined. The sequences were compared with GenBank sequences by using BLAST (1), and the best matches are indicated in Table 4. Most of the negative-signal-producing strains characterized were identified as Pseudomonas spp.; one was a Janthinobacterium sp.
TABLE 4.
Identification of negative signaling strains
| Strain | Genusa and species or strain | Accession no.b | Scorec | E valuec (% sim.) |
|---|---|---|---|---|
| PU-15 | Pseudomonas NZ081 | AF388206 | 2,755 | 0.0 (96) |
| PU-43 | “Pseudomonas cedrella” | AF064461 | 981 | 0.0 (96) |
| PU-177 | Pseudomonas brennerii | AF268968 | 1,542 | 0.0 (98) |
| PU-295 | Janthinobacterium lividum | AY247410 | 1,322 | 0.0 (96) |
| PU-761 | Pseudomonas trivialis | AJ492831 | 1,491 | 0.0 (98) |
Genus was determined by a BLAST search with 16S ribosomal DNA fragments amplified as described in Materials and Methods.
The accession number for the closest GenBank match is given.
The raw score for an alignment and the expectation (E) value from the BLAST search with the percentage of sequence identity (sim.) are shown. The lower the E value, the more significant the score.
Preliminary characterization of negative signals.
In order to isolate AHLs, furanones, or cyclic dipeptides (known negative signals), cultures of several negative-signal-producing strains were grown and extracted with acidified ethyl acetate. None of the ethyl acetate extracts had a negative effect on expression of the phzB::lacZ fusion, whereas all of the aqueous phases retained the inhibiting activity (data not shown). Strain PU-15 was chosen for further characterization because it did not inhibit strain 30-84 or G. graminis var. tritici, the fungal pathogen used in later inhibition studies. Treatment with pronase, proteinase K, or heat had no effect on inhibitory activity (Table 5). Similar to extractions with acidified ethyl acetate, extractions with unacidified ethyl acetate, chloroform, or hexane failed to separate the inhibitory activity from the aqueous phase (data not shown).
TABLE 5.
Effect of various treatments on the ability of PU-15 CM to reduce phzB::lacZ expression in strain 30-84Z
| Treatment | β-Galactosidase activity |
|---|---|
| LB medium only | 389 ± 13a |
| None (CM only) | 11 ± 2 |
| Heat | 18 ± 3 |
| Pronase | 14 ± 2 |
| Proteinase K | 14 ± 3 |
| Aqueous phaseb | 22 ± 11 |
| Acidified ethyl acetate phaseb | 228 ± 5 |
Values are the averages of results of four replicates ± standard errors.
CM of PU-15 was extracted with acidified ethyl acetate, and the aqueous and the acidified ethyl acetate phases were separated. The acidified ethyl acetate phase had little effect on phzB::lacZ expression, whereas the aqueous phase remained highly inhibitory.
Effect of negative signal on phzI and phzR expression and effect of extra copies of phzI and phzR on negative signaling.
To determine whether the negative signals affected the expression of phzI or phzR, CM prepared from strain 30-84I1/I2 or PU-15 were inoculated with strains 30-84(pDW7311uidA containing a phzI::uidA fusion) and 30-84R (containing a phzR::lacZ fusion). β-Glucuronidase assays indicated that CM inoculated with PU-15 did not affect phzI expression (β-glucuronidase levels for 30-84I1/I2 and PU-15 were 135 ± 7 and 161 ± 9 units, respectively). However, β-galactosidase assays indicated that PU-15 CM reduced phzR expression by 39% (Miller units for strains 30-84I1/I2 and PU-15 were 288 ± 12 and 177 ± 16, respectively). CM from strains 30-84I1/I2 or PU-15 were also inoculated with strain 30-84Z(pLAF-2.7RVΔ3) to investigate whether extra copies of phzI in trans could overcome PU-15 negative signal inhibition. Expression of phzB in strain 30-84Z(pLAF-2.7RVΔ3) in CM prepared from strain PU-15 was similar to that of strain 30-84Z(pLAFR3) (Table 6), indicating that phzB inhibition by strain PU-15 was not reversed by additional copies of phzI. CM with the addition of extracted AHLs also did not overcome inhibition (data not shown). To determine whether additional copies of both phzR and phzI could reduce the effect of the negative signals on phzB expression, strains 30-84Z(pLAFR3) and 30-84Z(pLSP10-21) were inoculated into CM prepared from strains 30-84I1/I2, PU-15, PU-43, and PU-295. β-Galactosidase activity was determined after 24 h and indicated that additional copies of phzR and phzI increased phzB expression regardless of which strain the CM was prepared from (Table 6).
TABLE 6.
Effect of extra copies of phzI alone or phzR and phzI in trans on phzB expression by CM
| CM source | β-Galactosidase activity
|
||
|---|---|---|---|
| 30-84Z (pLAFR3)a | 30-84Z (pLAF-2.7RVΔ3)b | 30-84Z (pLSP10-21) | |
| 30-84I1/I2 | 161 ± 11 | 328 ± 30 | 1,055 ± 147 |
| PU-15 | 13 ± 3 | 17 ± 4 | 478 ± 66 |
| PU-43 | 44 ± 1 | ND | 1,299 ± 188 |
| PU-295 | 25 ± 6 | ND | 360 ± 35 |
Strain 30-84Z(pLAFR3) contains the vector alone, strain 30-84Z(pLAF-2.7RVΔ 3) contains phzI, and strain 30-84Z(pLSP10-21) contains phzR and phzI. Activity is given as average Miller units ± standard deviations of results for three replicates.
ND, not determined.
Negative-signal strains reduce pathogen inhibition by strain 30-84.
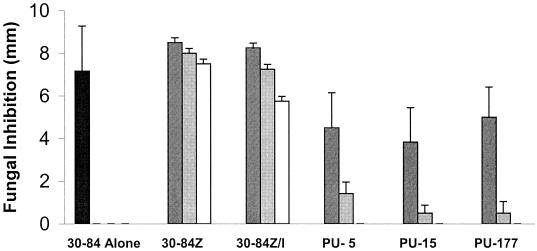
Several negative-signal-producing strains were tested for their ability to inhibit growth of G. graminis var. tritici by an in vitro plate inhibition assay. Each strain was grown separately to the same cell density and combined just prior to plating. Strain 30-84 was coinoculated with a control strain (30-84Z or 30-84Z/I) or with a negative-signal-producing strain (PU-5, PU-15, or PU-177) at a ratio of 75:25, 50:50, or 25:75 and compared to inoculation by each strain alone for the ability to inhibit growth of G. graminis var. tritici (Fig. 2). Strains 30-84Z and 30-84Z/I do not produce phenazines and do not inhibit G. graminis var. tritici growth. Strains PU-5, PU-15, and PU-177 were chosen, as they were the only strains tested that did not inhibit G. graminis var. tritici themselves. There was very little reduction in mycelial inhibition when strain 30-84 was grown in a mixture with a positive-signal-producing strain (30-84Z) or a neutral strain (30-84Z/I), even if strain 30-84 comprised only 25% of the starting mixture. However, the ability of strain 30-84 to inhibit the mycelial growth of G. graminis var. tritici was significantly reduced regardless of its ratio to the negative-signal-producing strain. In fact, fungal inhibition was reduced to less than 20% of wild-type inhibition or was completely abolished when strain 30-84 comprised 50 or 25% of the starting mixture, respectively. Data obtained by dilution plating of the final populations of the mixtures verified that strain 30-84 populations were not reduced by the negative-signal strains; in fact, strain 30-84 made up a larger than expected part of the final populations. For example, when introduced in a 50:50 starting mixture, the final ratio of strain 30-84/negative-signal-producing strain was always 25:1 or greater, and strain 30-84 always reached a final population density which was greater than 50% of the final density of strain 30-84 alone. Similarly, in a 25:75 starting mixture, the final ratio of strain 30-84/negative-signal-producing strain was always 8:1 or greater. These data demonstrate that the reduction in fungal inhibition was not due to the elimination of strain 30-84 from the mixture.
FIG. 2.
Negative communication reduces G. graminis var. tritici inhibition in vitro. Strains were grown separately, mixed in appropriate ratios, and spotted as described in Materials and Methods. Bars indicate fungal growth inhibition by strain 30-84 or mixtures of strain 30-84 and either strain 30-84Z (Phz− PhzI+), strain 30-84Z/I (Phz− PhzI−), or a negative-signal strain (PU-5, PU-15, or PU-177). Treatments are indicated as follows: black bars, wild-type strain 30-84; dark grey bars, 75:25 mix of strain 30-84 and the second strain indicated; light grey bars, 50:50 mix of strain 30-84 and the second strain indicated; white bars, 25:75 mix of strain 30-84 and the second strain indicated. Strains 30-84Z, 30-84Z/I, PU-5, PU-15, and PU-177 had no fungal inhibition alone (data not shown). Error bars indicate standard errors of results for six replicates.
DISCUSSION
Many gram-negative bacterial species utilize AHL signals to regulate the expression of genes that produce products involved in bacterium-host interactions (4, 16, 25, 34). The biological control bacterium P. aureofaciens strain 30-84 utilizes AHL signals as part of a complex regulatory network to control the production of phenazine antibiotics. Phenazine production by strain 30-84 is necessary for rhizosphere persistence and biological control by this strain (22, 26). It has been shown previously that the wheat rhizosphere community contained a subpopulation of bacteria that positively influenced phenazine gene expression in situ in strain 30-84 (26). We have now identified a second subpopulation from the same wheat rhizosphere community that interfered with phenazine production in strain 30-84. CM produced from selected wheat rhizosphere isolates decreased phenazine production and expression of the phenazine biosynthetic genes in strains 30-84 and 30-84Z, respectively. These results are consistent with the hypothesis that these strains are producing an extracellular signal that inhibited phenazine gene expression at the transcriptional level. There was no effect on the growth of strain 30-84, because it reached similar cell densities in CM produced from the negative strains and in CM produced from the control strain 30-84I1/I2. These data indicate that the inhibitory effect is caused by an extracellular signal and is not an effect of cellular stress. A similar relationship was observed between bacteria and the alga Delisea pulchra, which produces a halogenated furanone that inhibited transcription of lux-like genes in bacteria but had no other adverse effects on growth (15, 21).
We PCR amplified a portion of the 16S rRNA gene from five of the negative-signal-producing strains. Four were identified as different Pseudomonas species, and the fifth (PU-295) was identified as a member of the genus Janthinobacterium, a subdivision of Chromobacterium (7). There were fluorescent and nonfluorescent members among the Pseudomonas isolates. These data were included primarily as an indication that the negative-signal strains are not identical rather than as an absolute identification.
We chose one strain, PU-15, for more detailed analysis. Preliminary studies showed that the PU-15 signal is heat stable, small, and unaffected by pronase or proteinase K, indicating that it is either nonproteinaceous or a small peptide. The negative-acting signals from all strains tested were not extractable by ethyl acetate or other nonpolar solvents. This is of interest because all other published quorum-sensing inhibitors, including the furanones, diketopiperazines, and AHLs, readily partition into ethyl acetate (15, 17, 23). These inhibitors are hypothesized to work by competition with the AHL signal for binding of LuxR-like activators. Although the presence of additional copies of phzI did increase phzB::lacZ expression in strain 30-84Z when grown in strain 30-84I1/I2 CM as expected, there was no increase in phzB::lacZ expression in strain 30-84Z when grown in CM prepared from strain PU-15 or PU-43 (Table 6). CM prepared from strain PU-15 had no affect on phzI expression but caused a 39% decrease in phzR expression. Whether this decrease is biologically relevant is unclear. However, extra copies of phzR and phzI did overcome the negative-signal effects (Table 6). Current hypotheses for the mechanism of action of the negative signal include (i) competitive binding to PhzR (inhibitory long-chain AHLs are not displaced by native AHL signals [23]), (ii) inhibition of phzR expression, (iii) negative effects on phzR mRNA stability, and (iv) degradation of the endogenous strain 30-84 AHL signal analogous to the role of AiiA in Bacillus spp. (8) or AiiD in Ralstonia spp. (19). However, the fact that signal was present in culture supernatants and was not degraded by protease treatments does not support the last hypothesis. Additional work is necessary to clarify the mechanism of inhibition.
We have shown that negative communication can abolish or severely limit strain 30-84's ability to inhibit the fungus G. graminis var. tritici when negative communicators are present at equal or greater numbers in vitro. Even when strain 30-84 is present in greater numbers than the negative communicators, inhibition levels are inconsistent (Fig. 2). Future studies will examine this interaction in situ on wheat roots.
Because the negative-signal strains used in this work were isolated from the same ecosystem as strain 30-84, negative cross-communication could have important implications in phenazine production by strain 30-84 in the rhizosphere. P. aureofaciens also contains another quorum-sensing system, CsaR/CsaI, that is involved in controlling cell surface properties (39). Thus, positive and negative signals produced by other members of the wheat rhizosphere community could have multiple effects on the lifestyle of strain 30-84. Since strain 30-84 exists as a member of a multispecies community in the rhizosphere, positive and negative cross-communication among the members of this community could be essential to the temporal, spatial, and functional development of this community. Current research is aimed at elucidating the roles of phenazine production and modulation of phenazine gene expression on rhizosphere community structure.
Acknowledgments
We thank James Nabhan and Scott Coyle for technical assistance in this work. We also thank Scott Chancey and Cheryl Whistler for helpful conversations during the design and completion of these studies and Derong Liu and Krishna Maddula for critical comments.
This work was supported by U.S. Department of Agriculture NRICGP grant 2001-02684 (to L.S.P.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, R. A., A. R. Eriksson, R. Heikinheimo, A. Mae, M. Pirhonen, V. Koiv, H. Hyytiainen, A. Tuikkala, and E. T. Palva. 2000. Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expR(Ecc). Mol. Plant-Microbe Interact. 13:384-393. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, W. D., and J. B. Robinson. 2002. Disruption of bacterial quorum sensing by other organisms. Curr. Opin. Biotechnol. 13:234-237. [DOI] [PubMed] [Google Scholar]
- 4.Beck von Bodman, S., W. D. Bauer, and D. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 5.Beck von Bodman, S., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chancey, S. T., D. W. Wood, and L. S. Pierson III. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lay, J., P. Segers, and M. Gillis. 1978. Intra- and intergenic similarities of Chromobacterium and Janthinobacterium ribosomal ribonucleic acid cistrons. Int. J. Syst. Microbiol. 28:154-168. [Google Scholar]
- 8.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumenyo, C. K., A. Mukherjee, W. Chun, A. K. Chatterjee. 1998. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur. J. Plant Pathol. 104:569-582. [Google Scholar]
- 10.Figurski, K. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flavier, A. B., L. M. Ganova-Raeva, M. A. Schell, and T. P. Denny. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:7089-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signaling. Nat. Rev. Mol. Cell. Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentzer, M., L. Eberl, J. Nielsen, and M. Givskov. 2003. Quorum sensing: a novel target for the treatment of biofilm infections. BioDrugs 17:241-250. [DOI] [PubMed] [Google Scholar]
- 17.Holden, M. T. G., S. R. Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. C. Salmond, G. S. A. B. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 18.Kroes, I., P. Lepp, and D. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, Y-H., J.-L. Xu, J. Hu, L.-H. Wang, S. L. Ong, J. R. Leadbetter, and L.-H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum sensing enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 20.Loh, J., E. A. Pierson, L. S. Pierson, G. Stacy, and A. Chatterjee. 2002. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 5:285-290. [DOI] [PubMed] [Google Scholar]
- 21.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 22.Mazzola, M., R. J. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson III. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum-sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierson, E. A., D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson III. 1998. Interpopulation signaling via N-acyl-homoserine lactone among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 11:1078-1084. [Google Scholar]
- 27.Pierson, L. S., III, and L. S. Thomashow. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant-Microbe Interact. 5:330-339. [DOI] [PubMed] [Google Scholar]
- 28.Pierson, L. S., III, K. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Staskawicz, B., J. D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 33.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16s ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, K., S. G. Hughes, and R. A. Jefferson. 1992. The Escherichia coli gus operon: introduction and expression of the gus operon in E. coli and the occurrence and use of gus in other bacteria, p. 7-22. In S. R. Gallagher (ed.), GUS protocols: using the gus gene as a reporter of gene expression. Academic Press, Inc., San Diego, Calif.
- 36.Wisniewski-Dye, F., and J. A. Downie. 2002. Quorum-sensing in rhizobium. Antonie Leeuwenhoek 81:397-407. [DOI] [PubMed] [Google Scholar]
- 37.Wood, D., and L. S. Pierson III. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]
- 38.Wood, D. W., F. Gong, M. M. Daykin, P. Williams, and L. S. Pierson III. 1997. N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179:7663-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Z., and L. S. Pierson III. 2001. A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 67:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]