Abstract
Phenotypically, Bacillus atrophaeus is indistinguishable from the type strain of Bacillus subtilis except by virtue of pigment production on certain media. Several pigmented variants of B. subtilis have been reclassified as B. atrophaeus, but several remain ambiguous in regard to their taxonomic placement. In this study, we examined strains within the American Type Culture Collection originally deposited as Bacillus globigii, B. subtilis var. niger, or Bacillus niger using 16S rRNA gene sequencing and amplified fragment length polymorphism (AFLP) analysis to determine the level of molecular diversity among these strains and their relationship with closely related taxa. The 16S rRNA gene sequences revealed little variation with one base substitution between the B. atrophaeus type strain ATCC 49337 and the other pigmented bacilli. AFLP analysis produced high-quality DNA fingerprints with sufficient polymorphism to reveal strain-level variation. Cluster analysis of Dice similarity coefficients revealed that three strains, ATCC 31028, ATCC 49760, and ATCC 49822, are much more closely related to B. atrophaeus than to B. subtilis and should be reclassified as B. atrophaeus. A very closely related cluster of B. atrophaeus strains was also observed; this cluster was genetically distinct from the type strain. The level of variation between the two groups was approximately the same as the level of variation observed between members of the two B. subtilis subspecies, subtilis and spizizenii. It is proposed that the cluster of strains typified by ATCC 9372 be designated a new subspecies, B. atrophaeus subsp. globigii.
Bacillus atrophaeus is a gram-positive, aerobic, endospore-forming, rod-shaped bacterium whose description is virtually identical to that of Bacillus subtilis except for the production of a pigment on media containing an organic nitrogen source (12). Many of the isolates belonging to this species were previously classified as Bacillus subtilis var. niger, Bacillus niger, or even earlier as Bacillus globigii. Several of these strains are used in industry as sterilization control organisms or sources of restriction endonucleases (4, 14, 18), but specifically within the biodefense research and testing community, some of these strains are used extensively as nonpathogenic surrogates for Bacillus anthracis (16, 17). Isolates used in this latter capacity are still commonly referred to by their historical designation of B. globigii or simply BG.
Nakamura first proposed the species B. atrophaeus after examining a number of pigmented and nonpigmented strains of B. subtilis (12). Using DNA-DNA reassociation measurements, multilocus enzyme electrophoresis, and pigment production, he demonstrated that a subgroup of the pigmented strains differed significantly from the other pigmented and nonpigmented strains typified by B. subtilis and hence warranted a new species designation. The remaining pigmented strains were considered true variants of B. subtilis. The isolates included in the Nakamura study were reclassified in publicly available culture collections. Recently, Fritze and Pukall (5) examined two additional strains, ATCC 9372 and ATCC 51189, and on the basis of ribotyping and DNA-DNA reassociation measurements, demonstrated the need to reclassify them as B. atrophaeus. These reclassifications have resulted in some confusion within the biodefense research community, since the taxonomic placement of other isolates used as BG remains ambiguous.
Molecular variability among members of the B. atrophaeus species has yet to be examined, and to our knowledge, no comparative analyses have been performed with an analysis or technique capable of resolving genetic variability at the strain level as well as the species level. In this study, we report the molecular characterization of several B. atrophaeus strains and other selected pigmented and nonpigmented Bacillus species using amplified fragment length polymorphism (AFLP) analysis. This highly discriminatory genetic fingerprinting technique has been evaluated against traditional methodologies as a tool for bacterial taxonomy (9) and has shown utility in detecting molecular variability in very closely related bacterial strains (6-8, 10, 11). A cluster of very closely related strains of B. atrophaeus is identified, and a proposal for a subspecies designation is made.
MATERIALS AND METHODS
Bacterial cultures.
Bacillus strains used in this study are listed in Table 1 and were purchased from the American Type Culture Collection (ATCC) (Manassas, Va.) or obtained from the Bacillus Genetic Stock Center (BGSC) (Ohio State University, Columbus). All cultures were initially recovered in nutrient broth (Difco) incubated at 30°C and then streaked for isolation on nutrient agar. Single colonies were used to inoculate nutrient broth for DNA isolation and glycerol stock production.
TABLE 1.
Bacillus strains used in this study
| Current Bacillus species | Sourcea | Strainb | Depositor designation | Other designation(s) |
|---|---|---|---|---|
| B. atrophaeus | ATCC | ATCC 49337 | Bacillus subtilis var. niger Smith et al. | NRS 213 (CCUG 28524, CIP 107159, DSM 7264, IFO 15539, JCM 9070, LMG 16797) |
| ATCC | ATCC 9372 | Bacillus subtilis var. niger Smith et al. | NRS 1221A (CIP 77.18, DSM 675, IFO 13721, IFO 16183, NCIB 8058) | |
| ATCC | ATCC 51189 | Bacillus subtilis (Ehrenberg) Cohn | NCTC 10073 (Camp Detrick, DSM 2277, NCIB 8649, NRCB-467) | |
| ATCC | ATCC 6537 | Bacillus niger Migula | ||
| ATCC | ATCC 7972 | Bacillus niger Migula | NRS 650 | |
| B. circulans | ATCC | ATCC 4516 | Bacillus globigii Kral | NRS 313 |
| B. subtilis | ATCC | ATCC 49822 | Bacillus globigii Migula | |
| ATCC | ATCC 49760 | Bacillus globigii Migula | ||
| ATCC | ATCC 31028 | Bacillus globigii Migula | FD 6404 | |
| ATCC | ATCC 6633 | Bacillus subtilis | NRS 231 (BUCSAV 425, Boots 218, CCM 1999, CIP 52.62, DSM 347, IAM 1069, JCM 2499, NCIB 8054, NCTC 10400, NCTC 6752, PCI 219, WHO 9) | |
| BGSC | PY79 | Bacillus subtilis subsp. subtilis | Bacillus 168 derivative | |
| B. pumilus | ATCC | ATCC 27142 | Bacillus pumilus Meyer and Gottheil | E601 (CIP 77.25, NCIMB 10692, NCTC 10327) |
ATCC, American Type Culture Collection, Manassas, Va.; BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus.
The B. atrophaeus type strain is ATCC 49337. The strain used as a simulant at the U.S. Army Dugway Proving Ground is B. atrophaeus 9372 commonly called BG. All the Bacillus strains except PY79, ATCC 6633, and ATCC 27142 were chosen for this study on the basis of having some indicated relationship to B. globigii or B. subtilis var. niger in catalogues, such as the ATCC catalogue.
DNA isolation.
Genomic DNA was isolated by a nonphenol methodology using a PUREGENE kit (Gentra, Minneapolis, Minn.) and the manufacturer's protocol for gram-positive bacteria. Preparations were quantitated spectrophotometrically by measuring absorbance at 260 nm and stored at −20°C until use.
16S ribosomal DNA (rDNA) sequencing.
A 500-bp fragment of the 16S rRNA gene was amplified from genomic DNA from each isolate by PCR and sequenced using the reagents and protocol supplied in the MicroSeq 500 bacterial sequencing kit (Applied Biosystems, Foster City, Calif.). The full-length 16S rRNA gene was amplified and sequenced using the MicroSeq Full Gene 16S rDNA bacterial sequencing kit (Applied Biosystems). Sequencing reaction products were analyzed with an ABI PRISM 310 genetic analyzer outfitted with a 50-cm-long capillary tube filled with POP-6 polymer (Applied Biosystems).
AFLP analysis.
Twenty nanograms of purified genomic DNA from each isolate was digested with EcoRI and MseI (New England Biolabs, Inc. Beverly, Mass.). EcoRI and MseI adaptor pairs (Applied Biosystems) were ligated to restriction fragments according to the manufacturer's protocol. The ligated DNA samples were diluted 20-fold, and 4 μl of the diluted DNA was amplified by PCR using EcoRI+0 and MseI+0 primers. The reaction mixture was diluted 20-fold, and 1.5 μl was subsequently used in a second amplification with each of 16 primer combinations (EcoRI plus G/MseI plus N, EcoRI plus C/MseI plus N, EcoRI plus G/MseI plus CN, EcoRI plus C/MseI plus CN, where N = A, T, C, or G) contained in the EcoRI/MseI AFLP microbial kit (Applied Biosystems). Products were diluted 10-fold in a solution of Gene-Scan 500 ROX Size Standard/Hi-Di formamide (Applied Biosystems) mixed at a ratio of 1:20 prior to separation and detection of fragments on an ABI PRISM 3100 genetic analyzer outfitted with an array of 36-cm-long capillaries filled with POP-4 polymer (Applied Biosystems).
Data analysis.
16S rRNA gene sequence data were analyzed using Sequencher analysis software (Gene Codes Corp., Ann Arbor, Mich.). Sequence alignments were performed using Jellyfish software version 3.0 (Lab Velocity Inc., San Francisco, Calif.). BLASTn sequence similarity searches (1) were performed on the National Center for Biotechnology Information website. AFLP data generated from the ABI PRISM 3100 genetic analyzer were analyzed using GelCompar II software package (Applied Maths, Kortrijk, Belgium). To standardize analysis of AFLP data in our laboratory using GelCompar II, several parameters were determined and subsequently used to analyze all of the data. Reproducibility and baseline variability were determined by repeating AFLP analysis with a single strain on four separate occasions. The following comparison parameters were established by determination of the minimum values for each that resulted in a 100% similarity value among the four replicates: optimization (0.2%), position tolerance (0.5%,) and change toward the end of fingerprints (0.1%). Fragment-matching analyses were performed on comparisons of profiles from all the strains selectively amplified with a particular primer set. Matched fragment classes that did not contain fragment peaks representing a minimum area of 5% of the total relative area for each fingerprint were eliminated from the comparison. A composite data set of fragment classes taken from each of the different primer set results was used to perform a cluster analysis. Dice similarity coefficients and unweighted paired group mathematical average method (UPGMA) were employed. The reliability of internal branches of the dendrogram generated by cluster analysis was measured by bootstrap analysis with 1,000 samplings.
Nucleotide sequence accession numbers.
The partial 16S rDNA sequences for strains ATCC 31028, ATCC 49760, ATCC 49822, ATCC 6537, ATCC 7972, and ATCC 51189 are deposited in GenBank under the sequential accession numbers AY379767 through AY379772.
RESULTS
16S rRNA gene sequencing.
The first 500 bp of the 16S rRNA gene was sequenced from the nine Bacillus strains listed in Table 1 deposited in the ATCC as B. globigii, B. niger, and B. subtilis var. niger. A comparison revealed that these sequences were virtually identical with the exception of Bacillus circulans strain ATCC 4516. The B. circulans sequence shared 85% identity with the type strain of B. atrophaeus, ATCC 49337. Among the other eight strains, a single base substitution was observed at position 266 using the E. coli 16S rRNA gene as a numbering reference. The substitution, a cytosine for a thymine, exists only in strain ATCC 49337. No additional variation was observed in the full-length 16S rRNA gene sequenced from strains ATCC 49337 and ATCC 9372. The full-length 16S rRNA gene was sequenced from strain PY79, a derivative of B. subtilis 168. A comparison of PY79 with ATCC 49337 sequences revealed 10 substitutions in approximately 1,400 bases or 99.3% identity with the type strain of B. atrophaeus.
Standardization of AFLP data analysis.
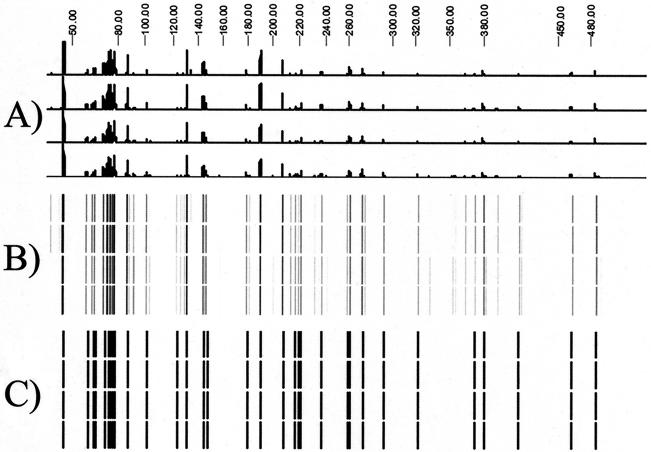
AFLP analysis was performed using genomic DNA isolated from all strains listed in Table 1 and 16 different selective primer sets. Two of the primer sets, EcoRI plus C/MseI plus CC and EcoRI plus C/MseI plus CA, yielded the best quality patterns and were selected for use in replicate AFLP runs. The best-quality pattern was defined as the selective primer sets that generated 30 to 100 DNA fragments within the size range of 35 and 500 bases and revealed the most polymorphism among the strains compared. The reproducibility of the fingerprint patterns generated from four replicate runs representing two separate DNA isolations of B. atrophaeus ATCC 49337 was greater than 98% (Fig. 1). The minimum settings for comparison parameters that resulted in a 100% similarity value among the four replicates were 0.2% lane optimization, 0.5% fragment position tolerance, and 0.1% change toward the end of the fingerprint. As shown in Fig. 1C, several less intense fragments that would have been difficult to judge were eliminated from the data set due to fragment class filtering settings.
FIG. 1.
Standardization of AFLP data. (A) Electropherogram generated by GeneScan analysis software on the ABI PRISM 3100 genetic analyzer. (B) Digitized image generated by GelCompar II software using data from the electropherogram. (C) Fragment-calling results from GelCompar II software using the parameters stated in Materials and Methods. The scale across the top of the figure represents fragment size (in bases).
Species-level variation.
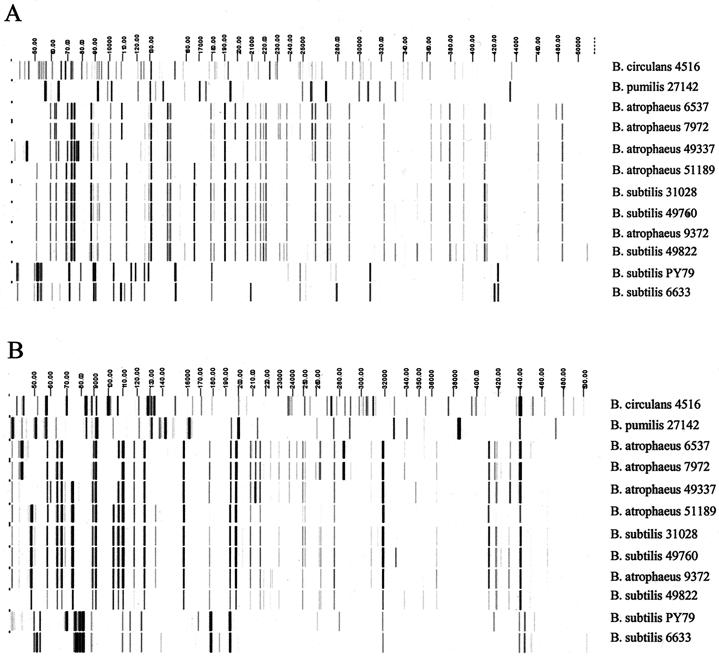
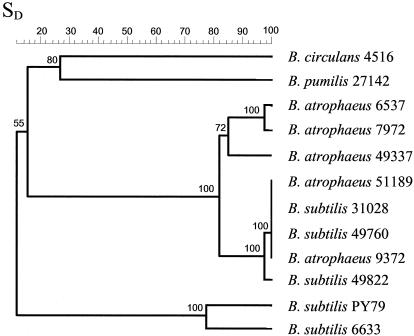
AFLP analysis using two primer set combinations generated a total of 164 scorable fragments among the taxa examined (Fig. 2). The fingerprint patterns from the various species were very different from one another, with only one fragment shared among all taxa and less than 30% similarity between members of any two species when cluster analysis was performed using Dice similarity coefficients (Fig. 3). Fingerprints from three B. subtilis strains, ATCC 49822, ATCC 31028, and ATCC 49760, were clustered with the type strain of B. atrophaeus with a similarity of 81%. These same strains were only 12% similar to B. subtilis strains BGSC PY79 and ATCC 6633, clearly indicating a much closer phylogenetic relationship to B. atrophaeus than to B. subtilis.
FIG. 2.
Digitized AFLP patterns of Bacillus taxa generated using primer sets EcoRI plus C/MseI plus CA (A) and EcoRI plus C/MseI plus CC (B). Across the top of each image is the fragment size scale (in bases). The Bacillus species and strain designations for each isolate are indicated to the right of each profile. All strains are ATCC strains, except for B. subtilis strain PY79.
FIG. 3.
UPGMA dendrogram generated from Dice similarity coefficients (SD) in percentages among Bacillus taxa based on 164 AFLP fragments. Bootstrap values calculated from 1,000 replications are shown at each internal branch. All strains are ATCC strains, except for B. subtilis strain PY79.
Strain-level variation.
AFLP fingerprinting revealed sufficient variation for use in differentiating subspecies of B. subtilis (Fig. 3). Sixteen polymorphisms were detected among 43 scorable fragments between the two strains PY79 and ATCC 6633. Cluster analysis indicated a 77% similarity between PY79 and ATCC 6633. Among the isolates that clustered with the B. atrophaeus type strain (ATCC 49337), there were a total of 72 scorable fragments including 31 polymorphisms. Cluster analysis revealed a very monomorphic group of the pigmented Bacillus strains that were nearly indistinguishable under the AFLP conditions used in this study (Fig. 3). These isolates included strains ATCC 51189 and ATCC 9372 that were recently reclassified as B. atrophaeus as well as three strains, ATCC 49822, ATCC 49760, and ATCC 31028, that were classified as B. subtilis strains in the ATCC. The AFLP fingerprints of these strains revealed 21 polymorphisms from a total of 66 scorable fragments compared to the type strain of B. atrophaeus, ATCC 49337 and cluster analysis indicated 81% similarity. Little variation was detected between strains ATCC 6537 and ATCC 7972. There were three polymorphic fragments detected in the two isolates, resulting in a 97% similarity by cluster analysis; however, there were 15 polymorphisms observed between these isolates and strain ATCC 49337 from 63 scorable fragments.
DISCUSSION
In this study, we have examined the genetic variation among isolates of B. atrophaeus and other related taxa using 16S rRNA gene sequencing and AFLP analysis. In light of the reclassification of several of these strains as B. atrophaeus, the ambiguous placement of other uncharacterized strains in use in industrial and research laboratories is an issue of concern. This is especially true in the case of interlaboratory comparisons of data in the development and validation of instrumentation or methodologies for biological warfare agent identification or decontamination.
The 16S rRNA gene sequence data proved useful in determining the proper placement of three Bacillus strains as B. atrophaeus rather than B. subtilis. The resolution of this methodology, however, was not high enough to examine genetic variability beyond the species level. In fact, even at the species level, the variability detected in 16S rDNA gene sequences of B. subtilis and B. atrophaeus is unusually low at 99.3% sequence identity. Stackebrandt and Goebel (15) have noted that the relationship observed between 16S rRNA homology and DNA-DNA reassociation is not linear and that reassociation values as low as 25% can exist between organisms with very high (99.8%) rRNA sequence homology. Interestingly, two examples can be found within the Bacillus genus where there is high 16S rRNA sequence homology, but the taxa are recognized as separate species or subspecies, namely, Bacillus anthracis, Bacillus cereus, Bacillus mycoides, and Bacillus thuringensis (2, 3, 11) and B. subtilis subsp. subtilis and B. subtilis subsp. spizizenii (13).
AFLP analysis was used successfully to discriminate between related taxa of Bacillus.
Use of two primer set combinations revealed sufficient polymorphism to easily distinguish between species. Across the different taxa, a similarity level of 30% or lower in our analysis was indicative of a difference in species. However, the utility of these data in determining meaningful phylogenetic relationships between species is less clear. Our cluster analysis indicated a closer relationship between B. atrophaeus and B. circulans than between B. atrophaeus and B. subtilis. This result is contradictory to data from a study measuring the evolutionary distance between these species using 16S rRNA sequences (2).
Three of the strains included in this study, ATCC 49760, ATCC 49822, and ATCC 31028, were clearly more closely related to B. atrophaeus than to B. subtilis and should be reclassified as members of the former species. The level of similarity between bona fide members of B. subtilis and these isolates was well below the similarity levels between the other species examined. These isolates have remained members of B. subtilis simply due to their omission from molecular characterization studies performed with other pigmented strains of B. subtilis.
AFLP analysis was useful in discriminating between members of the two recognized subspecies of B. subtilis. Cluster analysis indicated a similarity of 77% between the subspecies and provided a basis upon which the variation among B. atrophaeus strains was compared. Interestingly, a very monomorphic group of pigmented strains was revealed that was closely related to the type strain of B. atrophaeus, yet was also plainly distinct. Cluster analysis indicated a similarity level near that observed between the subspecies of B. subtilis, suggesting that the difference is significant. We propose that strains ATCC 49760, ATCC 49822, ATCC 30128, ATCC 9372, and ATCC 51189 represent members of a subspecies of B. atrophaeus genetically distinct from the type strain ATCC 49337. We also propose that the subspecies be named globigii to realign the name of this group with the historical and present use of its members throughout the biodefense research and industrial communities.
Comparison of strains ATCC 51189, ATCC 9372, and ATCC 49337 by automated ribotyping revealed nearly identical fingerprints with similarity coefficients of greater than or equal to 92% (5). Our 16S rRNA gene sequence data are consistent with this observation; however, AFLP analysis of these strains revealed significant variation between these isolates. This suggests that under the conditions reported, automated ribotyping is sufficient for determining species-level variation but does not have the resolution necessary to detect strain-level variation among members of B. atrophaeus.
Acknowledgments
We thank Paul J. Jackson, Biosciences Division, Los Alamos National Laboratory, for a critical review of the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, B. cereus, and related species on the basis of reverse transcriptase sequencing of the 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 4.de Oliveira, D. C., and J. Pinto Tde. 2002. Study of sterilizing effectivity of different ethylene oxide gaseous mixtures using CFCs and HCFCs (Oxyfume 12R and 2002R). PDA J. Pharm. Sci. Technol. 56:242-247. [PubMed] [Google Scholar]
- 5.Fritze, D., and R. Pukall. 2001. Reclassification of bioindicator strains Bacillus subtilis DSM 675 and Bacillus subtilis DSM 2277 as Bacillus atrophaeus. Int. J. Syst. Evol. Microbiol. 51:35-37. [DOI] [PubMed] [Google Scholar]
- 6.Grady, R., D. Blanc, P. Hauser, and J. Stanley. 2001. Genotyping of European isolates of methicillin-resistant Staphylococcus aureus by fluorescent amplified-fragment length polymorphism analysis (FAFLP) and pulsed-field gel electrophoresis (PFGE) typing. J. Med. Microbiol. 50:588-593. [DOI] [PubMed] [Google Scholar]
- 7.Grady, R., M. Desai, G. O'Neill, B. Cookson, and J. Stanley. 1999. Genotyping of epidemic methicillin-resistant Staphylococcus aureus phage type 15 isolates by fluorescent amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 37:3198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huys, G., R. Coopman, P. Janssen, and K. Kersters. 1996. High-resolution genotypic analysis of the genus Aeromonas by amplified fragment length polymorphism fingerprinting. Int. J. Syst. Bacteriol. 46:572-580. [DOI] [PubMed] [Google Scholar]
- 9.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as an new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 10.Janssen, P., K. Maquelin, R. Coopman, I. Tjernberg, P. Bouvet, K. Kersters, and L. Dijkshoorn. 1997. Discrimination of Acinetobacter genomic species by amplified fragment length polymorphism fingerprinting. Int. J. Syst. Bacteriol. 47:1179-1187. [DOI] [PubMed] [Google Scholar]
- 11.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura, L. K. 1989. Taxonomic relationship of black-pigmented Bacillus subtilis strains and a proposal for Bacillus atrophaeus sp. nov. Int. J. Syst. Bacteriol. 39:295-300. [Google Scholar]
- 13.Nakamura, L. K., M. S. Roberts, and F. M. Cohan. 1999. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 49:1211-1215. [DOI] [PubMed] [Google Scholar]
- 14.Penna, T. C., P. G. Mazzola, and A. M. Silva Martins. 2001. The efficacy of chemical agents in cleaning and disinfection programs. BMC Infect. Dis. 1:16. [Online.] http://www.biomedcentral.com/1471-2334/1/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 16.Stratis-Cullum, D. N., G. D. Griffin, J. Mobley, A. A. Vass, and T. Vo-Dinh. 2003. A miniature biochip system for detection of aerosolized Bacillus globigii spores. Anal. Chem. 75:275-280. [DOI] [PubMed] [Google Scholar]
- 17.Turnbough, C. L. 2003. Discovery of phage display peptide ligands for species-specific detection of Bacillus spores. J. Microbiol. Methods 53:263-271. [DOI] [PubMed] [Google Scholar]
- 18.U.S. Pharmacopeia. 2000. Biological indicator for ethylene oxide sterilization, paper strip, p. 231-232. In The United States Pharmacopeia/The National Formulary, USP 24-NF19. U.S. Pharmacopeia, Rockville, Md.