Abstract
In this study, the chloroplast (cp) genome sequences from three early diverged leptosporangiate ferns were completed and analyzed in order to understand the evolution of the genome of the fern lineages. The complete cp genome sequence of Osmunda cinnamomea (Osmundales) was 142,812 base pairs (bp). The cp genome structure was similar to that of eusporangiate ferns. The gene/intron losses that frequently occurred in the cp genome of leptosporangiate ferns were not found in the cp genome of O. cinnamomea. In addition, putative RNA editing sites in the cp genome were rare in O. cinnamomea, even though the sites were frequently predicted to be present in leptosporangiate ferns. The complete cp genome sequence of Diplopterygium glaucum (Gleicheniales) was 151,007 bp and has a 9.7 kb inversion between the trnL-CAA and trnV-GCA genes when compared to O. cinnamomea. Several repeated sequences were detected around the inversion break points. The complete cp genome sequence of Lygodium japonicum (Schizaeales) was 157,142 bp and a deletion of the rpoC1 intron was detected. This intron loss was shared by all of the studied species of the genus Lygodium. The GC contents and the effective numbers of co-dons (ENCs) in ferns varied significantly when compared to seed plants. The ENC values of the early diverged leptosporangiate ferns showed intermediate levels between eusporangiate and core leptosporangiate ferns. However, our phylogenetic tree based on all of the cp gene sequences clearly indicated that the cp genome similarity between O. cinnamomea (Osmundales) and eusporangiate ferns are symplesiomorphies, rather than synapomorphies. Therefore, our data is in agreement with the view that Osmundales is a distinct early diverged lineage in the leptosporangiate ferns.
Keywords: chloroplast genomes, Diplopterygium glaucum, early diverged leptosporangiate ferns, inversion mutations, Lygodium japonicum, Osmunda cinnamomea, rpoC1 intron loss
INTRODUCTION
Comparative chloroplast (cp) genomic studies provide an invaluable source of information for understanding plant evolution and plant phylogeny. Therefore, the cp genome is the most widely studied genome when compared to the two other genomes found in plant cells. Approximately 400 cp genome sequences for land plants are available from a public database, but the majority of them belonged to seed plants (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?opt=plastid&taxid=3193). The cp genomes hold numerous important evolutionary features, including structural changes, gene content differences, and base substitution patterns.
Structural changes in the cp genome, such as gene rearrangements (Chumley et al., 2006; Tangphatsornruang et al., 2010; Wu et al., 2007), gene/intron losses or duplications (Guisinger et al., 2011; Hiratsuka et al., 1989; Jansen et al., 2007), and small inversions (Kim and Lee, 2004; Yi and Kim, 2012) are well known at the genus, family, or ordinal levels of seed plants. Therefore, the genome evolution and phylogenetic relationships of seed plants are relatively well understood. However, the cp genome studies in ferns are limited to just a few lineages.
One of the distinct features of cp genomes is its high levels of adenosine and thiamine (AT) content (Sablok et al., 2011; Smith, 2009). However, a relatively wide range of AT content variation was reported for a number of different plant lineages (Smith, 2009). The GC content differences in the cp genomes usually correlate well with codon usage bias. The effective number of codons (ENCs) represents a simple way to measure synonymous codon usage bias and is independent of coding region length and amino acid composition (Wright, 1990). Therefore, the comparative ENC values may show a broad spectrum of base usage patterns among major lineages of plant groups.
Ferns are an important plant group for the understanding plant evolution because of the long evolutionary history and the complicated phylogenetic relationships (Pryer et al., 2004). The extant ferns are composed of one monophyletic class and 11 monophyletic orders (Pryer et al., 2009). Since the physical map of the Osmunda cinnamomea cp genome was known (Palmer and Stein, 1982; 1986), many researchers tried to understand the ferns cp genome evolution. Recently, the complete cp genome sequences of four orders of eusporangiate ferns were analyzed, and the data aided in understanding the evolutionary history of eusporangiate ferns (Grewe et al., 2013; Karol et al., 2010). In leptosporangiate ferns, the six complete cp genome sequences have been reported. These are Alsophila spinulosa (Gao et al., 2009), Adiantum capillus-veneris (Wolf et al., 2003), Cheilanthes lindheimeri, Pteridium aquilinum subsp. aquilinum (Wolf et al., 2011), Lygodium japonicum, and Marsilea crenata (Gao et al., 2013). In addition, the incomplete sequences were also used for the reconstruction of the fern trees (Wolf et al., 2010). Although the comparative cp genome studies of eusporangiate ferns and leptosporangiate ferns were published, it is still difficult to understand cp genome evolution from all fern lineages because there is a lack of cp genome data for the early diverged fern lineages. This appears as missing links in data.
In order to provide the data in the missing lineages, we report three complete cp genome sequences from the early diverged leptosporangiate ferns in this paper. Two are newly reported groups (Osmundales and Gelicheniales) and one (Schizaeales) is a previously reported group. Using these data, we address the following two questions about cp genome evolution of early diverged leptosporangiate ferns: (i) which of the cp genome structures are more similar to that of basal Osmundales, and (ii) whether or not Osmundales really have an intermediate-type cp genome that is between eusporangiate and leptosporangiate ferns.
Osmundales consists of a monophyletic family, three genera, and ca. 20 species (Smith et al., 2006), but it includes more than 150 fossil species (Tidwell and Ash, 1994). Many researchers consider the Osmundales to be closely related to eusporangiate ferns (Pryer et al., 2001; 2004; Schneider et al., 2004; Schuettpelz and Pryer, 2007; Wolf et al., 1995). Osmun-dales also have been considered as intermediate taxa between eusporangiate and leptosporangiate ferns based on their external appearance, and anatomical and meristem characteristics (Cross, 1931a; 1931b; Freeberg and Gifford Jr, 1984; Gifford Jr, 1983). Using fossil records, Osmundaceae could be traced back to the Late Permian period, but the genus Osmunda was known from the Late Triassic period. O. cinnamomea seemed to exist since the Late Cretaceous period (Taylor et al., 2009) and was identified as a sister species to the rest of the Osmundaceae (Metzgar et al., 2008; Yatabe et al., 1999). Considering the morphological characters and phylogenetic relationship of ferns, the cp genome of O. cinnamomea could be regarded as an ancestral type from leptosporangiate ferns.
Gleicheniales consists of three families, 10 genera, and ca. 140 species, and most of the species are members of Gleicheniaceae (Smith et al., 2006). Gleicheniaceae is considered as an old lineage originating from the Permian (Pryer et al., 2004; Taylor et al., 2009). We report the cp genome sequence of Diplopterygium glaucum, which was a synonym of Gleichenia japonica (Iwatsuki et al., 1995), and was widely distributed in the Asian tropics.
Schizaeales consists of three families, four genera, and ca. 155 species (Smith et al., 2006). The oldest Schizaeaceae fossil originated from the Jurassic period (Taylor et al., 2009), and Schizaeales diverged from the core leptosporangiate ferns in the Permian (Pryer et al., 2004). The genus Lygodium is considered as a basal group in the Schizaeales (Schuettpelz and Pryer, 2007), and the cp genome of Lygodium japonicum had two large inversions and gene deletions (Gao et al., 2013; Wolf et al., 2010). We report the cp genome sequences of a Korean population of L. japonicum.
In this study, the complete cp genome sequences of O. cinnamomea and D. glaucum filled the evolutionary gap between eusporangiate and leptosporangiate ferns and gave us information, such as gene/intron losses, inversions, codon usage bias, and patterns of repeating units in early diverged leptosporangiate ferns, thus allowing us to understand cp genome evolution in these interesting phylogenetic groups. In addition, knowing the complete cp genome sequence of L. japonicum is helpful to understanding the cp genome evolution between Chinese and Korean populations.
MATERIALS AND METHODS
DNA extraction, sequencing, and assembling
O. cinnamomea (KUS 2006-0338), D. glaucum (KUS 2000-0022), and L. japonicum (KUS 2007-0451) were collected in Korea. All voucher specimens were kept in Korea University’s Herbarium (KUS). The genomic DNAs of O. cinnamomea and D. glaucum were isolated from the fresh leaves by the CTAB method (Doyle and Doyle, 1987) and purified by ultracentrifugation in cesium chloride/ethidium bromide gradients. We designed common primer sets based on the cp genome sequence in ferns using known complete cp genome sequences. Using the primers, we amplified the 5–10 kb overlapping cp genome fragments using TaKaRa LA Taq by PCR. PCR products were sequenced by Big-Dye chemistry and ABI3730XL. For L. japonicum, the chloroplasts were isolated by sucrose step-gradient methods (Palmer, 1986), and the cp genome was isolated using 5x lysis buffer (Jansen et al., 2005). The cp genome of L. japonicum was sequenced using GS-FLX 454 at Macrogen Co. (Korea). A total of 108,241 sequence reads were generated. A total of 4,430 reads were fully incorporated into the assembly and 2,595 reads were partially incorporated. There were 400 contigs over 500 bp, and the largest contig was 13,610 bp. Gaps were filled by PCR. All sequenced contigs were de novo assembled using Geneious 6.1.2 (Kearse et al., 2012). Gene annotations were performed by DOGMA (Wyman et al., 2004) and tRNAscan (Lowe and Eddy, 1997). Then, the exact positions of all genes were determined by local BLAST searches using the gene database of ferns obtained from NCBI.
Phylogenetic analyses and comparative sequence analyses of cp genomes
Thirty-five cp genome sequences were used for the phylogenetic analysis (Table 1). We sampled all of the published complete cp genome sequences from monilophytes (14), lycophytes (4), and bryophytes (5), and eight selected species from spermatophytes. Two charophytes were included as out-groups. In addition, two unpublished monilophytes sequences were also included in these taxon samplings (H.-T. Kim and K.-J. Kim, unpublished data). Eighty-nine genes, including 84 protein coding genes and five ribosomal RNA genes, were aligned using MUSCLE program (Edgar, 2004), and the phylogenetic trees were constructed using four different tree building methods. First, the maximum parsimony (MP) tree was generated by PAUP (Swofford, 2003) under the options of equal character weighting, random taxon addition, and TBR branch swapping options. Gaps were treated as missing. Second, the neighbor joining (NJ) tree was generated with Geneious 6.1.7 using the HKY genetic distance model. Third, for the maximum likelihood (ML) tree, we selected the optimal model with Modeltest 3.7 (Posada and Crandall, 1998). The ML tree was evaluated by the GTR + I + G model using RAxML (Stamatakis, 2006; Stamatakis et al., 2008) that is performed using the CIPRES Science Gateway (Miller et al., 2010). The strengths of all of the internal branches in MP, NJ, and ML analyses were evaluated by 1,000 bootstrap replications. Fourth, the Bayesian inference (BI) tree was reconstructed by Mrbayes under the following conditions: nst = 6, rates = invgamma, Ngen = 500,000 and samplef = 100, using the CIPRES Science Gateway (Miller et al., 2010).
Table 1.
The list of complete chloroplast genome sequences and rpoC1 sequences
| Target | Taxa | Group | GenBank |
|---|---|---|---|
| Phylogenetic analysis | Arabidopsis thaliana | Spermatophytes | NC000932 |
| Panax chinseng | Spermatophytes | NC006290 | |
| Nymphaea alba | Spermatophytes | NC006050 | |
| Amborella trichopoda | Spermatophytes | NC005086 | |
| Cycas revoluta | Spermatophytes | NC020319 | |
| Ginkgo biloba | Spermatophytes | NC016986 | |
| Welwitschia mirabilis | Spermatophytes | NC010654 | |
| Pinus koraiensis | Spermatophytes | NC004677 | |
| Adiantum capillus-veneris | Polypodiales(core leptosporangiate ferns) | NC004766 | |
| Cheilanthes lindheimeri | Polypodiales(core leptosporangiate ferns) | NC014592 | |
| Pteridium aquilinum subsp. aquilinum | Polypodiales(core leptosporangiate ferns) | NC014348 | |
| Alsophila spinulosa | Cyatheales(core leptosporangiate ferns) | NC012818 | |
| Marsilea crenata | Salviniales(core leptosporangiate ferns) | KC536646 | |
| Lygodium japonicum (K.-J. Kim et al. KUS 2006-0338) | Schizaeales (early diverged leptosporangiate ferns) | KF225593* | |
| Diplopterygium glaucum (C.-H. Kim et al. KUS 2000-0022) | Gleicheniales (early diverged leptosporangiate ferns) | KF225594* | |
| Osmunda cinnamomea (H.-W. Kim et al. KUS 2007-0451) | Osmundales (early diverged leptosporangiate ferns) | KF225592* | |
| Angiopteris evecta | Marattiales (eusporangiate ferns) | NC008829 | |
| Ophioglossum californicum | Ophioglossales (eusporangiate ferns) | NC020147 | |
| Mankyua chejuensis | Ophioglossales (eusporangiate ferns) | NC017006 | |
| Psilotum nudum 1 | Psilotales (eusporangiate ferns) | NC003386 | |
| Psilotum nudum 2 | Psilotales (eusporangiate ferns) | KC117179 | |
| Equisetum arvense 1 | Equisetales (eusporangiate ferns) | NC014699 | |
| Equisetum arvense 2 | Equisetales (eusporangiate ferns) | JN968380 | |
| Equisetum hyemale | Equisetales (eusporangiate ferns) | NC020146 | |
| Huperzia lucidula | Lycophytes | NC006861 | |
| Isoetes flaccida | Lycophytes | NC014675 | |
| Selaginella moellendorffii | Lycophytes | NC013086 | |
| Selaginella uncinata | Lycophytes | AB197035 | |
| Anthoceros formosae | Bryophytes | NC004543 | |
| Syntrichia ruralis | Bryophytes | NC012052 | |
| Physcomitrella patens subsp. patens | Bryophytes | NC005087 | |
| Marchantia polymorpha | Bryophytes | NC001319 | |
| Aneura mirabilis | Bryophytes | NC010359 | |
| Chaetosphaeridium globosum | Charophytes | NC004115 | |
| Chara vulgaris | Charophytes | NC008097 | |
| rpoC1 intron analysis | Lygodium japonicum (K.-J. Kim et al. TC 2010-0136) | Schizaeales (early diverged leptosporangiate ferns) | KF225595* |
| Lygodium polystachyum (K.-J. Kim et al. TL 2008-1701) | Schizaeales (early diverged leptosporangiate ferns) | KF225596* | |
| Vandenboschia striata (K.-J. Kim et al. TC 2010-1362) | Hymenophyllales (early diverged leptosporangiate ferns) | KF225597* | |
| Lygodium flexuosum (K.-J. Kim et al. TL 2008-1886) | Schizaeales (early diverged leptosporangiate ferns) | Not sequenced | |
| Lygodium microphyllum (K.-J. Kim et al. TL 2008-1661) | Schizaeales (early diverged leptosporangiate ferns) | Not sequenced | |
| Lygodium salicifolium (K.-J. Kim et al. TL 2008-1772) | Schizaeales (early diverged leptosporangiate ferns) | Not sequenced |
Asterisk on the GenBank accession numbers indicate newly reported sequences in this paper.
The cp genomes modifications, such as gene/intron gains or losses, inversion events, and the anticodon changes, were treated as binary characters. A total of 30 variable evolutionary events were recorded from the fern lineages. Next, the character states were plotted on the ML tree topology in order to deduce the evolutionary direction of these characteristics. The evolutionary directions were accounted on the ACCTRAN criteria on the parsimony analysis using PAUP (Swofford, 2003).
The complete cp genome sequences of 194 species of land plants were used to analyze the GC contents of coding sequences in the cp genome (Supplementary Table 1). All cp genome sequences were obtained from NCBI Organelle Genome Resources. The GC contents of the entire coding gene (GCall), first position (GC1), second position (GC2), and third position (GC3), and the effective numbers of codons (ENCs) (Wright, 1990) were calculated using Acua 1.0 (Vetrivel et al., 2007). We also analyzed dispersed repeats using REPuter (Kurtz et al., 2001). Then, each repeat sequence was sorted by similarity. These repeat sequences were reanalyzed using a DNA pattern search (http://www.geneinfinity.org/sms/sms_DNApatterns.html#) for calculating the exact numbers of repeat sequences in the complete cp genome (Kim et al., 2009; Yi et al., 2012).
Analysis of rpoC1 intron loss
Five species of the genus Lygodium and one accession of Vandenboschia striata were collected from Laos, China and Korea for rpoC1 intron loss analysis. All specimens were kept in the KUS (Table 1). Their genomic DNAs were obtained using the same method as described for O. cinnamomea and D. glaucum. The primer set was designed based on the consensus rpoC1 sequence among 15 fern cp genomes. The forward primer ‘FrpoC1 exon1’ (5′-GAAAGCCYAGTVTATTGCGA-3′) was located at the end of exon1, and the reverse primer ‘FrpoC1 exon2’ (5′-ATGCARACGAATRGCRCGTCC-3′) was in the middle of exon2 in rpoC1. We amplified and sequenced the region. The exon/intron of rpoC1 was annotated by DOGMA, and the rpoC1 partial sequences were aligned with 14 full fern rpoC1 sequences by MUSCLE alignment program (Edgar, 2004). We also amplified and sequenced the region for L. japonicum as a reference, even though the species was subjected to the completed sequencing of the cp genome.
RESULTS
Phylogenetic analysis of ferns and related groups
The aligned sequences of 89 cp genes from 35 taxa consisted of 94,790 bp. Among them, 31,312 sites (33.0%) were constant, 10,740 sites (11.3%) were parsimony-uninformative, and 52,738 sites (55.7%) were parsimony-informative. Figure 1 shows the ML tree topology with ML and MP bootstrapping support values and Bayesian probability. The MP, ML, NJ, and BI analyses showed largely concordant tree topologies, except on the two nodes leading to lycophytes and Equisetales. First, the lycophytes was a sister group to the euphyllophytes (spermatophytes + monilophytes) in the ML and BI trees (Fig. 1A). However, the lycophytes was a sister group to the spermatophytes in the MP and NJ trees (Fig. 1B). In addition, ML and MP boot strap values prefer to the lycophytes + spermatophytes clade. The ML values between two topologies are not significantly different for this large data set (LM = −1,285,886 versus LM = −1,285,889). Second, the Equisetales was a sister group to all other members of monilophytes in ML, MP, and NJ trees, as shown in Fig. 1A. In addition, the bootstrap values of ML, NJ, and MP analyses prefer to the Equisetales + other members of monilophytes clade. However, only the BI tree prefer to the Equisetales + (Psilotales + Ophioglossales) clade, as shown in Fig. 1C. The ML values between two topologies are not significantly different for this large data set (LM = −1,285,886 vs LM = −1,285,874). We also constraint to the alternative tree topologies on both of the nodes to be the lycophytes + spermatophytes clade and the Equisetales + (Psilotales + Ophioglos-sales) clade, as shown in Fig. 1D. The LM values increased from −1,285,855 to −1,285,921 in the analysis
Fig. 1.
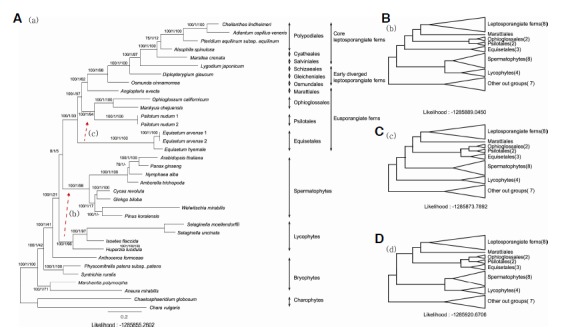
Phylogenetic tree of ferns and related groups. The best ML tree was constructed using RAxML under the GTR + I + G base substitution model (A). The three consecutive numeric values on each internal node in (A) tree indicate the ML bootstrap support percentage, Bayesian probability, and MP bootstrap support percentage, respectively. Two alternative suboptimal tree topologies (B, C) that are observed frequently in MP, BI, and NJ analyses, were also generated using the topology constraint analyses option of RaxML using the same base substitution assumption. The lycophytes was a sister group of the euphyllophytes (spermatophytes + monilophytes) in the (A) tree, while the lycophytes was a sister group to the spermatophytes in the (B) tree. The Equisetales was a sister group to all other members of monilophytes in the (a) tree, while the Equisetales was a sister clade to the Psiotales and Ophioglossales in the (C) tree. The (D) tree topology represents the combination of the topology of the (B) and (C) trees.
Comparison of the cp genome structures among three leptosporangiate ferns
The physical maps of cp genomes from three early diverged leptosporangiate ferns are shown in Fig. 2, and the three newly completed sequences were deposited in the NCBI database under the Nos. KF 225592–225594. The O. cinnamomea cp genome sequence was 142,812 bp in length. Large single copy (LSC), small single copy (SSC), and inverted repeats (IRs) were 100,294 bp, 22,300 bp, and 10,109 bp, respectively (Table 2). The gene orders and IR-LSC boundaries of O. cinnamomea are similar to that of Equisetum arvense. The D. glaucum cp genome sequence was 151,007 bp in length (KF 225594) and consisted of an LSC (99,857 bp), SSC (21,982 bp), and two IRs (14,584 bp). The D. glaucum cp genome had a 9.7 kb inversion between the trnL-CAA and trnV-GCA when compare to O. cinnamomea. The ndhB exon2 and trnL-CAA was duplicated in the IR region, and trnV-GCA moved to the LSC from the IR. The L. japonicum cp genome sequence was 157,142 bp (KF 225593) and it was composed of an LSC (85,432 bp), SSC (21,634 bp), and two IRs (25,038 bp).
Fig. 2.
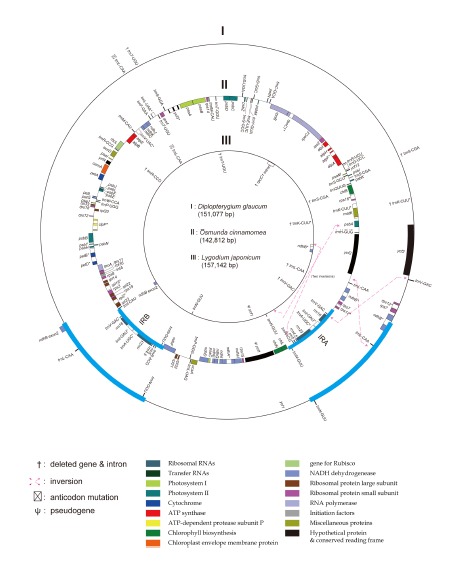
The cp gene maps of three early diverged leptosporangiate ferns. When compared to the middle circle of Osmunda cinnamomea (Osmundales), the differences of the gene orders and gene/intron contents are marked on the outer circle for Diplopterygium glaucum (Gleicheniales) and on the inner circle for Lycodium japonicum (Schizaeales). The red broken arrows indicate the inversion mutations when compare to the O. cinnamomea. The IR region (blue line) is short in O. cinnamomea and shows gene arrangements similar to that of eusporangiate ferns. Gene names with asterisk(s) indicate on or two intron containing gene.
Table 2.
The length of quadripartite chloroplast genome of three early diverged leptosporangiate ferns
| Taxa | LSC(bp) | IR(bp) | SSC(bp) | Total(bp) |
|---|---|---|---|---|
| Lygodium japonicum | 85432 | 25038 | 21634 | 157142 |
| Diplopterygium glaucum | 99857 | 14584 | 21982 | 151007 |
| Osmunda cinnamomea | 100294 | 10109 | 22300 | 142812 |
Comparison of the gene/intron contents among leptosporangiate ferns
A total of 130 genes were identified in the O. cinnamomea cp genome, and they consisted of 84 protein-coding genes, eight rRNA genes, and 38 tRNA genes (Supplementary Table 2). Among them, four rRNA and five tRNA genes were duplicated in two IR regions. The anticodon sequence of the trnK gene was changed from UUU to CUU. In addition, the ycf1 gene was a pseudogene because of a frameshift mutation. Five genes had alternative start codons, such as ACG or GTG (Table 3). A total of 128 genes were identified in the cp genome of D. glaucum. The genes consisted of 85 protein-coding genes, eight rRNA genes, and 35 tRNA genes. Four rRNA and five tRNA genes were duplicated in two IRs. The trnS-CGA, trnT-UGU, and trnK-UUU genes were lost, and the anticodon sequence of the trnL gene between trnF and rps4 was changed from UAA to CAA. Thirty three genes had internal stop codons, and 19 genes had alternative start codons (Table 3).
Table 3.
Potential and detected RNA editing sites in chloroplast genome of ferns
| Group | Taxon | No. of alternative start codons | No. of genes with internal stop codons | Maximum no. of internal stop codons in gene | No. of RNA editing sitesa |
|---|---|---|---|---|---|
| Core leptosporangiate ferns | Adiantum capillus-veneris | 25 | 18 | 4 | 349 |
| Cheilanthes lindheimeri | 26 | 22 | 4 | - | |
| Pteridium aquilinum subsp. Aquilinum | 29 | 25 | 8 | - | |
| Alsophila spinulosa | 22 | 30 | 10 | - | |
| Marsilea crenata | 28 | 21 | 3 | - | |
| Early diverged leptosporangiate ferns | Lygodium japonicum* | 21 | 17 | 2 | - |
| Diplopterygium glaucum* | 19 | 33 | 15 | - | |
| Osmunda cinnamomea* | 5 | 0 | 0 | - | |
| Eusporangiate ferns | Angiopteris evecta | 1 | 0 | 0 | - |
| Psilotum nudum | 0 | 0 | 0 | - | |
| Ophioglossum californicum | 7 | 1 | 1 | ||
| Mankyua chejuensis | 7 | 3 | 1 | - | |
| Equisetum arvense | 2 | 0 | 0 | - | |
| Equisetum hyemale | 1 | 0 | 0 | - |
The numbers of RNA editing sites were reported by Wolf et al. (2004).
Patterns of GC contents and ENCs in early diverged leptosporangiate ferns
We compared the GC contents of the cp genome coding sequence of bryophytes (8 spp.), Lycopodiopsida (4 spp.), Polypodiopsida (14 spp.), gymnosperms (26 spp.), and angiosperms (142 spp.; Supplementary Table 1). The GC content ranged from 29.5 to 39.2% in bryophytes, from 36.8 to 54.4% in Lycopodiopsida, from 33.6 to 42.4% in Polypodiopsida, from 35.1 to 40.0% in gymnosperms, and from 34.4 to 41.3% in angiosperms. We analyzed the GC content for each codon position and the ENCs using a box plot for each taxonomic group. In the GC position-plot, almost all data were distributed near the regression line, and the slope of GC3 was twice as high as the slope of GC1 and GC2 (Fig. 3A). The GC3 showed wider variation than the GC1 and the GC2 (Fig. 3B). The median value of GC3 showed a little variation among seed plants. However, the range of GC3 in Polypodiopsida showed substantial variation. The GC3 value seemed to increase from eusporangiate ferns to leptosporangiate ferns. The ENCs showed a similar distribution pattern when compare to the GC3 values. The ENCs of seed plants were concentrated between 45 and 50, but the ENCs of Polypodiopsida ranged from 41 to 54 (Fig. 3B).
Fig. 3.
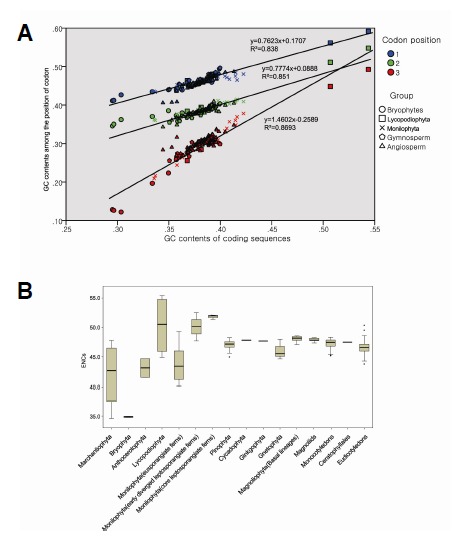
GC contents and the effective numbers of codons (ENCs). (A) The scatter diagram of the GC1, GC2, and GC3 against all GC contents. The regression lines and the equations are shown on the diagram. First (blue), second (green), and third (red) codon positions, and the taxonomic groups are distinguished by colors and symbols (five different symbols). (B) The boxplot of the GC contents by codon position (the upper diagram) and the ENCs (the lower diagram) for taxonomic groups. Seed plant lineages usually show small ranges of variation, while the fern lineages, especially the fern allies, show a wide range of variation both in GC contents and ENCs. The core leptosporangiate ferns show higher ENCs than early diverged leptosporangiate ferns.
Repeat sequences in the cp genomes of early diverged leptosporangiate ferns
The D. glaucum cp genome contained more than 100 dispersed repeat sequences. Most of them were located around tRNA genes, especially between trnL-CAA and rrn16. The cp genome of D. glaucum had a 9.7 kb inversion mutation between trnL-CAA and trnV-GAC. As a result, the position of trnL-CAA was moved to the IR region near the rrn16 gene. The repeat sequences between trnL-CAA and rrn16 usually had a repeating backbone sequence of GGAC-NNNN-AATCC. The sequence was repeated 43 times in the intergenic spacer (IGS) region between trnL-CAA and rrn16, and 31 of them were AGGAC-NNN-AAATCCT. The 15 bp sequence was similar to the tRNA anticodon loop sequences of trnF-GAA and trnC-GCA (Fig. 4). The first 5 bp and the last 7 bp in the sequence were conserved, and the middle 3 bp sequence were variable. The IGS region between trnL-CAA and ndhB also contained many repeat sequences. TCNATGTAGAAA was repeated 10 times, and GAAATAGTAGGGGTTGACATT was repeated four times in this IGS. The O. cinnamomea cp genome contained fewer repeat sequences than D. glaucum. GGAC-NNNN-AATCC was detected 11 times, with six of them located between trnL-CAA and ycf2, but AGGAC-NNN-AAATCCT was not repeated in D. glaucum. The L. japonicum cp genome contained the smallest number of repeat sequence. The GGAC-NNNN-AATCC sequence was repeated six times, with four of the repeats being located between the trnR-ACG and ndhB genes. AGGACNNN-AAATCCT was repeated four times and three of them located between the trnR-ACG and ndhB genes. Tandem repeats over 20 bp in length were also detected in all three investigated species. The D. glaucum and L. japonicum cp genomes contained only AT dinucleotide repeats, while the O. cinnamomea cp genome contained di-, hexa-, 7-, 12-, and 17-nucleotide repeats.
Fig. 4.
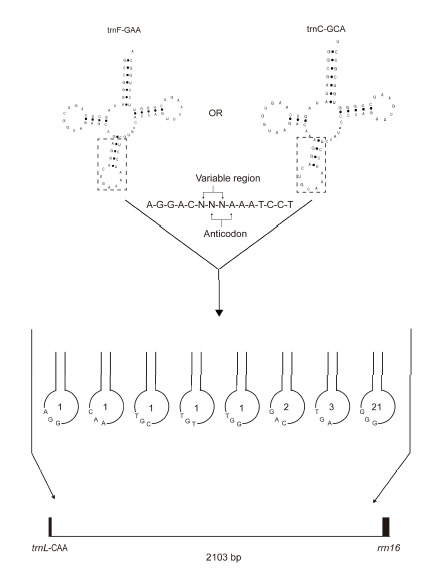
The IGS region between trnL and rrn16 in D. glaucum. A 15 bp sequence is repeated 31 times, and the repeating unit is similar to the anticodon loop sequences of tRNA. The number in the circles indicates the number of duplications in each repeating unit.
Intraspecific cp genome difference of L. japonicum and rpoC1 intron loss in the genus Lygodium
The L. japonicum cp genome from a Korean population was 118 bp shorter than the Chinese population (KC 536645). The difference between two populations were due to tandem repeats between psbM and trnD-GUC in LSC and between ndhB and trnR-ACG in IR. A total of 19 single nucleotide polymorphisms (SNPs) were detected between the two populations. In addition, one small inversion was detected between rpl32 and trnP-GGG.
RpoC1 intron loss was described in the L. japonicum cp genome (Gao et al., 2013). We extended the intron survey of rpoC1 to other species of Lygodium (Schizaeales) and Vandenboschia (Hymenophyllaes) using PCR and sequencing methods. The amplified rpoC1 sequences from five species of the genus Lygodium were about 960 bp, but it was over 1.5 kb in Vandenboschia. The amplified region included a whole rpoC1 intron and the 5′ end of rpoC1 exon 2. We generated the sequences from all amplified products. However, background noise and double peaks at both ends were present in three sequences of the genus Lygodium, so these sequences were excluded from further analyses. Therefore, the sequences from L. japonicum (China), L. poystachyum, and V. striata were aligned with the 14 complete cp genome sequences from ferns (Fig. 5). All surveyed species of the genus Lygodium lost the rpoC1 intron, while all other fern lineages have the rpoC1 intron.
Fig. 5.
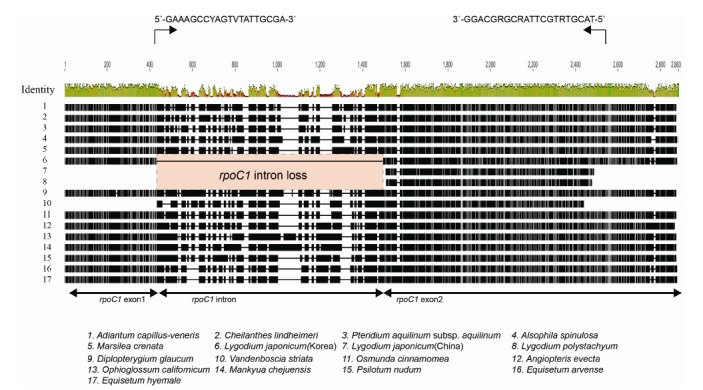
Alignments of rpoC1 gene regions in ferns. We designed a primer set at the end of exon 1 and in the middle of exon 2 (indicated by black arrows) in order to test the presence or the absence of the rpoC1 intron. The intron is lost only in the genus Lygodium. All eusporangite ferns and all other leptosporangiate ferns contained the intron.
DISCUSSION
Phylogenetic relationship of monilophytes
Monilophytes consist of four orders of eusporangiate ferns and a clade of leptosporangiate ferns (Smith et al., 2006). The phylogenetic relationships among the four eusporangiate ferns were uncertain even though most of the recent data indicate they are paraphyletic assemblages. Specifically, the phylogenetic position of Equisetales is largely different among the data sets, and this relationship remains to be resolved (Karol et al., 2010; Pryer et al., 2001). Our phylogenetic tree developed based on the most comprehensive cp genome data so far placed the Equisetales at the most basal position among the members of monilophytes, even though they did not show a large ML value difference (Fig. 1). The branch length leading to the Equisetum is relatively long when compared to other monilophyte lineages, and it is comparable with the branch lengths of Selaginella and Welwitschia. In contrast, several recent molecular phylogenetic analyses based on cp gene sequences placed the Equisetales either as a sister group to marattioid ferns (Pryer et al., 2001; 2004) or as a sister group to Psilotales (Karol et al., 2010). We believe that the tree differences may due to the taxon sampling for the long branched taxa. The Equisetum lineages were recognized as a distinct phylum for a long time because of the distinct morphological characteristics of both sporophytes and gametophytes (Bold et al., 1987). Their ancestry shows a long evolutionary history that dates back to the Devonian period, and they flourished during the Carboniferous periods in fossil history (Good and Taylor, 1975; Schweitzer, 1972). The Equisetum is the only extant lineages of the group. Addition of more complete cp genome sequences from eusporangiate ferns and basal leptosporangiate ferns will be helpful to resolve the different phylogenetic hypotheses. Our data support the monophyly of leptosporangiate ferns. We added two new complete cp genome sequences from early diverged leptosporangiate ferns: Ophioglossum (Ophioglossales) and Diplopterygium (Gleicheniales). Osmundales was the first diverged leptosporangiate fern in the tree, and this result concordant to the previous studies (Pryer et al., 2004; Schuettpelz and Pryer, 2007). In addition, Geicheniales was placed between Osmundales and Schizaeales. The addition of two early diverged leptosporangiate ferns helped us to understand not only cp genome evolution, but also the evolution of leptosporangiate fern groups. Our phylogenetic analyses also suggest that two alternative positions of Lycophytes, either as a sister group to euphyllophytes or as a sister group to spermatophytes, are not significantly different in terms of ML values and other support values.
Cp genome structure evolution in early diverged leptosporangiate ferns
Several cp genome structural modifications, including gene/intron loss and inversion, have been reported for various ferns (Gao et al., 2009; 2013; Hasebe and Iwatsuki, 1990; Wolf et al., 2003; 2010). However, most of these studies were focused on the core leptosporangiate ferns. The complete cp DNA sequences from three early diverged leptosporangiate ferns provide us with new information on the evolution of the cp genome and the phylogenetic relationships of ferns. Figure 6 shows the genome evolutionary history on the phylogenetic tree. The coding gene losses occurred mainly for eusporangiate ferns. In contrast, the tRNA gene losses and anticodon substitutions usually occur on non-Osmundalean leptosporangiate ferns. Large inversions among IR-LSC are characteristic of the early diverged leptosporangiate ferns.
Fig. 6.
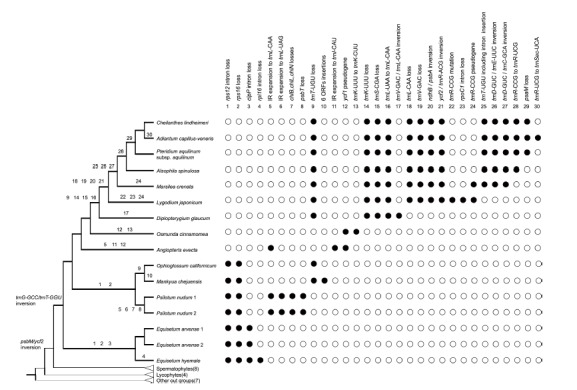
The major evolutionary changes of cp genomes among fern lineages. The gene/intron gains or losses, inversion events, and the anti-codon changes were plotted on the abbreviated phylogenetic tree of ferns as shown on Fig. 1A. The evolutionary events were accounted for using the ACCTRAN criteria during the parsimony analysis using MacClade program. The solid and empty circles indicate the presence and the absence of each character states, respectively.
The inversion between trnL-CAA and trnV-GCA in the cp genome of D. glaucum is interesting. Due to this inversion, the position of trnL-CAA is moved from the LSC to an IR. In addition, many repeat sequences are located in the IGS between rrn16 and trnL-CAA. The repeating AGGAC-NNN-AAATCCT backbone sequence makes up the repeating sequences within this region. The repeat sequence has also been detected in L. japonicum. This 15 bp backbone sequence was also reported in the IGS between trnT-UGU and trnR-ACG in Alsophila spinulosa (Gao et al., 2009). However, the sequence was not detected in other core leptosporangiate ferns. Therefore, we hypothesize that the 15 bp repeat occurred widely in early diverged leptosporangiate ferns and then the repeat sequences were degraded due to slipped mispairing (Moore, 1983).
RpoC1 intron loss was detected only from the genus Lygodium (Fig. 5) in fern lineages. RpoC1 intron losses occur in various plant lineages. They are recognized as a synapomorphic characteristic of the subfamily Cactoideae of Cactaceae (Wallace and Cota, 1996) and the tribes Drosanthemeae and Ruschieae of Aizoaceae (Thiede et al., 2007). However, the intron losses occur independently even within a single genus depending on the species (Downie et al., 1996). Similar to rpoC1 intron losses in angiosperm, the analyses of the rpoC1 intron loss in ferns will help to indicate some taxonomic grouping. The primer set we developed in this study work well for fern species. So far, we tested the limited samples of ferns and found intron loss only from Lygodium. All species of Lygodium share this intron loss. This survey needs to expand to more fern species.
The patterns of GC contents by codon position and the ENCs of ferns are different from those of seed plants (Fig. 3). Early diverged ferns show low GC contents and ENCs when compared to the recently originated group. They also show a wide range of variation in GC and ENC values, while seed plants were similar to each other. The difference among groups may be due to the sampling error because many cp genome sequences are reported in seed plants, but only fifteen cp genome sequences are reported in ferns. Nevertheless, the value of GC3 and ENCs are notably different between ferns and seed plants. Furthermore, the GC3 and ENCs values are markedly different between the early diverged leptosporangiate and the core leptosporangiate ferns. We need more information about the cp genome sequences from ferns in order to address this question properly.
The molecular characteristics of the O. cinnamomea cp genome
Osmundales have several common characteristics with eusporangiate ferns. However, it is normally recognized as a member of leptosporangiate ferns based on other morphological characteristics. However, the cp genome of Osmunda is notably different from other leptosporangiate ferns in the following characters: 1) The cp genome structure is similar to that of E. arvense, which is eusporangiate fern; 2) the gene/intron losses that occurred in non-Osmundalean leptosporangiate ferns do not occur in the cp genome of O. cinnamomea; and 3) RNA editing sites in the cp genome are frequently predicted or detected in leptosporangiate ferns, but almost no RNA editing sites occur in the O. cinnamomea cp genome.
Molecular characteristics are frequently used to indicate specific taxonomic groups. The large inversion between psbM and ycf2 is recognized as a synapomorphic characteristic of seed plants and ferns (Raubeson and Jansen, 1992). In addition, a 9-bp insertion in rps4 is also a monophyletic characteristic of ferns (Pryer et al., 2001). Based on molecular characters, leptosporangiate ferns can be divided into two groups: Osmunda-lean ferns and non-Osmundalean ferns. Osmundales is also differentiated from other leptosporangiate ferns by echina-bearing tubercles on the surfaces of spores (Tryon and Lugardon, 1991). Therefore, it is reasonable to distinguish non-Osmundalean leptosporangiate ferns from Osmundalean ferns, or vice versa.
CONCLUSION
The complete cp DNA sequences from three major lineages of basal leptosporangiate ferns provide us a substantial information not only on evolution of the cp genomes and also on the phylogenetics of fern lineages. Our phylogenetic analysis, which was based on the largest numbers of complete cp genomes of Monilophytes so far, showed the paraphyly of the eusporangiate ferns. The Equisetales was the sister group to all other members of monilophytes. The results were consistent for the majority of the other analyses. In contrast to the paraphyly of eusporangiate ferns, the leptosporangiate ferns from a monophyletic group. Within the eusporangiate ferns, the cp genome structures, gene/intron contents, and RNA editing sites of Osmunda cinnamomea (Osmundales) were similar to that of the eusporangiate ferns. Therefore, these cp genome characteristics in Osmunda are symplesiomorphic conditions. Several lines of morphological and anatomical data, both from sporophyte and gametophyte of Osmunda, also support the intermediate or the distinctive natures of Osmundalean ferns from other leptosporangiate ferns. Therefore, the Osmundalean ferns can be recognized as living fossil lineages from leptosporangiate ferns. The GC contents and ENCs in ferns vary significantly when compared to that of seed plants. The both values in the early diverged leptosporangiate ferns showed intermediate levels between eusporangiate and core leptosporangiate ferns. The cp genome of Diplopterygium glaucum (Gleicheniales) has a large unique inversion between the trnL-CAA and trnV-GCA genes. Several repeated sequences were detected around the inversion break points. The cp genome of Lygodium japonicum (Schizaeales) showed rpoC1 intron loss, which is shared among all Lygodium species.
Supplementary
Acknowledgments
This research was supported by a research grant (#KRF-2010-0011796) from the Korea Research Foundation and by the Eco-Innovation project of the Ministry of Environment (416-111-007) from KEITI to Ki-Joong Kim. A Ph.D. fellowship to HTK was granted by the graduate student education program from NIBR (2012-2013). DNA materials used in this study were deposited in and are available from the Plant DNA Bank of Korea (PDBK).
Note. Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bold H.C., Alexopoulos C.J., Delevoryas T. Morphology of Plants and Fungi. 4th ed. New York: Harper and Row; 1987. [Google Scholar]
- Chumley T.W., Palmer J.D., Mower J.P., Fourcade H.M., Calie P.J., Boore J.L., Jansen R.K. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Cross G.L. Embryology of Osmunda cinnamomea. Bot. Gaz. 1931a;92:210–217. [Google Scholar]
- Cross G.L. Meristem in Osmunda cinnamomea. Bot. Gaz. 1931b;91:65–76. [Google Scholar]
- Downie S.R., Llanas E., Katz-Downie D.S. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA’s. Syst. Bot. 1996;21:135–151. [Google Scholar]
- Doyle J.J., Dolye J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg J.A., Gifford E.M., Jr The root apical meristem of Osmunda regalis. Am. J. Bot. 1984;71:558–563. [Google Scholar]
- Gao L., Yi X., Yang Y.X., Su Y.J., Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol. Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Wang B., Wang Z.W., Zhou Y., Su Y.J., Wang T. Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biol. Evol. 2013;5:1403–1407. doi: 10.1093/gbe/evt099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford E., Jr Concept of apical cells in bryophytes and pteridophytes. Annu. Rev. Plant Physiol. 1983;34:419–440. [Google Scholar]
- Good C.W., Taylor T.N. The morphology and systematic position of calamitean elater-bearing spores. Geosci. Man. 1975;11:133–139. [Google Scholar]
- Grewe F., Guo W., Gubbels E.A., Hansen A.K., Mower J.P. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol. Biol. 2013;13:8. doi: 10.1186/1471-2148-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Hasebe M., Iwatsuki K. Chloroplast DNA from Adiantum capillus-veneris L., a fern species (Adiantaceae); clone bank, physical map and unusual gene localization in comparison with angiosperm chloroplast DNA. Curr. Genet. 1990;17:359–364. [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C.R., Meng B.Y., et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K., Yamazaki T., Boufford D., Ohba H. Flora of Japan. Vol. I, Pteridophyta and Gymnospermae. Tokyo: Kodansha; 1995. [Google Scholar]
- Jansen R.K., Raubeson L.A., Boore J.L., dePamphilis C.W., Chumley T.W., Haberle R.C., Wyman S.K., Alverson A.J., Peery R., Herman S.J., et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Depamphilis C.W., Leebens-Mack J., Muller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K., et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol K.G., Arumuganathan K., Boore J.L., Duffy A.M., Everett K.D., Hall J.D., Hansen S.K., Kuehl J.V., Mandoli D.F., Mishler B.D., et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.J., Lee H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Park C.W., Kim K.J. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol. Cells. 2009;27:365–381. doi: 10.1007/s10059-009-0047-6. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar J.S., Skog J.E., Zimmer E.A., Pryer K.M. The paraphyly of Osmunda is confirmed by phylogenetic analyses of seven plastid loci. Syst. Bot. 2008;33:31–36. [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; in Gateway Computing Environments Workshop (GCE), 2010 (IEEE)
- Moore G.P. Slipped-mispairing and the evolution of introns. Trends Biochem. Sci. 1983;8:411–414. [Google Scholar]
- Palmer J.D. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. [Google Scholar]
- Palmer J.D., Stein D.B. Chloroplast DNA from the fern Osmunda cinnamomea: physical organization, gene localization and comparison to angiosperm. Curr. Genet. 1982;5:165–170. doi: 10.1007/BF00391801. [DOI] [PubMed] [Google Scholar]
- Palmer J.D., Stein D.B. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986;10:823–833. [Google Scholar]
- Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pryer K.M., Schneider H., Smith A.R., Cranfill R., Wolf P.G., Hunt J.S., Sipes S.D. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature. 2001;409:618–622. doi: 10.1038/35054555. [DOI] [PubMed] [Google Scholar]
- Pryer K.M., Schuettpelz E., Wolf P.G., Schneider H., Smith A.R., Cranfill R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am. J. Bot. 2004;91:1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Pryer K.M., Smith A.R., Rothfels C. Polypodiopsida Cronquist, Takht. & Zimmerm. 1966. Ferns. Version 14, January 2009 (under construction) 2009 http://tolweb.org/Polypodiopsida/20615/2009.01.14 in The Tree of Life Web Project, http://tolweb.org/ [Google Scholar]
- Raubeson L.A., Jansen R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255:1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- Sablok G., Nayak K.C., Vazquez F., Tatarinova T.V. Synonymous codon usage, GC(3), and evolutionary patterns across plastomes of three pooid model species: emerging grass genome models for monocots. Mol. Biotechnol. 2011;49:116–128. doi: 10.1007/s12033-011-9383-9. [DOI] [PubMed] [Google Scholar]
- Schneider H., Schuettpelz E., Pryer K.M., Cranfill R., Magallon S., Lupia R. Ferns diversified in the shadow of angiosperms. Nature. 2004;428:553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- Schuettpelz E., Pryer K.M. Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon. 2007;56:1037–1050. [Google Scholar]
- Schweitzer H.-J. Die Mitteldevon-Flora von Lindlar (Rheinland). 3. Filicinae-Hyenia elegans Kräusel & Weyland. Palaeontographica Abteilung B. 1972:154–175. [Google Scholar]
- Smith D.R. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 2009;71:627–639. doi: 10.1007/s11103-009-9545-3. [DOI] [PubMed] [Google Scholar]
- Smith A.R., Pryer K.M., Schuettpelz E., Korall P., Schneider H., Wolf P.G. A classification for extant ferns. Taxon. 2006;55:705–731. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. PAUP; Phylogenetic Analysis Using Parsimony, version 4.0 b10. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Tangphatsornruang S., Sangsrakru D., Chanprasert J., Uthaipaisanwong P., Yoocha T., Jomchai N., Tragoonrung S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.L., Taylor T.N., Krings M. Paleobotany: The Biology and Evolution of Fossil Plants. Burlington, Massachusetts: Elservier Ltd.; 2009. [Google Scholar]
- Thiede J., Schmidt S.A., Rudolph B. Phylogenetic implication of the chloroplast rpoC1 intron loss in the Aizoaceae (Caryophyllales) Biochem. Syst. Ecol. 2007;35:372–380. [Google Scholar]
- Tidwell W.D., Ash S.R. A review of selected Triassic to Early Cretaceous ferns. J. Plant Res. 1994;107:417–442. [Google Scholar]
- Tryon A., Lugardon B. Spores of the Pteridophyta. New York: Springer-Verlag; 1991. [Google Scholar]
- Vetrivel U., Arunkumar V., Dorairaj S. ACUA: a software tool for automated codon usage analysis. Bioinformation. 2007;2:62–63. doi: 10.6026/97320630002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R.S., Cota J.H. An intron loss in the chloroplast gene rpoC1 supports a monophyletic origin for the subfamily Cactoideae of the Cactaceae. Curr. Genet. 1996;29:275–281. doi: 10.1007/BF02221558. [DOI] [PubMed] [Google Scholar]
- Wolf P.G., Pryer K.M., Ueda K., Ito M., Sano R., Gastony G., Yokoyama J., Manhart J., Murakami N., Crane E. Fern phylogeny based on rbcL nucleotide sequences. Am. Fern J. 1995;85:134–181. [Google Scholar]
- Wolf P.G., Rowe C.A., Sinclair R.B., Hasebe M. Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris. L. DNA Res. 2003;10:59–65. doi: 10.1093/dnares/10.2.59. [DOI] [PubMed] [Google Scholar]
- Wolf P.G., Rowe C.A., Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Wolf P.G., Roper J.M., Duffy A.M. The evolution of chloroplast genome structure in ferns. Genome. 2010;53:731–738. doi: 10.1139/g10-061. [DOI] [PubMed] [Google Scholar]
- Wolf P.G., Der J.P., Duffy A.M., Davidson J.B., Grusz A.L., Pryer K.M. The evolution of chloroplast genes and genomes in ferns. Plant Mol. Biol. 2011;76:251–261. doi: 10.1007/s11103-010-9706-4. [DOI] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wu C.S., Wang Y.N., Liu S.M., Chaw S.M. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol. Biol. Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yatabe Y., Nishida H., Murakami N. Phylogeny of Osmundaceae inferred from rbcL nucleotide sequences and comparison to the fossil evidences. J. Plant Res. 1999;112:397–404. [Google Scholar]
- Yi D.K., Kim K.J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 2012;7:e35872. doi: 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D.K., Lee H.L., Sun B.Y., Chung M.Y., Kim K.J. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analyses with other three asterids. Mol. Cells. 2012;33:497–508. doi: 10.1007/s10059-012-2281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


