Abstract
Autophagy targets cytoplasmic cargo to a lytic compartment for degradation. Autophagy-related (Atg) proteins, including the transmembrane protein Atg9, are involved in different steps of autophagy in yeast and mammalian cells. Functional classification of core Atg proteins in plants has not been clearly confirmed, partly because of the limited availability of reliable assays for monitoring autophagic flux. By using proUBQ10-GFP-ATG8a as an autophagic marker, we showed that autophagic flux is reduced but not completely compromised in Arabidopsis thaliana atg9 mutants. In contrast, we confirmed full inhibition of auto-phagic flux in atg7 and that the difference in autophagy was consistent with the differences in mutant phenotypes such as hypersensitivity to nutrient stress and selective autophagy. Autophagic flux is also reduced by an inhibitor of phosphatidylinositol kinase. Our data indicated that atg9 is phenotypically distinct from atg7 and atg2 in Arabidopsis, and we proposed that ATG9 and phosphatidylinositol kinase activity contribute to efficient autophagy in Arabidopsis.
Keywords: concanamycin A, GFP-ATG8 processing assay, pexophagy, plant autophagy, wortmannin
INTRODUCTION
In diverse eukaryotic species, autophagy targets a portion of the cytoplasm to a lytic compartment for bulk degradation (Yang and Klionsky, 2010). The best-characterized type of autophagy is macroautophagy (hereinafter referred to as auto-phagy), which is initiated by the formation of a membrane cisterna called the phagophore. A phagophore is an expanded membrane sac that sequesters cytoplasmic constituents and matures into the autophagosome, a double-membraned cytoplasmic compartment. In yeast and plant cells, the outer membrane of the autophagosome fuses with the vacuolar membrane. In the vacuole, the inner membrane and its cargo are called an autophagic body, which is rapidly degraded by vacuolar hydrolases.
A set of Autophagy-related (Atg) genes is responsible for autophagy in yeast and is conserved in higher eukaryotes. Homology-based reverse genetic studies have revealed the function of autophagy in plants (Li and Vierstra, 2012). On the basis of the classification of conserved yeast and mammalian Atg genes involved in phagophore initiation and expansion (Yang and Klionsky, 2010), it was proposed that core ATG homologs in Arabidopsis are classified into four groups (Kim et al., 2012): (i) ATG1 kinase complex containing ATG13 (Suttangkakul et al., 2011) and ATG11 (Li et al., 2014); (ii) phosphatidylinositol (PI) 3-kinase complex containing VPS34 and ATG6 (Liu et al., 2005); generating phosphatidylinositol 3-phosphate (PI3P), which may be enriched in autophagic membrane (Zhang et al., 2011); (iii) ATG8 and ATG12 conjugation system containing ATG7 (Doelling et al., 2002), ATG4 (Yoshimoto et al., 2004), ATG5 (Thompson et al., 2005), ATG10 (Phillips et al., 2008), and ATG12 (Chung et al., 2010); and (iv) transmembrane protein ATG9 (Hanaoka et al., 2002; Inoue et al., 2006) and ATG2-ATG18a complex (Inoue et al., 2006; Xiong et al., 2005). All Arabidopsis mutants defective in ATG8 and ATG12 conjugation showed the same degree of phenotype (Chung et al., 2010). For example, atg7, atg5, atg10, atg4a atg4b double, and atg12a atg12b double mutants were unable to survive 12 days of darkness, while more than 50% of wild-type seedlings were still recovered after the dark treatment (Chung et al., 2010). During post-germinative growth, atg5 and atg7 mutant hypocotyls also showed similar stabilization of peroxisomal proteins (Kim et al., 2013). However, only a few studies have revealed phenotypic differences among multiple classes of Arabidopsis atg mutants (Hofius et al., 2009; Inoue et al., 2006; Suttangkakul et al., 2011; Wang et al., 2011).
Yeast Atg9 and its mammalian homolog ATG9 are thought to act early in autophagosome formation, but detailed mode of action may differ between two homologues (Zavodszky et al., 2013). Yeast Atg9 is required for autophagy (Noda et al., 2000), whereas mammalian ATG9 appears to be important but not strictly required for this process (Orsi et al., 2012). Yeast Atg9 interacts with Atg2 and with PI3P-binding Atg18 (Wang et al., 2001), all of which are localized at the edge of the phagophore (Suzuki et al., 2013). In fact, recent studies suggested that Atg1, Atg2, and Atg18 are all interconnected and regulate Atg9 traf fickin in yeast (Zavodszky et al., 2013). In contrast, interactions of mammalian ATG9 with ATG2 and ATG18 were not reported, and the relation between ATG9 and ATG2-ATG18 remains unclear.
Arabidopsis atg9-1 was described as the first plant atg mutant (Hanaoka et al., 2002), although it is not clear whether this mutant is deficient for autophagy. Wild type (WT) and mutants homozygous for the second allele, atg9-2, accumulated cytoplasmic inclusions in the vacuole when they were treated with protease inhibitors (Inoue et al., 2006). In contrast, atg2 and atg5 mutants accumulated much less cytoplasmic inclusions in the vacuole, suggesting that autophagy in Arabidopsis roots requires ATG2 and ATG5 but not ATG9 (Inoue et al., 2006). Thus, it appears that Arabidopsis ATG9 and ATG2 belong to different classes of core ATG genes. To confirm the functional classification of Arabidopsis ATG genes, further information must be obtained from comparative analysis of atg mutants, which must be validated by assays for autophagy (Klionsky et al., 2012). However, the dispensability of ATG9 for autophagy in Arabidopsis was not confirmed by analysis with the auto-phagic marker GFP-ATG8, which was used to demonstrate that autophagy is inhibited in atg7 (Chung et al., 2010), atg5 (Chung et al., 2010), and atg2 (Wang et al., 2011).
Autophagic flux is defined as the dynamic process of auto-phagosome synthesis, delivery of autophagic substrates to the lytic compartment, and degradation of autophagic substrates inside the lytic compartment (Mizushima et al., 2010). Determination of autophagic flux is important because a steady-state level of autophagic markers may be increased not only by induction of autophagy but also by a block in a later step such as fusion or vacuolar degradation (Klionsky et al., 2012). Concanamycin A (CA), an inhibitor of vacuolar proton pumps (Matsuoka et al., 1997), is useful for monitoring autophagic flux in plants. Treatment with CA leads to increased vacuolar pH and inactivation of acid hydrolases in the vacuole. The objective of this study was to identify phenotypic difference between atg7, atg9, and atg2 mutants. By using autophagic markers and inhibitors, we found that atg9 mutants are phenotypically distinct from atg7 and atg2 and exhibited reduced autophagic flux when compared to WT.
MATERIALS AND METHODS
Plant materials and growth conditions
TAIR Accession numbers of Arabidopsis genes mentioned are as follows: ATG2, AT3G19190; ATG5, AT5G17290; ATG7, AT5G45900; ATG8a, AT4G21980; ATG9, AT2G31260. Arabidopsis thaliana T-DNA insertional mutants atg2-1 (Inoue et al., 2006), atg5-1 (Thompson et al., 2005), atg7-2 (Chung et al., 2010), atg9-3 (SALK_128991), and atg9-4 (SALK_145980) were previously described or obtained from the Arabidopsis Information Resource (TAIR, http://arabidopsis.org). Transgenic marker lines expressing GFP-ATG8a under the control of the cauliflower mosaic virus 35S promoter (Thompson et al., 2005) and Arabidopsis UBQ10 promoter (Kim et al., 2013) were designated pro35S-GFP-ATG8a and proUBQ10-GFP-ATG8a, respectively.
Surface-sterilized Arabidopsis seeds were stored at 4°C for 3 days and incubated at 20–22°C under a 16-h light/8-h dark photoperiod. For liquid culture, seedlings were incubated in a 24-well plate containing Murashige-Skoog (MS) liquid medium supplemented with 1% (w/v) sucrose (Kim et al., 2013). Nitrogen supply to the seedlings was limited by changing the medium to liquid MS-N containing 1% sucrose (Chung et al., 2009). For solid culture, seeds were germinated on MS solid medium with 1% sucrose (Kim et al., 2013). A carbon starvation experiment was performed as described previously (Chung et al., 2010).
Immunoblot analysis
Seeds were germinated on the solid medium for analysis of isocitrate lyase; hydroponically grown seedlings were used for all other analyses. Dissected hypocotyls (for isocitrate lyase; Ettinger and Harada, 1990) or whole seedlings (other analysis) were homogenized in Laemmli buffer and clarified by centrifugation at 16,000 × g for 10 min. Protein samples were separated by 12% SDS-PAGE. Immunoblot analysis was performed as described previously (Kim et al., 2013) except that anti-GFP antibodies (Roche) were diluted to 1:2000.
Microscopy
Nine-day-old root tissues from the maturation zone were analyzed using a Zeiss 510 laser scanning confocal microscope (Carl Zeiss). GFP was excited with a 488-nm excitation line and a BP500-530IR emission filter was used for detection. Z-sections through the central vacuole were acquired at 1-μm intervals to distinguish autophagic bodies from transvacuolar strands.
RESULTS
ATG9 is not strictly essential for autophagy but affects autophagic flux
To investigate the requirement of ATG9 for autophagy in Arabidopsis, we used two atg9 alleles, designated atg9-3 and atg9-4. The atg9-3 and atg9-4 alleles have a T-DNA insertion in exon 6 and in the 5′ end of exon 8, respectively, of the single-copy ATG9 gene (Supplementary Fig. S1). Reverse transcriptase (RT)-PCR analysis detected no ATG9 transcript flanking the T-DNA insertion in atg9-3 and atg9-4, indicating that they are null for functional ATG9 transcripts (Supplementary Fig. S1D).
We initially crossed the atg9 mutants with transgenic plants carrying the pro35S-GFP-ATG8a transgene (Thompson et al., 2005), a popular marker for the autophagosome and autophagic bodies. However, we observed a marked reduction in GFP signal in the progeny produced from the crosses, compared to GFP from the original pro35S-GFP-ATG8a transgenic line (data not shown). The reduction likely resulted from transgene silencing, because the presence of cauliflower mosaic virus 35S promoter in T-DNA tagging mutants often represses trans-gene expression driven by the same promoter (Daxinger et al., 2008). Thus, we decided to use an alternative marker, proUBQ10-GFP-ATG8a (Kim et al., 2013), which contains the promoter of Arabidopsis UBQ10 gene instead of the 35S promoter. We crossed the proUBQ10-GFP-ATG8a transgenic plants with atg9-3 and atg9-4, and their F2 individuals were genotyped to obtain atg9 mutants containing the transgene. The proUBQ10-GFP-ATG8a seedlings with WT, atg7-2, atg9-3, or atg9-4 background were incubated in liquid medium containing either CA or dimethylsulfoxide (DMSO) as a solvent control. CA is frequently used to stabilize vacuolar GFP-ATG8a puncta, which are interpreted as autophagic bodies (Yoshimoto et al., 2004). Confocal microscopy of DMSO-treated WT and atg9 roots (Fig. 1A) revealed a diffuse GFP-ATG8a signal in the cytoplasm, except for the occasional puncta that may show phagophores or autophagosomes. The CA-treated WT vacuole contained numerous GFP-ATG8a-labeled autophagic bodies, while the atg7-2 vacuole did not contain GFP-ATG8a puncta, as previously reported (Chung et al., 2010). GFP-ATG8a puncta were detected in the vacuole of CA-treated atg9 seedlings, although they were not as abundant as in the WT (Fig. 1A).
Fig. 1.
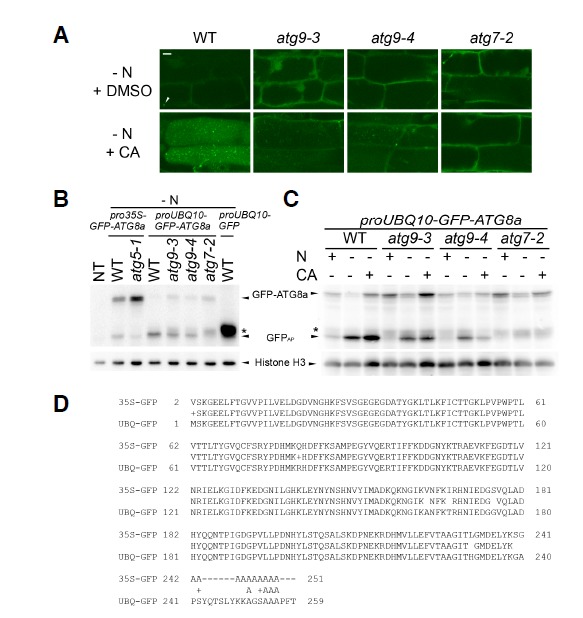
Autophagic marker analysis of atg7-2 and atg9 mutants. Hydroponically grown 7-d-old Arabidopsis seedlings were further incubated in either nitrogen (N)-sufficient or -deficient medium for 2 days before analysis. (A) Subcellular localization of GFP-ATG8a in N-limited wild-type (WT), atg9-3, atg9-4, and atg7-2 roots. Eight-day-old proUBQ10-GFP-ATG8a seedlings with indicated genotypes were treated with either 1 μM concanamycin A (CA, second row) or an equal volume of dimethylsulfoxide (DMSO, first row) for 16 h, and analyzed by confocal microscopy. Arrowhead shows a phagophore-like signal. Representative images were chosen from at least five biological replicates. Scale bar = 10 μm. (B) and (C) GFP-ATG8 processing assay of atg9 and atg7 mutants. Non-transgenic WT (NT), pro35S-GFP-ATG8a, proUBQ10-GFP, and proUBQ10-GFP-ATG8a transgenic seedlings of indicated geno-types were incubated in N-sufficient liquid medium for 7 days and then incubated in N-deficient medium for 2 days before immunoblot analysis (B). In (C), N-supplied or -limited seedlings were treated with CA (+) or DMSO (-) for 16 h prior to protein extraction. Immunoblot analysis by using anti-GFP (upper panel) or anti-histone H3 (lower panel; loading control) antibodies was performed. Arrowheads show the protein bands corresponding to GFP-ATG8a, GFPAP, and histone H3. Asterisk indicates the position of protein bands corresponding to GFPIP which shows slightly slower mobility than that by GFPAP. Representative immunoblots were selected from at least three independent experiments. (D) Comparison of two autophagic marker lines, pro35S-GFP-ATG8a (upper lines, 35S-GFP) and proUBQ10-GFP-ATG8a (lower lines, UBQ-GFP), by pairwise alignment of N-terminal amino acid sequences that were predicted from GFP-ATG8a coding sequences in two transgenic constructs.
To monitor autophagic flux in atg9 seedlings, we carried out the GFP-ATG8 processing assay (Klionsky et al., 2012). This assay is routinely used to study autophagy in yeast and is based on the fact that GFP-ATG8 in the vacuole is processed to generate free GFP which is more stable than GFP-ATG8. Autophagy-deficient mutants typically lack free GFP, whereas the inhibition of vacuolar degradation results in accumulation of free GFP and GFP-ATG8. Although the assay can be used for plant cells (Chung et al., 2010; Suttangkakul et al., 2011), there is a potential problem with this method. In atg5-1 mutants expressing the conventional autophagic marker pro35S-GFP-ATG8a (Thompson et al., 2005), we often detected a faint protein band (Fig. 1B, lane 3) that co-migrated with the free GFP band in the WT extract (Fig. 1B, lane 2). Interestingly, when we used proUBQ10-GFP-ATG8a for anti-GFP immunoblot analysis (Figs. 1B and 1C), we identified three protein bands with different mobility in SDS-PAGE: one band at the predicted size of GFP-ATG8a, and two faster bands similar to the size of free GFP moiety. The WT had the fastest band, atg7-2 had a smear band with slightly slower mobility (indicated by asterisks in Figs. 1B and 1C), and atg9-3 and atg9-4 included both. In the WT, the intensity of the fastest protein band was increased by nitrogen limitation and further by CA treatment. These increases suggest that this band represents a processed GFP species that is more stable than GFP-ATG8a in the vacuole and is thus a true indicator of autophagic flux. This protein species was designated GFPAP (for autophagy-dependent, processed GFP). In contrast, the slower band appeared to be associated with inefficient autophagy, as it was rarely detectable in the WT but more noticeable in atg9-3, atg9-4, and atg7-2; this protein species was designated GFPIP (for autophagy-independent, processed GFP). Although GFPIP level was slightly increased by nitrogen limitation in atg7 and atg9 mutants, it was not increased by CA (Fig. 1C, smear bands indicated by asterisk), showing that GFPIP is not associated with the vacuole and cannot be considered an indicator of autophagic flux.
Compared to WT, atg9 seedlings contained a lower level of GFPAP (Figs. 1B and 1C), indicating reduced autophagic flux in the mutants. The atg9 mutants showed a WT-like increase in GFPAP in response to nitrogen limitation, but unlike the WT, CA treatment had only a minor effect on GFPAP levels in these mutants (Fig. 1C). Since CA mainly acts after the fusion of the autophagosome with the vacuole, this result suggested that ATG9 acts in an early step before the fusion event.
We further tested the hypothesis that GFPAP is a product of vacuolar degradation. Tamura et al. (2003) reported that GFP tagged with a C-terminal vacuolar-targeting signal is processed in the vacuole, and that a processed GFP moiety was stabilized when plants were incubated in the dark. Consistent with our hypothesis, the abundance of GFPAP, but not of GFPIP, was increased when proUBQ10p-GFP-ATG8a plants were incubated in the dark (Fig. 2).
Fig. 2.
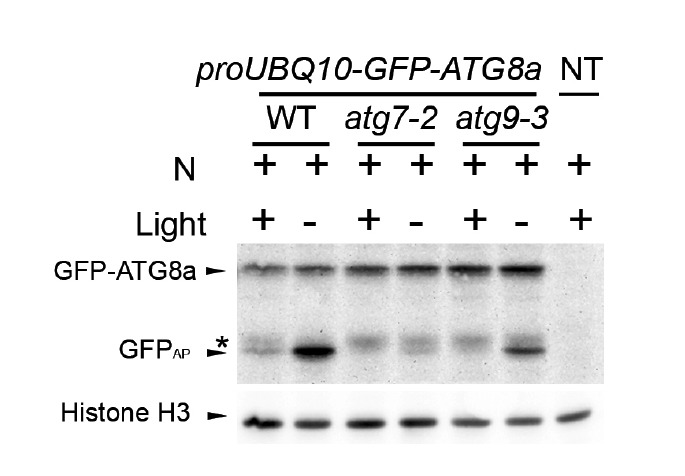
Response of autophagic marker lines to light deprivation. Autophagic marker lines proUBQ10-GFP-ATG8a with indicated genotypes were incubated in N-sufficient liquid medium for 8 days. At day 8, a plate containing the seedlings was wrapped with aluminum foil (- light) and another plate was left illuminated (+ light). After 24 h, proteins were extracted from the seedlings and immunoblot analysis was performed using anti-GFP (upper panel) or anti-histone H3 (lower panel, loading control) antibodies. Arrowheads show the protein bands corresponding to GFP-ATG8a, GFPAP, and histone H3. Asterisk indicates the position of protein bands corresponding to GFPIP which shows slightly slower mobility than that by GFPAP. NT, non-transgenic control.
Phenotypes of atg9 differ from those of atg7 and atg2
We compared atg7, atg2, and atg9 for hypersensitivity to nutrient limitation, which is the canonical phenotype of autophagy-defective Arabidopsis mutants (Doelling et al., 2002; Hanaoka et al., 2002; Inoue et al., 2006). As expected from our analysis of autophagy, atg7-2 and atg2-1 showed hypersensitivity to fixed carbon limitation (Chung et al., 2010), but atg9 seedlings did not (Fig. 3A). Phenotypic differences between atg9 and other core atg mutants were also seen for phenotypes associated with pexophagy, a selective type of autophagy preferentially targeting peroxisomes for degradation (Kim et al., 2013). Degradation of isocitrate lyase (ICL), a peroxisomal matrix enzyme, was shown to be delayed in atg7 and atg5 hypocotyls (Kim et al., 2013). Delayed degradation was also confirmed in atg2-1 hypocotyls (Fig. 3B), consistent with a recent report that another atg2 allele suppressed defects in peroxisomal functions (Farmer et al., 2013). ICL degradation was also delayed in atg9 hypocotyls (Fig. 3B), although atg7-2 and atg2-1 contained a higher level of ICL at day 9 than atg9 did. These phenotypic data were consistent with our observation that autophagic flux was fully compromised in atg7 and atg2 but not in atg9.
Fig. 3.
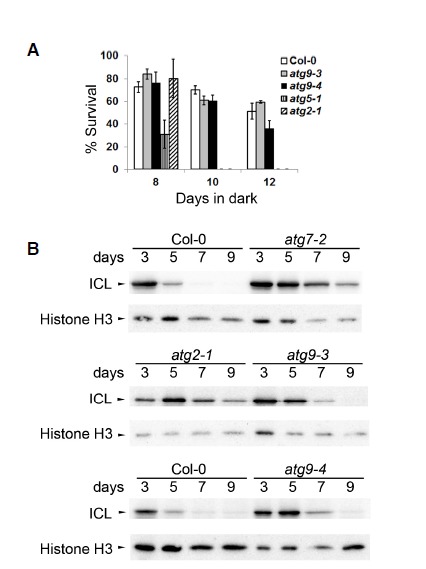
Phenotypes of atg7-2, atg2-1, and atg9 mutants. (A) Carbon starvation phenotype was assessed using a light-deprivation experiment (Chung et al., 2010). Two-week-old seedlings were deprived of light for the indicated number of days. Percentage of survival was calculated from self-fertilized progenies collected from ≥ 4 parental plants. Mean ± S.E. (N = 4 or 5 sibling populations) (B) Peroxisomal protein degradation in hypocotyls of the indicated genotypes was assessed by immunoblot analysis using anti-isocitrate lyase (ICL, upper panels) and anti-histone H3 (lower panels; loading control) antibodies. Representative immunoblots were selected from at least three independent experiments.
Wortmannin blocks autophagic flux
To determine whether PI 3-kinase activity is required for autophagy in plant cells, we tested the effect of wortmannin (WM), an inhibitor of mammalian and plant PI 3-kinases (Matsuoka et al., 1995). We incubated pro35S-GFP-ATG8a transgenic seedlings in liquid medium containing CA for various durations; the effect of CA on the level of free GFP was prominent after 16 h of incubation (Supplementary Fig. S2A). Next, we incubated the transgenic plants in liquid medium containing either DMSO or CA and various amounts of WM for 16 h and found that 10–30 μM WM dampened the effect of CA on free GFP (Supplementary Figs. S2B and S2C). However, only a slight reduction in free GFP was seen in transgenic seedlings treated with 30 μM WM alone for 16 h, compared to DMSO control (Supplementary Fig. S2B) and no treatment control at 0 h (Supplementary Fig. S2D). These results suggested that PI 3-kinase activity is required for either vacuolar targeting of GFP-ATG8a or for stabilization of free GFP by CA.
To gain insight into the relationship between different core ATG complexes in plants, we further investigated the effects of CA and WM on the autophagic markers in various mutant backgrounds. We compared confocal images of proUBQ10-GFP-ATG8a transgenic roots of WT, atg7-2, atg9-3, and atg9-4 background that were treated with DMSO, CA, WM, or the combination of CA and WM (Fig. 4A). The effect of CA in WT and atg9 was markedly diminished by treatment with WM, indicating that efficient delivery of GFP-ATG8a to the vacuole relies on PI 3-kinase activity. In WM-treated WT and atg9 cells, most GFP-ATG8a signal appeared to be diffuse in the cytosol and to occasionally form cytoplasmic foci, but almost no signal was detected in atg9 vacuoles. No GFP-ATG8a puncta were detected in the vacuoles of autophagy-defective negative control atg7-2 treated with CA and WM.
Fig. 4.
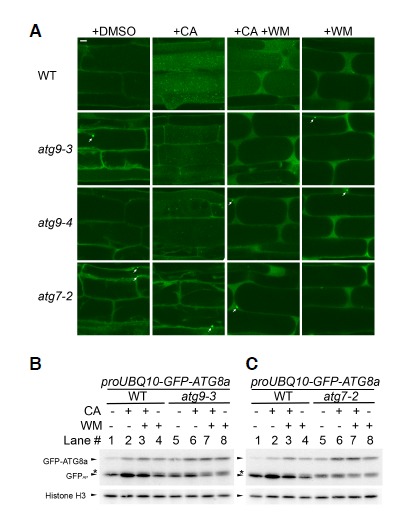
Autophagic flux is blocked using wortmannin (WM). Nitrogen supply was limited to seedlings for 2 days prior to analysis. (A) Confocal microscopy of GFP-ATG8a transgenic roots treated with dimethylsulfoxide (DMSO), concanamycin A (CA), WM, or both. GFP-ATG8a transgenic roots from wild-type (WT, first row), atg9-3 (second row), atg9-4 (third row), and atg7-2 (fourth row; negative control) background. Eight-day-old seedlings were further incubated for 16 h in liquid medium containing DMSO (first column), 1 μM CA (second column), 1 μM CA and 30 μM WM (third column), or 30 μM WM (fourth column). Representative images were chosen from at least five biological replicates. Scale bar = 10 μm. (B, C) Immunoblot analysis of GFP-ATG8a transgenic seedlings of WT, atg9-3, or atg7-2 treated with DMSO, WM, CA, or WM and CA, as in (A). Immunoblots were reacted with anti-GFP (upper panels) or with anti-histone H3 (lower panels; loading control) antibodies. Arrowheads identify the protein bands corresponding to GFP-ATG8a, GFPAP, and histone H3. Asterisk indicates the position of GFPIP. A minus (-) sign represents no treatment, while a plus (+) sign means treatment with CA or WM. Representative immunoblots were selected from at least three independent experiments.
We also investigated the effect of WM on autophagic flux. The GFP-ATG8 processing assay was performed using crude extracts prepared from whole seedlings that were treated with DMSO, CA, WM, or combination of CA and WM (Figs. 4B and 4C). We not only confirmed the absence of GFPAP in atg7-2 (Fig. 4C, lanes 5–8) and the presence of both GFPAP and GFPIP in atg9 (Fig. 4B, lanes 5–7), but also observed a small amount of GFPIP in WM-treated WT samples (Figs. 4B and 4C; lanes 3 and 4), suggesting that WM blocked a step in autophagy. In the WT, WM treatment increased the level of GFP-ATG8a but not that of GFPAP (Figs. 4B and 4C; lanes 1 and 4), while CA treatment enhanced the intensity of both GFP-ATG8a and GFPAP (Figs. 4B and 4C; lanes 1 and 2). WM treatment also suppressed the effect of CA on GFPAP accumulation (Figs. 4B, C; lanes 2 and 3). Combined with the confocal microscopy data, these immunoblot data indicated that WM interferes with the targeting of cytoplasmic GFP-ATG8a-PE to the vacuole for processing to GFPAP and then full degradation. GFPAP was largely depleted in atg9-3 seedlings by WM treatment alone (Fig. 4B, lanes 5 and 8), although CA stabilized a trace amount of GFPAP (Fig. 4B, lanes 7 and 8). The additional inhibition by WM in atg9 is consistent with the notion that ATG9 belongs to a different class from the autophagy-specific PI 3-kinase complex in Arabidopsis. In conclusion, the collective data confirmed that WM-sensitive PI 3-kinase activity and ATG9 contribute to efficient autophagic flux, especially in early autophagic process prior to the fusion of the autophagosome with the vacuole (Fig. 4).
DISCUSSION
In this study, we compared different classes of core ATG proteins in Arabidopsis, by using genetic analysis, confocal microscopy, and the GFP-ATG8a processing assay. Our data confirmed that mutations affecting the ATG8 conjugation system and ATG2-ATG18 complex resulted in more severe phenotypes than did atg9 (Figs. 1–4). ATG9 and PI 3-kinase activity may act at different stages of Arabidopsis autophagy, since atg9 mutants retained sensitivity to WM (Fig. 4).
We developed an Arabidopsis GFP-ATG8 processing assay that can separate autophagy-dependent and -independent pools of free GFP processed from GFP-ATG8a. The former pool, named GFPAP, which appeared to accumulate in the vacuole, was stabilized by CA and increased in response to N limitation. Thus, GFPAP is an indicator of autophagic flux. The nature of the autophagy-independent pool (GFPIP) is not clear, but GFPIP may be related to reduction or blockage in autophagic flux. This speculation is based on our observation of GFPIP in atg7 and atg9 seedlings (Figs. 1B, 1C, 2, 4B, and 4C).
Although the pro35S-GFP-ATG8a transgene (Thompson et al., 2005) has been popular as an autophagic marker in Arabidopsis, there are drawbacks to its use. We crossed the pro35S-GFP-ATG8a transgenic line with atg9 mutants to obtain atg9 mutants expressing the transgene but later found that the transgene was silenced in mutant progeny, probably because of the presence of the 35S promoter (Daxinger et al., 2008). We used an alternative autophagic marker line, proUBQ10-GFP-ATG8a (Kim et al., 2013), in which GFP-ATG8a showed a lower level of accumulation (Fig. 1B). The calculated molecular weight of free GFP moieties predicted from pro35S-GFPATG8a and proUBQ10-GFP-ATG8a, were 27.8 and 28.9 kDa, respectively. The 1.1-kDa difference is mostly caused by the C-terminal residues of the GFP moiety, where the pro35S-GFP-ATG8a transgene has a polyalanine linker and the proUBQ10-GFP-ATG8a transgene has a different linker derived from a recombination site in the vector system (Fig. 1D). The explanation for why only proUBQ10-GFP-ATG8a resulted in the separation of GFPAP from GFPIP may lie in the differential C-terminal sequences. For example, the polyalanine linker may cause a structurally weak junction (amino acids 242 to 251 of upper sequence in Fig. 1D) that is susceptible to attack by extravacuolar proteolytic activity, which may also attack at amino acids 254 to 256 of GFP-ATG8a derived from proUBQ-GFP-ATG8a (Fig. 1D, lower sequence) and generate a slightly longer GFP moiety, that is, GFPIP. Only in autophagy-competent plants, GFPAP may form by further digestion to ∼240th amino acid by vacuolar proteases.
The difference in mobility between GFPAP and GFPIP is small. Thus, when the proUBQ10-GFP-ATG8a line is used for the GFP-ATG8 processing assay, it is recommended that protein samples be run with a reference sample (e.g., atg9 or WM-treated WT extract) in SDS-PAGE. In addition, CA may be useful for confirming normal autophagic flux, as recommended previously (Klionsky et al., 2012). For example, we detected reduced autophagic flux in atg9 that would not have been revealed without CA treatment (Figs. 1C and 4B).
Although WM and CA are useful as inhibitors of early and late steps in autophagy, respectively, it is important that they be used with caution. Since CA indirectly blocks the action of vacuolar hydrolases by inhibiting vacuolar proton pumps, it took 16 h of CA treatment for autophagic cargo to be markedly stabilized (Supplementary Fig. S2A). In addition, we treated Arabidopsis seedlings with liquid medium containing CA and WM-uptake efficiency by plant tissues is variable under such conditions (e.g., shoot tissues may be less affected by CA and WM than root tissues). This partial uptake may explain our observation that GFPAP is only slightly increased by treatment of WT with CA and WM (Figs. 1 and 4) but significantly increased by dark treatment (Fig. 2).
We showed that Arabidopsis ATG9 is not strictly required for autophagy but contributes to efficient autophagic flux and that atg9 phenotypes are distinct from those of atg2 and atg7. Our observation is consistent with that in previous studies of the atg9-2 allele (Inoue et al., 2006) but contrasts with that in an earlier study of the atg9-1 allele (Hanaoka et al., 2002). The atg9-1 allele has a Wassilewskija genetic background, while atg9-2, atg9-3, and atg9-4 alleles have a Columbia background. In addition, atg9-1 has a large deletion spanning exons 4–10 near its T-DNA insertion (see Supplementary Fig. S1A). Hyper-sensitivity to carbon limitation was observed in atg9-1 mutants (Hanaoka et al., 2002), while we did not note a marked difference from the WT (Fig. 3). We do not know the cause of this phenotypic difference; the difference may suggest that the C-terminal half of ATG9 is dispensable for its function if a partial ATG9 is produced from atg9-2, atg9-3, and atg9-4 (see Supplementary Fig. S1A). Alternatively, variation in genetic background may explain the phenotypic differences among mutants. Analysis of autophagic flux in atg9-1 will provide valuable information to help clarify this issue.
Supplementary
Acknowledgments
We thank John Harada for kindly providing the anti-ICL antisera. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (2011-0010683) and by a grant from the Next-Generation Bio-Green 21 Program (No. PJ009004), Rural Development Administration, Republic of Korea.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Chung T., Suttangkakul A., Vierstra R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149:220–234. doi: 10.1104/pp.108.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- Daxinger L., Hunter B., Sheikh M., Jauvion V., Gasciolli V., Vaucheret H., Matzke M., Furner I. Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 2008;13:4–6. doi: 10.1016/j.tplants.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- Ettinger W.F., Harada J.J. Translational or post-translational processes affect differentially the accumulation of isocitrate lyase and malate synthase proteins and enzyme activities in embryos and seedlings of Brassica napus. Arch. Biochem. Biophys. 1990;281:139–143. doi: 10.1016/0003-9861(90)90423-v. [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell. 2013;25:4085–4100. doi: 10.1105/tpc.113.113407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jorgensen L.B., Jones J.D., Mundy J., Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–1652. doi: 10.1093/pcp/pcl031. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kwon C., Lee J.H., Chung T. Genes for plant autophagy: functions and interactions. Mol. Cells. 2012;34:413–423. doi: 10.1007/s10059-012-0098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H.N., Kim S.H., Shin K.D., Chung T. Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell. 2013;25:4956–4966. doi: 10.1105/tpc.113.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D. AUTOPHAGY-RELATED (ATG)11 plays a critical role in general autophagy and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26:788–807. doi: 10.1105/tpc.113.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Talloczy Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Bassham D.C., Raikhel N.V., Nakamura K. Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 1995;130:1307–1318. doi: 10.1083/jcb.130.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Higuchi T., Maeshima M., Nakamura K. A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell. 1997;9:533–546. doi: 10.1105/tpc.9.4.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D.J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., Tooze S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. doi: 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H., Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Kim J., Huang W.P., Abeliovich H., Stromhaug P.E., Dunn W.A., Jr, Klionsky D.J. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011;68:74–87. doi: 10.1111/j.1365-313X.2011.04669.x. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42:535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Vicinanza M., Rubinsztein D.C. Biology and trafficking of ATG9 and ATG16L1, two proteins that regulate autophagosome formation. FEBS Lett. 2013;587:1988–1996. doi: 10.1016/j.febslet.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li S., Zhou L.Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011;68:1081–1092. doi: 10.1111/j.1365-313X.2011.04761.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


