Abstract
An ∼32-kDa protein (albusin B) that inhibited growth of Ruminococcus flavefaciens FD-1 was isolated from culture supernatants of Ruminococcus albus 7. Traditional cloning and gene-walking PCR techniques revealed an open reading frame (albB) encoding a protein with a predicted molecular mass of 32,168 Da. A BLAST search revealed two homologs of AlbB from the unfinished genome of R. albus 8 and moderate similarity to LlpA, a recently described 30-kDa bacteriocin from Pseudomonas sp. strain BW11M1.
Ruminococcus albus and Ruminococcus flavefaciens are important cellulose-degrading bacteria in the rumen (6). R. albus has been reported to outnumber R. flavefaciens in defined laboratory cocultures (4, 16, 17, 22) and in the rumens of sheep (25) and dairy cattle (28). Several reports suggested that predominance of R. albus may be due to its production of bacteriocin-like substances (4, 16, 17), but none of these agents had been purified. The objective of this study was to purify and characterize bacteriocin-like substances produced by R. albus.
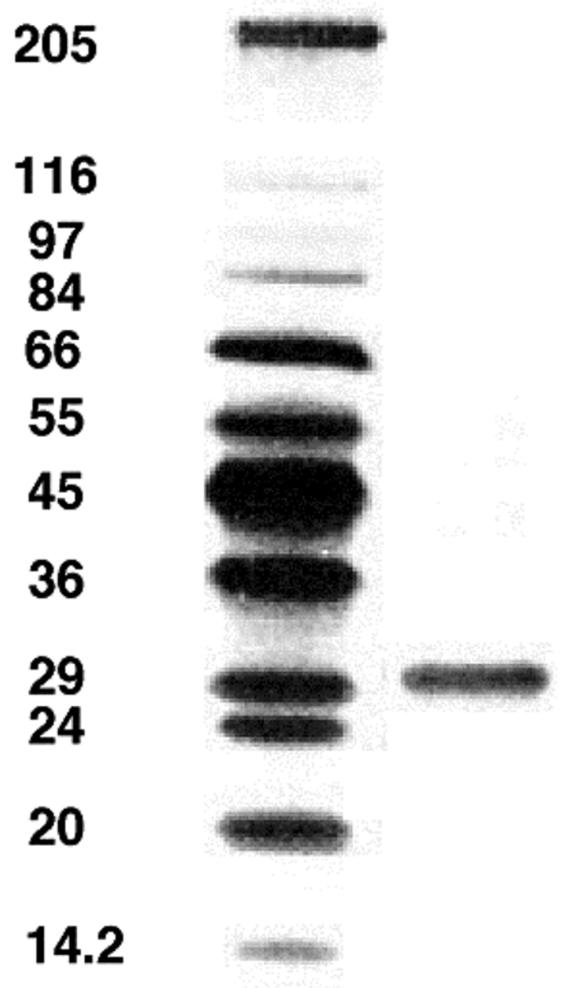
Culture supernatants of R. albus 7 grown in modified Dehority medium (27) with cellulose or cellobiose as a fermentable carbohydrate inhibited growth of R. flavefaciens FD-1 when assayed by a plate culture assay (4). Because most of the bacteriocins purified from gram-positive bacteria have low molecular masses (<10 kDa), we examined filtrates of R. albus 7 cultures for inhibitory activity toward R. flavefaciens. Cultures grown in 10 liters of modified Dehority medium with cellobiose were passed through a 0.2-μm-pore-size hollow fiber cartridge, and the filtrate was passed through a 10-kDa-molecular-mass-cutoff cartridge. Substantial inhibitory activity was observed in the nominal 10-kDa filtrate, but surprisingly this filtrate contained a 32-kDa protein (referred to here as albusin B) that was ultimately shown to account for most of the activity. Although the retentate from the 10-kDa cartridge also contained an inhibitory protein of 32 kDa, the mixture of components in the retentate was far more complex than that in the filtrate. The simple protein profile of the filtrate facilitated purification of albusin B, which was accomplished by a process that also included ammonium sulfate precipitation (40 to 60% saturation range) and a BioGel P6 gel filtration column (Table 1). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis revealed a single protein band (32 kDa) from the P6 column (Fig. 1). While the purification was facilitated by the 10-kDa-molecular-mass-cutoff ultrafiltration step, the complete protocol resulted in only a 4.7% activity yield and an estimated purification of 220-fold. Because some of the inhibitory activity in culture supernatants was due to a second inhibitor (albusin A, which will be described elsewhere), whose concentration declined during the course of the purification, the extent of purification of albusin B must be regarded as a lower bound.
TABLE 1.
Purification of the bacteriocin albusin B from R. albus 7
| Purification step | Total protein (mg)a | Total activity (AU)b | Sp act (AU/mg) | Yield (%) |
|---|---|---|---|---|
| Supernatant (10 liters) | 1,400 | 3,920 | 2.8 | 100 |
| 4× concn of 10-kDa filtrate | 11.1 | 343 | 31 | 8.8 |
| 40 to 60% (NH4)2SO4 precipitation | 1.6 | 205.8 | 128.6 | 5.3 |
| P6 column (6-kDa cut off) | 0.3 | 185.2 | 617.3 | 4.7 |
Protein concentration was analyzed by the MicroBCA protein assay kit (Pierce), using lysozyme as the standard.
AU, activity unit. Activity units are defined as the reciprocal of the highest dilution at which growth inhibition was detectable and are expressed on a per-milliliter basis.
FIG. 1.
SDS-polyacrylamide gel of protein standards (left lane; molecular masses [in kilodaltons] are on the left) and the bacteriocin albusin B (right lane), following its elution from the BioGel P6 column (Table 1). The image was obtained by scanning the dried gel directly with an Envisions flatbed scanner into Adobe Photoshop 7.0 (operating on Macintosh OS9.0) and converting the colored image to grayscale prior to saving it as a TIFF image.
All seven R. flavefaciens strains tested (FD-1, B34b, 17, C94, B1c45, B146, and R13e2) were sensitive to albusin B, but other ruminal species tested (Fibrobacter succinogenes S85, Selenomonas ruminantium D, and Streptococcus bovis JB-1) and the nonruminal species Escherichia coli ZK126 and Bacillus subtilis AD623 were not affected. Inhibitory activity of albusin B was lost upon treatment with Streptomyces griseus protease or porcine pancreatin or by boiling for 15 min. On the basis of its apparent protein nature and moderate specificity against a species related to the producing strain, the inhibitor fits the definition of a bacteriocin, according to Klaenhammer (12).
After electrotransfer from the SDS-polyacrylamide gel to a polyvinylidene difluoride membrane, the 32-kDa protein band was subjected to N-terminal sequencing (performed at the Protein Chemistry Laboratory, University of Texas Medical Branch, Galveston), which yielded the sequence AVISVNTVVDAKNGNADLVQGKFY. An oligonucleotide probe (5′-GGIAAYGCIGAYCTIGTICAGGG-3′), designed on the basis of the N-terminal sequence and codon usage by R. albus (codon usage database [www.kazusa.or.jp/codon]), was synthesized and 5′ digoxigenin labeled by Sigma-Genosys. The probe hybridized to a 1-kb HincII fragment of the R. albus 7 chromosome. HincII fragments of approximately 1 kb were cloned into the pGEM-3Z vector by using standard protocols (21), and two positive clones were detected by Southern hybridization screening. Sequencing data showed that the two cloned fragments were identical but opposite in orientation. A partial open reading frame (ORF) was present in the cloned sequence, designated albB, that encoded 16 amino acids (aa) (DAKNGNADLVQGKFY) matching the peptide sequence obtained by N-terminal sequencing (AVISVNTVVDAKNGNADLVQGKFY).
Because the cloned sequence did not contain the nucleotide sequence for the first 9 aa, gene-walking PCR (18) was performed (using primers described in Table 2) to obtain the 5′ sequence of albB. This procedure yielded a total of 284 nucleotides of sequence upstream of the cloned region. Sequencing revealed an ORF for albB predicted to encode a 336-aa protein. This protein has a predicted cleavage site between Ala (position 46) and Ala (position 47) that would yield a mature bacteriocin with predicted molecular mass of 32,168 Da and a predicted pI of 7.70. The amino acid composition of the putative mature 290-aa bacteriocin displayed a relatively hydrophilic content, with 24 (8.3%) acidic (D and E), 28 (9.7%) basic (K and R), 83 (28.6%) hydrophobic (A, I, F, L, W, and V), and 105 (36.2%) polar (N, C, Q, S, T, and Y) residues. On the basis of its size, its relatively hydrophilic character, and its pI near neutrality, albusin B appears to belong to class III of the bacteriocins defined by Klaenhammer (12).
TABLE 2.
Primers used for gene-walking PCRa
| Primer | Sequence (5′-3′) |
|---|---|
| Arbitrary primers | |
| arb1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
| arb2 | GGCCACGCGTCGACTAGTAC |
| arb6 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC |
| Specific primers | |
| Internal | |
| P101 | CGCACTTTGTACCGCCTCTGTTC |
| P111 | CGACTGCGTTACCAAGTACAGCAC |
| P201 | CAACGAGATGCAACAAACTAAAATC |
| P211 | GGAACAATGAAGAAGGTCATCA |
| External | |
| P102 | GAGGTTGCCGTCGTTTTG |
| P112 | CCTGTTGCTGCCAAAGTGCTG |
| P202 | CCGTAATTTCCAAGCAGAGCATTT |
| P212 | TCATCAGCACTTTGGCAGCAAC |
The primer pairs arb1-P101 and arb2-P102 were used in the first gene-walking PCR run, and arb1-P111 and arb2-P112 were used in the second run, to obtain one DNA strand upstream of the cloned sequence b19. arb1-P201 and arb2-P202 were used in the first run, and arb1-P211 and arb2-P212 were used in the second run, to obtain the complementary strand. The primer arb6 was sometimes substituted for arb1 if the latter failed to yield a product.
Leader peptides are common for bacteriocins from gram-positive bacteria (7), and there is a predicted 46-aa leader peptide preceding the N terminus of the putative mature albusin B. For smaller bacteriocins such as lantibiotics (the class I bacteriocins described by Klaenhammer [12]), the leader peptide sequences are generally shorter (24 to 30 aa) (7). There are no generalized lengths for the leader peptide from larger class III bacteriocins (7). Possible functions of the leader peptide include stabilizing a prepeptide during translation, keeping the molecule biologically inactive against the producing strain, maintaining the specific conformation of the prepeptide during processing, and assisting with the translocation of the prepeptide by specific transport systems (8).
The AGGAAG sequence preceding the translation start by seven nucleotides represents a likely ribosome-binding site, as is typically found in gram-positive bacteria (15). Putative promoter TAAACT (−10) and TAAAAT (−35) regions were found, with 16 bp between them, which is 1 bp less than the consensus distance, and they differ considerably from the consensus sequences TATAAT (−10) and TTGACA (−35). A similar putative promoter sequence was reported in an endoglucanase gene from R. albus AR67 (26), but this promoter was shown to be functional in E. coli and not in R. albus AR67. Other studies of RNA transcribed in E. coli have also shown the occurrence of multiple transcription initiation sites for genes from the ruminal bacteria Prevotella ruminicola (14) and Butyrivibrio fibrisolvens (13).
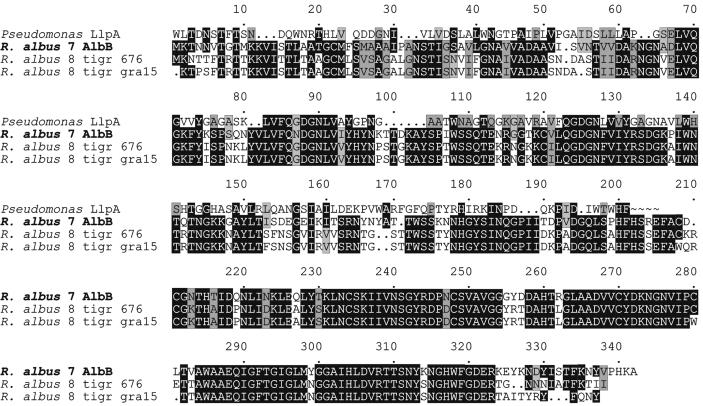
A BLAST search (2) revealed that the deduced mature protein sequence encoded by albB from R. albus 7 displayed 69% identity to the amino acid sequence predicted from two segments of the unfinished genome of R. albus 8 (Fig. 2). The latter, preliminary sequence data were obtained from The Institute of Genomic Research website (www.tigrblast.tigr.org/ufmg). The N-terminal region (aa 1 to 158) of the deduced AlbB protein sequence displayed 27% identity and 46% similarity to a C-terminal region of LlpA, a recently described bacteriocin (putidacin) from a strain of Pseudomonas putida isolated from the rhizospheres of banana plants (19) (Fig. 2). The llpA gene product has not been purified, but the gene has been heterologously expressed in E. coli, resulting in bacteriocin activity in the recombinant strain. Similarities between AlbB and LlpA include size (290 and 276 aa, respectively), sensitivity to heat, relatively hydrophilic character, and the presence of putative mannose-binding domains. Significant alignments (22 to 27% identity) between the N-terminal region of AlbB and a number of lectins and proteins known to bind mannose were also obtained. The sequence similarities of both AlbB and LlpA to these lectins appear to be localized in discrete regions, the so-called monocot mannose binding lectin domains (19). Such domains have been identified in a variety of functionally different proteins produced by phylogenetically diverse, primarily gram-negative bacteria (see reference 19 for a discussion).
FIG. 2.
Multiple alignment of the predicted amino acid sequence of the albB gene product from R. albus 7 with those of a C-terminal portion of LlpA (a 30-kDa bacteriocin encoded by llpA from Pseudomonas sp. strain BW11M1; GenBank accession no. AY118112) (19) and two putative AlbB homologs predicted from the unfinished genome of R. albus 8. Numbers above the sequence indicate the total number of residues. The predicted amino acid sequences for R. albus 8 are from contigs of the unfinished genome obtained from the TIGR website (see the text). Amino acid identity and similarity (1) are indicated with black and gray shading, respectively.
Because weak sequence similarity was noted between AlbB and mannose-binding proteins, and because mannose is a major component of the glycocalyces of both R. albus 7 and R. flavefaciens FD-1 (27), attempts were made to determine if the inhibitory activity of albusin B could be attenuated by exposure to mannose or to glucomannan, a plant polysaccharide containing a backbone of mannosyl units. Neither d-mannose nor Salep glucomannan (0.2 mg/ml, the latter generously provided by J. M. Hackney [5]) attenuated the inhibitory activity of R. albus extracts in the plate culture assay, and the activity of purified albusin B (15 μg) was not removed by exposure to 4 mg of d-mannose or glucomannan. Likewise, Parret et al. (19) have reported that LlpA does not bind mannose. While the importance, if any, of the putative mannose-binding domains in these bacteriocins remains unclear, it appears that these two proteins represent a previously unrecognized group of antimicrobial proteins. Although there are substantial differences between the two bacteriocins (e.g., the lack of a leader sequence in LlpA), their presence in two phylogenetically distant bacterial species from very different environments suggests that this group of bacteriocins may have wide taxonomic distribution.
Purification of only three ruminal bacteriocins has been reported (10, 11, 30), all from noncellulolytic species. All are relatively small (<10-kDa), membrane-active proteins that appear to belong to class I or II, as defined by Klaenhammer (12). Albusin B, the inhibitory agent described in this study, has a relatively high molecular mass (>30 kDa), is relatively hydrophilic, and has a narrow range of antibacterial activity-characteristics typical of class III bacteriocins (12). Albusin B from R. albus 7 resembles the unpurified bacteriocins from R. albus strains 7, MO2a, and MO3g (3) in that they are all heat labile. A BLAST search of the unfinished genome of a related strain, R. albus 8, revealed two sequences similar to that of albB from R. albus 7. The R. albus 8 albB homologs have similar-sized ORFs, and their amino acid sequence identity with albB was 69%, much higher than the similarities of albusin B with mannose-binding proteins. R. albus 8 has been reported to produce a bacteriocin-like agent that inhibits R. flavefaciens FD-1 (16). Although this agent has not been purified, it appears to differ from albusin B described here in that it is heat stable. Thus, the preliminary genomic data suggest that R. albus 8 produces an additional, possibly heat-labile bacteriocin similar to AlbB produced by R. albus 7.
It is widely believed that bacteriocins can have a significant impact on microbial populations in natural environments, and bacteriocins have been suggested as an alternative to the use of antibiotics in animal agricultural operations (9, 20, 24, 29). The former notion is based primarily upon laboratory studies with defined mixed cultures. No experimental data have yet related bacteriocin production and the population sizes of individual bacteriocin-producing species to those of competing, bacteriocin-sensitive species. R. albus has been shown to be more abundant than R. flavefaciens in defined mixed culture on cellulosic substrates in both batch (17) and continuous (4) culture modes. R. albus has also been shown to be more abundant than R. flavefaciens in both the ovine (25) and bovine (28) rumen. Because R. albus has a lower maximum growth rate and poorer affinity for cellodextrin products of cellulose hydrolysis (23), the ability of this species to produce bacteriocins might be expected to play a major role in determining its success against R. flavefaciens in the rumen. However, the relative population sizes of R. albus and R. flavefaciens in the bovine rumen are closely correlated with one another (28), suggesting that both species have a preference for similar environmental conditions in the dynamic ruminal environment. Further research is necessary to determine if albusin B (or any other bacteriocin) in the rumen is present at concentrations sufficient to provide a selective advantage to R. albus in vivo, either in the rumen as a whole or in microenvironments near its site of production.
Nucleotide sequence accession number.
The complete albB sequence has been deposited in the GenBank database under accession number AF469209.
Acknowledgments
This research was supported by project 3655-21000-033-00D of the Agricultural Research Service, U.S. Department of Agriculture. Funding for the R. albus genome sequencing effort is provided by the U.S. Department of Agriculture through a grant to the North American Consortium for Genomics of Fibrolytic Rumen Bacteria.
We thank M. A. Cotta and B. A. Dehority for bacterial strains; C. L. Odt for technical assistance; and H. Goodrich-Blair, W. R. Kenealy, D. A. Kunz, J. B. Russell, and H. J. Strobel for useful discussions. We also thank A. H. A. Parret and R. DeMot for communicating results prior to publication and for useful suggestions.
Mention of specific commercial products is for informational purposes only and does not imply an endorsement or warranty of these products over other products not mentioned.
REFERENCES
- 1.Altschul, S. F. 1991. Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol. 219:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, W. W., and B. A. Dehority. 1999. Production of Ruminococcus flavefaciens growth inhibitor(s) by Ruminococcus albus. Anim. Feed Sci. Technol. 77:61-71. [Google Scholar]
- 4.Chen, J., and P. J. Weimer. 2001. Competition among three predominant ruminal cellulolytic bacteria in the absence or presence of non-cellulolytic bacteria. Microbiology 147:21-30. [DOI] [PubMed] [Google Scholar]
- 5.Hackney, J. M., R. H. Atalla, and D. L. van der Hart. 1994. Modification of crystallinity and crystalline structure of Acetobacter xylinum cellulose in the presence of water-soluble β-1,4-linked polysaccharides. Int. J. Biol. Macromol. 16:215-218. [DOI] [PubMed] [Google Scholar]
- 6.Hungate, R. E. 1966. The rumen and its microbes. Academic Press, New York, N.Y.
- 7.Jack, R. W., J. R. Tagg, and B. Bay. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James, F., C. Lazdunski, and F. Pattus (ed.). 1992. Bacteriocins, microcins and lantibiotics. Springer-Verlag, New York, N.Y.
- 9.Kalmokoff, M. L., F. Bartlett, and R. M. Teather. 1996. Are ruminal bacteria armed with bacteriocins? J. Dairy Sci. 79:2297-2306. [DOI] [PubMed] [Google Scholar]
- 10.Kalmokoff, M. L., D. Lu, M. F. Whitford, and R. M. Teather. 1999. Evidence for production of a new lantibiotic (Butyrivibriocin OR79A) by the ruminal anaerobic Butyrivibrio fibrisolvens OR79: Characterization of the structural gene encoding butyrivibriocin OR79A. Appl. Environ. Microbiol. 65:2128-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalmokoff, M. L., and R. M. Teather. 1997. Isolation and characterization of a bacteriocin (butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B. fibrisolvens. Appl. Environ. Microbiol. 63:394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:9-86. [DOI] [PubMed] [Google Scholar]
- 13.Lin, L.-L., E. Rumbak, H. Zappa, J. A. Thomson, and D. R. Woods. 1990. Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a β-glucosidase. J. Gen. Microbiol. 136:1567-1576. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita, O., J. B. Russell, and D. B. Wilson. 1990. Cloning and sequencing of a Bacteroides ruminicola B14 endoglucanase gene. J. Bacteriol. 172:3620-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin, J. R., C. L. Murray, and J. C. Rabinowitz. 1981. Unique features in the ribosome binding site sequence of the Gram-positive Staphylococcus aureus β-lactamase gene. J. Biol. Chem. 256:11283-11291. [PubMed] [Google Scholar]
- 16.Odenyo, A. A., R. I. Mackie, D. A. Stahl, and B. A. White. 1994. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: development of probes for Ruminococcus species and evidence for bacteriocin production. Appl. Environ. Microbiol. 60:3688-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odenyo, A. A., R. I. Mackie, D. A. Stahl, and B. A. White. 1994. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: pure-culture studies with cellulose and alkaline peroxide-treated wheat straw. Appl. Environ. Microbiol. 60:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker, J. D., P. S. Rabinovitch, and G. C. Burmer. 1991. Targeted gene walking polymerase chain reaction. Nucleic Acids Res. 19:3055-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parret, A. H. A., G. Schoofs, P. Proost, and R. DeMot. 2003. Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J. Bacteriol. 185:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell, J. B. 1984. Factors influencing competition and composition of the rumen bacterial flora, p. 313-345. In F. M. C. Gilchrist and R. I. Mackie (ed.), Herbivore nutrition in the subtropics and tropics. The Science Press, Craighall, South Africa.
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, New York, N.Y.
- 22.Shi, Y., C. L. Odt, and P. J. Weimer. 1997. Competition for cellulose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 63:734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi, Y., and P. J. Weimer. 1996. Utilization of individual cellodextrins by three predominant ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 62:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teather, R. M., and R. J. Forster. 1998. Manipulating the rumen microflora with bacteriocins to improve ruminant production. Can. J. Anim. Sci. 78:57-69. [Google Scholar]
- 25.Van Gylswyk, N. O. 1970. The effect of substituting a low-protein hay on the cellulolytic bacteria in the rumen of sheep and on the digestibility of cellulose and hemicellulose. J. Agric. Sci. (Cambridge) 74:169-180. [Google Scholar]
- 26.Vercoe, P. E., and K. Gregg. 1995. Sequence and transcriptional analysis of an endoglucanase gene from Ruminococcus albus AR67. Anim. Biotechnol. 6:59-71. [Google Scholar]
- 27.Weimer, P. J., A. H. Conner, and L. F. Lorenz. 2003. Solid residues of Ruminococcus cellulose fermentations as components of wood adhesive formulations. Appl. Microbiol. Biotechnol. 63:29-34. [DOI] [PubMed] [Google Scholar]
- 28.Weimer, P. J., G. C. Waghorn, C. L. Odt, and D. R. Mertens. 1999. Effect of diet on population of three species of ruminal cellulolytic bacteria in lactating dairy cows. J. Dairy Sci. 82:122-134. [DOI] [PubMed] [Google Scholar]
- 29.Wells, J. E., D. O. Krause, T. D. Callaway, and J. B. Russell. 1997. A bacteriocin-mediated antagonism by ruminal lactobacilli against Streptococcus bovis. FEMS Microbiol. Ecol. 22:237-243. [Google Scholar]
- 30.Whitford, M. F., M. A. McPherson. R. J. Forster, and R. M. Teather. 2001. Identification of bacteriocin-like inhibitors from rumen Streptococcus spp. and isolation and characterization bovicin 255. Appl. Environ. Microbiol. 67:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]