Abstract
A bacterial strain, PS4040, capable of degrading polycyclic aromatic hydrocarbons for use as the sole carbon source was isolated from oily-sludge-contaminated soil. The 16S rRNA gene showed 98.8% homology to that of Leclercia adecarboxylata. Comparative molecular typing with the clinical strain of L. adecarboxylata revealed that there were few comigrating and few distinct amplimers among them.
Metabolic diversity of bacterial flora is a well-established phenomenon, and a consequence of this diversity is the degradation of various biohazardous or persistent anthropogenic compounds by microbial activities. Among these recalcitrant compounds, polycyclic aromatic hydrocarbons (PAHs) represent a unique class of petroleum hydrocarbons due to their pyrogenic nature and the complexity of the assemblages in which they occur (9). Although many genera of microorganisms have the ability to degrade these recalcitrant compounds and use them as a source of carbon or energy, such phenomena are not commonly encountered in enteric bacteria (3).
The enteric bacteria in the family Enterobacteriaceae are mainly regarded as inhabitants of animal guts (3). The ability of this group to degrade high-molecular-weight PAH compounds appears to be an unusual feature, as this phenomenon has been associated with typical soil bacteria. However, very few reports have indicated utilization of aromatic compounds by enterobacteria, particularly those of the genera Klebsiella, Enterobacter, Escherichia, and Hafnia (3, 5, 7). Although there are several reports of bioremediation of high-molecular-weight PAHs, research pertaining to biodegradation of these substances by enteric bacteria has been relatively rare (3, 9).
In this article we report the isolation of an enteric bacterial strain, PS4040, which can degrade the high-molecular-weight, four-benzene-ring PAH pyrene for use as a sole source of carbon. Phenotypic profiling and sequence analysis identified the strain as Leclercia adecarboxylata. We report for the first time the degradation of PAHs by L. adecarboxylata.
Source, enrichment, and isolation of bacteria.
PAH-degrading bacterial strains were isolated from subsurface soil collected from an oily-sludge storage pit at the Digboi oil refinery, situated in the northeastern region of India (27°15′N, 98°15′E). The site has a history of petroleum hydrocarbon contamination of over 100 years. A defined mineral salt medium (MSM) (11) was used for isolation and enrichment of PAH-degrading bacteria. An oily-sludge-contaminated soil sample (10 g) was used to inoculate 200 ml of MSM containing pyrene (200 mg/liter), and the culture was incubated at 30°C on a rotary shaker (200 rpm) for 7 days. After 10 such cycles of enrichment, 1 ml of the culture was diluted 108-fold and the diluted culture was plated on MSM agar containing pyrene (200 mg/liter). The nine bacterial colonies that gave a zone of clearance on pyrene-coated MSM agar plates were further purified on MSM agar with pyrene as the carbon and energy source.
Sequencing of 16S rRNA gene and phylogenetic analysis.
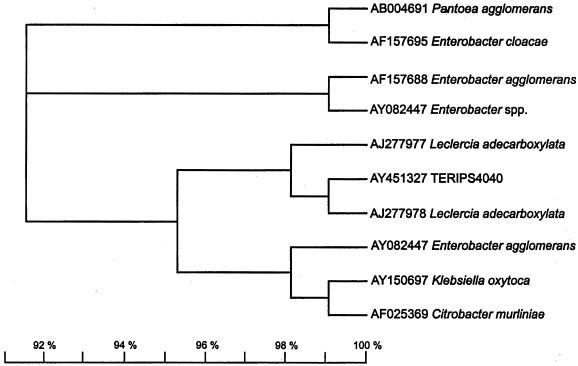
Sequencing of the full-length 16S rRNA genes of the nine PAH-degrading bacterial isolates was done as described previously (2). Alignment of the 16S rRNA gene sequences of these isolates with sequences obtained by doing a BLAST search of the National Center for Biotechnology Information (NCBI), Ribosomal Database Project (RDP), and Microseq (Microseq Analysis software version 1.40, Microseq 16S rRNA Sequence Database version 1.01; PE Applied Biosystems, Inc.) databases revealed up to 98 or 99% similarity to different bacterial species. The highest degree of identity was shown by strain PS4040; it was revealed to be 98.8% identical to L. adecarboxylata in a BLAST search. The 16S rRNA gene sequence of isolate PS4040 differs by only 1.2% from the most closely related sequence, that of L. adecarboxylata, when examined with the RDP SIMILARITY_RANK program (12). A phylogenetic tree, constructed by using the TREEVIEW program (15), illustrates the phylogenetic relatedness of strain PS4040 to the selected Enterobacteriaceae strains obtained from GenBank (NCBI) and RDP (Fig. 1). For further validation of the tree, the sequences were aligned using CLUSTALW(19) and the alignments were analyzed using PAUP, version 3.0 (16), and neighbor-joining (SEQBOOT) programs obtained from the PHYLIP package (4). The results supported the positioning of strain PS4040 in the phylogenetic tree over the alternative positions in a majority of the bootstrap resamplings, indicating a distinct lineage within the Enterobacteriaceae.
FIG. 1.
16S rRNA gene phylogenetic analysis of PS4040 and other members of the family Enterobacteriaceae. The tree was constructed by the TREEVIEW program after an initial analysis of the sequences by using the PAUP program and SEQBOOT software.
Degradation of PAH compounds by strain PS4040 under aerobic conditions.
Growth of the selected bacterial strain PS4040 was verified by demonstrating an increase in bacterial cell protein concentration concomitant with a decrease in the PAH concentration. Strain PS4040 was grown in batch cultures in 500-ml flasks containing 200 ml of MSM supplemented with pyrene (200 mg/liter) as the sole carbon source. The flasks were inoculated with 5% (vol/vol) inoculum (108 CFU/ml) and were incubated at 30°C in the dark on a rotary shaker (200 rpm) for 20 days. At specific intervals, one flask was taken out and 1 ml of culture was withdrawn for protein estimation by the protocol described by Itzaaki and Gill (8). The residual pyrene from the same culture flask was extracted first with an equal volume of toluene and then twice with an equal volume of chloroform. Evaporation of solvents was done under a gentle nitrogen stream in a fume hood. The residual pyrene was dissolved in 5 ml of acetone, and 1 μl of the resultant solution was analyzed by gas chromatography (GC; Hewlett-Packard 5890 series II gas chromatograph fitted with flame ionization detectors and a 30-m-long DB 5.625 column [internal diameter, 0.25 μm; film thickness, 0.25 μm]).
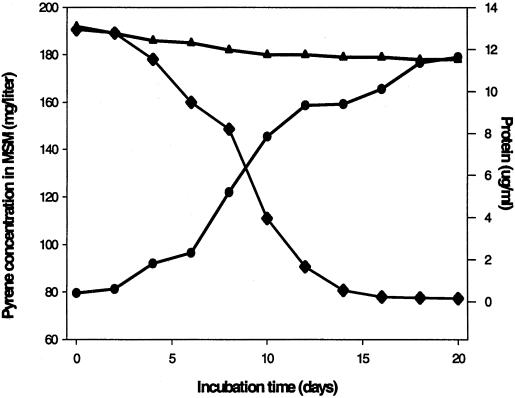
The concentration of pyrene in the culture decreased from 200 to 77 mg/liter, indicating 61.5% degradation of pyrene by strain PS4040, in 20 days (Fig. 2). The bacterial cell protein concentration increased from 0.4 μg/ml at the time of inoculation to a maximum of 10.3 μg/ml at day 15. There was no disappearance of pyrene in the uninoculated control flasks (Fig. 2). Strain PS4040 was also examined for its ability to degrade naphthalene, catechol, anthracene, fluorene, and fluoranthene when used separately as a sole source of carbon under conditions similar to those described above. GC analysis revealed 73.2, 53.1, 40.6, and 47.6% degradation of catechol, naphthalene, fluorene, and fluoranthene, respectively, by strain PS4040 in 20 days, while anthracene did not support growth (data not shown).
FIG. 2.
Degradation of pyrene by L. adecarboxylata PS4040 grown in MSM with pyrene as the sole carbon and energy source. Growth was monitored by estimation of bacterial cell protein (•), while pyrene concentration (♦) was determined by GC analysis of the residual pyrene in culture broth. The pyrene concentration in uninoculated flasks (▴) is also shown.
Extensive research pertaining to the degradation of PAHs has resulted in documentation of a diverse microflora that can either mineralize PAHs or use them as a sole source of carbon and energy (6, 9, 22). The ability of an enteric bacterium to degrade high-molecular-weight PAH compounds appears to be an uncommon finding, as there have been no reports indicating the ability of any enteric bacterial strain to degrade high-molecular-weight PAHs such as pyrene.
L. adecarboxylata strains have been reported to cause a few pathogenic manifestations in humans (1, 17, 18, 24) and to have mercury resistance genes (13), but there have been no reports indicating that this bacterium is capable of degrading PAHs. As discussed by Diaz et al. (3), upon analyzing the ecology of enteric bacteria it can be seen that an enteric bacterium may easily encounter aromatic compounds. Although PAH compounds may not be expected to be normal substrates for enteric bacteria, there is evidence that competition for nutrients results in selection of substrates and shifts in nutrient preference (20). Manonmani et al. (14) also reported a role for substrate concentration in adaptation of, and acquisition of degradative ability by, bacterial flora. Since strain PS4040 was isolated from a site with a contamination history of over 100 years, organic pollutants can be postulated to have exerted effects leading to adaptation of this strain to utilize specific recalcitrant compounds as sources of carbon or energy.
Comparison of strain PS4040 with a clinical strain of L. adecarboxylata by phenotypic tests and molecular typing.
Isolate PS4040 is a short, rod-shaped, motile, gram-negative bacterium. It produces yellowish, circular (2-mm-diameter), convex colonies that have an odor similar to that of Escherichia coli when grown on Luria-Bertani (LB) agar plates for 18 h. The substrate utilization pattern generated on Biolog GN2 plates (10) (Biolog Inc., Hayward, Calif.) identified strain PS4040 as L. adecarboxylata. Biolog tests were done for a clinical strain of L. adecarboxylata (a kind gift from Mario Vaneechoutte, Ghent University Hospital, Ghent, Belgium). Resistance to antibiotics was determined by growing the isolate in LB broth containing different concentration of antibiotics (5 to 40 μg/ml). A comparative representation of the substrate utilization pattern and antibiotic profile is shown in Table 1.
TABLE 1.
Phenotypic comparison and antibiotic profiles of strain PS4040 and the clinical strain of L. adecarboxylata
| Positive for both strains | Negative for both strains | Positive for PS4040, negative for clinical strain | Negative for PS4040, positive for clinical strain |
|---|---|---|---|
| Catalase | Voges-Proskauer | Galactose | Rhamnose |
| β-Glucosidase | α-Ketobutyric acid | Maltose | Cellobiose |
| Glucose | Glutaric acid | Cyclodextrin | Aconitic acid |
| Lactose | Sorbitol | Histidine | Citric acid |
| Mannitol | Erythritol | Phenylalanine | Dextrin |
| Sucrose | Xylitol | Ampicillin (50 μg/ml) | Inositol |
| Fructose | Citrate utilization | Vancomycin (10 μg/ml) | |
| Arabinose | Urease | ||
| Malonic acid | Gelatinase | ||
| Gluconic acid | Oxidase | ||
| Glycerol | H2S production | ||
| Leucine | Ornithine | ||
| Kanamycin (30 μg/ml) | Proline | ||
| Streptomycin (30 μg/ml) | d- and l-serine | ||
| Gentamycin (15 μg/ml) | |||
| Chloromphenical (20 μg/ml) |
Molecular typing of strain PS4040 was done so that a whole-genome comparison with the clinical strain of L. adecarboxylata could be performed using tRNA intergenic spacer length polymorphism (ILP) analysis, as described by Welsh and McClelland (23), and repetitive-element PCR (Rep-PCR)-based DNA fingerprinting with primer sets for repetitive-element (enterobacterial repetitive intergenic consensus, repetitive extragenic palindromic, and BOXAIR) sequences as described elsewhere (2, 23). Rep-PCR and tRNA ILP have been extensively used for strain-level differentiation (1, 2, 21). PCR-based DNA fingerprinting by tRNA ILP and Rep-PCR yielded unique fingerprint patterns for strain PS4040 and the clinical strain of L. adecarboxylata. tRNA ILP resulted in nine distinct bands for strain PS4040, whereas there were six distinct amplimers for the clinical strain. When comparative tRNA ILP analysis of strain PS4040 and the clinical strain of L. adecarboxylata was performed, four of the amplimers comigrated while there was nine distinct amplimers in both strains. ERIC-PCR showed four amplimers for strain PS4040 and seven amplimers for the clinical strain of L. adecarboxylata. Comparison of the banding patterns revealed that two amplimers comigrated while eight unique amplimers resulted for both strains. Similarly, REP-PCR and BOXAIR-PCR respectively generated eight and six amplimers for strain PS4040 but nine and three amplimers for the clinical strain. Comparative analysis of REP-PCR and BOXAIR-PCR banding patterns showed four and two common amplimers for the clinical strain and three and six distinct amplimers for PS4040, respectively. Thus, banding patterns with few comigrating amplimers and few distinct amplimers were observed for both strains. This indicated a specific strain-level genomic difference between these two strains, as discussed previously in several reports (2, 21).
In summary, an enteric bacterium, L. adecarboxylata, with the rare ability to degrade PAHs as a sole source of carbon and energy was isolated, identified, and characterized. It is genotypically different from a clinical strain of L. adecarboxylata. Although isolated from a pyrene-enriched culture, it can degrade other two- and three-benzene-ring PAH compounds. This study provides new insights into the degradation of PAH compounds by enteric bacteria and demonstrates a need for further biochemical and genetic studies of PAH metabolism and transformation
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain PS4040 has been deposited in the GenBank database under accession number AY451327.
Acknowledgments
We are thankful to R. K. Pachauri, Director General, TERI, and to T. P. Singh, Director, TERI School of Advanced Studies, for providing the infrastructure for carrying out the present study. We express our gratitude to Mario Vaneechoette, Ghent University Hospital, Ghent, Belgium, for providing clinical isolates of L. adecarboxylata. Thanks are due to Neeti Chauhan and Theo Smiths (EPFL, Lausanne, Switzerland) for their contributions to the phylogenetic analysis.
We also thank the Department of Biotechnology, Government of India, for funding the research.
REFERENCES
- 1.Baere, T., G. Wauters, A. Huylenbroeck, G. Claeys, R. Peleman, G. Verschraegen, D. Allemeersch, and M. Vaneechoutte. 2001. Isolation of Leclercia adecarboxylata from a patient with a chronically inflamed gallbladder and from a patient with sepsis without focus. J. Clin. Microbiol. 39:1674-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya, D., P. M. Sarma, S. Krishnan, S. Mishra, and B. Lal. 2003. Evaluation of the genetic diversity among some strains of Pseudomonas citronellolis isolated from oily sludge-contaminated sites. Appl. Environ. Microbiol. 69:1435-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz, E., A. Fernandez, M. A. Prieto, and J. L. Garcia. 2001. Bioremediation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 5.Grant, D. J. W. 1967. Kinetic aspects of the growth of Klebsiella aerogenes with some benzenoid carbon sources. J. Gen. Microbiol. 46:213-224. [DOI] [PubMed] [Google Scholar]
- 6.Heitkamp, M. A., J. P. Freeman, D. W. Miller, and C. E. Cerniglia. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ijah, J. J. 1998. Studies on relative capabilities of bacterial and yeast isolates from tropical soil in degrading crude oil. Waste Manag. 18:293-299. [Google Scholar]
- 8.Itzaaki, R. F., and D. M. Gill. 1964. A micro biuret method for estimating proteins. Anal. Biochem. 9:401-410. [DOI] [PubMed] [Google Scholar]
- 9.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konapka, A., L. Oliver, and R. F. Turco. 1998. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 35:103-115. [DOI] [PubMed] [Google Scholar]
- 11.Lal, B., and S. Khanna. 1996. Mineralization of [14C]octacosane by Acinetobacter calcoaceticus S30. Can. J. Microbiol. 42:1225-1231. [Google Scholar]
- 12.Larsen, N., G. J. Olsen, B. L. Maidek, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The Ribosomal Database Project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebert, C. A., J. Wireman, T. Smith, and A. O. Summers. 1997. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manonmani, H. K., D. H. Chandrashekaraiah, N. S. Reddy, C. D. Elcey, and A. A. M. Kunhi. 2000. Isolation and acclimation of a microbial consortium for improved aerobic degradation of α-hexachlorohexane. J. Agric. Food Chem. 48:4341-4351. [DOI] [PubMed] [Google Scholar]
- 15.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 10:41-43. [DOI] [PubMed] [Google Scholar]
- 16.Swofford, D. L. 1991. PAUP: phylogenetic analysis using parsimony. Illinois Natural History Survey, Champaign.
- 17.Tamura, K., R. Shkazaki, Y. Kasoko, and E. Yoshizaki. 1986. Leclercia adecarboxylata gen. nov., comb. nov., formerly known as Escherichia adecarboxylata. Curr. Microbiol. 13:157-158. [Google Scholar]
- 18.Temesgen, Z., D. R. Toal, and F. R. Cockerill III. 1997. Leclercia adecarboxylata infections: case report and review. Clin. Infect. Dis. 25:79-81. [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 21.Versalovic, J., F. J. de Bruijn, and J. R. Lupski. 1998. Repetitive sequences based PCR (Rep-PCR) DNA fingerprinting of bacterial genome, p. 437-454. In F. J. de Bruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes: physical structure and analysis. Chapman and Hall, New York, N.Y.
- 22.Vila, J., Z. Lopez, J. Sabate, C. Minguillon, A. M. Solanas, and M. Grifoll. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring PAH. Appl. Environ. Microbiol. 67:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsh, J., and M. McClelland. 1991. Genomic fingerprints produced by PCR with consensus tRNA primers. Nucleic Acids Res. 19:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo, P. C., E. Y. Cheung, K. Leung, and K. Yuen. 2001. Identification by 16S ribosomal RNA gene of an Enterobacteriaceae species with ambiguous biochemical profile from a renal transplant recipient. Diagn. Microbiol. Infect. Dis. 39:85-93. [DOI] [PubMed] [Google Scholar]