Abstract
Bacterial processes in soil, including biodegradation, require contact between bacteria and substrates. Knowledge of the three-dimensional spatial distribution of bacteria at the microscale is necessary to understand and predict such processes. Using a soil microsampling strategy combined with a mathematical spatial analysis, we studied the spatial distribution of 2,4-dichlorophenoxyacetic acid (2,4-D) degrader microhabitats as a function of 2,4-D degrader abundance. Soil columns that allowed natural flow were percolated with 2,4-D to increase the 2,4-D degrader abundance. Hundreds of soil microsamples (minimum diameter, 125 μm) were collected and transferred to culture medium to check for the presence of 2,4-D degraders. Spatial distributions of bacterial microhabitats were characterized by determining the average size of colonized soil patches and the average number of patches per gram of soil. The spatial distribution of 2,4-D degrader microhabitats was not affected by water flow, but there was an overall increase in colonized patch sizes after 2,4-D amendment; colonized microsamples were dispersed in the soil at low 2,4-D degrader densities and clustered in patches that were more than 0.5 mm in diameter at higher densities. During growth, spreading of 2,4-D degraders within the soil and an increase in 2,4-D degradation were observed. We hypothesized that spreading of the bacteria increased the probability of encounters with 2,4-D and resulted in better interception of the degradable substrate. This work showed that characterization of bacterial microscale spatial distribution is relevant to microbial ecology studies. It improved quantitative bacterial microhabitat description and suggested that sporadic movement of cells occurs. Furthermore, it offered perspectives for linking microbial function to the soil physicochemical environment.
The spatial distribution of soil bacteria has rarely been considered in microbial ecology, particularly not at the microscale. However, its importance in soil functioning can be grasped from simple observations. Soil porosity is variable depending on the soil type, but it commonly represents about 50% of the soil volume. If the total bacterial abundance is 108 cells per cm3 of soil and the cell volume is 1 μm3, soil bacteria occupy only 0.1 mm3 of the 500 mm3 of pores in 1 cm3 of soil. This comparison is even more striking when potential bacterial occupancy is normalized to the surface area in soil. Nevertheless, the influence of bacterial spatial distribution on soil biological functions has not been adequately studied. Although most microbiological research is carried out on the macroscale (grams of soil), bacterial cells interact with their environment at the microscale, which is particularly important in microbial ecology studies. Information on the spatial distribution of microorganisms at the microscale is scarce, and microhabitats are poorly defined (16). In contrast, the spatial distribution of macroscopic organisms is routinely measured as it determines the frequency of encounters with food and other organisms. The frequency of encounters between bacteria and their substrates and among bacteria may be an important parameter driving bacterial activity in soils. Encounters depend on how bacteria are distributed in the bulk soil. At the microscale, do the bacteria coalesce in a few spots, or are they evenly distributed in a soil layer? The lack of quantitative data on the spatial distribution of bacteria at the microhabitat scale (16, 17) is due to limitations of sampling and sample-processing methods. Most previous studies on spatial patterns of bacterial microcolonies were based on microscopic observations (16, 17, 18, 22), which focused on two-dimensional distribution.
A better understanding of the spatial organization of bacterial microhabitats and the probability of encounters could improve our understanding of the spatial variability of bacterial activity and its spatial response to changing environmental conditions and thus could lead to improved mathematical models (2). It might also lead to development of more efficient bioremediation techniques (21), based on increasing the probability of bacterial encounters with substrates. Furthermore, bacterial spatial distribution at the microscale might also influence horizontal gene transfers, since such transfers are dependent on cell-to-cell or cell-to-free DNA contacts.
Grundmann et al. (15) and Dechesne et al. (8) proposed a method that combines a soil microsampling strategy and a mathematical spatial analysis to characterize the spatial distribution of bacterial microhabitats at the microscale. At the microhabitat level, NO2− oxidizers were more spread out than NH4+ oxidizers, which probably resulted in efficient NO2− interception (15). Moreover, different bacterial types present at the same density in soil might have significantly different microscale spatial distributions, and 2,4-dichlorophenoxyacetic acid (2,4-D) degraders might be more spread out than NH4+ oxidizers (8).
The objectives of this work were to identify and characterize changes in the spatial distribution of a bacterial group in soil at the microscale during changes in the population density and to link degradation efficiency and microscale distribution. To do this, 2,4-D degraders were used as a model bacterial group. The organic herbicide 2,4-D is commonly used as a model compound to study the degradation of chlorinated aromatic compounds in soils (13). Populations of 2,4-D degraders are present at low densities in pristine soils, and the abundance of these organisms increases after addition of 2,4-D (24). In this study, experiments were carried out by using unsaturated repacked soil columns under flow conditions comparable to the conditions encountered in the field (e.g., during a heavy rain or field irrigation). The spatial distribution of 2,4-D degrader microhabitats was determined by using the microscale spatial analysis method (15, 8).
MATERIALS AND METHODS
Soil and soil columns.
The soil used was a sandy clay loam (Alfisol, typic hapludalf) in a cultivated field planted with corn (28) (La Côte Saint André, Isère, France) with no history of 2,4-D application. Soil samples were obtained at a depth interval from 0 to 30 cm and were sieved (pore size, 4 mm). The soil was then kept at 15°C and prevented from drying out (gravimetric water content, 15%) in order to maintain its physical structure and to preserve microorganisms. The soil at a depth of 0 to 30 cm had an organic matter content of 2.5%, a C/N molar ratio of 11, a high level of coarse elements (40% of the particles were more than 2 mm in diameter), a bulk density of 1.34 g cm−3, and a pH of 7.0 (25). The water-holding capacity of the soil was 25.8 g of H2O per 100 g (dry weight) (32).
The soil columns consisted of an Altuglass (clear acrylic plastic) cylinder that was 36 cm long and had an inside diameter of 5 cm. The soil columns were prepared by sequentially introducing 80-g portions of humid soil, each of which was packed down in the same manner. This procedure resulted in a dry bulk density of 1.46 ± 0.02 g cm−3 and a porosity of 0.45 ± 0.01 in the columns. A metallic grid (pore size, 1 mm) at the top of the column allowed even spreading of the input solution throughout the column cross section, and a uniform one-dimensional flow was achieved throughout the columns. All experiments were performed at 22°C.
Column experiments.
Column experiments were performed under unsaturated conditions (the volumetric water content was less than the saturation volumetric water content) and with a steady-state water flow (the Darcy flux and the volumetric water content were constant). An input solution (flow rate, 3 cm3 h−1) was applied to the top of each column by using a piston pump (307 model; Gilson), while the output tubing was connected to a fraction collector. At the water flow rate used, the gravimetric water content of the columns was 26.5% ± 0.1%. The columns were percolated with water until steady-state flow conditions were reached. Then a pulse of Br− (used as a flow tracer) and 2,4-D, which is also anionic at the soil pH (pKa 2.64) (4), was applied. The input concentrations of Br− (KBr; Rectapur; Prolabo) and 2,4-D (sodium salt; Sigma Aldrich Chemicals) were both 250 mg liter−1. The electrical conductivity of the input solutions was adjusted with KCl to 415 μS cm−1 (corresponding to the ionic strength of the groundwater at the field site). Water was injected after each pulse of Br− and 2,4-D until no tracer or 2,4-D remained in the output samples.
The following different column experiments were performed: percolation with water (control columns C, five replicates) and injection of one to three pulses of Br− and 2,4-D (one pulse [columns IP, three replicates], two pulses [columns IIP, three replicates], and three pulses [column IIIP, one replicate]). Each pulse volume was equal to one-half the volume of water in the soil column.
Hydrodynamic properties and degradation efficiency.
Hydrodynamic parameters were estimated by fitting the experimental Br− breakthrough curves (i.e., the curves describing the solute concentrations in output samples as a function of time) with the MIM (mobile-immobile) transport model, where solute transport is described by taking into account the fact that part of the fluid is immobile and does not participate in the flow process (7). The apparent dispersion coefficient (in square centimeters per second), the solute exchange coefficient (in hours−1), and the immobile water content (in cubic centimeters per cubic centimeter) were determined (23). A preliminary study (data not shown) indicated that 2,4-D adsorption on this soil is reversible and that it slightly delays solute transport but does not affect the quantity available for transport. Therefore, the 2,4-D mass balance between the injected pulse and the output breakthrough curve was used to calculate the percentage of 2,4-D degraded.
2,4-D and Br− analysis.
Concentrations of 2,4-D were determined by high-performance liquid chromatography (LC-10S; Shimadzu) by using a flow rate of 0.15 cm3 s−1 (isocratic mode) and a 5-μm Hypersil C18 (n-octadecyl) column (250 by 4.6 mm). The eluant was ethanol-water (70:30, vol/vol) acidified with H3PO4 (final concentration, 0.6 mM). The detector (LC-455; Waters) was set to 228 nm.
Concentrations of Br− were determined by capillary electrophoresis (Quanta 4000; Waters). Separation was performed by using a fused silica capillary column (75 μm by 60 cm). Injections were realized by elevating the sample at 10 cm for 40 s. Reverse UV detection was performed at 254 nm (Hg lamp).
Bacterial counts.
Heterotrophic culturable bacteria and 2,4-D degraders were enumerated in the original soil prior to column packing, in control columns (columns C), and in columns amended with 2,4-D (columns IP, IIP, and IIIP). Bacterial enumeration was performed in triplicate. Five-gram soil samples were placed in 50 ml of sterile 0.8% NaCl. A 10-fold dilution series in 0.8% NaCl was prepared from each 50-ml soil sample solution. Then 100-μl portions of appropriate dilutions were plated in triplicate on 10% tryptic soy agar (3) supplemented with cycloheximide (200 mg liter−1) to estimate the soil heterotrophic bacterial population, and 500-μl portions of appropriate dilutions were transferred to 2 ml of Loos culture medium (27) (eight replicates) containing (per liter) 0.166 g of 2,4-D (sodium salt), 0.05 g of yeast extract, 0.016 g of bromocresol purple (sodium salt), 0.02 g of K2HPO4, and 0.005 g of KH2PO4 (pH 7.0) to enumerate 2,4-D degraders by the most-probable-number method (37). The presence of 2,4-D degraders following acidification of the culture medium (27) was determined by a color change from purple to yellow. The incubation times were 5 days for heterotrophic bacteria and 28 days for 2,4-D degraders. Both media were incubated at 28°C in the dark.
The detection threshold of Loos medium (i.e., the number of 2,4-D degraders required to cause the color change of the medium) was estimated. A pure culture of Ralstonia eutropha JMP134(pJP4), a well-studied 2,4-D-degrading strain (10), was grown overnight, and the density of this bacterium was determined by 4′,6′-diamidino-2-phenylindole (DAPI) staining (31) and by using spread plates to check for culturable cells. The density was then adjusted to 40, 8, or 4 CFU ml−1 with 0.8% NaCl. After this, 0.5 ml of each dilution was added to 2 ml of Loos medium (96 replicates) in order to inoculate an average of 20, 4, and 2 cells per well. The number of positive wells (as described above) was recorded after 28 days of incubation in the dark at 28°C. Concurrently, the theoretical numbers of positive wells expected for each dilution were calculated by using the binomial distribution and the hypothesis that a single 2,4-D degrader could induce the color change to yellow.
Soil microsampling.
Soil microsampling and spatial analysis were carried out with the original soil and soil from the depth interval from 2 to 3 cm in control columns (columns C) and columns amended with 2,4-D (columns IP, IIP, and IIIP). The sampling strategy (15) was based on random sampling of soil microsamples of definite sizes, which were then checked for the presence of 2,4-D degraders. Natural soil aggregates were separated with a sterile scalpel blade. Then soil microsamples were taken one by one with a sterile Pasteur pipette, and their sizes were determined with a grid gauge. A sample was referred to as size 125 when its diameter was completely inscribed in a square of the grid gauge that was 125 μm on each side; sizes 250, 500, 1000, and 2000 were determined similarly. A total of 144 size 125 microsamples, 144 size 250 microsamples, 144 size 500 microsamples, 96 size 1000 microsamples, and 96 size 2000 microsamples were taken. Each of the samples was transferred to 2 ml of Loos medium (27) in a 24-well microtiter plate, which was incubated for 28 days at 28°C. Some tests (data not shown) performed with Loos medium without 2,4-D showed that false-positive responses could be obtained (light yellow instead of bright yellow) for the largest microsamples (sizes 1000 and 2000). Consequently, only bright yellow wells were considered positive.
Spatial analysis.
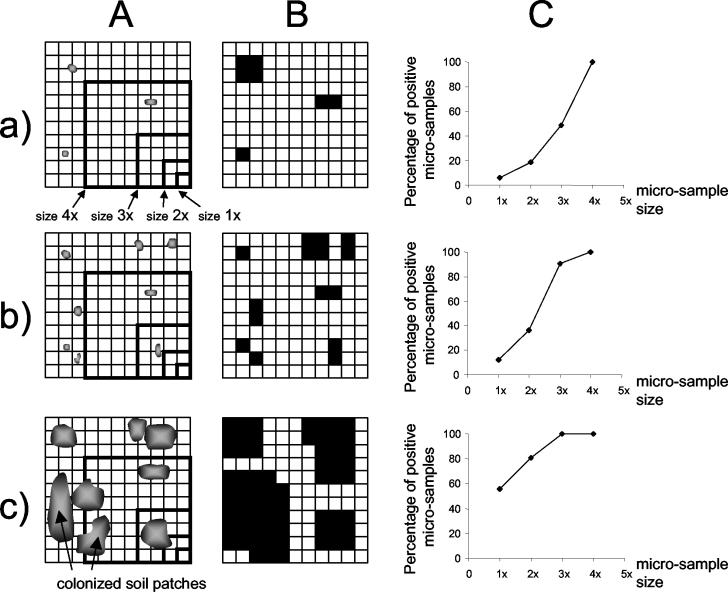
The experimental results were interpreted by plotting the percentage of microsamples positive for the presence of 2,4-D degraders as a function of the microsample size (Fig. 1C). Visual analysis of a set of two-dimensional theoretical distributions (Fig. 1) showed that both the number and the size of soil patches colonized by bacteria determined the resulting curves. Quantitative spatial analysis (8) was used to determine which theoretical spatial distributions of colonized soil patches could correspond to the observed experimental percentages of positive microsamples (i.e., the curves shown in Fig. 1C). The soil was simulated as a three-dimensional cubic grid. Each elementary unit of the grid was a cube with 125-μm sides (125 μm is the diameter of the smallest soil microsamples). Each elementary unit was considered positive if it harbored at least one bacterium. Based on previous results (15, 22), only clustered distributions (random distribution of clusters of positive elementary units) were tested. These clusters referred to aggregates in the simulated grid and corresponded to colonized soil patches in the real soil.
FIG. 1.
Principles of the spatial analysis. (A) Distribution of soil patches colonized by bacteria in a two-dimensional grid with an indication of the four sizes of microsamples used. (B) Same distribution after the test for the presence of bacteria. The black and white elementary units represent positive and negative results, respectively. (C) Corresponding curves obtained after sampling, showing the percentages of positive microsamples as a function of the four microsample sizes. Different types of distribution of bacteria are shown in rows a, b, and c.
Each theoretical spatial distribution was characterized by three parameters: (i) the percentage of elementary units that harbored 2,4-D degraders, (ii) the number of colonized patches per gram of soil, and (iii) the diameter of colonized patches. For each column experiment, the spatial analysis yielded several theoretical distributions compatible with the experimental data (maximum type I error for the acceptance of a theoretical distribution, 0.05). Therefore, for each experiment, the results are presented below on a graph (x axis, diameter of colonized patches; y axis, number of patches per gram of soil) as an area corresponding to the range of compatible theoretical distributions. If two experiments did not share any compatible theoretical distributions (i.e., there were nonoverlapping areas of compatible distributions), their microhabitat spatial patterns were significantly different (maximum type I error, 0.025).
The diameters of the colonized patches of the simulated distribution in the grid were corrected to compensate for the difference in bulk density between the microsamples and the soil columns (1.8 and 1.4 g cm−3). Below, the results are expressed as the average diameter of colonized patches (in millimeters) and the average number of colonized patches per gram of soil. A detailed discussion of the methodology has been presented elsewhere (8, 15).
RESULTS
Bacterial counts and 2,4-D degradation efficiency.
The Loos medium allowed detection of 2,4-D degraders at the single-cell level, and the theoretical binomial distribution fit the results obtained for experimental detection of serially diluted R. eutropha JMP134 suspensions (P > 0.9).
There was no difference in 2,4-D degrader abundance (P > 0.05) between the original soil and the control columns, except for column Cb, in which there was a 1-order-of-magnitude increase (Table 1). Conversely, an increase in 2,4-D degrader abundance was observed in columns amended with 2,4-D; the level increased from 102 degraders per g (dry weight) of soil to 2 × 105 degraders per g (dry weight) of soil after one pulse of 2,4-D and to 106 degraders per g (dry weight) of soil after two or three pulses. The density of heterotrophic bacteria (107 CFU per g [dry weight] of soil) was not affected (P > 0.05) by the addition of 2,4-D (Table 1).
TABLE 1.
Hydrodynamic and microbiological characteristics of original soil, soil from control columns, and soil from columns amended with one, two, or three pulses of 2,4-D
| Treatment | Soila | Apparent dispersion coefficient (cm2 h−1) | Solute exchange coefficient (h−1) | Fraction of mobile water | No. of heterotrophs per g [dry wt] of soil | No. of 2,4-D degraders per g [dry wt] of soil | % of 2,4-D degraded | Determination of spatial distributionb |
|---|---|---|---|---|---|---|---|---|
| Original soil | S | NRc | NR | NR | 3.6 × 107 | 1.9 × 102 | NR | + |
| Control column | Ca | NDd | ND | ND | 2.7 × 107 | 3.1 × 102 | + | |
| Cb | ND | ND | ND | 4.0 × 107 | 3.7 × 103 | + | ||
| Cc | ND | ND | ND | ND | ND | NR | + | |
| Cd | ND | ND | ND | ND | ND | + | ||
| Ce | ND | ND | ND | ND | ND | + | ||
| Column amended | IP1a | 0.05 | 0.03 | 0.83 | 3.8 × 107 | 2.0 × 105 | 25 | + |
| with one pulse | IP1b | ND | ND | ND | ND | 5.6 × 105 | ND | + |
| IP1c | ND | ND | ND | ND | ND | ND | + | |
| Column amended | IIP1a | 0.05 | 0.03 | 0.83 | ND | ND | 46 | − |
| with two pulses | IIP2a | 0.22 | 0.03 | 0.82 | 5.7 × 107 | 5.5 × 106 | 97 | + |
| IIP2b | ND | ND | ND | 1.4 × 107 | 9.4 × 105 | 75 | + | |
| IIP2c | ND | ND | ND | 3.8 × 107 | 1.1 × 106 | 98 | + | |
| Column amended | IIIP1 | 0.05 | 0.03 | 0.83 | ND | ND | 50 | − |
| with three pulses | IIIP2 | 0.22 | 0.03 | 0.83 | ND | ND | 59 | − |
| IIIP3 | 0.22 | 0.03 | 0.83 | 3.8 × 107 | 6.2 × 106 | 73 | + |
S, original soil; C, soil from control column; IP, IIP, and IIIP, soil from columns amended with one, two, and three pulses of 2,4-D, respectively. The first, second, and third pulses are indicated by 1, 2, and 3, respectively. The suffixes a, b, c, d, and e indicate replicates.
+, spatial microsampling performed; −, spatial microsampling not performed.
NR, not relevant.
ND, not determined.
There was a systematic increase in the percentage of 2,4-D degraded with successive 2,4-D pulses; the percentage increased from 46 to 97% between the first and second pulses in column IIPa, from 50 to 59% between the first and second pulses in column IIIP, and from 50 to 73% between the first and third pulses in column IIIP (Table 1). The hydrodynamic properties (namely, the apparent dispersion coefficient, the mobile fraction, and the solute exchange coefficient) were similar for the various columns used (Table 1). Under these reproducible experimental conditions, bacterial activity varied significantly among column replicates, and the first pulse was degraded twice as much in two columns (46% for column IIP1a and 50% for column IIIP1) than in another column (25% for column IP1a) (Table 1). In columns IIP2a and IIIP3 (Table 1), which had similar bacterial densities (5.5 × 106 and 6.2 × 106 degraders g−1; P > 0.05), 97% of the injected 2,4-D and 73% of the injected 2,4-D were degraded, respectively. This indicates that factors other than bacterial abundance or hydrodynamic properties were involved in the degradation.
Frequency of 2,4-D degraders in microsamples.
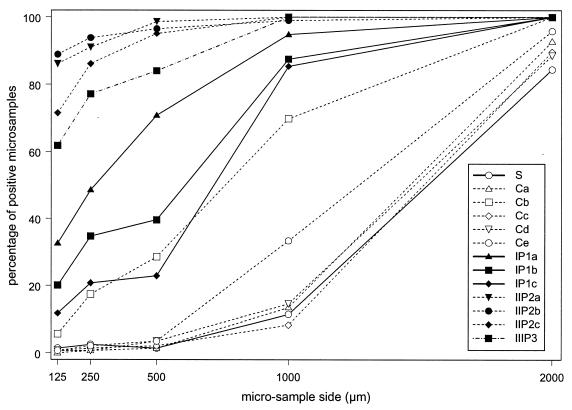
Figure 2 shows the percentages of positive microsamples as a function of the microsample sizes. Interestingly, the curves associated with each type of experiment are ordered as a function of 2,4-D degrader abundance. The results for the control columns (five replicates) were similar and resembled the results for the original soil. Conversely, the percentages of positive microsamples were higher (P < 0.05) for soil percolated with 2,4-D than for the original soil (Fig. 2), indicating that a larger number of microsamples were occupied by 2,4-D degraders. The 2,4-D percolation thus resulted in an increase in the abundance together with spread of 2,4-D degraders. The comparison of the original soil and control columns showed that this spread was not due to the water flow.
FIG. 2.
Experimental results of the spatial analysis: percentages of microsamples that were positive for the presence of 2,4-D degraders as a function of microsample size for the different column treatments. S, original soil; C, soils from control columns; IP1, IIP2, and IIIP3, soils after one, two, and three pulses of 2,4-D, respectively. The suffixes a, b, c, d, and e indicate replicates of the same treatment.
It should be noted that control column Cb, which had a higher 2,4-D degrader abundance than the other control columns, showed higher frequencies (Fig. 2).
Spatial analysis.
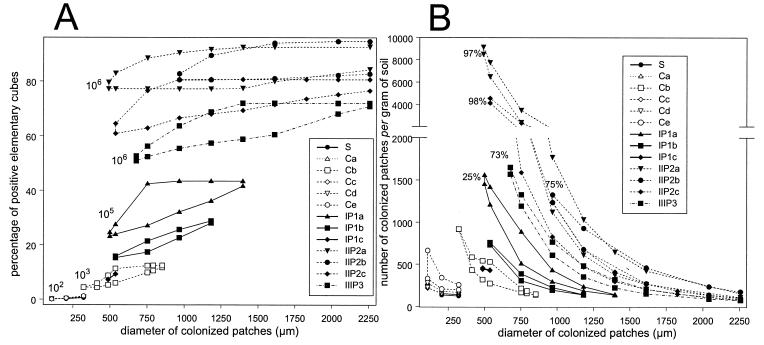
The spatial distributions of 2,4-D degrader microhabitats were characterized further (8). For each experiment, a range of possible spatial distributions was identified, the characteristics of which are represented by areas in Fig. 3. The arrangement of the areas associated with each column was ordered following the increase in 2,4-D degrader abundance. Although acceptance of theoretical distributions was based on large confidence intervals (8), there was limited overlap between the areas corresponding to the different treatments. This means that the calculated spatial characteristics for the different treatments were significantly different (P ≤ 0.025). Consequently, the spatial patterns of 2,4-D degrader microhabitats in soil were different for different 2,4-D degrader abundances.
FIG. 3.
Theoretical distributions compatible with experimental data. Each experiment is represented by an area corresponding to the range of compatible theoretical distributions. Nonoverlapping areas indicate that theoretical distributions are significantly different (P ≤ 0.025). (A) Percentage of positive elementary units in the grid simulating the soil as a function of the diameter of soil patches colonized by 2,4-D degraders. The order of magnitude of 2,4-D degrader densities per gram of soil are indicated next to the results. (B) Number of colonized patches per gram of soil as a function of the diameter of soil patches colonized by 2,4-D degraders. The percentages of 2,4-D degraders are indicated next to the results. S, original soil; C, soils from control columns; IP1, IIP2, and IIIP3, soils after one, two, and three pulses of 2,4-D, respectively. The suffixes a, b, c, d, and e indicate replicates of the same treatment.
The spreading out of 2,4-D degraders caused by 2,4-D amendment could have resulted from an increase in the diameter of the colonized patches and/or from the creation of new colonized patches. There was an increase in the diameter of colonized patches between the original soil and the soil amended with 2,4-D, from 0.1 to 0.3 mm in diameter to 0.5 to 2.25 mm in diameter, when the 2,4-D degrader density increased from 102 to 106 cells per g (dry weight) of soil (Fig. 3A). Conversely, no conspicuous trend was observed for the number of colonized patches per gram of soil; either stability or an increase in the number was possible (Fig. 3B). Indeed, the range of possible numbers of patches per gram of soil varied from 150 to 900 for the control columns to 150 to 1,650 for the columns amended with 2,4-D except columns IIP2a and IIP2c. The areas of accepted distributions for the latter two columns appeared to be atypical (Fig. 3B), and the data revealed a range for the number of patches per gram of soil that was larger than the range for the other columns. These two columns also showed the highest percentages of 2,4-D degraded (97 and 98%, respectively) (Table 1).
Estimation of spatial isolation.
The spatial analysis allowed evaluation of the spatial isolation of 2,4-D degrader microhabitats within the soil. Estimates of the average distance between a colonized patch and its closest neighbor were deteremined with a spatial resolution of 125 μm (smallest sample size) (8); the values were as high as more than 900 μm for control columns (102 CFU g−1), whereas they ranged from 100 to 700 μm after one 2,4-D pulse (105 CFU g−1). After two or three 2,4-D pulses, the 2,4-D degrader microhabitat network was very dense since the colonized patches coalesced.
DISCUSSION
The microscale spatial distribution of bacteria has been hypothesized to be a key parameter in soil, because the frequency of encounters between bacteria and substrates is a determining factor for bacterial activity. Here we used a method in which a soil microsampling strategy and statistical spatial analysis were combined to characterize the distribution of 2,4-D degrader microhabitats as a function of 2,4-D degrader abundance. Successive soil amendment with 2,4-D was used to increase 2,4-D degrader abundance.
The spatial analysis indicated that there was a significant change (P ≤ 0.025) in the spatial distribution of the 2,4-D degraders at the microhabitat scale when the abundance of 2,4-D degraders increased. This observation was based on reliable comparisons as hydrodynamic conditions were controlled and the column setting was reproducible. The soil volume occupied by 2,4-D degrader microhabitats increased from less than 1% to more than 50% when the abundance increased from 102 2,4-D degraders g−1 (control columns) to 106 2,4-D degraders g−1 (columns amended with two or three 2,4-D pulses). However, these values are overestimates due to the size of the smallest microsamples used (125 μm on a side), which is probably larger than the microhabitat volume, as discussed previously (15). The increase in the volume occupied by 2,4-D degrader microhabitats far exceeded the increase in volume occupied by cells if the cells stayed in the form of tight microcolonies. Indeed, if bacteria were organized in microcolonies, the increase in volume occupied by cells when the bacterial density increased from around 102 cells per g (dry weight) of soil to around 106 cells per g (dry weight) of soil could not be detected by using microsamples with 125-μm sides, because when 106 1-μm3 cells stay clustered together, they are inscribed in a cube with 100-μm sides.
The spatial analysis indicated that the spreading out of 2,4-D degraders during 2,4-D percolation was characterized by an overall increase in the sizes of colonized patches; colonized microsamples were dispersed in the soil when the degrader densities were low (102 cells per g [dry weight] of soil) but clustered in patches that were more than 0.5 mm in diameter at higher densities (Fig. 3A). For low abundances, there was strong spatial isolation within the 2,4-D degrader community with a large average distance between colonized patches (>900 μm) and low estimated average number of 2,4-D degraders per colonized patch (<2 2,4-D degraders). A similar spatial isolation was observed previously (36) for an agrobacterial community at the same level (103 CFU g−1); 35 of the 42 microsamples (with 500-μm sides) which harbored agrobacteria were colonized by a single Agrobacterium cell. In the case of higher abundances of 2,4-D degraders, the isolation was reduced as the distance between colonized patches decreased (from 900 μm to contact). Nevertheless, the estimated average number of cells per positive microsample remained low (less than 5 cells at densities less than 106 CFU g−1 and 11 to 28 cells at densities greater than 106 CFU g−1). The low average microscale cell density agrees with the observation that in soil bacterial cells occur singly or in microcolonies consisting of two or three cells (26).
Cell movement involved in the spread of 2,4-D degrader microhabitats can be related to other evidence of microscale cell movement in soil. El Balkhi et al. (12) used electron microscopy to examine thin slices of soil percolated with saccharose and observed the disappearance of bacterial colonies and the appearance of isolated bacteria together with an increase in the number of bacteria. In a study of agrobacteria (36), individuals of a clone could be separated by 1 cm, suggesting that bacterial movement at this scale is common in soils. Several mechanisms could explain the spreading observed here. The hypothesis that there is passive bacterial movement with water flow is unlikely as water flow did not induce changes in the spatial distribution of 2,4-D degraders and very few 2,4-D degraders were found at the outlet of the column (data not shown). Furthermore, the use of repacked soil columns limited the passive movement of bacteria due to a reduction in the number of macropores (6). Two nonexclusive hypotheses may explain the spreading of 2,4-D degraders: active movement and plasmid transfer.
Active movement of cells is the most probable hypothesis as one of the dominant 2,4-D-degrading bacteria isolated from the soil after 2,4-D percolation, Variovorax sp., is motile (data not shown). Furthermore, the water content in the column experiments was favorable for active movement of cells (30). It has recently benn shown (19) that R. eutropha JMP134(pJP4) cells are chemotactically attracted to 2,4-D and that this chemotaxis is induced by growth on 2,4-D and is plasmid encoded. Furthermore, the greater hydrophobicity of newly divided cells than of older cells, as shown for Escherichia coli (1), could explain the detachment of cells. In the case of 2,4-D degraders, it has been suggested that at an early time, bacteria are less firmly attached to soil particles than the remainder of the bacterial community is (20).
2,4-D degradation is often plasmid encoded (5, 11, 24, 35), and conjugation, which could explain soil adaptation to 2,4-D degradation (33), could also contribute to the spreading process. A recent study by Newby et al. (29) provided evidence that there is pJP4 plasmid transfer to an indigenous soil microbial population. Dejonghe et al. (9) showed that a high transconjugant abundance could be reached in some cases. Thus, conjugation was not exclusive to other potential processes, although it required contact between cells and it is known that a small percentage of the soil matrix surface is colonized (14).
More generally, one can ask what major factors drive bacterial spatial patterns in soil. Spatial patterns described for different bacterial types with similar high densities (106 CFU g−1) corresponded to spread-out distributions; these bacterial types included NO2− oxidizers with stable population levels (15) and 2,4-D degraders after growth (this study). This suggests that bacterial abundance plays a role. If it does not, it should be possible to find a bacterial type that is present at a high level but occupies only a few densely colonized patches. The role of the bacterial type was suggested by the comparison of NH4+ oxidizers and 2,4-D degraders, which displayed significantly different patterns in the same soil for similar levels (8). It is important to characterize these bacterial spatial patterns as they could be involved in bacterial activity in soil and could influence biodegradation efficiency at the macroscale. Indeed, for the same 2,4-D degrader density, greater spreading of degraders through the soil volume was associated with a higher percentage of 2,4-D degraded. We suggest that spatial spreading of bacteria increased the probability of encountering the substrate. This could correspond to increased interception potential, leading to improved degradation.
Young and Ritz (38) suggested that structural heterogeneity and biological heterogeneity coexist and are connected in soil. In particular, it has been recognized that spatial isolation may control community structure involved in biological processes relevant to preservation of biodiversity and bioremediation (34). In this respect, spatial isolation refers to relative connectivity within the soil and usually depends on water content and/or soil structure (34). In this study, we estimated the potential spatial isolation due to distances between members of a bacterial group. The distances changed with time, favoring encounters between cells of different taxa and eventually gene transfer. Sporadic cell movements could counteract the effect of spatial isolation. Beyond the mechanism involved in the cell movements, the following question arises concerning the locations of new microhabitat settlements: do they happen at random or deterministically, driven by soil surface quality or substrate location? In this study, the spatial pattern of a bacterial community interacting with a soluble substrate could be characterized, but further work needs to be performed to determine if similar patterns can be found for different species and sampling sites or different soil structures. The microscale used in this study seems to be useful for determining the complex relationships among bacterial localization, bacterial activities, and the physicochemical environment in soil.
REFERENCES
- 1.Allison, D. G., D. J. Evans, M. R. W. Brown, and P. Gilbert. 1990. Possible involvement of the division cycle in dispersal of Escherichia coli from biofilms. J. Bacteriol. 172:1667-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arah, J. R. M. 1990. Modelling spatial and temporal variability of denitrification. Biol. Fertil. Soils 9:71-77. [Google Scholar]
- 3.Balestra, G. M., and I. J. Misaghi. 1997. Increasing the efficiency of the plate counting method for estimating bacterial diversity. J. Microbiol. Methods 30:111-117. [Google Scholar]
- 4.Bekbölet, M., O. Yenigün, and I. Yücel. 1999. Sorption studies of 2,4-D on selected soils. Water Air Soil Pollut. 111:75-88. [Google Scholar]
- 5.Bhat, M. A., M. Tsuda, K. Horiike, M. Nozaki, C. S. Vaidynathan, and T. Nakazawa. 1994. Identification and characterization of a new plasmid carrying genes for degradation of 2,4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl. Environ. Microbiol. 60:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo, M. V., C. Masson, and C. W. Greer. 1997. The influence of moisture on microbial transport, survival and 2,4-D biodegradation with a genetically marked Burkholderia cepacia unsaturated soil column. Biodegradation 8:87-96. [DOI] [PubMed] [Google Scholar]
- 7.Coats, K. H., and B. D. Smith. 1964. Dead-end pore volume and dispersion in porous media. Soc. Pet. Eng. J. 3:49-52. [Google Scholar]
- 8.Dechesne, A., C. Pallud, D. Debouzie, J. P. Flandrois, T. M. Vogel, J. P. Gaudet, and G. L. Grundmann. 2003. A novel method for characterizing the microscale 3-D spatial distribution of bacteria in soil. Soil Biol. Biochem. 35:1537-1546. [Google Scholar]
- 9.Dejonghe, W., J. Goris, S. El Fantoussi, M. Göfte, P. De Vos, W. Verstraete, and E. M. Top. 2000. Effect of dissemination of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation plasmids on 2,4-D degradation and on bacterial community structure in two different soil horizons. Appl. Environ. Microbiol. 66:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Don, R. H., and J. M. Pemberton. 1981. Properties of six degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Don, R. H., and J. M. Pemberton. 1985. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J. Bacteriol. 161:466-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Balkhi, M., F Mangenot, J. Proth, and G. Kilbertus. 1978. Influence de la percolation d'une solution de saccharose sur la composition qualitative et quantitative de la microflore bactérienne d'un sol. Soil Sci. Plant Nutr. 24:15-25. [Google Scholar]
- 13.Estrella, M. R., M. L. Brusseau, R. S. Maier, I. L. Pepper, P. J. Wierenga, and R. M. Miller. 1993. Biodegradation, sorption, and transport of 2,4-dichlorophenoxyacetic acid in saturated and unsaturated soils. Appl. Environ. Microbiol. 59:4266-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, R. C. 1988. Micro-environments of soil micro-organisms. Biol. Fertil. Soils 6:189-203. [Google Scholar]
- 15.Grundmann, G. L., A. Dechesne, F. Bartoli, J. L. Chassé, J. P. Flandrois, and R. Kizungu. 2001. Simulation of the spatial distribution of micro-habitat of NH4+- and NO2−-oxidizing bacteria in soil. Soil Sci. Soc. Am. J. 65:1709-1716. [Google Scholar]
- 16.Harris, P. J. 1994. Consequences of spatial distribution of microbial communities in soil, p. 239-247. In K. Ritz, J. Dighton, and K. Giller (ed.), Beyond the biomass. John Wiley and Sons, Chichester, United Kingdom.
- 17.Hattori, T. 1973. Microbial life in the soil. An introduction. Marcel Dekker, New York, N.Y.
- 18.Hattori, T. 1988. Soil aggregates as microhabitats of microorganisms. Rep. Inst. Agric. Res. Tohoku Univ. 37:23-36. [Google Scholar]
- 19.Hawkins, A. C., and C. S. Harwood. 2002. Chemotaxis of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 68:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holben, W. E., B. M. Schroeter, V. G. M. Calabrese, R. H. Olsen, J. K. Kukor, V. O. Biederbeck, A. E. Smith, and J. M. Tiedje. 1992. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 58:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holden, P. A., and M. K. Firestone. 1997. Soil microorganisms in soil cleanup: how can we improve our understanding? J. Environ. Qual. 26:32-40. [Google Scholar]
- 22.Jones, D., and E. Griffith. 1964. The use of thin sections for the study of soil microorganisms. Plant Soil 20:232-240. [Google Scholar]
- 23.Jury, W. A., and K. Roth. 1990. Transfer functions and solute movement through soils, theory and applications. Birkhaüser Verlag, Basel, Switzerland.
- 24.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kengni, L., G. Vachaud, J. L. Thony, R. Laty, B. Garino, H. Casabianca, P. Jame, and R. Viscogliosi. 1994. Field measurements of water and nitrogen losses under irrigated maize. J. Hydrol. 162:23-46. [Google Scholar]
- 26.Kilbertus, G. 1980. Etude des microhabitats contenus dans les agrégats du sol. Leur relation avec la biomasse bactérienne et la taille des procaryotes présents. Rev. Ecol. Biol. Sol 17:543-557. [Google Scholar]
- 27.Loos, M. A. 1975. Indicator media for microorganisms degrading chlorinated pesticides. Can. J. Microbiol. 21:104-107. [DOI] [PubMed] [Google Scholar]
- 28.Netto, A. M., R. A. Pieritz, and J. P. Gaudet. 1999. Field study on the local variability of soil water content and solute concentration. J. Hydrol. 215:23-37. [Google Scholar]
- 29.Newby, D. T., K. L. Josephson, and I. L. Pepper. 2000. Detection and characterization of plasmid pJP4 transfer to indigenous soil bacteria. Appl. Environ. Microbiol. 66:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papendick, R. I., and G. S. Campbell. 1981. Theory and measurement of water potential, p. 1-22. In J. F. Pau, W. R. Gardner, and L. F. Elliott (ed.), Water potential relations in soil microbiology. Soil Science Society of America special publication 9. Soil Science Society of America, Madison, Wis.
- 31.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 32.Ranjard, L., A. Richaume, L. Jocteur-Monrozier, and S. Nazaret. 1997. Response of soil bacteria to Hg(II) in relation to soil characteristics and cell location. FEMS Microbiol. Ecol. 24:321-331. [Google Scholar]
- 33.Rensing, C., D. T. Newby, and I. L. Pepper. 2002. The role of selective pressure and selfish DNA in horizontal gene transfer and soil microbial community adaptation. Soil Biol. Biochem. 34:285-296. [Google Scholar]
- 34.Treves, D. S., B. Xia, J. Zhou, and J. M. Tiedje. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45:20-28. [DOI] [PubMed] [Google Scholar]
- 35.Vallaeys, T., F. Persello-Cartieaux, N. Rouard, C. Lors, G. Laguerre, and G. Soulas. 1997. PCR-RFLP analysis of 16S rRNA, tfdA and tfdB genes reveals a diversity of 2,4-D degraders in soil aggregates. FEMS Microbiol. Ecol. 24:269-278. [Google Scholar]
- 36.Vogel, J., P. Normand, J. Thioulouse, X. Nesme, and G. L. Grundmann. 2003. Relationship between spatial and genetic distance in Agrobacterium spp. in 1 cubic centimeter of soil. Appl. Environ. Microbiol. 69:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woomer, P. A. 1994. Most probable number counts, p. 59-79. In Methods of soil analysis, part 2. Microbiological and biochemical properties. SSSA Book Series no. 5. Soil Science Society of America, Madison, Wis.
- 38.Young, I. M., and K. Ritz. 1998. Can there be a contemporary ecological dimension to soil biology without a habitat? Soil Biol. Biochem. 30:1229-1232. [Google Scholar]