Abstract
Root mat of cucumbers and tomatoes has previously been shown to be caused by Agrobacterium radiobacter strains harboring a root-inducing Ri plasmid (pRi). Nine other pRi-harboring α-Proteobacteria have subsequently been isolated from root mat-infected crops. Fatty acid profiling and partial 16S rRNA sequence analysis identified three of these strains as being in the genus Ochrobactrum, five as being in the genus Rhizobium, and one as being in the genus Sinorhizobium. An in vitro pathogenicity test involving inoculation of cucumber cotyledons was developed. All pRi-harboring α-Proteobacteria induced typical root mat symptoms from the cotyledons. Average transformation rates for rhizogenic Ochrobactrum (46%) and Rhizobium (44%) strains were lower than those observed for rhizogenic A. radiobacter strains (64%). However, individual strains from these three genera all had transformation rates comparable to those observed from cotyledons inoculated with a rhizogenic Sinorhizobium strain (75%).
Since the early 1990s, hydroponic cucumber and tomato crops in the United Kingdom and some other European countries have been affected by a disorder known as root mat. In affected crops symptoms are expressed as extensive root proliferation within rockwool cubes and slabs with, in severe cases, losses in marketable yield. An earlier outbreak of root mat in the 1970s, in soil and straw bed cucumber crops, had been associated with the presence of Agrobacterium biovar 1 strains (Agrobacterium radiobacter). In the late 1990s, surveys of affected crops and subsequent host tests with Agrobacterium strains isolated during the surveys showed that the disease in cucumbers, and a similar disorder in tomatoes, was caused by A. radiobacter strains harboring a root-inducing Ri plasmid (pRi) (32, 33).
Root mat symptoms are induced following transfer and expression of a segment of pRi DNA (T-DNA) into the plant cell genome. Unlike some other tumor-inducing Ti plasmids (pTi) and pRi, which have two distinct T-DNA fragments, the T-DNA of a root mat-associated (RMA) pRi comprises a single continuous T-DNA fragment (24). The T-DNA of pRi and pTi in transformed plant tissues encode genes that induce cell proliferation and also synthesis genes for unusual amino acid derivatives or sugar-phosphodiesters. These compounds are termed opines and are catabolized by genes also present on the infecting pRi or pTi (12). The opine associated with RMA pRi has been termed cucumopine (5).
The taxonomy of Agrobacterium is confused. The majority of Agrobacterium species form a phylogenetic clade that may include some related organisms, such as Rhizobium galegae, while a number of pRi- or pTi-containing strains, known as Agrobacterium rhizogenes or biovar 2, form a group that is phylogenetically within the genus Rhizobium (see Fig. 2). Although a proposal has recently been made to incorporate all Agrobacterium species into the genus Rhizobium (37), we have retained the established nomenclature because the majority of Agrobacterium species, including the recognized root mat pathogen A. radiobacter biovar 1, are phylogenetically distinct from Rhizobium spp.
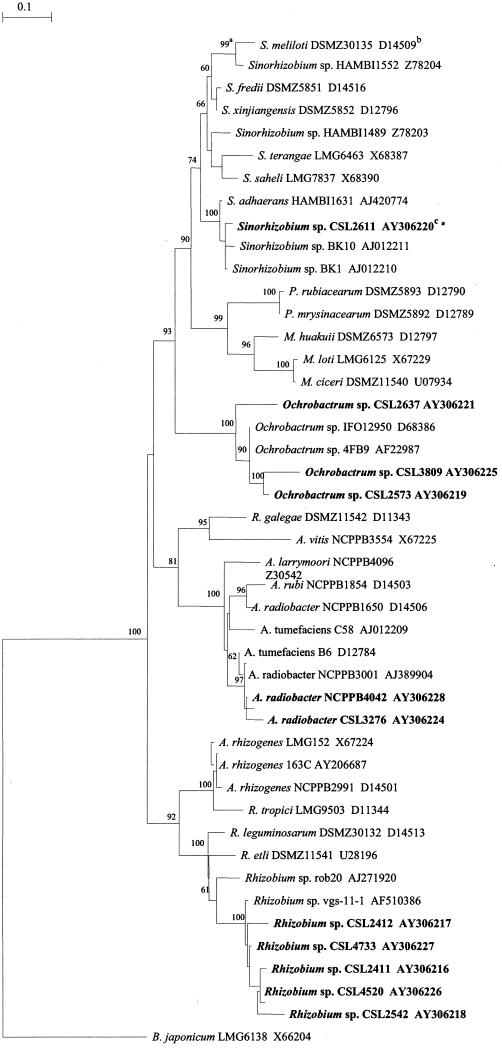
FIG.2.
Neighbor-joining phylogeny for partial 16S rRNA sequences (alignment length, 1,050 bp) showing relationships between rhizogenic α-Proteobacteria isolated from RMA cucumber and tomato crops as well as a panel of representative strains. Footnotes: a, numbers on the dendrogram represent the percentages of grouping confidence calculated by bootstrap analysis (100 bootstrap replicates); b, GenBank accession numbers; c, strains in bold are the nine rhizogenic α-Proteobacteria strains plus two rhizogenic A. radiobacter strains isolated from RMA cucumber crops.
During the surveys of the late 1990s, bacterial strains had been isolated from cucumber and tomato root samples plated onto either nutrient dextrose medium or an Agrobacterium biovar 1 semiselective medium (23) and had been identified by fatty acid profiling (26), which is known to differentiate many taxa within Agrobacterium and other genera of the α-Proteobacteria (28). Strains with fatty acid profiles similar to those obtained from known A. radiobacter were tested by the PCR protocols of Haas et al. (11), which provide a tentative discrimination between pTi and pRi. Strains with fatty acid profiles not consistent with known A. radiobacter strains were stored (at −80°C) but not tested for the presence of the plasmid, as it was assumed that rhizogenic A. radiobacter strains were the causal agent of the disorder.
Experimental transfer of Agrobacterium pTi via conjugation to recipient α-Proteobacteria in other genera has been reported previously (13, 27, 31). Hooykaas et al. (13) reported that Rhizobium leguminosarum biovar trifolii transconjugants were able to induce tumors on inoculated host plants.
This paper reports the isolation, from RMA cucumber and tomato crops, of pRi-harboring α-Proteobacteria which do not belong to the genus Agrobacterium. An in vitro hairy root culture assay was modified to act as a rapid host test to determine if these strains possessed the ability to induce typical root mat symptoms on inoculated cucumber cotyledons.
MATERIALS AND METHODS
Detection of pRi DNA in putative non-Agrobacterium strains.
Over 300 putative non-Agrobacterium strains, isolated from affected cucumber and tomato crops since 1997, were removed from a cryogenic (−80°C) storage system (Protect Bacterial Preservers; Technical Service Consultants, Heywood, Lancashire, United Kingdom) and grown overnight on nutrient agar at 28°C. DNA was extracted from resulting cultures by heating a 100-μl suspension of each culture (6 min, >95°C). A supernatant was obtained from this suspension by centrifugation (14,500 rpm, 3 min) (Eppendorf MiniSpin plus; Eppendorf AG, Hamburg, Germany) and used as template in two PCR assays, the conventional PCR of Haas et al. (11), testing for the virD2 region found in all pTi and pRi, and the rol TaqMan PCR (35), testing for a gene commonly found in the T-DNA of RMA pRi.
Strains positive for both PCR assays were tested by the fliG TaqMan assay (36), an assay specific to Agrobacterium spp., using the original DNA extract as template. Strains were then reidentified by fatty acid profiling (26). Strains were cultured at 28°C for 24 ± 1 h on Trypticase soy agar, and saponification, methylation, and purification of fatty acid methyl esters was carried out in a single test tube. Fatty acid methyl esters were separated by the MIDI microbial identification system (Newark, N.J.) utilizing a Hewlett Packard HP 6890 series gas chromatograph. Profiles were compared with those stored in the MIDI TSBA 40 and in-house National Collection of Plant Pathogenic Bacteria (NCPPB; Central Science Laboratory [CSL], Sand Hutton, York, United Kingdom) libraries. The former comprises a wide range of species of aerobic bacteria, the latter comprises taxa of plant-pathogenic bacteria and is based on strains housed in the NCPPB.
Strain characterization by 16S rRNA sequence analysis.
A DNA extract was prepared from rhizogenic strains which did not have fatty acid profiles consistent with known Agrobacterium species and had tested negative in the fliG PCR assay with a genomic DNA extraction kit (Wizard genomic DNA purification kit; Promega Corporation, Madison, Wis.). These extracts were used for partial 16S rRNA sequencing. Sequencing was done directly from a 1,050-bp PCR product amplified by the primers fA (5′-GGAGAGTTAGATCTTGGCTCAG) and rG (5′-CCCCACCTTCCTCTCGGCTTATC) (2). The PCR product was purified (Wizard SV Gel and PCR Clean-Up System; Promega Corporation) and sent to the University of York Technology Facility for sequencing. The PCR product was sequenced from the forward (fA) and reverse (rG) primers to produce a consensus sequence for the 1,050-bp product. The resulting sequence was compared to similar sequences in the GenBank database via a BLAST search.
In vitro pathogenicity test.
The cucumber hairy root culture protocol of Shi et al. (25) was adapted as follows. Cucumber seeds (cv. Jessica) were surface disinfected overnight in a 5% plant preservative mixture solution (Plant Cell Technology Inc., Washington, D.C.). Disinfected seeds were germinated on hormone-free Murashige and Skoog basal medium (MS) (17) containing 3 g of Phytagel (Sigma-Aldrich Ltd., Poole, Dorset, United Kingdom) per liter at 25°C, 16 h of light. Seedlings were harvested 6 to 10 days postgermination. Cotyledons were removed and cut at the basal end of the leaf (approximately 1-cm cut width) and cultured overnight on hormone-free MS medium at 25°C. Disks were then inoculated with a liquid MS medium containing the bacterial strain to be tested (5-μl loop of overnight culture in 10 ml) by floating the disks on this suspension in a petri dish. After 45 min the disks were placed back on the original MS medium for 48 to 72 h at the original culture conditions. Disks were then transferred onto MS (plus 100 mg of cefotaxime/liter) medium, and root formation was induced at 28°C, 16 h of light. Disks were inoculated with a panel of rhizogenic A. radiobacter strains and all other α-Proteobacteria strains shown to harbor pRi DNA. In all protocols disks were inoculated with liquid MS medium and a rhizogenic A. radiobacter strain (NCPPB 4042) to serve as negative and positive controls, respectively. Plates were observed for the presence of roots every 3 to 4 days.
Extraction of root DNA and analysis by PCR.
To demonstrate the validity of the root culture assay and to indicate that roots, induced from inoculated disks and showing root mat morphology, did contain transformed tissue, DNA was extracted from 0.2 g of root material induced from a cucumber cotyledon inoculated with the rhizogenic A. radiobacter NCPPB 4042 strain (5 weeks after inoculation) by using a DNA extraction kit (Wizard magnetic DNA purification system for food; Promega Corporation). This extract was used as template in the Agrobacterium fliG and the pRi T-DNA rol PCR assays as well as the internal control COX TaqMan PCR assay (34), designed within the published sequence (22) of the constitutive cytochrome oxidase (COX) plant gene.
Nucleotide sequence accession numbers.
The 16S rRNA sequence data cited in this study were deposited in GenBank under accession numbers AY306216 to AY306221 and AY306224 to AY306228.
RESULTS
Identification of rhizogenic non-Agrobacterium spp.
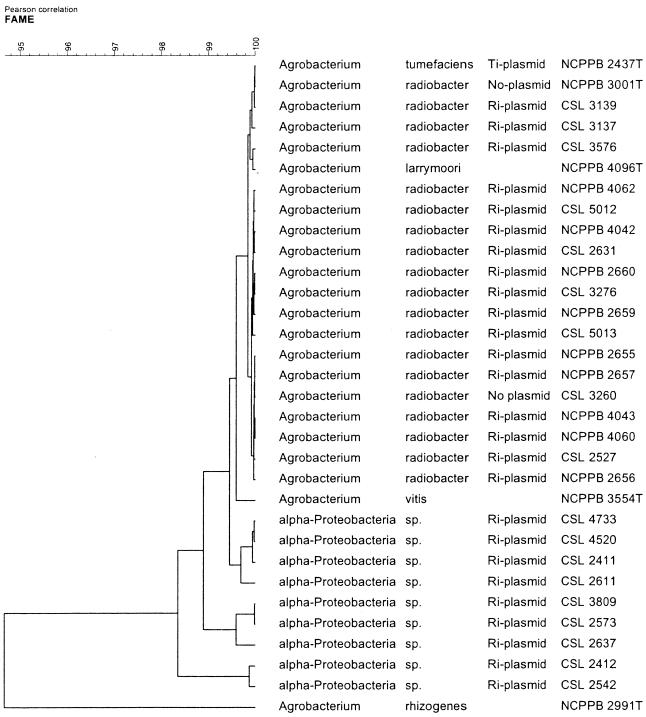
Nine strains were isolated that tested positive in each of the pRi PCR assays, tested negative in the fliG assay, and had fatty acid profiles that indicated they were not in the genus Agrobacterium (Table 1). Phylogenetic analysis of fatty acid data indicated that although these strains do not belong to a known Agrobacterium species, they were closely related to the genus Agrobacterium and were thus termed α-Proteobacteria strains (Fig. 1).
TABLE 1.
Characterization and pathogenicity of pRi-harboring α-Proteobacteria and representative Agrobacterium strains isolated from RMA cucumber and tomato crops
| Strain(s) | Identificationa | Source | PCRb result for:
|
Cucumber hairy root culturec | ||
|---|---|---|---|---|---|---|
| fliG | virD2 | rol | ||||
| CSL 2411 | Rhizobium sp. | Cucumber | − | + | + | Positive (5/11, 45.5%) |
| CSL 2412 | Rhizobium sp. | Cucumber | − | + | + | Positive (2/14, 14.3%) |
| CSL 2542 | Rhizobium sp. | Cucumber | − | + | + | Positive (4/11, 36.4%) |
| CSL 2573 | Ochrobactrum sp. | Cucumber | − | + | + | Positive (8/12, 66.6%) |
| CSL 2611 | Sinorhizobium sp. | Cucumber | − | + | + | Positive (9/12, 75%) |
| CSL 2637 | Ochrobactrum sp. | Cucumber | − | + | + | Positive (3/12, 25%) |
| CSL 3809 | Ochrobactrum sp. | Tomato | − | + | + | Positive (5/11, 45.5%) |
| CSL 4520 | Rhizobium sp. | Cucumber | − | + | + | Positive (9/13, 69.2%) |
| CSL 4733 | Rhizobium sp. | Cucumber | − | + | + | Positive (7/13, 53.8%) |
| CSL 3260 | A. radiobacter | Cucumber | + | − | − | Negative (0/9) |
| All rhizogenic Agrobacterium strains | A. radiobacter | Cucumber | + | + | + | Positive (47/74, 63.5%) |
Identification based on fatty acid profiling and 16S rRNA sequence analysis.
For more information on procedures for fliG PCR, see Weller et al. (36); for virD2 PCR, see Haas et al. (11); for rol PCR, see Weller and Stead (35).
Positive results are from the total number of discs surviving in culture for at least 14 days.
FIG. 1.
UPGMA dendrogram for fatty acid methyl ester data from a panel of RMA A. radiobacter strains, representative Agrobacterium spp., and rhizogenic α-Proteobacteria isolated from RMA crops.
Strain characterization by partial 16S rRNA sequencing.
Partial 16S rRNA sequence analysis of the nine rhizogenic α-Proteobacteria strains indicated that, according to a BLAST search on the GenBank database, these strains were not in the genus Agrobacterium. 16S rRNA sequences of representative α-Proteobacteria were obtained and edited so that the first 11,00 bp of sequence from the position of the fA primer could be aligned with the 16S sequences of the rhizogenic strains, including two pRi-harboring A. radiobacter strains (CSL 3276 and NCPPB 4042) which had been similarly edited. Phylogenetic analysis of these sequences showed that none of the nine rhizogenic α-Proteobacteria strains belonged to the genus Agrobacterium, with strains falling into two main clusters. One cluster of five strains (CSL 2411, 2412, 2542, 4520, and 4733) formed a clade with two Rhizobium spp., and one cluster of three strains (CSL 2573, 2637, and 3809) formed a clade with two Ochrobactrum spp. One other strain (CSL 2611) formed a clade with several Sinorhizobium spp. The two RMA Agrobacterium strains were placed within a clade of other Agrobacterium biovar 1 strains as expected (Fig. 2).
In vitro pathogenicity test.
Results from the in vitro host tests on cucumber cotyledon leaves are summarized in Table 1. Symptoms were first observed as callus formation at the midrib of the cut leaf edge, usually within 5 to 14 days postinoculation. Roots then developed from the callus tissue within a further 5 to 14 days. Roots were thick in appearance, with irregular branching (Fig. 3). Secondary callus formation along the cut edge, and sometimes on the leaf surface at the site of forceps damage, was also observed. Roots again initiated from these calli. Root induction from noninoculated controls and disks inoculated with an Agrobacterium sp. (CSL 3260) not possessing an Ri plasmid was occasionally observed. However, callus tissue was not associated with such disks and roots were thin in appearance, with regular branching, similar in appearance to roots observed in noninoculated cucumber cotyledons of a previous study (25). Because a proportion of cotyledons did not survive the initial tissue culture steps, leaf disks were not considered negative unless they had survived at least 14 days postinoculation. Positives were only those disks from which roots had developed from callus tissue. Occasional disks, inoculated with rhizogenic strains, on which callus tissue was observed but from which roots were not induced were considered negative.
FIG. 3.
Rooting from cucumber cotyledon inoculated with rhizogenic A. radiobacter strain (NCPPB 4042) 12 days postinoculation.
Transformation efficiencies varied between species, with rhizogenic Agrobacterium strains having an average rate of nearly 64%, compared with rates of strains from the Rhizobium and Ochrobactrum clades which averaged nearly 44% and 46%, respectively. The single rhizogenic Sinorhizobium sp. had a transformation efficiency of 75%, although individual rhizogenic Agrobacterium, Rhizobium, and Ochrobactrum strains also had similarly high rates.
Analysis of root culture tissue by PCR.
A DNA extract, prepared from root tissue induced from a cotyledon inoculated with the rhizogenic A. radiobacter strain NCPPB 4042, tested positive in each of the fliG, rol, and COX PCR assays, indicating that both plant and bacterial DNA was present in the DNA extract and implying that the cefotaxime in the MS medium suppressed rather than eliminated A. radiobacter. A DNA extract was prepared from a pure culture of NCPPB 4042. From this extract a 10-fold dilution series was prepared and used as template in individual fliG, rol, and COX PCRs. Mean (two replicates) threshold cycle (Ct) values, a measure of the quantity of DNA present within each sample, from this dilution series were compared with those obtained from the root DNA extract (Table 2). There is a linear relationship between Ct values for rol and fliG in the dilution series, whereas for the root tissue the rol threshold is reached 4.17 cycles earlier than would be predicted by this relationship. From this fact it can be estimated that the abundance of rol relative to fliG is elevated 16.3-fold in the root tissue in comparison to that of the bacterial culture, under the assumption that bacterial DNA was extracted equally from both bacterial and root cultures. It is probable that much of this increase is attributable to transformation of plant tissue with T-DNA (which carries rol but not fliG). However, it is also possible that the copy number of pRi might be higher in the bacteria that have colonized the leaf disk. Changes in pTi copy number of up to eightfold have been reported for Agrobacterium sp. in response to an autoinducer (20). The calculations above necessitate fliG Ct data. As none of the nine α-Proteobacteria strains are detected by the fliG assay (Table 1), it is not possible to extend this assay to root material induced by these strains, as it could not be proven that any increase in rol signal is not solely caused by an abundance of residual bacteria. As the roots induced by these strains showed morphology typical of those induced by rhizogenic A. radiobacter and were also induced from callus tissue, it was assumed that these roots were also transformed.
TABLE 2.
Quantitative PCR demonstrating transformation of plant tissue as well as persistence of bacteriaa
| DNA extract (dilution) | Mean Ct value from PCRb of:
|
||
|---|---|---|---|
| COX | fliG | rol | |
| Root (undiluted) | 24.64 | 21.32 | 18.50 |
| NCPPB 4042 (undiluted) | —c | 16.06 | 17.19 |
| NCPPB 4042 (10−1) | — | 19.30 | 20.72 |
| NCPPB 4042 (10−2) | — | 22.89 | 24.20 |
| NCPPB 4042 (10−3) | — | 25.97 | 27.52 |
Ct values are from three separate TaqMan assays performed on a root DNA extract obtained from a cucumber cotyledon inoculated with NCPPB 4042 and a dilution series obtained from a pure culture of NCPPB 4042.
For information on procedures for COX PCR, see Weller et al. (34); for fliG PCR, see Weller et al. (36); for rol PCR, see Weller and Stead (35).
—, no fluorescence above threshold was detected.
DISCUSSION
To the best of our knowledge, this is the first report of the isolation direct from environmental samples of non-Agrobacterium strains which harbor an Ri plasmid. RMA crops show extensive root proliferation (32, 33). The presence of large populations of pRi-harboring A. radiobacter strains and quantities of cucumopine will create more favorable conditions in such a rhizosphere for both conjugal transfer and maintenance of pRi in recipient bacterial strains than those found for the spread of pTi in crown gall-affected plants, where volumes of transformed tissues and populations of pTi-harboring Agrobacterium strains are generally lower.
Initial fatty acid and PCR analysis identified nine pRi-harboring strains which had profiles that differed from those of a panel of RMA A. radiobacter strains and five Agrobacterium type strains. These strains appeared to be related to the genus Agrobacterium, and thus it was unclear whether these strains belonged to a previously undescribed Agrobacterium species or were in fact genuinely not Agrobacterium strains. A phylogenetic tree constructed from partial 16S rRNA sequence data showed that the rhizogenic strains were placed in three places within the α-Proteobacteria group. Five closely related strains showed closest homology with two Rhizobium strains (rob20 and vgs-11). Three other strains formed a clade with two Ochrobactrum strains (4FB9 and IFO 12950), and an individual strain was placed within the Sinorhizobium cluster. The tree, based on aligned sequences of around 1,100 bp in length, bears close comparison with similar trees constructed by using entire gene (1,400 to 1,500 bp) 16S rRNA sequence data (8, 27).
Although the sole use of the 16S rRNA gene for phylogenetic placement of α-Proteobacteria has recently been questioned (30), the sequence of some 70% of this gene, together with the results obtained from the fliG PCR and fatty acid profiling, allows these nine strains to be confidently placed outside the genus Agrobacterium and into the genera identified. However, further work would be needed in order to assign each of these strains to individual species.
Two PCR assays were employed to demonstrate the presence of pRi in these strains, one targeted to the conserved virD2 gene (11) found in all pTi and pRi and one targeted to the T-DNA of pRi commonly associated with RMA crops (35). We have assumed that positive results in both of these assays and the subsequent results in the in vitro pathogenicity test indicate the presence of pRi in the nine α-Proteobacteria strains, although the presence of pRi was not demonstrated by other techniques, such as electrophoretic isolation of plasmids (14).
Teyssier-Cuvelle et al. (27) report the use of a conjugal transfer system, consisting of a counter-selectable polyauxotrophic derivative of the Agrobacterium sp. C58 strain and a highly transferable pTi (pSTiEGK), which demonstrated the transfer of pTi to recipient Sinorhizobium spp. in soil microcosm experiments. One of the strains isolated in our study (CSL 2611) had very close 16S rRNA sequence similarity to these recipient strains (BK1 and BK10). This transfer system has subsequently been used to demonstrate that pTi can be transferred to other members within the Rhizobiaceae group, including Ochrobactrum strains, but the more taxonomically remote genus Bradyrhizobium members could not accept the plasmid (X. Nesme, personal communication). These results support the observations of our study, i.e., pRi was not detected in strains outside α-Proteobacteria.
The complete nucleotide sequence of the mikimopine-type Ri plasmid pRi1724 has been determined previously (16) and has been shown to have close similarity to the cucumopine-type plasmid pRi2659, a known RMA pRi (32). The conjugal transfer (tra) system of pRi1724 shows closer similarity to the tra system of the sequenced Sinorhizobium Sym plasmid pNGR234 (6) and to some plasmids in R. leguminosarum (29) than to the tra systems in several Ti plasmids (16). The plasmid replication genes of Agrobacterium plasmids are also related to those of rhizobia (21). A Rhizobium symbiotic plasmid (pSymG100) has been shown to belong to the same incompatibility class as the Agrobacterium Ri plasmid (pRi1855) (18), indicating that a self-transmissible RMA pRi could disseminate to other α-Proteobacteria. Large quantities of cucumopine in the rhizosphere are likely to favor conjugation of pRi. Conjugation of pTi has been shown to be induced by the presence of other opines (7).
The small sample size of isolated rhizogenic α-Proteobacteria means it is not possible to determine the full taxonomic range of pRi transfer in RMA rhizospheres. The nine strains were, in effect, isolated by accident during a process designed to isolate Agrobacterium strains. As many α-Proteobacteria are uncultivable on the media used in this study, many potential transconjugants may not have been isolated. Indeed, only cultivable strains were analyzed in this report, and as more than 99% of bacteria present in environmental samples are generally considered to be uncultivable (1), we cannot be certain that pRi cannot disseminate outside α-Proteobacteria.
Rhizogenic A. radiobacter strains have nearly always been associated with RMA crops (32). The extent of symptom expression on whole plants induced by other pRi-harboring α-Proteobacteria in the absence of A. radiobacter strains has not been determined in this study. The cucumopine opine exuded from transformed tissues will likely create a bias favoring cucumopine-catabolizing bacteria (10, 19), and genes for cucumopine catabolism will be present on cucumopine-type pRi. Thus, the presence of cucumopine in the root mat rhizosphere would confer an advantage on pRi-harboring α-Proteobacteria even if they were unable to transform root tissues.
All isolated pRi-harboring α-Proteobacteria induced root mat symptoms following in vitro inoculation of cucumber cotyledons. The pathogenicity of such strains, especially in a natural environment, will not be determined solely by the acquisition of pRi. Chromosomal virulence genes, encoding functions such as bacterial attachment to the plant cell or exopolysaccharide production, are a necessary component of the Agrobacterium transformation mechanism (9). Similar genes, necessary for the establishment of the symbiotic relationship between nodulating α-Proteobacteria spp. and root tissues, have been detected within the Sinorhizobium meliloti chromosome (4, 15), implying that suitable chromosomal elements exist within α-Proteobacteria to ensure pRi-mediated transformation is possible by transconjugant strains. Experimental transfer of a pTi to R. leguminosarum biovar trifolii resulted in this strain becoming pathogenic (13), although transfer of another pTi to S. meliloti did not result in a pathogenic strain despite vir gene induction and T-DNA formation (31). This failure was possibly due to poor attachment of the bacteria to the plant cells or failure of T-DNA transfer from the bacterium and indicates that transfer of pRi may not always result in pathogenic α-Proteobacteria strains in root mat rhizospheres.
Although the in vitro test is limited in that leaf, rather than root, tissue is inoculated, average transformation rates were higher for rhizogenic A. radiobacter strains than for rhizogenic Rhizobium and Ochrobactrum strains. However, individual strains from both of these genera, plus the rhizogenic Sinorhizobium strain, showed higher transformation rates comparable to those seen with individual A. radiobacter strains. This implies that although a randomly selected pRi-harboring A. radiobacter strain is more likely to induce extensive symptoms, strains exist outside the Agrobacterium genus that are also likely to be effective pathogens.
This study has been undertaken as part of a larger project to select biocontrol agent(s) that can control root mat in hydroponic crops. Ri plasmid transfer to other α-Proteobacteria from A. radiobacter complicates this choice. A biocontrol agent that specifically targets A. radiobacter may only lead to root mat being induced by non-Agrobacterium strains harboring pRi. Although we have only shown in vitro induction of root mat symptoms by such strains, the numbers and genetic diversity of potential pRi recipients suggest that there is a high probability that efficient non-Agrobacterium root mat inducers exist. Thus, biocontrol agent(s) with a broad range of action and protection may be the best avenue to success. Even if no efficient non-Agrobacterium pathogens exist, rhizogenic α-Proteobacteria will play an important epidemiological role in the disease. Avirulent Agrobacterium strains are ubiquitous both on hydroponic cucumber nurseries (32) and in nature (3), and thus pRi-harboring non-Agrobacterium species may act as reservoirs of pRi.
Acknowledgments
This study was funded by the Department of Environment, Food, and Rural Affairs (DEFRA), Horticultural Crop Sciences Unit, project no. HH 2308SPC.
We thank Celina Whalley of the University of York for DNA sequencing.
REFERENCES
- 1.Amman, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bala, A., P. Murphy, and K. E. Giller. 2002. Occurrence and genetic diversity of rhizobia nodulating Sesbania sesban in African soils. Soil Biol. Biochem. 34:1759-1768. [Google Scholar]
- 3.Bouzar, H., and L. W. Moore. 1987. Isolation of different Agrobacterium biovars from a natural oak savannah and tallgrass prairie. Appl. Environ. Microbiol. 53:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., L. Hung, V. Puvanesarajah, G. Stacey, D. A. Ozga, J. A. Leigh, and E. W. Nester. 1987. Common loci for Agrobacterium tumefaciens and Rhizobium meliloti exopolysacchride synthesis and their roles in plant interactions. J. Bacteriol. 169:2086-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davioud, E., A. Petit, M. E. Tate, M. H. Ryder, and J. Tempe. 1988. Cucumopine-a new T-DNA encoded opine in hairy root and crown gall. Phytochemistry 27:2429-2433. [Google Scholar]
- 6.Freiburg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaunt, M. W., S. L. Turner, L. Rigottier-Gois, S. A. Lloyd-Macgilp, and J. P. W. Young. 2001. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 51:2037-2048. [DOI] [PubMed] [Google Scholar]
- 9.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyon, P., A. Petit, J. Tempe, and Y. Dessaux. 1993. Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol. Plant-Microbe Interact. 6:92-98. [Google Scholar]
- 11.Haas, J. H., L. W. Moore, W. Ream, and S. Manulis. 1995. Universal PCR primers for detection of phytopathogenic Agrobacterium strains. Appl. Environ. Microbiol. 61:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, S. B., I. Hwang, Y. Dessaux, P. Guyon, K. S. Kim, and S. K. Farrand. 1997. A T-DNA gene required for agropine biosynthesis by transformed plants is functionally and evolutionary related to a Ti plasmid gene required for catabolism of agropine by Agrobacterium strains. J. Bacteriol. 179:4831-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooykaas, P. J. J., P. M. Klapwijk, M. P. Nuti, R. A. Schilperoort, and A. Rörsch. 1977. Transfer of the Agrobacterium tumefaciens TI plasmid to avirulent agrobacteria and to Rhizobium ex planta. J. Gen. Microbiol. 98:477-484. [Google Scholar]
- 14.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marks, J. R., T. J. Lynch, J. E. Karlinsey, and M. F. Thomashow. 1987. Agrobacterium tumefaciens virulence locus pscA is related to the Rhizobium meliloti exoC locus. J. Bacteriol. 169:5835-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriguchi, K., Y. Maeda, M. Satou, N. S. N. Hardayani, M. Kataoka, N. Tanaka, and K. Yoshida. 2001. The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J. Mol. Biol. 307:771-784. [DOI] [PubMed] [Google Scholar]
- 17.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15:473-497. [Google Scholar]
- 18.O'Connell, M. P., M. F. Hynes, and A. Pühler. 1987. Incompatibility between a Rhizobium Sym plasmid and a Ri plasmid of Agrobacterium. Plasmid 18:156-163. [DOI] [PubMed] [Google Scholar]
- 19.Oger, P., A. Petit, and Y. Dessaux. 1997. Genetically engineered plants producing opines alter biological environment. Nat. Biotechnol. 15:369-372. [DOI] [PubMed] [Google Scholar]
- 20.Pappas, K. M., and S. C. Winans. 2003. A LuxR type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 48:1059-1073. [DOI] [PubMed] [Google Scholar]
- 21.Palmer, K. M., S. L. Turner, and J. P. W. Young. 2000. Sequence diversity of the plasmid replication gene repC in the Rhizobiaceae. Plasmid 44:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Quinones, V., S. Zanlungo, L. Holuigue, S. Litvak, and X. Jordana. 1995. The cox1 initiation codon is created by RNA editing in potato mitochondria. Plant Physiol. 108:1327-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroth, M. N., J. P. Thompson, and D. C. Hildebrand. 1965. Isolation of Agrobacterium tumefaciens-A. radiobacter group from soil. Phytopathology 55:645-647. [Google Scholar]
- 24.Serino, G., D. Clerot, J. Brevet, P. Costantino, and M. Cardarelli. 1994. rol genes of Agrobacterium rhizogenes cucumopine strain: sequence, effects and pattern of expression. Plant Mol. Biol. 26:415-422. [DOI] [PubMed] [Google Scholar]
- 25.Shi, H. P, L. Li, and R. C Pan. 1998. Genetic transformation of Cucumis sativus by Agrobacterium rhizogenes. Acta Botanica Sinica 40:470-473. [Google Scholar]
- 26.Stead, D. E., J. E. Sellwood, J. Wilson, and I. Viney. 1992. Evaluation of a commercial microbial identification system based on fatty acid profiles for rapid, accurate identification of plant pathogenic bacteria. J. Appl. Bacteriol. 72:315-321. [Google Scholar]
- 27.Teyssier-Cuvelle, S., C. Mougel, and X. Nesme. 1999. Direct conjugal transfers of Ti plasmid to soil microflora. Mol. Ecol. 8:1273-1284. [DOI] [PubMed] [Google Scholar]
- 28.Tighe, S. W., P. de Lajudie, K. Dipietro, K. Lindstrom, G. Nick, and B. D. W. Jarvis. 2000. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock microbial identification system. Int. J. Syst. Evol. Microbiol. 50:787-801. [DOI] [PubMed] [Google Scholar]
- 29.Turner, S. L., K. A. L. Knight, and J. P. W. Young. 2002. Identification and analysis of rhizobial plasmid origins of transfer. FEMS Microbiol. Ecol. 42:227-234. [DOI] [PubMed] [Google Scholar]
- 30.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindstrom, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Veen, R. J. M., H. den Dulk-Ras, R. A. Schilperoort, and P. J. J. Hooykaas. 1989. Ti plasmid containing Rhizobium meliloti are non-tumourogenic on plants, despite proper virulence gene induction and T-strand formation. Arch. Microbiol. 153:85-89. [Google Scholar]
- 32.Weller, S. A., D. E. Stead, T. M. O'Neill, D. Hargreaves, and G. M. McPherson. 2000. Rhizogenic Agrobacterium biovar 1 strains and cucumber root mat in the UK. Plant Pathol. 49:43-50. [Google Scholar]
- 33.Weller, S. A., D. E. Stead, T. M. O'Neill, and P. S. Morley. 2000. Root mat of tomato caused by rhizogenic strains of Agrobacterium biovar 1 in the UK. Plant Pathol. 49:799. [Google Scholar]
- 34.Weller, S. A., J. G. Elphinstone, N. C. Smith, N. Boonham, and D. E. Stead. 2000. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66:2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weller, S. A., and D. E. Stead. 2002. Detection of root mat associated Agrobacterium strains from plant material and other sample types by post-enrichment TaqMan PCR. J. Appl. Microbiol. 92:118-126. [DOI] [PubMed] [Google Scholar]
- 36.Weller, S. A., S. A. Simpkins, D. E. Stead, A. Kurdziel, H. Hird, and R. J. Weekes. 2002. Identification of Agrobacterium spp. present within Brassica napus seed by TaqMan PCR-implications for GM screening procedures. Arch. Microbiol. 178:338-343. [DOI] [PubMed] [Google Scholar]
- 37.Young, J. M., L. D. Kuykendall, E. Martinez-Romero, A. Kerr, and H. Sawada. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Microbiol. 51:89-103. [DOI] [PubMed] [Google Scholar]