Abstract
Diversity and community structure of aerobic methane-oxidizing bacteria in the littoral sediment of Lake Constance was investigated by cloning analysis and terminal restriction fragment length polymorphism (T-RFLP) fingerprinting of the pmoA gene. Phylogenetic analysis revealed a high diversity of type I and type II methanotrophs in the oxygenated uppermost centimeter of the sediment. T-RFLP profiles indicated a high similarity between the active methanotrophic community in the oxic layer and the inactive community in an anoxic sediment layer at a 10-cm depth. There were also no major changes in community structure between littoral sediment cores sampled in summer and winter. By contrast, the fingerprint patterns showed substantial differences between the methanotrophic communities of littoral and profundal sediments.
Aerobic methane-oxidizing bacteria (methanotrophs) play an important role in controlling methane fluxes from anoxic environments to the atmosphere (11, 19). In freshwater sediments, the activity of methanotrophs is largely restricted to a narrow layer at the oxic-anoxic interface, where methane and oxygen gradients overlap (5, 28, 33).
In the case of Lake Constance, where oxygen penetrates only a few millimeters into the sediment (16, 26; S. Gerhardt, A. Brune, and B. Schink, unpublished data), 90% of the methane produced in the profundal zone (<1 mmol of CH4 m−2 day−1) is oxidized aerobically (16, 39). The situation is different in the much more productive littoral zone (5 to 95 mmol of CH4 m−2 day−1), where a large proportion of methane is lost through ebullition (39). Nevertheless, most of the methane diffusing upwards is also oxidized by methanotrophs (6).
Methanotrophs in littoral sediments are exposed to environmental conditions that differ considerably from those in profundal sediments. The littoral zone is subject to diurnal and annual cycles of light and temperature that also influence other environmental variables, e.g., oxygen status and methane production. In Lake Constance, the oxic-anoxic interface in littoral sediment cores shifts several millimeters between darkness and daylight conditions and methane production differs by 90 mmol of CH4 m−2 day−1 between summer and winter (36, 39).
Other important characteristics differentiating the littoral sediments from the profundal sediments are irregular disturbances due to wave action or changes in the water level. Together with sedimentation, they are responsible for the burial of microbiota, including methanotrophs, in the deeper, anoxic layers of the sediment. Although methanotrophs cannot be metabolically active under such conditions, a previous study has shown a large potential for aerobic methane oxidation in the anoxic zone of littoral sediments of Lake Constance (6).
Although it is suggestive that such conditions should favor a methanotrophic community in littoral sediments different from that in profundal sediments, previous studies concentrated on profundal sediments and its oxygenated zone (2, 3, 10, 11); there is presently no study of diversity and community structure of methanotrophs in littoral sediments of a freshwater lake. In a culture-independent analysis, employing the pmoA gene (encoding the α subunit of the particulate methane monooxygenase) as a molecular marker (11, 24), we investigated the methanotrophic communities in littoral sediment of Lake Constance.
Lake Constance is a warm monomictic lake that is oxic down to the sediment (4). The study sites were located in the bay Obere Güll (littoral, 2-m depth) and at the northern shore between Birnau and Nussdorf (profundal, 90-m depth). Samples for clone libraries and terminal restriction fragment length polymorphism (T-RFLP) analysis were taken in December 2001 (littoral, ∼4°C). Additional samples for T-RFLP analysis were taken in August and September 2002 (littoral, ∼20°C; profundal, ∼4°C). All samples were collected with a modified sediment corer as described by Tessenow et al. (38), using Plexiglas tubes of 37 cm in length and 8 cm in diameter.
Construction of clone libraries.
DNA was extracted from sediment of the uppermost centimeter sampled in winter (1 g [fresh weight]). For clone library A, extraction with the Nucleo Spin Food kit (Macherey-Nagel) was preceded by bead mill homogenization (31) in Nucleo Spin Food lysis buffer (containing proteinase K). Clone library B was derived from the same core by using the beat-beating protocol described by Lueders and Friedrich (31).
Extracted DNA (1 to 5 ng) was used for amplification of pmoA fragments (531 bp) with the primer pair A189f and A682r (23) and recombinant Taq polymerase (MBI Fermentas). Amplification was initiated by denaturation at 95°C for 4 min and proceeded in two phases: (i) a 6-cycle touchdown program (1 min at 92°C, 1 min at 62°C, decreasing 1°C per cycle, and 45 s at 72°C) and (ii) 25 cycles of a standard amplification program at a 56°C annealing temperature. The final extension step was at 72°C for 5 min. PCRs resulted in two amplicons, one matching the predicted size of 531 bp and one approximately 100 bp longer.
Clone libraries were generated from the PCR products by using a TA cloning kit (Invitrogen). pmoA fragments from 105 randomly selected clones (44 and 61 clones from clone libraries A and B, respectively) were amplified by toothpick PCR using recombinant Taq polymerase (MBI Fermentas), analyzed by RFLP with MspI (1.5 U; MBI Fermentas), and grouped according to their restriction patterns.
Phylogenetic analysis of the littoral community.
For at least half of the clones of each RFLP group, both strands were sequenced. Sequences were checked for chimeras by dividing them into two partial sequences of equal length and subjecting the whole sequence and the two partial sequences to separate BLAST searches (1; http://www.ncbi.nlm.nih.gov/BLAST/). If the relatives within such a set of sequences differed, the sequences were excluded from further analysis. In total, 10 chimeras were identified. Sequences of clones resulting from the longer PCR product did not show any affiliation with pmoA sequences according to the results of BLAST searches and were excluded from further analysis.
Sequences were aligned by using the ARB software package (version 2.5b; [http://www.arb-home.de]), followed by a manual correction of the alignment when necessary. A phylogenetic tree of deduced PmoA amino acid sequences was reconstructed based on distance matrices, using the Fitch algorithm (27) (Fig. 1). The use of alternative tree construction methods, such as neighbor joining (35) and maximum-likelihood algorithms, implemented in the ARB software package (evolutionary correction using models of Dayhoff et al. [12] and Jones et al. [25]) resulted in dendrograms with almost identical topology (data not shown).
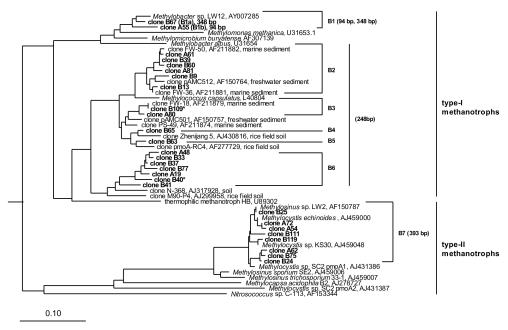
FIG. 1.
Phylogenetic tree based on deduced PmoA amino acid sequences, showing the positions of pmoA clones obtained in this study relative to cultured methanotrophs and pmoA clones of other studies. Clones are grouped into seven groups (B1 to B7) based on at least 90% nucleotide sequence identity. GenBank accession numbers are shown for sequences of cultured methanotrophs and clones of other studies. Numbers next to the clone groups represent the length of the respective T-RFs. Clones marked with an asterisk did not contain a restriction site. The bar indicates 10% sequence divergence. AmoA sequences of Nitrosomonas europaea, Nitrosomonas cryotolerans, and Nitrosospira multiformis were used as an outgroup.
The pmoA sequences obtained in this study were grouped by using a threshold of 90% sequence identity, taking into account the current concept that defines 16S rRNA gene sequences as belonging to the same species if they show >97% sequence identity (37) and the 3.5-times-higher nucleotide substitution rate of the pmoA gene (22). pmoA sequences originating from the two different DNA extraction methods clustered in the same clone groups, with the exception of clones B63 and B65 (Fig. 1). Altogether, seven clone groups were identified; six (B1 to B6) clustered within the family Methylococcaceae (type I methanotrophs), and one (B7) clustered within the family Methylocystaceae (type II methanotrophs). Clone group B1 was closely related (≥90% sequence identity) to Methylobacter spp., with Methylobacter sp. strain LW12 as the closest cultured relative from a freshwater sediment (3). Clone groups B2 to B5 were closely related to pmoA clones from profundal sediment of Lake Washington or soil studies but were not closely related to any pure culture. In contrast, most of the pmoA sequences of clone group B6 were not closely related to any partial pmoA gene of cultured methanotrophs or clones obtained in environmental studies. These pmoA sequences might represent so-far-unknown methanotrophs. Clone group B7 was closely related to Methylocystis and Methylosinus strains isolated in other studies (Table 1).
TABLE 1.
Relative abundance of methanotrophic groups based on frequencies of pmoA genes in clone librariesa
| Phylogenetic group | Clone group | No. of clones | Relative abundance (%) | Next relative (sequence identity) |
|---|---|---|---|---|
| Type I | B1 | 22 | 29 | Methylobacter sp. strain LW12 (3) (93-97%) |
| B2 | 5 | 7 | Clones FW-36 and FW-50 (32) and pAMC512 (11) (94-99%) | |
| B3 | 9 | 13 | Clone Fw-18 (32) (98-100%) | |
| B4 | 1 | 1 | Clone Zhenjiang 5 (GenBank accession no. AJ430816) (94%) | |
| B5 | 1 | 1 | Clone pmoA-RC4 (21) (94%) | |
| B6 | 10 | 13 | Clone N-368 (GenBank accession no. AJ317928) (88-94%) | |
| Type II | B7 | 27 | 36 | Methylocystis spp. (13, 22) (96-99%), Methylosinus sp. strain LW2 (11) (95-99%) |
Clone groups B1 to B7 contain the sequences obtained in this study and are based on at least 90% sequence identity within one group. The sequences of the clones are compared to their next phylogenetic relatives.
T-RFLP fingerprinting.
DNA was extracted by using a bead-beating protocol (31), and pmoA fragments were amplified as described above with two modifications: (i) primer A189f was labeled with a fluorescent dye (IRD 700, pentamethine carbocyanin; MWG), and (ii) FailSafe enzyme mix and PreMix B (Epicentre) were used because amplification with the recombinant Taq polymerase of MBI Fermentas in combination with the labeled A189f primer did not result in PCR products.
PCRs resulted in a single amplicon matching the predicted size of 531 bp. Amplicons were purified by using the MiniElute kit (Qiagen) and quantified photometrically. Approximately 40 ng of DNA was simultaneously digested with the restriction endonucleases AluI and MlsI (3 U each; MBI Fermentas). When amplified pmoA fragments from clones were analyzed, 5 ng of DNA was used. Digestions were carried out in a total volume of 20 μl for 3 h at 37°C. Digested amplicons were mixed with a loading dye containing formamide (stop solution; LI-COR) in a proportion of 5:3, denatured for 3 min at 95°C, and immediately placed on ice. Digested amplicons were electrophoretically separated on polyacrylamide gels (5.5%) at 1,200 V, 25 mA, 30 W, and 50°C using an automated DNA sequencer (model 4200; LI-COR) (signal gain, 64). Sizes of terminal restriction fragments (T-RFs) were calculated by comparison with molecular size markers (50 to 700 bp; LI-COR) and with T-RFs of sequenced pmoA clones as internal standards, using Gelscan Professional software (version 5.02; BioSciTec, Frankfurt, Germany).
In silico analysis of the clonal pmoA sequences showed that the combination of the restriction endonucleases AluI and MlsI gave the best resolution among the different clone groups; only clones B40 and B109 did not contain an AluI or MlsI restriction site. T-RFs of clones B67, A55, B60, and B24, representing clone groups B1a, B1b, B2-B6, and B7, were experimentally verified.
Comparison of different communities.
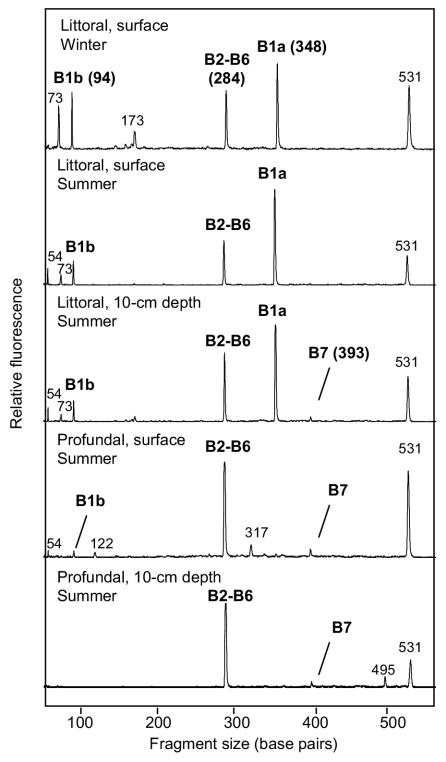
T-RFLP analyses were routinely performed with two independently sampled sediments from the same site, which always resulted in virtually identical profiles (data not shown). The T-RF profiles of the uppermost layer of littoral sediments sampled in winter and summer were similar with respect to both the presence and abundance of the major T-RFs (Fig. 2). Differences were visible in the presence and abundance of small, unaffiliated peaks. The profiles of sediments sampled at a 10-cm depth in summer were very similar to those of the uppermost layer. In all littoral sediment samples, the T-RF representing clone group B1a was most dominant, followed by the T-RFs representing clone groups B2 to B6 and B1b. By contrast, the T-RF representing clone group B7 was detected in only one of the littoral sediment samples collected in summer at a 10-cm depth, where it constituted only a very small peak (Fig. 2). These results indicate a stable community of the major methanotrophic groups throughout the year. This observation is supported by the comparison of the pmoA clone libraries from sediment sampled in winter (this study) to a clone library obtained from the same location during summer (8), where the phylotypes from all libraries clustered in the same groups (details not shown). Temporally stable communities of type II methanotrophs were also previously observed in the rice field ecosystem (15, 20), although the communities of type I methanotrophs changed in these studies.
FIG. 2.
Comparison of different pmoA-based T-RFLP profiles obtained from littoral sediment cores sampled in winter and from littoral and profundal sediment cores sampled in summer at the sediment surface (0 to 1 cm) and at a 10-cm depth. Identified peaks are marked with the represented clone group. The peak at 531 bp represents the undigested pmoA fragment.
T-RF profiles of profundal sediments sampled at the sediment surface and at a 10-cm depth were also quite similar (Fig. 2), despite minor differences in small, unaffiliated peaks. In contrast to littoral sediment samples, however, they were dominated by the T-RFs representing clone groups B2 to B6, whereas the T-RF representing clone group B1a or B1b was completely absent or close to the detection limit. As in the littoral samples, the T-RF representing clone group B7 had only a minor-peak height. It is reasonable to assume that differences in the detected methanotrophic communities of the littoral and profundal sediments reflect the different environmental conditions in the two habitats.
Some of the minor T-RFs, e.g., at 73 and 173 bp, could not be affiliated with the obtained pmoA sequences. These unaffiliated T-RFs differed from the hypothetical restriction sites of the chimeric sequences in the clone libraries, and although rarefaction analysis (17) indicated that the number of sequenced clones was sufficient to describe methanotrophic diversity at the species level (≥90% sequence identity [data not shown]), we must conclude that not all pmoA sequences present in the sediment are represented in the clone library.
All T-RF profiles contained a T-RF that had the length of the undigested PCR product (531 bp). Extending the incubation time to 16 h and increasing the enzyme activity to 10 U did not result in a reduction of this peak, indicating that it did not result from an incomplete digestion. It is possible that this peak represents pmoA genes like those of clones B40 and B109, which are not cut by the restriction enzymes used in this study, but it might also be a pseudo-T-RF (14). Pseudo-T-RFs are PCR artifacts resulting from the inability of restriction enzymes to cut at restriction sites that are situated in the single-stranded part of partially single-stranded PCR products (14).
Comparison of the relative abundance of pmoA clones in the clone libraries to their corresponding T-RFs in the T-RFLP analysis showed an apparent discrepancy. Type II methanotrophs (clone group B7), which constituted the largest fraction of clones in the clone libraries (Table 1), were close to the detection limit in the T-RFLP profiles of all samples obtained from littoral and profundal sediments. Instead, the profiles were dominated by T-RFs of type I methanotrophs, with Methylobacter-like methanotrophs forming the most abundant group. Such discrepancies have also been observed in other studies on 16S rRNA genes (9, 24, 30), and there is currently no simple explanation for this phenomenon. Both methods are subject to a potential PCR bias, which can be excluded only by PCR-independent approaches, e.g., fluorescent in situ hybridization with group-specific 16S rRNA probes (18). Nevertheless, the apparent predominance of type I methanotrophs suggested by the results of the T-RFLP analysis is in agreement with the results obtained for methanotrophs in profundal sediments of Lake Washington, where type I methanotrophs were also shown to dominate the methanotrophic community, using slot-blot hybridizations and polar lipid fatty acid analysis (2, 10). Interestingly, profundal sediments of Lake Washington are dominated by Methylomonas-like methanotrophs, whereas littoral sediments of Lake Constance appear to be dominated by Methylobacter-like methanotrophs.
Profundal sediments of Lake Constance have been shown to contain methanotrophs up to a depth of 32 cm, albeit at decreasing density (34). The oxic-anoxic interface, which sets the lower activity margin of methanotrophs (7), is located in the uppermost centimeter in both profundal and littoral sediments of Lake Constance (16, 26, 39). Nevertheless, spontaneous methane oxidation occurs up to a depth of 7.5 cm, and even in deeper sediment layers, methane oxidation of apparently dormant methanotrophs can be stimulated by the addition of oxygen (34). It seemed therefore reasonable to expect a shift in the methanotrophic communities over depth owing to the ability to survive anoxia and starvation. However, T-RFLP analysis did not detect any differences in the structure of the methanotrophic communities between the uppermost sediment layer, where oxygen and methane gradients overlap, and an anoxic sediment layer at a 10-cm depth. This was true for littoral sediments and for profundal sediments (Fig. 2). T-RFLP profiles describe the diversity of methanotrophs at the DNA level, independent of the expression of methanotrophic activity. Since methanotrophs cannot be metabolically active in the absence of oxygen, the results of this study add strong support to the notion that all methanotrophic populations present in the top layer survive burial and anoxic conditions for a considerable time period. Similar results were obtained with Wadden Sea sediment, where fluorescent in situ hybridization analysis with rRNA-targeted oligonucleotides revealed a similar relative abundance of the major phylogenetic bacterial groups throughout the redox gradient (29).
Nucleotide sequence accession numbers.
The pmoA nucleotide sequences obtained in this study have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession numbers AY488060 to AY488086.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 454).
We thank Dirk Schmitt-Wagner, Ulrich Stingl, and Markus Egert for helpful discussions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Auman, A. J., and M. E. Lidstrom. 2002. Analysis of sMMO-containing Type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517-524. [DOI] [PubMed] [Google Scholar]
- 3.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäuerle, E., and U. Gaedke. 1998. Foreword by the guest editors. Adv. Limnol. 53:V-VII. [Google Scholar]
- 5.Bender, M., and R. Conrad. 1994. Methane oxidation activity in various soils and freshwater sediments: occurence, charateristics, vertical profiles and distribution on grain size fractions. J. Geophys. Res. 99:16531-16540. [Google Scholar]
- 6.Bosse, U., P. Frenzel, and R. Conrad. 1993. Inhibition of methane oxidation by ammonium in the surface layer of a littoral sediment. FEMS Microbiol. Ecol. 13:123-134. [Google Scholar]
- 7.Brune, A., P. Frenzel, and H. Cypionka. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691-710. [DOI] [PubMed] [Google Scholar]
- 8.Bussmann, I., M. Pester, A. Brune, and B. Schink. 2004. Preferential cultivation of type-II methanotrophic bacteria from littoral sediments (Lake Constance). FEMS Microbiol. Ecol., 47:179-189. [DOI] [PubMed]
- 9.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 11.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcull. 1978. A model of evolutionary change in proteins, p. 345-352 In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5. National Biomedical Research Foundation, Silver Spring, Md.
- 13.Dunfield, P. F., M. T. Yimga, S. N. Dedysh, U. Berger, W. Liesack, and J. Heyer. 2002. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41:17-26. [DOI] [PubMed] [Google Scholar]
- 14.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eller, G., and P. Frenzel. 2001. Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl. Environ. Microbiol. 67:2395-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frenzel, P., B. Thebrath, and R. Conrad. 1990. Oxidation of methane in the oxic surface layer of a deep lake sediment (Lake Constance). FEMS Microbiol. Ecol. 73:149-158. [Google Scholar]
- 17.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulledge, J., A. Ahmad, P. A. Steudler, W. J. Pomerantz, and C. M. Cavanaugh. 2001. Family- and genus-level 16S rRNA-targeted oligonucleotide probes for ecological studies of methanotrophic bacteria. Appl. Environ. Microbiol. 67:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson, R. S., and T. S. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henckel, T., P. Roslev, and R. Conrad. 2000. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2:666-679. [DOI] [PubMed] [Google Scholar]
- 21.Henckel, T., U. Jäckel, and R. Conrad. 2001. Vertical distribution of the methanotrophic community after drainage of rice field soil. FEMS Microbiol. Ecol. 34:279-291. [DOI] [PubMed] [Google Scholar]
- 22.Heyer, J., V. F. Galchenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 24.Horz, H.-P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 26.Kappler, A., M. Benz, B. Schink, and A. Brune. 2004. Electron shuttling via humic acids in microbial iron(III) reduction in a freshwater sediment. FEMS Microbiol. Ecol. 47:85-92. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, M. 1983. The neutral theory of molecular evolution, p. 208-233. In M. Nei and R. K. Koehn (ed.), Evolution of genes and proteins. Sinauer, Sunderland, Mass.
- 28.Kuivila, K. M., J. W. Murray, A. H. Devol, M. E. Lidstrom, and C. E. Reimers. 1988. Methane cycling in the sediment of Lake Washington. Limnol. Oceanogr. 33:571-581. [Google Scholar]
- 29.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lueders, T., and M. W. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lueders, T., and M. W. Friedrich. 2002. Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl. Environ. Microbiol. 68:2484-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remsen, C. C., E. C. Minnich, R. S. Stephens, and L. Buchholz. 1989. Methane oxidation in Lake Superior sediments. J. Great Lakes Res. 15:141-146. [Google Scholar]
- 34.Rothfuss, F., M., Bender, and R. Conrad. 1997. Survival and activity of bacteria in a deep, aged lake sediment (Lake Constance). Microb. Ecol. 33:69-77. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbour-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. E vol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Schulz, S., and R. Conrad. 1995. Effect of algal decomposition on acetate and methane concentrations in the profundal sediment of a deep lake (Lake Constance). FEMS Microbiol. Ecol. 16:251-260. [Google Scholar]
- 37.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 38.Tessenow, U., T. Frevert, W. Hofgärtner, and A. Moser. 1975. Ein simultan schlieβender Wasserschöpfer für Sedimentkontaktwasser mit fotoelektrischer Selbstauslösung und fakultativen Sedimentstecher. Arch. Hydrobiol. Suppl. 48:438-452. [Google Scholar]
- 39.Thebrath, B., F. Rothfuss, M. J. Whiticar, and R. Conrad. 1993. Methane production in littoral sediment of Lake Constance. FEMS Microbiol. Ecol. 102:279-289. [Google Scholar]