DNA damage response (DDR) networks have long been noted to be implicated in cell death induced via ionizing radiation (1). These DNA damage sensing and signaling pathways establish control through cell cycle checkpoints, cellular senescence, and apoptosis (2). When functioning properly, DDR networks act as a barrier against tumor growth while maintaining genome integrity. New discoveries have unveiled specific roles of proteins in DDR networks, which may serve as potential therapeutic targets and sensitizers to ionizing radiation (3).
Unfortunately, although a clear connection has been established between dysfunctional DDR networks and malignancy, clinical trials targeting these pathways in the oncology realm have shown limited efficacy to date (4, 5). Lapsed regulation of DDR pathways in malignancy allows cells to bypass cellular checkpoints and progress through the cell cycle with stalled replication forks, incomplete DNA replication, and other forms of DNA damage (6). This genomic instability is propagated through cellular generations resulting in a neoplastic phenotype. A number of specific pathognomonic DDR defects have been identified in a number of cancers, including the mismatch repair protein MSH2 in colorectal cancer and the homologous recombination proteins BRCA1 and BRCA2 in breast and ovarian cancers (7, 8). Recent evidence suggests DDR mishaps may occur at an early stage in some precancerous lesions, double-strand break (DSB) markers such as nuclear gamma-H2AX are significantly elevated (9).
To further understand the role of DDR in malignancy, attention can be turned to the investigation of microRNAs (miRs), as another component of the DDR machinery in post-transcriptional gene regulation (10). miRs are small, non-coding RNA molecules that are complementary to one or more messenger RNA molecules (mRNA) (11). This specific pairing leads to the translational inhibition and degradation of the target mRNA. Global dysregulation of miRNAs is frequently observed in malignancy and patterns of dysregulation seem to be dependent on cancer type (12). More recently, it has been demonstrated that miR expression is regulated by DNA lesions and DDR proteins (13). It is suggested that miRs may play a regulatory role in an intermediary timeframe, in between rapid post-translational protein modifications and delayed transcriptional activation of target genes (14).
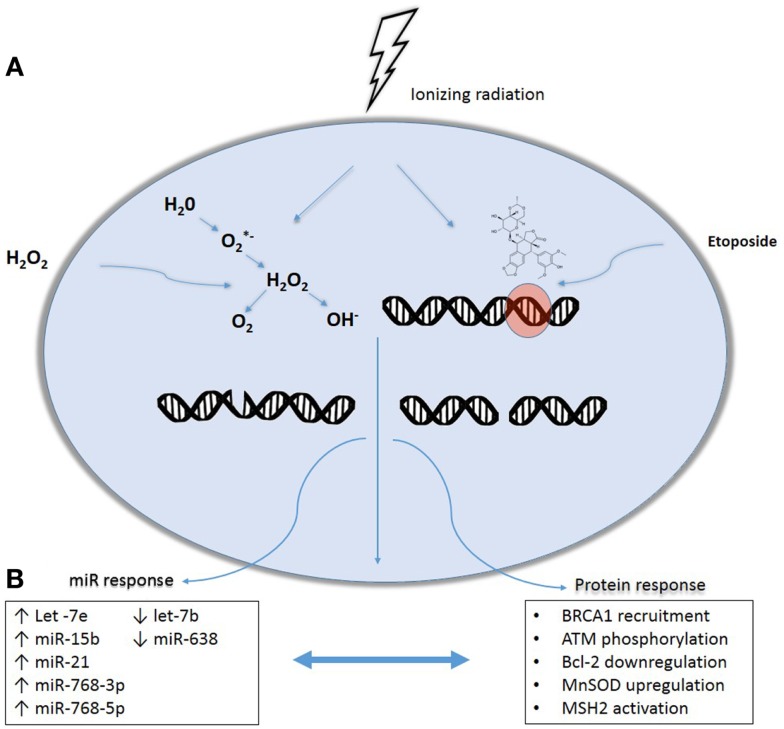
Our laboratory has previously shown that normal human fibroblasts exhibit unique miRNA signatures when exposed to exogenous agents that induce oxidative or genotoxic stress (15). A time course after exposure showed changes in 17 miR species following exposure to radiation, 23 after H2O2 treatment, and 45 after etoposide treatment. The miR signatures varied with direct (etoposide) and indirect (H2O2) effects (Figure 1). Eight miRs were altered specifically by radiation and etoposide, suggesting these might be used to discern direct DNA damage due to radiation. Alternatively, two miRs were altered with radiation and H2O2, suggesting these could comprise a signature of indirect DNA damage. These arrays did not demonstrate any significantly altered miRs that were unique to radiation alone. Interestingly, production of reactive oxygen species (ROS) increased with increasing doses of radiation. Additionally, pre-treatment with the thiol antioxidant cysteine decreased both ROS production and reversed the changes in the miRNA signature in response to irradiation.
Figure 1.
Effects of cytotoxic stressors on microRNAs and protein expression interplay. DNA damage may occur following exposure to genotoxic agents including ionizing radiation, etoposide, and hydrogen peroxide (H2O2) exposure. Ionizing radiation causes DNA damage both directly, by energetic disruption of DNA integrity, and indirectly, as a result of the formation of intracellular free radicals resulting in both double and single strand breaks. Similarly, DNA damage by H2O2 is produced by reactive oxygen species formation while etoposide generates double-strand breaks mimicking the direct DNA damage caused by radiation. (A) In response, damage-sensing repair molecules are recruited to the site of DNA damage, triggering multiple protein-mediated repair cascades. In parallel, DNA damage activates the processing of miRNA precursors, eventually leading to expression of a common miRNA signature produced in response to genotoxic stress. Post-transcriptional regulation of mRNAs mediated by these induced miRNAs plays a fundamental role in adjusting DDR machinery (B), with complex interplay between protein and miRNA expression.
The miRs affected in our study are reflective of more recent literature investigating individual miRs that are altered in response to DDR (16). In fact, they are implicated in more mechanistic studies dealing with homologous recombination, non-homologous end joining, and base excision repair (17, 18). Post-transcriptional regulation of mRNAs mediated by miRs plays a fundamental role in adjusting DDR machinery. miR-421 in neuroblastoma and HeLa cells downregulates ATM kinase, which is a crucial integrator of DNA DSBs repair machinery (19). Ectopic expression of miR-421 leads to S-phase cell cycle checkpoint changes and an increase in radiosensitivity. Although it has not been clearly demonstrated that miRs directly mediate the choice between homologous recombination and NHEJ-mediated repair of a DSB, evidence suggests that miRs are at least intimately involved by targeting factors that belong to a specific pathway. Expression of miR-182 directly downregulates BRCA1 and defers from homologous recombination (20). Alternatively, the expression of miR-101 and miR-34a would downregulate DNA-PKcs and p53 binding protein 1, respectively, impeding the NHEJ repair pathway (21, 22). Other miRNAs, such as miR-34, miR-521, miR-21, have been shown to regulate the expression of important DDR network proteins BCL2, manganese superoxide dismutase (MnSOD), and MSH2, respectively (23–25).
Due to the miRNA regulation of DDR machinery and to the clear connection between DDR dysregulation and a neoplastic phenotype, we believe miRs could define the relationship between cancer and DDR. Our laboratory’s studies suggest that miRs serve as integrators of the cellular response to ROS and DNA strand breaks, both of which are results of ionizing radiation. It is our opinion that further investigation of miR impact on cellular sensitivity to DNA-damaging agents could elucidate therapeutic targets to combat cancer, as miRs may provide the link between DDR and malignancy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was supported in part by the Kimmel Cancer Center’s NCI Cancer Center Support Grant P30 CA56036.
References
- 1.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. Philadelphia, PA: Lippincott Williams & Wilkins; (2012). [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem (2004) 73:39–85 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- 3.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science (2007) 316:1160–6 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- 4.O’Shaughnessy J, Schwartzberg LS, Danso MA, Rugo HS, Miller K, Yardley DA, et al. A randomized phase III study of iniparib (BSI-201) in combination with gemcitabine/carboplatin (G/C) in metastatic triple-negative breast cancer (TNBC). J Clin Oncol (2011) 29:201121502544 [Google Scholar]
- 5.Ma CX, Ellis MJC, Petroni GR, Guo Z, Cai S-R, Ryan CE, et al. A phase II study of UCN-01 in combination with irinotecan in patients with metastatic triple negative breast cancer. Breast Cancer Res Treat (2013) 137:483–92 10.1007/s10549-012-2378-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartkova J, Horejší Z, Koed K, Krämer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature (2005) 434:864–70 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- 7.Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell (1993) 75:1027–38 10.1016/0092-8674(93)90546-3 [DOI] [PubMed] [Google Scholar]
- 8.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell (2002) 108:171–82 10.1016/S0092-8674(02)00615-3 [DOI] [PubMed] [Google Scholar]
- 9.Gorgoulis VG, Vassiliou LVF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature (2005) 434:907–13 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- 10.Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell (2007) 26:745–52 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science (2001) 294:853–8 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A (2006) 103:2257–61 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNAs functions in the DNA damage response. Nat Rev Mol Cell Biol (2013) 14(3):181–9 10.1038/nrm3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai EC. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet (2002) 30:363–4 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- 15.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, DeGraff W, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One (2009) 4:e6377. 10.1371/journal.pone.0006377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pothof J, Verkaik NS, van Ijcken W, Wiemer EAC, Ta VTB, van der Horst GTJ, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J (2009) 28:2090–9 10.1038/emboj.2009.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res (2009) 69:1221–9 10.1158/0008-5472.CAN-08-2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A (2010) 107:5375–80 10.1073/pnas.0910015107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A (2010) 107:1506–11 10.1073/pnas.0907763107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell (2011) 41(2):210–20 10.1016/j.molcel.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo Y-Y, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One (2010) 5(7):e11397. 10.1371/journal.pone.0011397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kofman AV, Kim J, Park SY, Dupart E, Letson C, Bao Y, et al. microRNA-34a promotes DNA damage and mitotic catastrophe. Cell Cycle (2013) 12:3500–3511 10.4161/cc.26459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene (2007) 26:5017–22 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- 24.Josson S, Sung S-Y, Lao K, Chung LWK, Johnstone PAS. Radiation modulation of microRNA in prostate cancer cell lines. Prostate (2008) 68:1599–606 10.1002/pros.20827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci U S A (2010) 107:21098–103 10.1073/pnas.1015541107 [DOI] [PMC free article] [PubMed] [Google Scholar]