Abstract
Fusarium verticillioides, a pathogen of maize, produces a class of mycotoxins called fumonisins in infected kernels. In this study, a candidate regulatory gene, ZFR1, was identified in an expressed sequence tag library enriched for transcripts expressed by F. verticillioides during fumonisin B1 (FB1) biosynthesis. ZFR1 deletion mutants exhibited normal growth and development on maize kernels, but fumonisin production was reduced to less than 10% of that of the wild-type strain. ZFR1 encodes a putative protein of 705 amino acids with sequence similarity to the Zn(II)2Cys6 binuclear cluster family that are regulators of both primary and secondary metabolism in fungi. Expression of ZFR1 in colonized germ and degermed kernel tissues correlated with FB1 levels. Overexpression of ZFR1 in zfr1 mutants restored FB1 production to wild-type levels; however, FB1 was not restored in an fcc1 (Fusarium C-type cyclin) mutant by overexpression of ZFR1. The results of this study indicate that ZFR1 is a positive regulator of FB1 biosynthesis in F. verticillioides and suggest that FCC1 is required for ZFR1 function.
Fusarium verticillioides (Sacc.) Nirenburg (teleomorph: Gibberella moniliformis Wineland) is a phytopathogenic filamentous fungus that infects maize kernels, where it produces a group of mycotoxins known as fumonisins (24). Of the 15 fumonisin analogs isolated and characterized, fumonisin B1 (FB1) typically is found at the highest levels and is toxicologically the most important (22). Consumption of FB1-contaminated maize causes leukoencephalomalacia in equids, pulmonary edema in swine, and liver cancer in rats (16, 30, 40). Because of the potential health risks, guidelines for fumonisin levels in food have been established by the U.S. Food and Drug Administration and by other government agencies worldwide (11).
The initial studies on the structure of fumonisins indicated that these compounds are metabolites of a polyketide pathway (12). Proctor and coworkers confirmed this when they isolated and characterized the FUM1 gene, which encodes a polyketide synthase (26). Strains of F. verticillioides with a disrupted FUM1 gene fail to produce detectable levels of fumonisins (26). These researchers further established that FUM1 is one of 15 FUM genes clustered on chromosome 1 that are involved in fumonisin biosynthesis or possible self-protection from fumonisins (27). Included in the FUM cluster are genes encoding proteins similar to cytochrome P450 monooxygenases (FUM6, FUM12), dehydrogenases (FUM7, FUM13), an aminotransferase (FUM8), a dioxygenase (FUM9), a fatty acyl-coenzyme A synthetase (FUM10), a tricarboxylate transporter (FUM11), a peptide synthetase (FUM14), longevity assurance factors (FUM17, FUM18), and an ABC transporter (FUM19). Mutants with the clustered genes disrupted have aided in characterization of the fumonisin biosynthetic pathway. Butchko et al. (3) provided the first biochemical evidence directly linking a FUM gene (FUM13) to a specific reaction during fumonisin biosynthesis. The Fum13 protein, which has similarity to short-chain dehydrogenases/reductases, was found to catalyze the reduction of the C-3 carbonyl of the fumonisin backbone to a hydroxyl group.
A current focus of research is the molecular mechanisms that regulate fumonisin biosynthesis. For many fungal secondary metabolites, the structural genes responsible for biosynthesis are clustered, and specific regulatory genes have been found to reside in those gene clusters (reviewed by Keller and Hohn [14]). None of the 15 genes within the FUM cluster appears to have a regulatory function. However, two recently described genes (PAC1 and FCC1) appear to impact the regulation of fumonisin biosynthesis (10, 34). Experimental evidence showed that a PAC1 disruption mutant of F. verticillioides produced more FB1 than the wild-type strain and that only the mutant produced FB1 under alkaline conditions (10). The results indicated that Pac1 acts as a transcriptional repressor of fumonisin structural genes. F. verticillioides with a disrupted FCC1 gene fails to produce FB1 on maize kernels (34). The deduced product of FCC1 is similar to C-type cyclins, a class of proteins involved in the transcriptional activation or repression of genes associated with stress responses and development (7, 17, 19). Interestingly, the strain with FCC1 disrupted produces FB1 when cultured on synthetic media buffered at acidic pH. How acidic pH restores the ability of the fcc1 mutant to produce FB1 is unknown.
Here we describe the isolation of a putative regulatory gene and characterize its effect on FB1 biosynthesis. This gene, named ZFR1, encodes a polypeptide with significant homology to fungal proteins that contain a DNA binding motif consisting of a Zn(II)2Cys6 binuclear cluster (36). We introduced deletion mutations in the ZFR1 gene of F. verticillioides and show that the zfr1 mutants exhibited normal growth and conidiation when cultured on maize kernels, while FB1 biosynthesis was severely impaired. The results presented in this report support the hypothesis that ZFR1 is a positive regulator of genes involved in fumonisin biosynthesis by F. verticillioides.
MATERIALS AND METHODS
Strains and media.
F. verticillioides strains 7600 (M3125; Fungal Genetics Stock Center, Kansas City, Kans.) and FT536 (fcc1Δ mutant) were stored in 20% glycerin at −80°C. For inoculum, the strains were grown on potato dextrose agar (Difco Laboratories, Detroit, Mich.) at 28°C. For isolation of genomic DNA, fungi were grown in stationary YEPD medium (5 g of yeast extract per liter, 10 g of peptone per liter, 20 g of glucose per liter) at 28°C. For FB1 analysis and RNA isolation, fungi were grown on defined liquid (DL) medium (33) and cracked maize kernels as previously described (10).
Experiments that examined growth and fumonisin production by the wild-type, zfr1 deletion mutant, and ZFR1-rescued strains were conducted with DL medium, whole cracked maize kernels, and separated germ and degermed maize kernel fractions as previously described (32, 33). Comparisons of FB1 production on cracked whole maize kernels to expression profiles of ZFR1, FUM1, and FUM8 were performed as previously described (10). All experiments were conducted at least twice with the same results.
Nucleic acid isolation and analysis.
Bacterial plasmids were isolated with a Qiagen miniprep DNA purification system (Qiagen, Valencia, Calif.). Fungal genomic DNA was isolated by methods previously described (41). For Southern analysis, F. verticillioides genomic DNA was digested with EcoRI, size fractionated on a 0.7% agarose gel, and transferred to a nylon membrane (Nytran; Schleicher & Schuell, Keene, N.H.) by standard procedures (20). Total RNA was extracted by an acid-phenol extraction procedure described by de Vries et al. (8). For Northern blot analyses, 10 μg of RNA was separated by electrophoresis through a formaldehyde-denaturing gel and transferred to a nylon membrane as previously described (20). The F. verticillioides TUB2 (β-tubulin; GenBank accession number U27303; 43) was 32P labeled and hybridized as a loading control. F. verticillioides FUM1 (GenBank accession number AF155773; 26) and FUM8 (GenBank accession number AF155773; 30) were 32P labeled and hybridized together. All hybridization probes were labeled with the Prime-It II random prime labeling kit (Amersham Biosciences, Arlington Heights, Ill.). High-stringency hybridization was performed with 7% sodium dodecyl sulfate (SDS)-0.5 M sodium phosphate (pH 7.5)-10 mM EDTA (6) and followed by two 30-min washes, the first at room temperature in a solution of 2× SSC (0.3 M sodium chloride, 0.03 M sodium citrate) and 0.5% SDS and the second at 65°C in 0.2× SSC-0.5% SDS (20). Blots were exposed to a phosphorimaging screen and scanned with a Typhoon 9200 high-performance gel blot reader (Molecular Dynamics, Inc., Amersham Biosciences). Images were resized and adjusted for contrast with Adobe Photoshop software (Adobe Systems Incorporated, San Jose, Calif.).
Isolation of ZFR1.
A 45-kb cosmid clone, designated pZFRcos1, was identified by screening a genomic library of F. verticillioides. Standard protocols were used to plate and screen the library (20) with a 32P-labeled insert from expressed sequence tag (EST) clone wt_1_O18 (GenBank accession number CF452892).
Nucleotide sequence analysis.
The nucleic acid sequence of ZFR1 was obtained from plasmids pZFR-pst (6.1-kb) and pZFR-bam (6.3-kb). Both plasmids are subclones from cosmid pZFRcos1 (45-kb) and contain the full-length ZFR1 sequence. DNA sequencing reactions were performed by the Plant-Microbe Genomics Facility, Ohio State University, Columbus.
To obtain cDNA clones of ZFR1, total RNA was extracted from strain 7600 grown on cracked maize kernels for 7 days. Ten micrograms of total RNA was used in the first-strand cDNA synthesis reaction mixture with Superscript II RNase H- reverse transcriptase (Invitrogen, Carlsbad, Calif.) in a 20-μl reaction volume incubated at 42°C for 2 h. Regions of the ZFR1 cDNAs were amplified with the following PCR primer sets: zinc1 (5′-ATGCTCGTTGACCG-3′) and zinc2 (5′-GCTAAGCTCAGTAG-3′), based on the 5′ region (1.2-kb product), and zinc3 (5′-GTGAAGAACCAAAG-3′) and zinc4 (5′-ACGAGAACAGCTTTAG-3′), based on the 3′ region (1.4-kb product). The reaction conditions were the same as those described for the isolation of ZFR1. The PCR products were cloned into the pGEM T-easy vector and sequenced by the Purdue Genomics Core Facility, West Lafayette, Ind. All DNA sequences were analyzed and predicted amino acid sequences were deduced with MacDNASIS software (Hitachi Software Engineering America, Ltd., San Bruno, Calif.). Homology searches were conducted with the BLAST algorithm (1). The PSORTII program was used to analyze the deduced Zfr1 peptide sequence for predictions of subcellular localization (23). Multiple alignments were conducted with the ClustalW software (http://www.ch.embnet.org/software/ClustalW.html) (35).
Disruption of ZFR1.
The ZFR1 deletion vector (pZFR-Δ1) was constructed by insertion of a 1.4-kb HpaI fragment that contained a hygromycin resistance gene cassette from pCB1003 (Fungal Genetics Stock Center), into the EcoRV sites (405 bp apart in pZFR1-TA) that are located in the ZFR1 coding sequence. pZFR-Δ1 was amplified by PCR with primers zfrV5 (5′-ACTGGCAGTCTCTTCAG-3′) and zfrV3 (5′-CGCATGCGATTGTG-3′). A 2.4-kb product that consisted of the 1.4-kb hygromycin marker surrounded by ∼500 bp of ZFR1 flanking sequence was gel purified and used for fungal transformation.
A second ZFR1 deletion plasmid (pZFR-Δ2) was constructed from plasmid pHYG-TA, which consists of the 1.4-kb hygromycin resistance gene cassette from pCB1003 cloned into the pGEM T-easy cloning vector (Promega). Regions (500 bp) located at both ends of ZFR1 were cloned immediately up- and downstream of the hygromycin gene cassette contained within pHYG-TA. A 2.4-kb amplicon that contained the hygromycin gene cassette flanked by 500 bp of ZFR1 was amplified from pZFR-Δ2 by PCR with primers zfrD1 (5′-GATATACCTGCCTG-3′) and zfrD2 (5′-TCATCTCATGCAGCG-3′) and used for fungal transformation.
Protoplasts of strain 7600 were obtained and transformed as described by Proctor et al. (26). Transformants were selected on regeneration medium containing hygromycin B (Roche Molecular Biochemicals, Indianapolis, Ind.) at 60 μg/ml. Transformants were screened by PCR with primers that distinguished homologous crossover events. Primers zincD5 (5′-GGACTCTGTACTTGTTCG-3′) and h3P (5′-CGATAGTGGAAACCGACG-3′) produced a 500-bp DNA product from the homologous crossover at the 5′ end of the insertion DNA, and primers zincD3 (5′-GACCTGTGAGAGGTAG-3′) and h5P (5′-GATCAGAAACTTCTCGACAG-3′) produced a 1,020-bp product from the 3′ end. Conditions for PCR amplification were the same as those described for the isolation of ZFR1, with the exception of the annealing temperature (54°C).
Two deletion mutants (ZN27ss and zfr1Δ), one generated from each type of deletion vector construct, were complemented with pZFRsub-G418. This plasmid was constructed by insertion of a 4.3-kb DNA fragment that spans ZFR1 from 500 bp upstream of the ATG to 1.8 kb downstream of the translational stop codon into vector pUC-G418, which harbors a Geneticin resistance gene (21). The ZFR1 fragment used to construct pZFRsub-G418 was obtained by PCR from pZFRcos1 in a reaction mixture containing a high-fidelity polymerase (Pwo; Roche Molecular Biochemicals) with primers zincA (5′-CGAATCTGCGTAACG-3′) and zincB (5′-ATCTTGAGAGACACG-3′). Conditions for amplification were as follows: 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 54°C, and 2.5 min at 72°C. Geneticin-resistant transformants were selected on regeneration medium containing Geneticin (Sigma Chemical Co., St. Louis, Mo.) at 75 μg/ml. Analysis of the transformants with primer sets zincA-zincB and zincD3-zincD5 (described above) confirmed that the transformants contained a full-length ZFR1 gene (data not shown).
Overexpression of ZFR1.
The entire coding sequence of ZFR1 was amplified by PCR with primers ZFR-5p (5′-GAGTTACCATGGTTATGCTCGTTG-3′) and ZFR-3p (5′-TTCCTCCATGGTTTGCAGGTG-3′p) and a high-fidelity polymerase (Pwo polymerase; Roche Molecular Biochemicals); the resulting DNA fragment was cloned in frame immediately downstream of the GPDA promoter contained within pGPD-G418, a modified pNOM102 vector (28) that contains a 2.2-kb XbaI fragment from plasmid pSM334 that harbors a Geneticin resistance gene cassette (10). The resulting plasmid, pGPD-ZFR1, was used to transform both the zfr1 and fcc1 deletion mutants. Single conidia of Geneticin-resistant transformants were isolated, and integration of the pGPD-ZFR1 vector was confirmed by Southern blot analysis. Overexpression of ZFR1 was verified by Northern blot analysis.
Fumonisin analysis.
Fumonisins were extracted from cultures, and concentrations of FB1 were determined by high-pressure liquid chromatography (HPLC) as previously described (33). Briefly, fumonisins in samples were extracted overnight with acetonitrile-water (1:1, vol/vol). The extracts were passed through C18 solid-phase extraction columns (J & W Scientific, Folsom, Calif.). The fumonisins were eluted with 2 ml of acetonitrile-water (7:3, vol/vol), derivatized with o-phthaldehyde (Sigma Chemical Co.), and analyzed with an HPLC apparatus (Shimadzu Scientific Instruments, Inc., Kyoto, Japan) equipped with an analytical C18 column (150 by 4.6 mm) and a variable-wavelength spectrofluorometric detector (excitation wavelength, 335 nm; emission wavelength, 440 nm). FB1 was quantified by comparison with an FB1 standard (Sigma Chemical Co.).
Nucleotide sequence accession numbers.
The nucleic acid sequence and predicted amino acid sequence of ZFR1 were submitted to the GenBank database and assigned accession number AY493199.
RESULTS
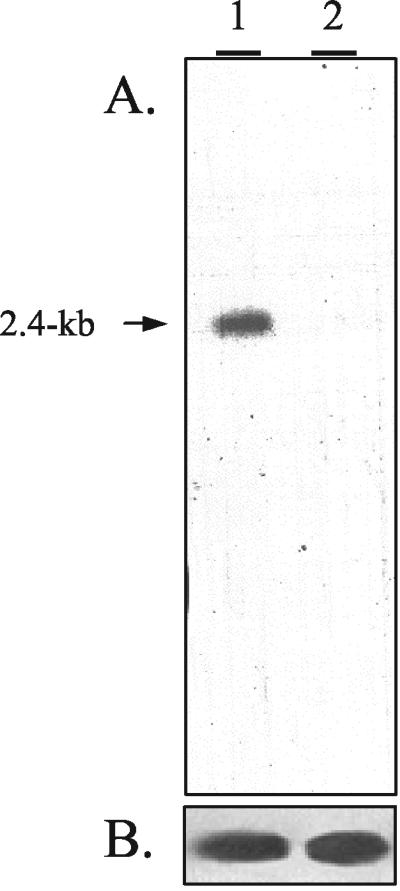
Isolation and characterization of ZFR1. A cDNA subtraction library was previously constructed from RNA isolated from an fcc1Δ mutant and a wild-type strain of F. verticillioides cultured on cracked maize kernels (34). One of the 655 ESTs sequenced from the library enriched with transcripts from the wild-type strain exhibited high similarity to genes encoding known transcription factors harboring a Gal4-like zinc finger DNA-binding motif (18). This EST (wt_1_O18) was used as a probe to verify differential expression via Northern blot analysis of total RNA extracted from maize kernels colonized by a wild-type or fcc1Δ mutant strain (Fig. 1). The same probe was used to screen a cosmid genomic library of F. verticillioides, and a single 45-kb cosmid designated pZFRcos1 was isolated. Sequence analysis of a subclone of pZFR1cos1, designated pZFRsub1, revealed the entire coding region of ZFR1. Comparison of the genomic sequence of ZFR1 to sequences from three overlapping cDNA clones revealed a single uninterrupted open reading frame that is predicted to encode a peptide of 705 amino acids. The deduced protein product of ZFR1 contains (at amino acid residues 157 to 187) a conserved zinc finger DNA binding domain similar to that of known transcription factors such as Gal4 of Saccharomyces cerevisiae (18) and AflR of Aspergillus flavus (42). Zfr1 also contains the peptide sequence RRKDPSCDACRERKVKC beginning at amino acid 152, which fits the pattern of a bipartite nuclear localization signal (23). Zfr1 exhibits the highest overall similarity (22%) to Acu-15 of Neurospora crassa (2) and exhibits 8% similarity to Gal4 and 10% similarity to AflR.
FIG. 1.
Northern blot analysis of wild type and fcc1 mutant cells cultured on cracked maize kernels. Total RNAs (10 μg) isolated from the wild-type strain (lane 1) and the fcc1Δ mutant strain (lane 2) cultured for 7 days were separated by electrophoresis in a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized with a 32P-labeled ZFR1-specific probe (A) and a 32P-labeled β-tubulin-specific probe (B).
Expression of ZFR1.
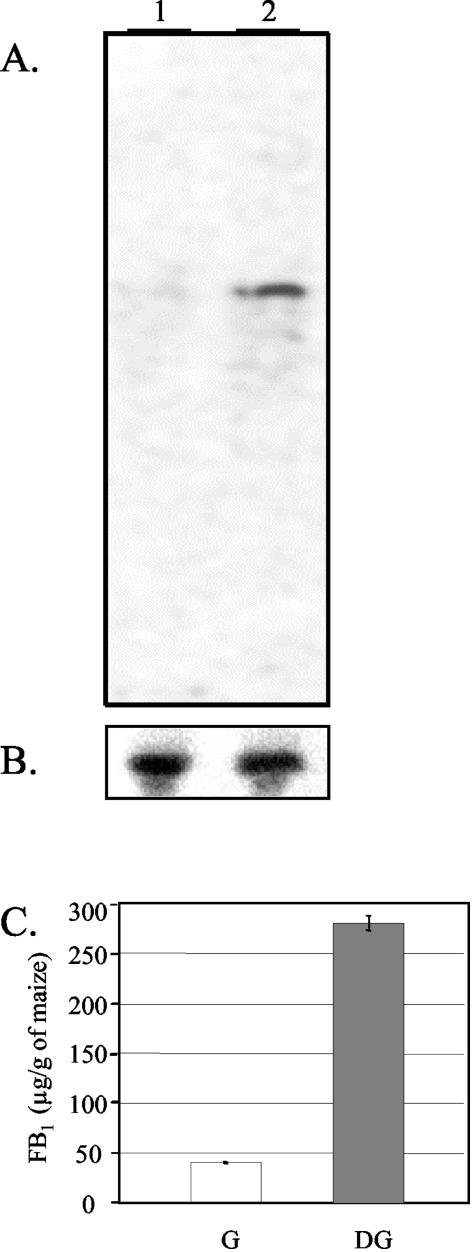
Northern analysis was conducted with total RNA of colonized germ and degermed tissues that were inoculated with strain 7600 and incubated for 7 days. When probed with ZFR1, a ∼2.4-kb band of hybridization was observed in the lane loaded with RNA isolated from the colonized degermed tissue (Fig. 2). No hybridization signal was detected for RNA isolated from colonized germ tissue (Fig. 2). FB1 production was considerably higher in the degermed kernel fraction (280 ± 20 μg of FB1/g of kernel tissue) than in the germ fraction (40 ± 2 μg of FB1/g of tissue) (Fig. 2C). Thus, ZFR1 transcript levels correlated with increased levels of fumonisin biosynthesis.
FIG. 2.
Northern blot analysis and FB1 production of the wild type cultured on separated maize kernel germ and degermed tissues. Total RNA (10 μg) isolated from the wild-type strain was cultured for 7 days on maize kernel germ (lane 1) and degermed (lane 2) fractions. The blot was probed with ZFR1 (A) and TUB2 (B) as a loading control. (C) Quantification of FB1 as determined by HPLC extracted from germ (G) and degermed (DG) kernel fractions. Averages of three repetitions are shown with error bars showing standard errors.
Disruption of ZFR1.
ZFR1 deletion mutants of strain 7600 were produced by homologous recombination. Of the 28 hygromycin-resistant transformants obtained, 1 contained a disrupted ZFR1 gene. The disrupted strain was designated ZN27ss. Southern blots probed with ZFR1 indicated a 1.0-kb larger band of hybridization in ZN27ss than observed for the wild type (Fig. 3). The increase in size corresponds to the replacement of 405 bp within ZFR1 (nucleotides +1590 to +1995 relative to the translational start codon, ATG) with the 1.4-kb hygromycin gene cassette. PCR products generated from the ends of the hygromycin cassette indicated that the disruption occurred by a homologous double-crossover event (data not shown).
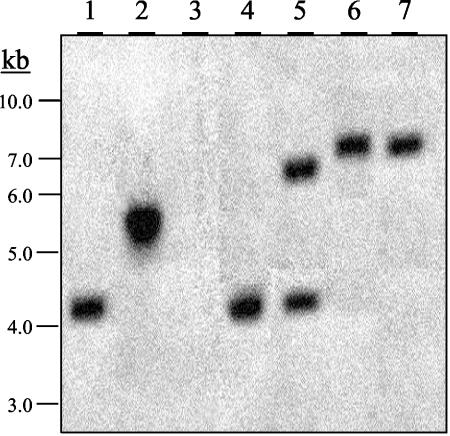
FIG. 3.
Analysis of the ZFR1 deletion and rescued strains. Shown is a Southern analysis of genomic DNAs (2 μg) from the wild-type (strain M-3125, lane 1), zn27ss (zfr1 deletion strain 1, lane 2), zfr1Δ (zfr1 deletion strain 2, lane 3), fcc1Δ (fcc1 deletion strain, lane 4), fcc1Δ GPD::ZFR1 (fcc1Δ mutant transformed with GPD::ZFR1, lane 5), zfr1Δ GPD::ZFR1 (zfr1 deletion strain 2 transformed with GPD::ZFR1, lane 6), and zfr1Δ ZFR1 (zfr1Δ strain transformed with ZFR1, lane 7) strains digested with EcoRI, separated by electrophoresis in a 0.7% agarose gel, transferred to a nylon membrane, and probed with a 32P-labeled DNA fragment of ZFR1. Molecular size standards are indicated on the left.
To rule out the possibility that ZN27ss may produce a functional (or partially functional) Zfr1 protein despite the gene disruption event described above, we constructed an additional vector (pZFR-Δ2) to delete nearly all of the coding sequences of ZFR1 (−50 to +1933 relative to ATG). Of the 41 hygromycin-resistant colonies resulting from the transformation of strain 7600 with pZFR-Δ2, two contained a deleted ZFR1 gene. The zfr1Δ mutants were not distinguishable from strain ZN27ss and exhibited the same fumonisin phenotype as strain ZN27ss on all of the substrates tested. Southern blot analysis of genomic DNAs isolated from both types of deletion mutations confirmed a homologous double-recombination event at the ZFR1 locus (Fig. 3).
Impact of ZFR1 on fumonisin biosynthesis.
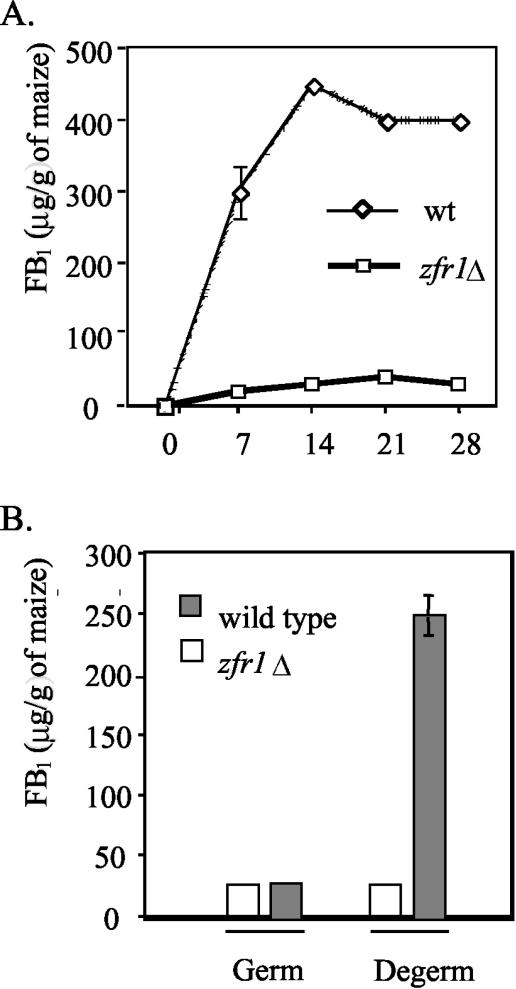
On whole cracked maize kernels, the wild-type strain produced 300 μg of FB1/g of maize after 7 days and 450 μg after 14 days (Fig. 4A). In contrast, a ZFR1 deletion strain, ZN27ss, produced 25 μg of FB1/g of maize after 7 days and 35 μg after 14 days of growth (Fig. 4A). FB1 biosynthesis was restored in ZN27ss and the zfr1Δ mutant when they were transformed with a plasmid (pZFR-G418) containing a full-length ZFR1 gene and a Geneticin resistance gene cassette. Southern analysis confirmed the presence of the ZFR1 gene in the ZN27ss and ZFRΔ2 genomes (Fig. 3). The ZFR1-rescued strains were designated ZN27-R and ZFRΔ2-R. ZN27-R produced the same amount of FB1 as the wild type over the duration of the time course (data not shown).
FIG. 4.
FB1 production by the wild type (wt) and the zfr1 deletion mutant on whole cracked maize kernels and separated maize kernel germ and degermed tissues. (A) Wild-type and zfr1Δ mutant (zfr1 deletion) strains cultured on whole cracked maize kernels for 28 days and analyzed for FB1 every 7 days. (B) Wild-type and zfr1Δ mutant strains cultured for 7 days on separated germ and degermed maize kernel tissue fractions and analyzed for FB1 production. All values are averages of three repetitions, and error bars indicate standard errors greater than 5% of the mean value.
FB1 production was also examined on separated maize-kernel germ and degermed tissue fractions. On degermed kernels, the wild-type strain produced high levels of FB1 (250 ± 25 μg/g of tissue), while on the germ fraction, lower levels of FB1 were produced (26 ± 2 μg/g of tissue) (Fig. 4B). In contrast, the zfr1 mutant exhibited reduced levels of FB1 production (25 μg/g of tissue) on the degermed kernels compared to either the wild type or the rescued strain, but it produced nearly the same amounts of FB1 on the germ tissue (Fig. 4B). Furthermore, the zfr1 mutant failed to produce detectable levels of FB1 when grown on DL medium while the wild-type and ZN27-R strains each produced in excess of 120 μg of FB1/ml of medium (Table 1). The zfr1 mutant produced mycelial mass equivalent to that of the control strains (Table 1). All of the experiments described above that included strains ZN27ss and ZN27-R were repeated to include strains ZFRΔ2 and ZFRΔ2-R, and the same results were obtained (data not shown).
TABLE 1.
Comparison of FB1 production by wild-type and zfr1Δ, and zfr1Δ-ZFR1 mutant strains on DL mediuma
| Strain | Mycelial massb | FB1c |
|---|---|---|
| Wild type | 1.36 ± 0.20 | 120 ± 10 |
| zfr1Δ | 1.38 ± 0.18 | NDd |
| zfr1Δ-ZFR1 | 1.30 ± 0.22 | 110 ± 20 |
FB1 was extracted from cultures grown for 14 days on DL medium (pH 5.6).
Dry weight (grams) of fungal tissue after vacuum filtration is shown. Each value represents the mean of three replicates ± the standard error.
Micrograms of FB1 per milliliter. Each value represents the mean of three replicates ± the standard error.
ND, none detected.
Overexpression of ZFR1.
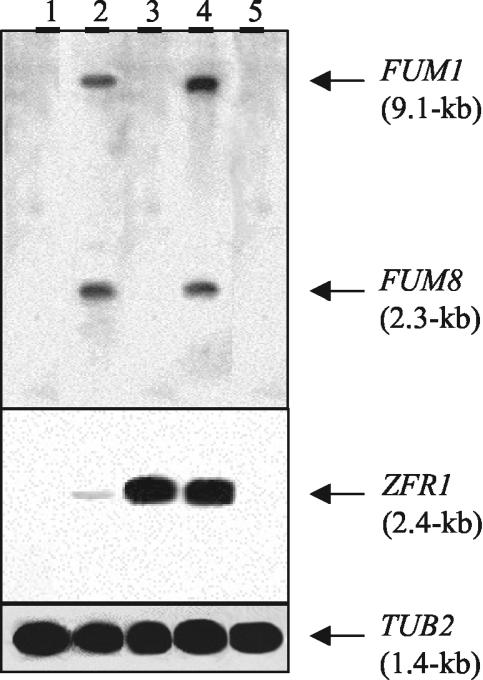
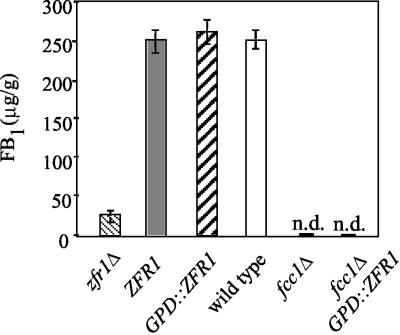
The entire coding sequence of ZFR1 was placed under the transcriptional control of the GPDA gene of A. nidulans (28) and introduced into the zfr1Δ and fcc1Δ mutant strains by transformation. Two transformants of each mutant were identified, and single-conidium isolates were obtained. Northern blot analysis confirmed that ZFR1 transcript was overproduced (Fig. 5). In the zfr1Δ transformant with constitutive expression of ZFR1, FB1 production was restored (Fig. 6), as well as transcription of FUM1 and FUM8 (Fig. 5). In contrast, constitutive expression of ZFR1 in the fcc1Δ mutant failed to restore FB1 biosynthesis (Fig. 6) or transcription of the FUM genes (Fig. 5).
FIG. 5.
Northern blot analysis of wild-type and ZFR1 overexpression strains. Total RNAs (10 μg) isolated from the zfr1Δ mutant (zfr1 deletion strain, lane 1), the wild type (lane 2), the fcc1Δ/GPD::ZFR1 mutant (fcc1Δ mutant strain transformed with a ZFR1 overexpression construct [GPD::ZFR1], lane 3), the zfr1Δ GPD::ZFR1 mutant (zfr1 deletion strain transformed with GPD::ZFR1, lane 4), and the fcc1Δ mutant (fcc1 deletion strain) extracted from cultures grown for 7 days on whole cracked maize kernels were separated by electrophoresis in a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized with 32P-labeled FUM1- and FUM8-specific probes (top), a 32P-labeled ZFR1-specific probe (middle), and a 32P-labeled β-tubulin-specific probe (bottom).
FIG. 6.
Effect of ZFR1 overexpression on FB1 production. The wild-type, zfr1Δ mutant (zfr1 deletion mutant), zfr1Δ-R mutant (zfr1Δ mutant transformed with ZFR1), zfr1Δ GPD::ZFR1 mutant (zfr1Δ mutant strain transformed with a GPD::ZFR1 overexpression construct), fcc1Δ mutant, and fcc1Δ GPD::ZFR1 mutant (fcc1Δ mutant transformed with a GPD::ZFR1 overexpression construct) strains were cultured for 7 days on whole cracked maize kernels and analyzed for FB1 production. All values are averages of three repetitions, and error bars indicate standard errors. n.d. = none detected.
DISCUSSION
The members of the Zn(II)2Cys6 binuclear cluster family of proteins are unique regulators of a wide range of processes in fungi, including primary and secondary metabolism (36). Among those described as positive regulators of secondary metabolism are AflR of A. flavus, Crg1 of Cercospora nicotianae, Pig1 of Magnaporthe grisea, and Cmr1 of Colletotrichum lagenarium. The gene that encodes AflR is the pathway-specific regulatory gene of aflatoxin biosynthesis (9). Crg1 appears to be involved in the activation of genes associated with production of and resistance to cercosporin (5). Pig1 and Cmr1 are involved in controlling gene expression leading to melanin biosynthesis (40). In the study presented here, a cDNA with sequence similarity to the Zn(II)2Cys6 binuclear cluster family of proteins was identified among >700 sequences obtained from a subtraction library enriched for F. verticillioides transcripts expressed during fumonisin biosynthesis on maize kernels. The EST was used to isolate a genomic cosmid clone that contained the entire gene designated ZFR1. F. verticillioides contains a single copy of ZFR1, which possesses a single uninterrupted open reading frame of 2,115 nucleotides and is predicted to encode a peptide of 705 amino acids. Sequence analysis of Zfr1 reveals a DNA binding motif at the N terminus of the deduced peptide characteristic of the Zn(II)2Cys6 binuclear cluster family.
Zfr1 exhibits the highest amino acid identity to the acetate regulatory proteins of A. nidulans and N. crassa (FacB and Acu-15, respectively) (2, 37). FACB and ACU-15 mutants fail to utilize acetate and therefore exhibit impaired growth when cultured on plates containing acetate as the sole carbon source (2, 37). Deletion of ZFR1 by homologous double-crossover transformation resulted in mutant strains that exhibited normal growth and conidiation on cracked maize kernels. The mutants grew at the same rate as the wild type on medium containing acetate as the sole carbon source (data not shown), indicating that ZFR1 is not orthologous to FACB or ACU-15. When cultured on DL medium otherwise conducive to FB1 production by the wild type, the ZFR1 mutants failed to produce detectable levels of FB1 although they produced equivalent mycelial mass. Despite data from extensive growth experiments on cracked maize kernels and DL medium that suggest that ZFR1 is not involved in fungal growth or development, zfr1 mutants were observed to produce very few conidial chains when cultured on defined agar plates such as Czapek's solution agar (10) and carnation leaf agar (34) compared to the wild-type and ZFR1-rescued strains (data not shown). Therefore, we cannot rule out the possibility that ZFR1 can conditionally influence conidiation. Interestingly, FB1 production by the ZFR1 mutants was less than 10% of the amount produced by the wild type when cultured on cracked maize kernels, and transcripts of FUM1 and FUM8 were not detectable in the mutants by Northern blot analysis. Fumonisin production was restored in the mutant strains when transformed with a functional copy of ZFR1. The observed effects of ZFR1 on fumonisin biosynthesis are analogous to that of AFLR in A. flavus and A. parasiticus (4, 42). Strains containing a mutated AFLR gene produce little aflatoxin, and transcription of the aflatoxin pathway genes is not detectable by Northern blot analysis.
Previous studies have shown that the greatest amount of FB1 is produced in the tissues of the degermed kernel (32). The concentration of FB1 produced by wild-type F. verticillioides cultured on the degermed kernel fraction, which is composed of the seed coat, aleurone layer, and endosperm, was nearly 10 times greater than that produced on colonized germ tissue, even though both kernel fractions supported equivalent growth of the fungus (32). In this study, expression of ZFR1 was greatest in the degermed kernel fraction. Furthermore, zfr1 mutants failed to produce higher levels of FB1 on degermed maize kernel fractions compared to the germ. These results indicate that elevated FB1 production on the degermed tissue fraction requires a functional ZFR1 gene.
A final question addressed by this study was if overexpression of ZFR1 can restore FB1 biosynthesis in an fcc1 mutant. ZFR1 was identified in a cDNA library that was constructed after subtractive hybridization with cDNAs from an fcc1 disruption mutant that fails to produce FB1 when cultured on cracked maize kernels (34). Northern blot analysis also indicated that no ZFR1 transcript was detectable in the fcc1 mutant. When grown on maize, two transformants of the fcc1 mutant that overexpressed ZFR1 failed to produce FB1 or detectable transcripts of FUM1 and FUM8. An explanation for the failure of constitutive ZFR1 expression to restore FB1 biosynthesis in the fcc1 mutant may be that FCC1 is required for transcription of the FUM genes, as well as ZFR1. Transcription of the FUM genes may also require activated Zfr1. Such a mechanism exists in yeast, where the C-type cyclin Srb11 (Ume3) is required for activation of the transcription factor Gal4, also a Zn(II)2Cys6 binuclear cluster protein (13). If a similar posttranslational activation mechanism exists in F. verticillioides, constitutive production of ZFR1 would not necessarily lead to production of the active protein without the corresponding Fcc1 (cyclin)-cyclin-dependent kinase interaction. In addition, constitutive expression of ZFR1 in the wild-type strain does not result in constitutive production of FB1 in culture (data not shown), supporting the hypothesis that additional factors are required for Zfr1 activity. Further experiments are required to confirm that Zfr1 is a direct regulator of fumonisin pathway gene expression and also to prove that Zfr1 interacts with Fcc1 and/or a cyclin-dependent kinase.
On the basis of the evidence presented in this report, we conclude that ZFR1 is a positive regulator of fumonisin biosynthesis. Because ZFR1 is predicted to encode a protein that contains a DNA binding motif similar to that found in other fungal transcription factors such as Gal4 and AflR, it remains to be determined if Zfr1 binds to specific sequences in the promoter regions of the FUM genes. Other questions concerning specificity are raised because ZFR1 does not reside within the FUM gene cluster, as there are several examples in fungi of close linkage of pathway-specific transcription factors to their respective structural genes. Furthermore, a DNA sequence with high similarity to that of ZFR1 is present within the genome of F. graminearum, a non-fumonisin-producing species that does not harbor FUM gene sequences, which reinforces the plausibility that Zfr1 may not be a specific regulator of fumonisin pathway genes. Future experiments will address these questions of specificity, as well as determine the involvement of Fcc1 in regulating Zfr1 activity.
Acknowledgments
We thank R. H. Proctor (National Center for Agricultural Utilization Research, USDA Agriculture Research Service, Peoria, Ill.) for generously providing a genomic library of F. verticillioides. We also thank Larry Dunkle and Jin-Rong Xu for helpful discussion and reviews of the manuscript.
Financial support was provided by USDA NRI Competitive Grants Program award 02-35201-11542.
Footnotes
Journal publication 17275 of the Purdue University Agricultural Research Program.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibbins, M., V. F. Crepin, N. J. Cummings, T. Mizote, K. Baker, K. H. Mellits, and I. F. Connerton. 2002. A regulator gene for acetate utilisation from Neurospora crassa. Mol. Genet. Genomics 267:498-505. [DOI] [PubMed] [Google Scholar]
- 3.Butchko, R. A., R. D. Plattner, and R. H. Proctor. 2003. FUM13 encodes a short chain dehydrogenase/reductase required for C-3 carbonyl reduction during fumonisin biosynthesis in Gibberella moniliformis. J. Agric. Food Chem. 51:3000-3006. [DOI] [PubMed] [Google Scholar]
- 4.Cary, J. W., K. C. Ehrlich, M. Wright, P.-K. Chang, and D. Bhatnagar. 2000. Generation of AFLR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:680-684. [DOI] [PubMed] [Google Scholar]
- 5.Chung, K. R., M. E. Daub, K. Kuchler, and C. Schuller. 2003. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 302:302-310. [DOI] [PubMed] [Google Scholar]
- 6.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, K. F., M. J. Mallory, J. B. Smith, and R. Strich. 1997. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 16:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries, S. C., J. Springer, and J. H. Wessels. 1982. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta 156:129-135. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty, J. E., and G. A. Payne. 1997. Overexpression of AFLR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl. Environ. Microbiol. 63:3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty, J. E., A. M. Pirttila, B. H. Bluhm, and C. P. Woloshuk. 2003. PAC1, a pH regulatory gene from Fusarium verticillioides. Appl. Environ. Microbiol. 69:5222-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Drug Administration Center for Food Safety and Applied Nutrition. 2001. Background paper in support of fumonisin levels in corn and corn products intended for human consumption. U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition Center for Veterinary Medicine, College Park, Md.
- 12.Gelderblom, W. C., K. Jaskiewicz, W. F. Marasas, P. G. Thiel, R. M. Horak, R. Vleggaar, and N. P. Kriek. 1988. Fumonisins—novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microbiol. 54:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirst, M., M. S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase Srb10/Cdk8. Mol. Cell 3:673-678. [DOI] [PubMed] [Google Scholar]
- 14.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 15.Keller, S. E., T. M. Sullivan, and S. Chirtel. 1997. Factors affecting the growth of Fusarium proliferatum and the production of fumonisin B1: oxygen and pH. J. Ind. Microbiol. Biotechnol. 19:305-309. [DOI] [PubMed] [Google Scholar]
- 16.Kriek, N. P. J., T. S. Kellerman, and W. F. O. Marasas. 1981. A comparative study of the toxicity of Fusarium verticillioides (= F. moniliforme) to horses, primates, pigs, sheep and rats. Onderstepoort J. Vet. Res. 48:129-131. [PubMed] [Google Scholar]
- 17.Kuchin, S., P. YeghiaYan, and M. Carlson. 1995. Cyclin-dependent protein-kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92:4006-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laughon, A., and R. F. Gesteland. 1984. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol. Cell. Biol. 4:260-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao, S. M., J. H. Zhang, D. A. Jeffrey, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. Vanvuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Marek, E. T., C. L. Schardl, and D. A. Smith. 1989. Molecular transformation of Fusarium solani with an antibiotic resistance marker having no fungal DNA homology. Curr. Genet. 15:421-428. [DOI] [PubMed] [Google Scholar]
- 22.Musser, S. M., and R. D. Plattner. 1997. Fumonisin composition in cultures of Fusarium moniliforme, Fusarium proliferatum, and Fusarium nygami. J. Agric. Food Chem. 45:1169-1173. [Google Scholar]
- 23.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, P. E., A. E. Desjardins, and R. D. Plattner. 1993. Fumonisins, mycotoxins produced by Furarium species: biology, chemistry, and significance. Annu. Rev. Phytopathol. 31:233-252. [DOI] [PubMed] [Google Scholar]
- 25.Park, D. L., and T. C. Troxell. 2002. U.S. perspective on mycotoxin regulatory issues. Adv. Exp. Med. Biol. 504:277-285. [DOI] [PubMed] [Google Scholar]
- 26.Proctor, R. H., A. E. Desjardins, R. D. Plattner, and T. M. Hohn. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27:100-112. [DOI] [PubMed] [Google Scholar]
- 27.Proctor, R. H., D. W. Brown, R. D. Plattner, and A. E. Desjardins. 2003. Co-expression of fifteen contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 38:237-249. [DOI] [PubMed] [Google Scholar]
- 28.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 29.Reinhardt, A., and T. Hubbard. 1998. Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res. 26:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross, P. F., P. E. Nelson, J. L. Richard, G. D. Osweiler, L. G. Rice, R. D. Plattner, and T. M. Wilson. 1990. Production of fumonisin by Fusarium moniliforme and Fusarium proliferatum isolates associated with equine leukoencephalomalacia and a pulmonary edema syndrome in swine. Appl. Environ. Microbiol. 56:3225-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo, J. A., R. H. Proctor, and R. D. Plattner. 2001. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 34:155-165. [DOI] [PubMed] [Google Scholar]
- 32.Shim, W.-B., J. E. Flaherty, and C. P. Woloshuk. 2003. Comparison of fumonisin B1 biosynthesis in maize germ and degermed kernels by Fusarium verticillioides. J. Food Prot. 66:2116-2122. [DOI] [PubMed] [Google Scholar]
- 33.Shim, W.-B., and C. P. Woloshuk. 1999. Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi. FEMS Microbiol. Lett. 177:109-116. [DOI] [PubMed] [Google Scholar]
- 34.Shim, W.-B., and C. P. Woloshuk. 2001. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 67:1607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 37.Todd, R. B., A. Andrianopoulos, M. A. Davis, and M. J. Hynes. 1998. FACB, the Aspergillus nidulans activator of acetate utilization genes, binds dissimilar DNA sequences. EMBO J. 17:2042-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuji, G., Y. Kenmochi, Y. Takano, J. Sweigard, L. Farrall, I. Furusawa, O. Horino, and Y. Kubo. 2000. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38:940-954. [DOI] [PubMed] [Google Scholar]
- 39.Wang, E., W. P. Norred, C. W. Bacon, R. T. Riley, and A. H. Merrill, Jr. 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266:14486-14490. [PubMed] [Google Scholar]
- 40.Wilson, T. M., P. F. Ross, D. L. Owens, L. G. Rice, S. A. Green, S. J. Jenkins, and H. A. Nelson. 1992. Experimental reproduction of ELEM—a study to determine the minimum toxic dose in ponies. Mycopathologia 117:115-120. [DOI] [PubMed] [Google Scholar]
- 41.Woloshuk, C. P., and G. A. Payne. 1994. The alcohol dehydrogenase gene ADH1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:670-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1995. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan, K., and M. B. Dickman. 1996. Isolation of a β-tubulin gene from Fusarium moniliforme that confers cold-sensitive benomyl resistance. Appl. Environ. Microbiol. 62:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]