Abstract
Since their initial discovery in samples from the north Atlantic Ocean, 16S rRNA genes related to the environmental gene clone cluster known as SAR202 have been recovered from pelagic freshwater, marine sediment, soil, and deep subsurface terrestrial environments. Together, these clones form a major, monophyletic subgroup of the phylum Chloroflexi. While members of this diverse group are consistently identified in the marine environment, there are currently no cultured representatives, and very little is known about their distribution or abundance in the world's oceans. In this study, published and newly identified SAR202-related 16S rRNA gene sequences were used to further resolve the phylogeny of this cluster and to design taxon-specific oligonucleotide probes for fluorescence in situ hybridization. Direct cell counts from the Bermuda Atlantic time series study site in the north Atlantic Ocean, the Hawaii ocean time series site in the central Pacific Ocean, and along the Newport hydroline in eastern Pacific coastal waters showed that SAR202 cluster cells were most abundant below the deep chlorophyll maximum and that they persisted to 3,600 m in the Atlantic Ocean and to 4,000 m in the Pacific Ocean, the deepest samples used in this study. On average, members of the SAR202 group accounted for 10.2% (±5.7%) of all DNA-containing bacterioplankton between 500 and 4,000 m.
The discovery that previously unidentified bacterioplankton 16S rRNA gene sequences predominate in the ocean's lower surface layer was one of the first pieces of evidence to suggest that marine bacterioplankton communities are stratified (8, 13, 47). The environmental gene clone SAR202 and close relatives were among the groups recovered from seawater in early investigations of bacterioplankton diversity at the Bermuda Atlantic time series study (BATS) site in the north Atlantic Ocean (13). Shortly thereafter, close relatives were detected in seawater samples from 1,000 m in the Atlantic Ocean and 3,000 m in the Pacific, rapidly extending the apparent range of this group of microorganisms throughout the mesopelagic zone and into the deep ocean (12).
Interestingly, SAR202 organisms and relatives are members of the Chloroflexi phylum, one of the 11 original phyla described by comparative 16S rRNA sequence analysis (45). The Chloroflexi line of descent is thought by many to have diverged early in the evolution of the domain Bacteria (29). Representatives of this phylum occupy a wide variety of habitats; Chloroflexi-related sequences have been identified in geothermal, soil, freshwater, marine, wastewater, and subsurface environments. In addition, the few cultivated representatives exhibit a diverse range of phenotypes, including anoxygenic photosynthesis (e.g., Oscillochloris and Chloroflexus) (23, 30), thermophilic organotrophy (Thermomicrobium) (21), and chlorinated hydrocarbon reduction (Dehalococcoides ethenogenes) (27). The phenotypic characteristics of the SAR202 clade of bacteria cannot be inferred from their phylogeny because of the diverse physiological traits exhibited by cultured representatives within this phylum (19, 33, 36).
Since their initial identification in BATS 250-m seawater, environmental gene clones related to the SAR202 cluster have been found in deep subsurface, soil, marine sponge, and freshwater environments (4, 7, 18, 41) and further sequences have been found in various seawater samples (2, 14, 46). While cultivation-independent rRNA gene cloning and sequencing results suggest that members of this diverse group are ubiquitous and potentially abundant in the marine environment, there are well-known sources of potential methodological bias that prohibit absolute cellular quantification from these data. Variable lysis efficiency between microbial cell types, variations in rRNA gene copy number, and PCR-induced biases and artifacts are just a few of the factors that confound and restrict quantitative estimates of abundance from gene clone library data (34, 37, 44). However, direct cell counts using fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes have been used to accurately count cells in natural samples (1, 6, 28).
In general, small, slow-growing microbial cells such as planktonic marine bacteria have traditionally been difficult to detect by FISH. Subsequently, various strategies have been used to decrease background noise and increase signal intensity and counting accuracy. Strategies have included the use of multiple fluorescently labeled oligonucleotide probes (25), signal amplification methods such as tyramide signal amplification (35), and unlabeled helper oligonucleotide probes (11). Our strategy has been to use multiple oligonucleotide probes that target different regions of the same 16S rRNA to produce an additive effect on signal intensity (28) and, in this case, to ensure that all available SAR202-related 16S ribosomal DNA (rDNA) sequences recovered from seawater were targeted by at least one probe.
The available data suggest that members of the SAR202 cluster are ubiquitous and that they may play an important role in lower-surface and deep-ocean biogeochemistry. However, no data about their physiology or cellular abundance are available. There are currently no cultured representatives of the SAR202 cluster or published quantitative abundance estimates. In this study, we used newly identified SAR202-related 16S rDNA sequences from marine bacterioplankton with published SAR202 cluster sequences recovered from a variety of environments to further resolve SAR202 phylogeny and to design oligonucleotide probes for quantitative FISH. We report SAR202 cluster cell counts from the BATS site in the Atlantic Ocean and the Hawaii ocean time series (HOT) site (station ALOHA) in the Pacific Ocean. In addition, depth profiles from coastal waters were obtained from five stations along the Newport hydroline (NH35 to NH127), extending from just off the Oregon coast to the edge of the north Pacific gyre.
MATERIALS AND METHODS
Sample collection.
North Atlantic Ocean seawater was collected at the BATS site (32°N, 64°W) from a total of 10 depths between 1 and 3,600 m. Surface samples (1 to 250 m) were collected on 5 February 2001, while samples from depths >250 m were collected on 6 February 2001. Central north Pacific Ocean samples were collected at station ALOHA (45°N, 158°W), the HOT study site, from a total of seven depths on 15 December 2002. Water from the eastern Pacific Ocean coastal transect was collected from various depths along the Newport hydroline (44°N) at stations NH15 (25°W), NH35 (53°W), NH55 (22°W), NH65 (36°W), NH85 (126°W), and NH127 (127°W). Five samples (1, 10, 30, 100, and 500 m) were collected on 7 May 2002, and five samples (20, 110, 600, 1,000, and 2,700 m) were collected on 8 May 2002 at station NH127. All samples were collected in Niskin bottles on conductivity, temperature, and density device rosettes and transferred to primary collection bottles. Atlantic subsample volumes of 500 ml were immediately fixed in filtered formalin at a final concentration of 10% and stored at −80°C for up to 6 months. Pacific subsample volumes of 10 to 250 ml were immediately fixed in filtered, buffered paraformaldehyde at a final concentration of 2% and stored at 4°C for 6 to 8 h. Fixed samples were filtered onto white 0.2 μm-pore-size polycarbonate filters (GE Osmonics, Minnetonka, Minn.), immediately placed in slide boxes containing silicon desiccant, and stored at −20°C.
Cloning.
Bacterial 16S rRNA gene clones from the original BATS 250-m clone library were prepared as described previously (13). In short, DNA was amplified from a mixed population of genomic DNA by PCR using primers specific for bacterial 16S rRNA genes. A clone library was constructed with the plasmid vector pCRII (Invitrogen, San Diego, Calif.) from the resulting PCR amplicon. The clones were assigned the prefix SAR, numbered discontinuously from 177 to 325, and stored at −20°C in Luria-Bertani (LB) broth containing 10% (wt/vol) glycerol. Two new SAR202-related clone sequences were identified in a clone library constructed from February 1992 BATS 200-m seawater (prefix D92). The D92 bacterial 16S rDNA library was prepared essentially as described above, but by a streamlined protocol for clone library analysis (42). rRNA genes were amplified from environmental DNA for cloning by PCR with Taq polymerase (Fermentas, Hanover, Md.) and variations of commonly used bacterial primers 8F (AGRGTTYGATYMTGGCTCAG) and 1492R (GGYTACCTTGTTACGACTT) (24). Amplifications were performed in a PTC-0200 thermocycler (MJ Research, Cambridge, Mass.) under the following conditions: 35 cycles of annealing at 55°C for 1 min, elongation at 72°C for 2 min, and denaturation at 94°C for 30 s. A single band of the predicted length was observed by agarose gel electrophoresis. The clone library was constructed with the pGEM-TEasy (Promega, Madison, Wis.) vector by following the manufacturer's instructions. Individual clones were numbered sequentially from D92-01 to D92-96.
Gene sequencing and phylogenetic analysis.
Complete 16S rRNA gene clone sequences were obtained and added to an aligned database of >12,000 homologous 16S rDNAs maintained with the ARB software package (26). Evolutionary distance, parsimony, and maximum-likelihood phylogenetic analysis methods were used in concert to identify robust phylogenetic relationships within the SAR202 cluster data set and were performed with the program PAUP*, version 4.0 beta 10 (39). The tree topology was inferred by maximum likelihood employing a heuristic search with a tree bisection-reconnection branch-swapping algorithm, a proportion of invariable sites of 0.2339, equal base frequencies, and a gamma distribution of rate heterogeneity at variable sites with a shape parameter of 0.6889 and four rate categories. Bootstrap proportions from 1,000 replicate resampled data sets were used to estimate the relative confidence in monophyletic groups and were determined by evolutionary-distance and parsimony methods. Likelihood ratio tests were used to select a substitution model for evolutionary distance calculations by employing the program Modeltest, version 3.06 (30a). The model selected was SYM+I+G (48), with the estimated proportion of invariable sites set to 0.2339, equal base frequencies, and a gamma distribution of rate heterogeneity at variable sites with a shape parameter of 0.6889 and four rate categories. Distance matrices from bootstrapped data sets were calculated with this model, and neighbor joining was used to generate trees for the bootstrap analysis. Parsimony analyses employed a heuristic search, tree bisection-reconnection, and a starting tree obtained by stepwise addition with random sequence addition. All sequences used in this analysis were >1,200 nucleotides in length; 914 nucleotide positions remained after masking out hypervariable and other ambiguously aligned regions from the alignment. In preliminary analyses, a range of bacterial phyla were employed as outgroups. The choice of outgroup did not influence the significant relationships shown in Fig. 1.
FIG. 1.
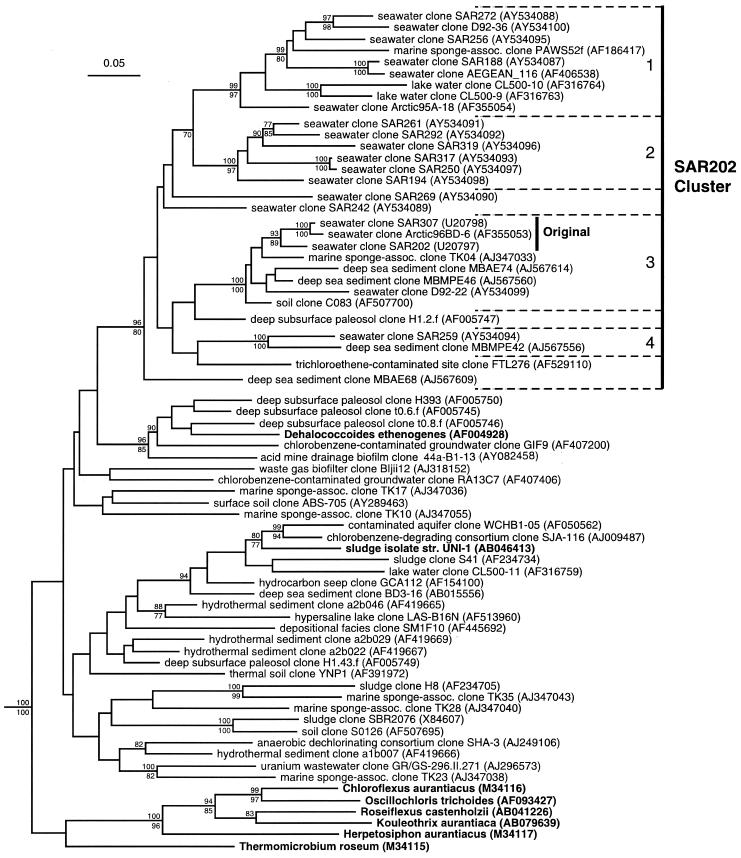
Phylogenetic tree of the SAR202 cluster and representatives of the phylum Chloroflexi. rRNA gene sequences from cultivated microorganisms are shown in boldface, while sequences derived from cultivation-independent studies are labeled with the environment from which they were derived and clone name. GenBank accession numbers are shown in parentheses. Nodes supported by bootstrap replicates >70% in evolutionary-distance (above) or parsimony (below) analyses are indicated. The scale bar corresponds to 0.05 substitutions per nucleotide position. Dashed brackets, subclusters (numbered 1 to 4). “Original” indicates the phylogenetic depth of the original SAR202 cluster (13).
FISH.
Hybridization reactions were performed essentially as described by Glöckner et al. (15) with the following modifications. Reactions were performed on one-quarter membrane sections at 37°C for 12 to 16 h in hybridization buffer (900 mM NaCl, 20 mM Tris [pH 7.4], 0.01% [wt/vol] sodium dodecyl sulfate [SDS], 35% formamide) and two Cy3-labeled oligonucleotide probes (SAR202-104R [GTTACTCAGCCGTCTGCC] and SAR202-312R [TGTCTCAGTCCCCCTCTG]) specific for members of the SAR202 cluster and designed with the ARB software package (26). Additionally, a control hybridization reaction was performed with a low-stringency buffer containing 15% formamide and a Cy3-labeled nonsense oligonucleotide (338F). All probes had a final concentration of 2 ng μl−1. Optimal hybridization stringency was achieved by washing the membranes in hybridization wash (70 [SAR202] or 150 [338F] mM NaCl, 20 mM Tris [pH 7.4], 6 mM EDTA, 0.01% SDS) for two 10-min intervals. An experimentally determined temperature of dissociation (Td) specific for the SAR202 probe suite (58.0°C) was used for all SAR202 hybridization reactions (see Fig. 2), and a low-stringency Td (50.0°C) was used for all 338F control hybridization reactions. Nucleic acid staining was achieved by transferring the membrane to a chilled (4°C) hybridization wash containing DAPI (4′,6′-diamidino-2-phenylindole) at a final concentration of 5 μg ml−1 for 10 min. The DAPI was rinsed for 2 min in a final hybridization wash chilled to 4°C. All reagents were filtered through a 0.2-μm-pore-size filter.
FIG. 2.
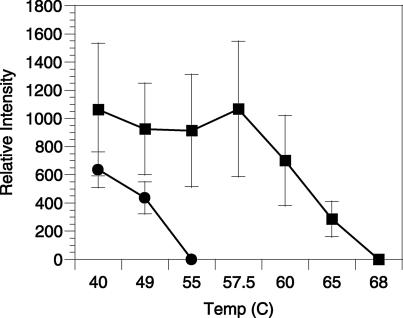
SAR202 probe pair dissociation curve. SAR202 cells from Oregon coast seawater (250 m; ▪) and an axenic SAR11 culture in exponential growth phase (•) hybridized to the SAR202 probe suite. Temperatures indicate various stringency conditions associated with the hybridization wash buffer.
Fluorescence microscopy.
After the filters were mounted in Citifluor (Ted Pella, Redding, Calif.), Cy3-positive and DAPI-positive cells were counted for each field of view with a Leica DMRB epifluorescence microscope equipped with a Hamamatsu ORCA-ER charge-coupled device digital camera, filter sets appropriate for Cy3 and DAPI, and Scanalytics IPLab, version 3.5.6, scientific imaging software. Consistent exposure times of 1 and 5 s were used for DAPI and Cy3 images, respectively. Cy3 images were manually segmented in IPLab and automatically made to overlie corresponding DAPI image segmentations in order to identify positive probe signals coincident with DAPI signals. Consistent size, morphology, and signal intensity criteria were used for all cell counts. Negative control counts were determined from the 338F hybridization using the same technique and subtracted from positive probe counts to account for objects detected with the Cy3 and DAPI filter sets in the absence of the positive probe set, such as autofluorescent cells.
Nucleotide sequence accession numbers.
Gene sequences were deposited in GenBank and given accession numbers AY534087 through AY534100.
RESULTS
A combination of methods were used to determine phylogenetic relationships among 16S rRNA gene sequences from members of the original SAR202 cluster (13); published relatives were identified by searching public nucleotide sequence databases (GenBank and the RDP-II), published reference sequences from other major subgroups of the Chloroflexi (33), and newly sequenced environmental gene clones recovered from the BATS study site in the north Atlantic Ocean. All of the analyses showed that the rRNA gene clones from pelagic marine bacterioplankton within the phylum Chloroflexi fell inside a single monophyletic cluster (Fig. 1), but the addition of newly identified clones greatly expanded the genetic diversity of this cluster relative to that based on the original observations (13). The first two full-length gene clones published in 1996, SAR202 and SAR307, are 94.9% similar. Currently, the most dissimilar Chloroflexi marine bacterioplankton gene clone sequences are 78.7% similar (D92-36 and SAR259 in Fig. 1). Within the Chloroflexi phylum, the closest relatives to the SAR202 cluster could not be identified with the 16S rRNA gene sequence data and analysis methods currently available.
Unlike clusters from other predominant groups of marine bacterioplankton, such as the SAR86 (31, 38) and Pelagibacter (SAR11) clusters (32, 33, 38), marine bacterioplankton environmental gene clones of the SAR202 cluster are not monophyletic; sequences retrieved from nonmarine and/or nonplanktonic communities are interspersed throughout the marine bacterioplankton clones. For example, environmental gene clones from freshwater bacterioplankton of Crater Lake, Oreg. (41), sponge symbionts from shallow marine environments (18), deep-sea sediments (unpublished data), and terrestrial soils (references 4 and 7 and unpublished data) are dispersed throughout the SAR202 cluster. Four subclusters within the SAR202 cluster were supported by high bootstrap proportions (Table 1; Fig. 1). While all four contained gene clones from marine bacterioplankton, only one was exclusively so (subcluster 2). In addition to marine bacterioplankton, subcluster 1 contained clones recovered from marine sponge and freshwater bacterioplankton communities, subcluster 3 contained clones from marine sponge, deep-sea sediment, and forest soil communities, and subcluster 4 contained a clone from a deep-sea sediment community. Several clones did not fall within the four monophyletic subgroups but instead formed independent lines of descent within the SAR202 cluster (e.g., clones SAR242, SAR269, and FTL256 in Fig. 1).
TABLE 1.
Probe specificity for members of the SAR202 cluster
| Clone | Source | No. of:
|
Reference or source | |
|---|---|---|---|---|
| SAR202-104R mismatches | SAR202-312R mismatches | |||
| Subgroup 1 | ||||
| SAR272 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR256 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR188 | Sargasso Sea seawater, 250 m | 1 | 0 | This study |
| D92-36 | Sargasso Sea seawater, 200 m | 0 | 0 | This study |
| AEGEAN_116 | North Aegean Sea seawater, 200 m | 0 | 0 | Unpublished |
| Arctic95A-18 | Arctic Ocean seawater, 500 m | 2 | 0 | 2 |
| PAWS52f | Sponge symbiont, 20-30 m, Pacific Ocean | 0 | 0 | 18 |
| CL500-9 | Freshwater Crater Lake, 500 m | 3 | 3 | 41 |
| CL500-10 | Freshwater Crater Lake, 500 m | 0 | 1 | 41 |
| Subgroup 2 | ||||
| SAR261 | Sargasso Sea seawater, 250 m | 1 | 0 | This study |
| SAR292 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR319 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR317 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR250 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR194 | Sargasso Sea seawater, 250 m | 0 | 1 | This study |
| Subgroup 3 | ||||
| SAR307 | Sargasso Sea seawater, 250 m | 0 | 0 | 13 |
| SAR202 | Sargasso Sea seawater, 250 m | 0 | 0 | 13 |
| D92-22 | Sargasso Sea seawater, 200 m | 1 | 0 | This study |
| Arctic96BD-6 | Arctic Ocean seawater, 500 m | 0 | 0 | 2 |
| TK04 | Sponge symbiont, 7-15 m, Mediterranean Sea | 0 | 0 | 18 |
| MBAE74 | Deep-sea sediment, Pacific Ocean | 3 | 0 | Unpublished |
| MBMPE46 | Deep-sea sediment, Pacific Ocean | 0 | 0 | Unpublished |
| C083 | Forest soil, Arizona | 0 | 0 | 7 |
| Subgroup 4 | ||||
| SAR259 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| MBMPE42 | Deep-sea sediment, Pacific Ocean | 4 | 0 | Unpublished |
| SAR269 | Sargasso Sea seawater, 250 m | 0 | 0 | This study |
| SAR242 | Sargasso Sea seawater, 250 m | 0 | 1 | This study |
| H1.2.f | Deep subsurface paleosol | 2 | 0 | 4 |
| MBAE68 | Deep sea sediment, Pacific Ocean | 2 | 0 | Unpublished |
| FTL276 | Trichloroethene-contaminated soil | 1 | 1 | Unpublished |
Two oligonucleotide probes were designed to target members of the SAR202 cluster. The probe SAR202-104R was designed to target a region corresponding to positions 104 to 121 of the Escherichia coli 16S rRNA. This probe matched perfectly 20 of 30 members of the SAR202 cluster possessing complete or nearly complete 16S rDNA sequences and 15 of19 marine bacterioplankton environmental gene clones in this cluster (Table 1). Outside of the SAR202 cluster, probe SAR202-104R matched perfectly the 16S rRNA gene sequence from the archaeaon Sulfolobus solfataricus (GenBank accession no. X90483) and closely related environmental gene clones and contained a single base mismatch with a wide variety of published 16S rRNA gene sequences, including those of several members of the SAR11 marine bacterioplankton cluster of the alpha Proteobacteria. The probe SAR202-312R was designed to target a region corresponding to positions 312 to 329 of the E. coli 16S rRNA. It matched perfectly 25 of 30 full-length members of the SAR202 cluster and 17 of 19 marine clones (Table 1). In addition, this probe matched perfectly 16S rRNA gene sequences from several members of candidate division OP11 (20) and had a minimum of two mismatches with all other known 16S rRNA gene sequences outside of the SAR202 cluster. Of 30 full-length, or nearly full-length, gene sequences within this cluster, only two (freshwater bacterioplankton clone CL500-9 and contaminated-soil clone FTL276 in Fig. 1) did not possess a target site that perfectly matched that of one of the two SAR202 cluster probes (Table 1). All 19 full-length marine bacterioplankton gene clone sequences within this cluster perfectly match at least one of the two SAR202 cluster probes.
Direct cell counts from the Atlantic and Pacific Oceans were obtained by hybridizing paraformaldehyde-fixed, filtered seawater samples with the two SAR202 cluster probes labeled with Cy-3. The Td of 58°C for cells hybridizing to the SAR202 probe pair was empirically determined from 100-m Oregon coast seawater (NH35). An axenic SAR11 cluster isolate (32), fortuitously exhibiting a single base mismatch to probe SAR202-104R, was used to evaluate the specificity of hybridization of this probe. SAR11 cells hybridized to the SAR202 probe pair showed a complete loss of probe-conferred fluorescence signal intensity between 49 and 55°C (Fig. 2). While it is known that base composition and rRNA secondary structure can affect in situ hybridization kinetics (9, 10), these results indicate that SAR11 cells containing the target sequence with a single base mismatch were excluded from counts reported in this study.
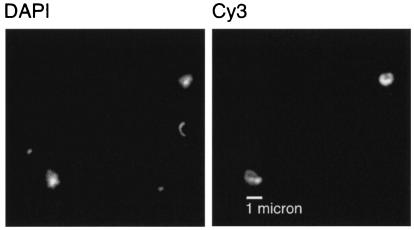
Additional confidence in the cell count measurements came from observations of the average morphology, size, and relative signal intensity of cells hybridizing to the SAR202 probe suite. Probe-positive cells had a coccoid morphology and were greater than 1 μm in diameter (Fig. 3) and unusually bright (1,067 ± 480 relative intensity units) compared to other pelagic bacterioplankton hybridizations (Fig. 2). Because of the distinctive size, morphology, and signal intensity of cells hybridizing to the SAR202 probe suite, there was very little ambiguity in the scoring of cells from below the upper ocean surface layer, where autofluorescent-cell counts are low.
FIG. 3.
FISH image of SAR202 cells from the Pacific Ocean (2,700 m). Identical fields of view show DNA-containing cells stained with DAPI and relatively large (cocci >1 μm in diameter) target cells stained with the SAR202 cluster probe pair labeled with Cy3. Images were obtained with a Hamamatsu ORCA-ER charge-coupled device digital camera.
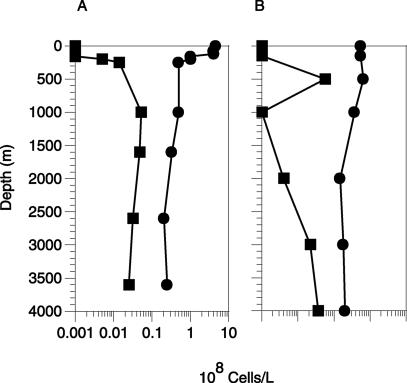
The overall abundance of SAR202 cells remained surprisingly constant below 500 m and accounted for an average of (3.0 ± 1.9) × 106 cells liter−1 in Atlantic (BATS) and Pacific (HOT) Ocean depth profiles (Fig. 4). On average, the SAR202 group accounted for 10.2% (±5.7%) of DAPI-stained cells present below the ocean surface layer. In surface waters, SAR202 cell counts were ≤1.0 × 106 cells liter−1, at or below the threshold of detection for surface waters. The threshold for accurate counting of the less-abundant bacterioplankton groups was higher in surface waters than in deep waters, due to the high autofluorescent-cell and particle counts associated with negative control probe hybridizations. Bulk nucleic acid hybridization data suggest that DNA from the SAR202 group decreases in surface waters (13), and surface cells (0 to 300 m) positive for both probe hybridization and DAPI always lacked green fluorescence (fluorescein isothiocyanate channel), indicative of chlorophyll autofluorescence. These data reinforce the ≥66% decrease in ocean surface layer SAR202 cells relative to the numbers in deeper waters suggested by the in situ hybridization data.
FIG. 4.
Group-specific FISH and prokaryotic-cell counts (DAPI-stained particles) in Atlantic and Pacific Ocean gyres. Shown are SAR202 (▪) and DAPI (•) cell counts for the north Atlantic Ocean at BATS sites (A) and in the central north Pacific Ocean at HOT sites (B). The Atlantic Ocean profile is a composite consisting of surface samples (1 to 250 m) and deep samples (1,000 to 3,600 m) taken from two different casts in February 2001.
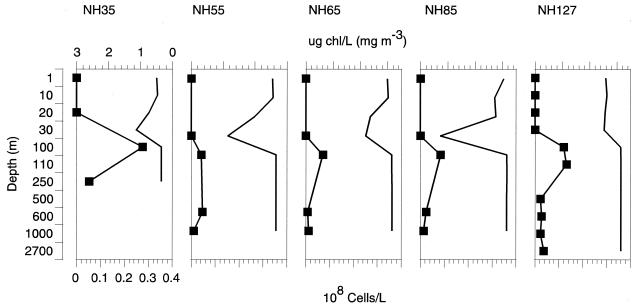
Depth profiles from stations along the Newport hydroline, which extended from the Oregon coast to the edge of the north Pacific gyre, showed a similar trend in the depth-specific distribution of the SAR202 group (Fig. 5). SAR202 cell counts were highest just below the deep chlorophyll maximum (DCM), reaching 27 × 106 cells liter−1 in the 100-m sample from station NH35 and accounting for an average of (12 ± 8.3) × 106 cells liter−1 just below the DCM. Average abundance values below SAR202 surface maximums declined to (2.5 ± 1.5) × 106 cells liter−1 but persisted throughout the water column to a maximum depth of 2,700 m at station NH127. These results confirm previous findings, showing a peak in relative SAR202 high-molecular-weight rRNA and 16S rRNA amplicon abundance just below the DCM (13) and extend their known range to depths throughout the mesopelagic zone and deep ocean.
FIG. 5.
Group-specific FISH of samples taken from off the Oregon coast. Squares, SAR202 depth profiles showing direct cell counts from NH35, NH55, NH65, NH85, and NH127; lines without squares, chlorphyll concentrations.
DISCUSSION
SAR202 is intriguing because of the apparently lengthy evolutionary history and extraordinary metabolic diversity of the phylum Chloroflexi and also because organisms with this diverse and complexly structured cluster resides in the deep pelagic zone of oceans and some lakes (12, 41). In this study we have added to the sparse information about the SAR202 cluster by identifying the associated cell morphology, providing accurate numbers of cells in the water column, and providing a detailed phylogeny for the group.
The data show that SAR202 cluster organisms occur throughout the mesopelagic zone, constituting about 10% of the microbial population there. They probably account for a somewhat larger proportion of deep-ocean microbial biomass, because they are larger than the average bacterioplankton cell (43). Their considerable abundance suggests an important role, but as yet no information about their metabolic activity has come to light. One aspect of the mesopelagic environment is the relatively constant availability of macronutrients (N and P), which are deficient in surface waters, where they likely drive competition among species (40). Energy for microbial metabolism is scarce in the deep ocean most of the time and mainly comes from the oxidation of recalcitrant organic compounds (semilabile dissolved organic carbon [DOC]), ammonium, and nitrite and from the metabolism of more-labile substrates originating from the indigenous fauna and sinking organic material (5, 22). The introduction of surface DOC to the upper mesopelagic zone by convective events associated with winter storms constitutes a large periodic input of DOC to the upper mesopelagic zone (3, 16, 17) and may sustain some elements of the microbial community that reside there.
The SAR202 cluster has been eclipsed because of interest in some of the more abundant bacterioplankton groups, but they occupy an important position in the bacterioplankton pantheon and will undoubtedly be a subject of keen interest as environmental genome sequences, environmental monitoring, and possibly cultures provide more information about this group.
Acknowledgments
We acknowledge Terah Wright, who identified several SAR202 clones from the BATS 250 m clone library, and Rachel Parsons, who prepared the BATS samples used for FISH.
This study was supported by the following grants from the National Science Foundation: An Oceanic Microbial Observatory (MCB-9977918) and Bacterial Activity in the NE Pacific (OCE-0002236).
Footnotes
This is HIMB contribution 1179 and SOEST contribution 6337.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Artic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson, C. A., H. W. Ducklow, and A. F. Michaels. 1994. Annual flux of dissolved organic carbon from the euphotic zone in the northwestern Sargasso Sea. Nature 371:405-408. [Google Scholar]
- 4.Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb. Ecol. 36:37-50. [DOI] [PubMed] [Google Scholar]
- 5.Cho, B. C., and F. Azam. 1988. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature 332:441-443. [Google Scholar]
- 6.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 7.Dunbar, J., B. M. Barns, T. O. Lawrence, and K. R. Cheryl. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field, K. G., D. Gordon, T. Wright, M. Rappé, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frischer, M. E., P. J. Floriani, and S. A. Nierzwicki-Bauer. 1996. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can. J. Microbiol. 42:1061-1071. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and A. A. Davis. 1997. Widespread Archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar. Ecol. Prog. Ser. 150:275-285. [Google Scholar]
- 13.Giovannoni, J. A., M. S. Rappe, K. L. Vergin, and N. L. Adar. 1996. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc. Natl. Acad. Sci. USA 93:7979-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giovannoni, S. J., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, Inc., New York, N.Y.
- 15.Glöckner, F. O., et al. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 16.Hansell, D. A., and C. A. Carlson. 1998. Deep ocean gradients in dissolved organic carbon. Nature 395:263-266. [Google Scholar]
- 17.Hansell, D. A., and C. A. Carlson. 2001. Biogeochemistry of total organic carbon and nitrogen in the Sargasso Sea: control by convective overturn. Deep Sea Res. II Top. Stud. Oceanogr. 48:1649-1667. [Google Scholar]
- 18.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, T. J., R. F. Ramaley, and W. G. Meinschein. 1973. Thermomicrobium, a new genus of extremely thermophilic bacteria. Int. J. Syst. Bacteriol. 23:28-36. [Google Scholar]
- 22.Karl, D. M., G. A. Knauer, and J. H. Martin. 1988. Downward flux of particulate organic matter in the ocean: a particle decomposition paradox. Nature 332:438-441. [Google Scholar]
- 23.Keppen, O. L., T. P. Tourova, B. B. Kuznetsov, R. N. Ivanovsky, and V. M. Gorlenko. 2000. Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria, and emended description of Oscillochloris and Oschillochloris trichoides in comparison with further new isolates. Int. J. Syst. Evol. Microbiol. 50:1529-1537. [DOI] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 25.Lee, S., C. Malone, and P. F. Kemp. 1993. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 101:193-201. [Google Scholar]
- 26.Ludwig, B. E., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Backleitner, and KH. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 27.Maymo-Gatell, X., Y.-T. Chien, J. Gossett, and S. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 28.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface microbial communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 29.Oyaizu, H., B. Debrunner-Vossbrinck, L. Mandelco, J. A. Studier, and C. R. Woese. 1987. The green non-sulfur bacteria: a deep branching in the eubacterial line of descent. Syst. Appl. Microbiol. 9:47-53. [DOI] [PubMed] [Google Scholar]
- 30.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 30a.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 31.Rappé, M. S., K. L. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 32.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 33.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 34.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schonhuber, W., B. Fuchs, S. Juretschko, and R. I. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiguchi, Y., H. Takahashi, Y. Kamagata, A. Ohashi, and H. Harada. 2001. In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol. 67:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, M. T., O. Béjà, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 2002. (PAUP)*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 40.Thingstad, T. F., and F. Rassoulzadegan. 1995. Nutrient limitation, microbial food webs, and biological C-pumps: suggested interactions in a P-limited Mediterranean. Mar. Ecol. Prog. Ser. 117:299-306. [Google Scholar]
- 41.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 42.Vergin, K. L., M. S. Rappe, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-940. [DOI] [PubMed] [Google Scholar]
- 43.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wintzingerode, F. V., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 45.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright, T. D. 1997. M.S. thesis. Oregon State University, Corvallis.
- 47.Wright, T. D., K. L. Vergin, P. W. Boyd, and S. J. Giovannoni. 1997. A novel δ-subdivision proteobacterial lineage from the lower ocean surface layer. Appl. Environ. Microbiol. 63:1441-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zharkikh, A. 1994. Estimation of evolutionary distances between nucleotide sequences. J. Mol. Evol. 9:315-329. [DOI] [PubMed] [Google Scholar]