Abstract
The 102 residue N-terminal extension of the HK97 major capsid protein, the delta domain, is normally present during the assembly of immature HK97 procapsids, but it is removed during maturation like well-known internal scaffolding proteins of other tailed phages and herpesviruses. The delta domain also shares other unusual properties usually found in other viral and phage scaffolding proteins, including its location on the inside of the capsid, a high predicted and measured α-helical content, and an additional prediction for the ability to form parallel coiled-coils. Viral scaffolding proteins are essential for capsid assembly and phage viability, so we tested whether the HK97 delta domain was essential for capsid assembly. We studied the effects of deleting all or parts of the delta domain on capsid assembly and on complementation of capsid-protein-defective phage, and our results demonstrate that the delta domain is required for HK97 capsid assembly.
Keywords: bacteriophage assembly, virus capsids, viral scaffolding proteins, viral protease incorporation, delta domain
Introduction
The assembly of many viral capsids requires the participation of proteins which are not present in the mature capsid (King and Casjens, 1974; Showe and Black, 1973). These are called scaffolding proteins and occur in two varieties, internal and external (Dokland, 1999; Fane and Prevelige, 2003; Prevelige and Fane, 2012). The tailed dsDNA phages (Casjens and Hendrix, 1988) and herpesviruses (Homa and Brown, 1997) utilize internal scaffolding proteins that are present during initial assembly, but they are removed before or during DNA packaging and final maturation of the capsid. In the absence of a functional scaffolding protein, major capsid proteins fail to assemble correctly, often making irregular structures called monsters or spirals (Cerritelli and Studier, 1996; Earnshaw and King, 1978; Hagen et al., 1976), tubes of capsid protein called polyheads (Howatson and Kemp, 1975; Kellenberger, 1980), or less frequently, smaller-than-normal complete capsid shells (Choi et al., 2006; Earnshaw and King, 1978; Thuman-Commike et al., 1998). Internal scaffolding proteins, as their name implies, occupy a significant space within procapsids and therefore must be removed before DNA can be fully packaged. This is often accomplished through the action of a capsid maturation protease, but scaffolding proteins sometimes exit through specialized ports in the capsid that close during maturation, as occurs for phage P22 (Prasad et al., 1993). Scaffolding proteins have also been found to have secondary structures that are dominated by α-helices (Morais et al., 2003; Tuma et al., 1996).
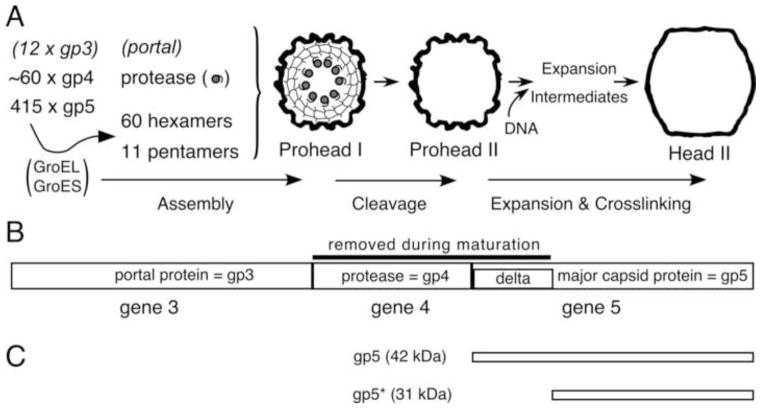
HK97 is a temperate dsDNA bacteriophage with a T=7 icosahedral capsid that does not have a separate scaffolding protein for capsid assembly (Duda et al., 1995b). Instead, the major capsid protein has a delta domain, a 102-amino-acid N-terminal extension, which is removed by the phage-encoded protease before DNA packaging. The term delta domain applies to parts of a major capsid protein (mcp) that are absent from mature capsids, originally applied to phage T4 (Tsugita et al., 1975). A schematic of HK97 capsid assembly and maturation is shown in Fig. 1. Assembly begins with the formation of hexamers and pentamers of the major capsid protein (Xie and Hendrix, 1995). These, collectively called capsomers, along with the portal protein and phage encoded protease, assemble into the immature Prohead I, with the delta domains in the interior of the capsid (Conway et al., 1995). Once assembly is complete, the protease is activated and cleaves the delta domain into peptides, and then cleaves itself (Duda et al., 2013). The liberated peptides appear to escape through holes in the shell (Conway et al., 1995; Duda et al., 1995b), leaving an empty capsid, Prohead II, which is ready for DNA packaging through the portal (Duda et al., 1995a). DNA packaging causes the capsid to expand and crosslink into the mature form, Head II.
Fig. 1. HK97 Capsid Assembly Pathway.
The E. coli chaperonin proteins GroEL and GroES assist in the folding and assembly of the mcp subunit into hexamers and pentamers. These, collectively called capsomers, assemble, along with the portal and the protease, into Prohead I. The active protease, present inside the capsid, cleaves the delta domains, then cleaves itself. The resulting peptides escape from the capsid, leaving an empty Prohead II shell. DNA is packaged through the portal and induces the capsid to transform, via a series of expansion intermediates, into the mature Head II. Head II is stabilized by covalent crosslinks and is ready to attach a tail to form a viable phage. B. HK97 capsid assembly genes with protein names. The HK97 protease substrates, including the delta domain are identified. C. The major forms of the HK97 major capsid protein.
Like viral scaffolding proteins, the HK97 delta domain is present during initial assembly, but absent in the mature structure (Conway et al., 1995; Duda et al., 1995b), suggesting that it may play similar crucial roles during the early steps of assembly. In most tailed-phage genomes, the scaffolding gene lies between the portal gene and the major capsid protein gene (Casjens and Hendrix, 1988; Hendrix and Duda, 1998). In the case of HK97, the delta domain, though fused to the major capsid protein, holds the same position in the genome. For a number of systems, including P22 (Greene and King, 1996, 1999; Weigele et al., 2005), P2 (Cerritelli and Studier, 1996), and the herpesviruses (Kennard et al., 1995), the respective scaffolding proteins bind to capsid interiors via their C-terminal ends, just as the HK97 delta domain is tied to the rest of the mcp by its C-terminus. In the absence of a detailed structure, the HK97 delta domain is predicted to be composed of three α-helical regions (Conway et al., 1995) with a short β-sheet near its C-terminus. The highly α-helical nature of the HK97 delta domain has been confirmed by Raman spectroscopy (Benevides et al., 2004), showing that it truly shares this common property of scaffolding proteins. Additionally, the sequence of the first two predicted α-helices of the delta domain give high scores when algorithms that calculate the probability of forming coiled-coils are applied (Conway et al., 1995). In herpesviruses, P22, T4, λ, and ϕ29, proper capsid assembly is dependent on the presence of the scaffolding protein (Earnshaw and King, 1978) (Ray and Murialdo, 1975) (Tatman et al., 1994) (Thomsen et al., 1994) (Keller et al., 1986) (Keller et al., 1988) (Hagen et al., 1976) (King et al., 1980). The HK97 delta domain shares many properties with other scaffolding proteins; therefore, one could hypothesize that the delta domain is required for HK97 capsid assembly. To test this, we made deletions of all or parts of the delta-domain segment of the HK97 major capsid protein gene and tested the assembly phenotypes of the mutants using plasmid expression studies. Our results confirm that the HK97 delta domain is essential for assembly.
Results and Discussion
Expression of the HK97 major capsid protein without the delta domain
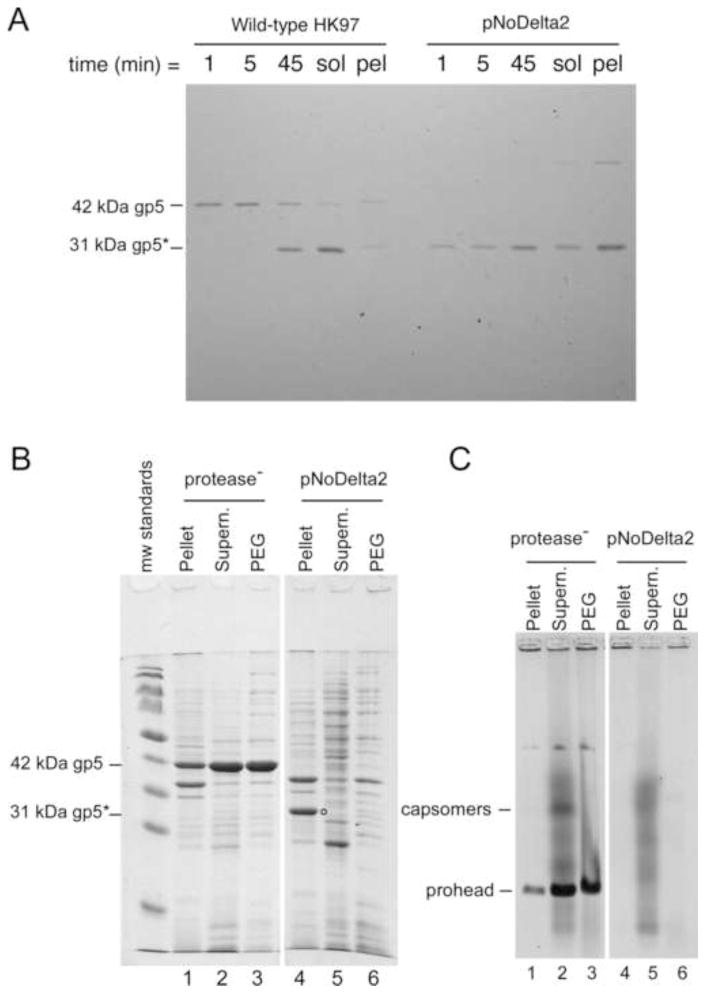
The HK97 maturation protease is not necessary for the full-length major capsid protein to assemble into Prohead I (Duda et al., 1995a) (Duda et al., 1995b) and appears to be incorporated into proheads by binding to the delta domain (Conway et al., 1995; Duda et al., 2013). This suggests that the protease would not be needed for assembly in the absence of the delta domain. Therefore, we deleted the protease and the entire delta domain from our standard expression plasmid to make a delta-less variant called pNoDelta1. In this plasmid, the Met start codon of HK97 major capsid protein, gp5, was moved from its normal position to just upstream of codon 104, which encodes the first residue of the mature HK97 major capsid protein. This plasmid failed to express any detectable protein (not shown), so we added the ribosome binding site and the first five codons from the efficiently expressed bacteriophage T7 major capsid protein (Olins et al., 1988) (Olins and Rangwala, 1989) to make plasmid pNoDelta2. This plasmid produced ample delta-less HK97 major capsid protein (Fig. 2). The proteins from pNoDelta2 and a control plasmid were expressed under radiolabelling conditions (Fig. 2A) and samples were collected 1, 5, and 45 minutes after label addition, along with the remaining soluble and pellet fractions of the culture. The wild-type control produced copious full-length 42 kDa mcp, which was also cleaved to the mature 31 kDa size during the experiment (Fig. 2A, left) indicating normal assembly and conversion to Prohead II. Plasmid pNoDelta2 expressed a protein that migrated similarly to the mature mcp at all time-points and at levels comparable to that of the wild-type protein (Fig. 2A, right). However, most of the protein was in the pellet fraction at the end of the experiment, suggesting that it is insoluble.
Fig. 2. Expression of the HK97 major capsid protein without the delta domain.
A. Radiolabelled proteins produced by the wild-type expression plasmid and plasmid pNoDelta2 were analyzed on a SDS polyacrylamide gel. The control plasmid synthesizes both the protease and the mcp, gp5. B. SDS gel analysis of high-level expression of pNoDelta2. Protease− plasmid (pVB) has a protease knockout mutation and produces Prohead I composed of 42 kDa gp5. pNoDelta2 produces mcp at the expected size in the pellet fraction only (labeled with circle). C. Fractions from the high-level expression were run on a 0.8% agarose gel, and stained. Prohead, head and capsomer bands are indicated. The pNoDelta2 mcp did not produce any distinct capsid assembly structures. The background smears of protein in the supernatant fractions are from cell proteins, which are also present when an empty plasmid vector is expressed.
In order to determine whether the truncated protein was capable of assembling into recognizable structures such as capsomers or proheads, we repeated the expression experiments using our high-level expression protocol and analyzed the pellet, supernatant, and PEG (polyethylene glycol precipitate) fractions (Fig. 2B, C). In this experiment, the control plasmid was a protease knockout plasmid (protease-) that expresses only the full-length HK97 major capsid protein. This plasmid produced a 42 kDa band as expected for uncleaved major capsid protein (lanes 1–3, Fig. 2B) in the SDS gel. The pNoDelta2 plasmid produced a band that migrated as expected for its size (indicated by a dot in lane 4 in Fig. 2B), but this band only appeared in the pellet fraction, confirming that the delta-less mcp is insoluble or, at least, highly aggregated. Protein samples from the pNoDelta2 plasmid also failed to produce any bands above background on a native agarose gel (lanes 4–6, Fig. 2C) in contrast to the control protease knockout plasmid (protease-), which produced clearly visible and soluble proheads and capsomer bands (lanes 1–3, Fig. 2C). Capsomers are hexamers and pentamers of the mcp, which migrate at the same position in these agarose gels (Xie and Hendrix, 1995). The migration behavior of HK97 prohead, capsomers and other assembled structures are well characterized (Dierkes et al., 2009; Duda et al., 1995a; Duda et al., 2013; Xie and Hendrix, 1995), so we are confident that we could detect significant production of soluble assembly products. The pellet fraction containing the bulk of the pNoDelta2 truncated mcp was examined by negative stain electron microscopy, and no recognizable assembled protein structures were found, only cell debris and aggregates of uncertain origin (data not shown). These results highlight the essential role that the HK97 delta domain plays in the successful folding and/or organized assembly of the HK97 major capsid protein, and we suggest that the delta domain may play a chaperone-like role in this process.
Complementation spot tests were used to test for biological function of mcp lacking the delta domain (Table 1). In these assays, mutant protein expressed from a plasmid is tested for its ability to complement phage with an amber mutation in the HK97 major capsid protein gene in order to produce viable phage particles. pNoDelta2 gave no detectable complementation, which is consistent with the biochemical results outlined above.
Table 1. Complementation of gene 4 and gene 5 amber mutant phage by deletion plasmids.
The complementation efficiency of the delta-domain deletion-mutant plasmids was compared to complementation of the wild-type expression plasmid using spot test scores to calculate a relative efficiency of plating (EOP), as described under Materials and Methods. The delta-domain deletion-plasmids were all reduced by 3 to 4 log10 in complementation efficiency in comparison to wild type.
| Plasmid | Wild-type relative EOP | Gene 5 amber relative EOP | Gene 4 amber relative EOP |
|---|---|---|---|
| Wild-type | 1* | 1* | 1* |
| pNoDelta2 | 1 | 0.0001 | n/a |
| Del1 | 1 | 0.001 | 1 |
| Del2 | 1 | 0.001 | 1 |
| Del3 | 1 | 0.001 | 1 |
| Del4 | 1 | 0.001 | 1 |
The asterisks (*) indicate that the wild-type plasmid scores were set to 1 and used as the denominator in EOP calculations. (n/a: not applicable)
Additional deletions within the delta domain
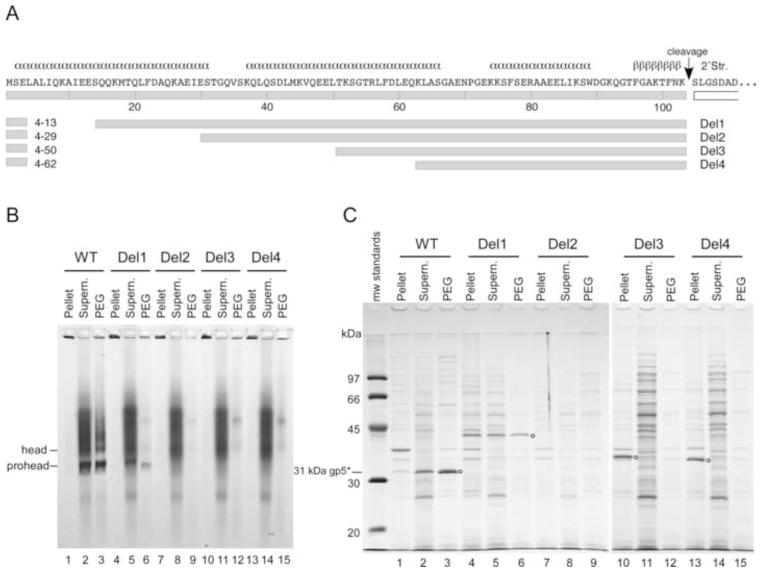
Several smaller deletions in the delta domain were also constructed to see if the capsid protein would retain partial function and perhaps provide more insight into the role of this domain. Deletions were made within a variant of our standard expression plasmid (Duda et al., 1995b). A unique SacI restriction endonuclease cleavage site in codon 4 of the major capsid gene served as one endpoint for all deletions. Four deletions were made (Fig. 3): Del1, removing 9 residues (from 4 to 13), Del2, removing 25 residues (from 4 to 29), Del3 removing 46 residues (from 4 to 50), and Del4, removing 58 residues (from 4 to 62). The locations of the deletions within the delta domain sequence are shown schematically in Fig. 3A. Del1 removes a portion of the predicted N-terminal α-helix of the delta domain; Del2 removes most of the first predicted α-helix; Del3 removes the first and half of the second predicted α-helix; Del4 removes the first and most of the second predicted α-helix.
Fig. 3. Analysis of four delta domain deletions of increasing length.
A. Delta domain sequence and deletion map showing predicted secondary structure. B. Mutant protein fractions from plasmids containing four different-sized deletions in the delta domain were analyzed using a 0.8% non-denaturing agarose gel. The wild-type plasmid produced distinct prohead and head bands, indicated in the figure. Del1 produced a prohead band in the supernatant and PEG fractions. The other deletion mutants did not show any distinct HK97 assemblies. C. SDS gel analysis of protein fractions of deletion mutants. Positions of wild-type and truncated mcp bands are indicated by small circles. The wild-type plasmid produced cleaved mcp (31 kDa). Del1 shows an uncleaved mcp band at the expected size for this deletion in all three fractions. Del2 did not show any detectable mcp. Del3 and Del4 showed mcp in the pellet fractions only, at the sizes expected for these deletions.
High-level expression experiments were again used to characterize the four additional delta domain deletion mutants (Fig. 3). On the non-denaturing agarose gel, the wild-type control produced prohead and head bands (lanes 1–3, Fig. 3B). The corresponding wild-type control lanes in the SDS gel showed cleaved mcp at the expected size of 31 kDa (lanes 1–3, Fig. 3C), indicating that the prohead band is indeed Prohead II and confirming successful assembly and cleavage of the delta domain. (In vivo, the wild-type plasmid produces only Prohead II, but the freeze/thaw step used to induce lysis also induces in vitro conversion of some of the Prohead II present to Head II(Duda et al., 1995b)). The mutants Del2, Del3, and Del4 did not produce any identifiable bands in the agarose gel and thus appeared to produce no soluble assembly products (lanes 7–15, Fig. 3B). Del2 did not produce a detectable mcp band in the SDS gel in any fraction, (lanes 7–10, Fig. 3C, confirmed by a repeat experiment), which suggests that this unnatural protein is very unstable in the host cells and is rapidly degraded. Del3 and Del4 each produced a distinct band in the SDS gel, which migrate at the positions expected for the sizes of the deletion proteins (indicated by circles next to lanes 10 and 14, Fig. 3C). However, each was found exclusively in the pellet fraction, which strongly suggests that Del3 and Del4 produced insoluble protein similar to the product from pNoDelta2. These results provide further support for the crucial role of the delta domain in the folding and solubility of the HK97 major capsid protein.
Del1 was the only deletion mutant that produced detectable bands in the agarose gel. These faint Del1 bands were found in both the supernatant and PEG fractions at the position expected for proheads (lanes 5 and 6, Fig. 3B), indicating that the Del1 protein is capable of assembling into proheads, although not as efficiently as the wild-type protein. The Del1 lanes in the SDS gel each showed a band of the appropriate mobility to be the uncleaved version of the Del1 protein (indicated by a circle next to lane 6, Fig. 3C) showing that cleavage did not occur for this mutant and that the proheads produced were a form of Prohead I. The pellet fraction of the SDS gel contained the highest concentration of the Del1 protein, indicating that the protein that did not assemble into Prohead I either misfolded and aggregated, or assembled abnormally into structures that were trapped in the pellet. This finding suggests that the Del1 mutant is defective in folding, even though it has retained some ability to assemble. The pellet fraction was not examined further.
Complementation tests were done to assess the functionality of the four N-terminal deletion mutants. All four mutants exhibited no significant complementation, indicating that they are all biologically dead (Table 1), including the Del1 mutant. None of the deletion mutants interfered with the growth of wild-type phage in the complementation test controls, showing that there were no dominant negative effects observed. Dominant negative effects have been observed with other HK97 mutants (Dierkes et al., 2009).
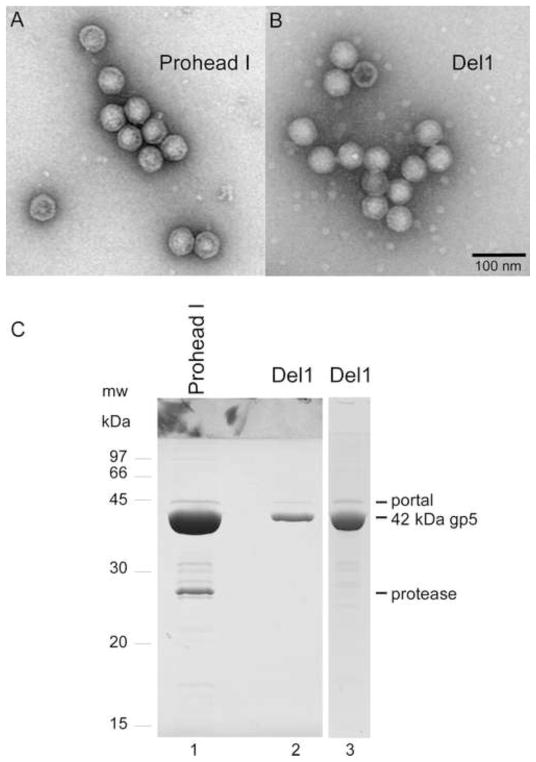
The lack of capsid protein cleavage for the Del1 mutant could be due to failure to incorporate the protease, or to a different kind of block to proteolysis. To test for Del1’s ability to co-assemble with the protease, the Del1 deletion was transferred to a plasmid that contains both the HK97 portal and an inactive variant of the HK97 protease with the active site mutation H65A (Duda et al., 2013). Following protein expression, the proheads were purified and compared to the proheads produced by an identical plasmid without the deletion. Electron microscopy showed that the two types of proheads were essentially identical in appearance in negative stain (Fig. 4A, B), indicating that the Del1 mutant made capsids of the proper size and shape. SDS gel analysis of the purified control proheads showed uncleaved major capsid protein, portal, and inactivated protease bands (Fig. 4C, lane 1), as expected. The yield of Del1 proheads was lower than wild-type, as indicated by the weaker mcp band observed when equivalent samples from the two preparations were compared (Fig. 4C, lane 2). A portal protein band was clearly visible in the Del1 sample, showing that the portal was incorporated. Strikingly, however, no protease band was detected in the Del1 sample, even when the total amount of protein loaded was increased to match the conditions where the inactive protease band was readily detectable in the control Prohead I (compare lanes 1 and 3 in Fig. 4C). These results show that the cleavage defect of the Del1 mutant is due to a failure to incorporate the HK97 protease, and further suggest that the N-terminal helix of the delta domain may participate in protease binding.
Fig. 4. Comparison of Del1 proheads (deleting residues 9–13) with wild-type proheads.
A and B. Electron micrographs of purified particles stained with 1% uranyl acetate. Both images were taken at the same magnification. The scale bar represents 100 nm. A. Wild-type Prohead I. B. Del1 proheads. C. SDS gel analysis of the composition of proheads: lane 1, Prohead I with portal and inactivated protease from plasmid pVP0-g4H65A; lanes 2 and 3, portal-containing Del1 proheads made in the presence of inactivated protease from plasmid pVP0-g4H65A-Del1.
The multiple essential roles of the HK97 delta domain
We have shown that the HK97 delta domain behaves like a scaffolding protein analog and is essential for assembly of the HK97 major capsid protein into a procapsid. The HK97 delta domain also appears to have chaperone-like qualities, because all large deletions in the delta domain produced unstable or insoluble protein. Deletion of only nine amino acids near the N-terminus in Del1 drastically reduced this protein’s solubility and interfered with assembly. Many internal viral scaffolding proteins have their N-terminal regions positioned toward the center of the capsid, which allow them to make intermolecular interactions (Dokland, 1999) (Parker et al., 1998) (Parker et al., 1997) (Wood et al., 1997). Similar interactions may be required in the delta domain, which could explain why the larger delta-domain deletions are unable to assemble capsomers into proheads. Furthermore, these interactions may involve coiled-coil interactions between delta domains. We believe that the highly α-helical nature of the HK97 delta domain may be important for delta-domain function by providing it with the ability to form such coiled-coil interactions with itself, and with other partners, including the protease and portal proteins.
In P22 procapsid assembly, mutations which block portal incorporation map to a near-C-terminal region of the scaffolding protein (Greene and King, 1996), showing that the multiple functions of a scaffolding protein map to different parts of the protein. While our current study did not reveal if the HK97 portal binds to any part of the delta domain, we did find that an intact N-terminal delta domain α-helix is required for protease incorporation, strongly suggesting that the protease may bind there. We have previously shown that the C-terminus of the HK97 protease contains the capsid targeting signal (Duda et al., 2013). The C-terminus of the protease is predicted to be largely α-helical, which together with the results reported here supports the idea that α-helical regions at the C-terminus of the HK97 protease may form coiled-coils with the N-terminal region of the delta domain to mediate its assembly into proheads (Duda et al., 2013). We have begun expanded studies of the HK97 delta domain that target specific residues and regions identified in analyses of aligned protein sequences of a larger set of phage delta domains. These studies will provide deeper insight into the specific roles of delta domains and scaffolding proteins in viral capsid assembly.
Materials and Methods
Bacterial strains, plasmids and plasmid construction
T7 promoter expression strain E. coli BL21(DE3)pLysS (Studier et al., 1990) was used for capsid protein expression and for complementation spot tests. E. coli stain DH10b was used for propagating plasmids. All plasmids were based on T7 expression vector pT7-5 (GenBank ACCESSION: AY230150). The wild-type HK97 expression plasmid, pV0 (pT7-Hd2.9) (Duda et al., 1995b) contains a part of HK97 gene 3 and all of genes 4 and 5. Plasmid pVB is pV0 with the protease gene inactivated (pT7-Hd2.9(fsBstBI)) (Duda et al., 1995b). Plasmid pV0SacI has a silent codon change that introduces a SacI site between codons 4 and 5 of gene 5. Plasmid pVP0 was made from pV0 by adding a genomic EcoRI fragment that contains the rest of the portal gene. Plasmid pVP0-g4H65A has a gene 4 mutation H65A that inactivates the protease (Duda et al., 2013). Plasmid pVP0 was used as the PCR template with Pfu polymerase (Stratagene) for making the no-delta plasmids. PCR products were purified by gel electrophoresis and pressure-extrusion (Gubin and Kincaid, 1998). The PCR products and plasmid pT7-5 were digested with EcoRI and HindIII, gel purified, ligated and transformed into E. coli DH10b cells. Del1, Del2, Del3, and Del4 were created in plasmid pV0SacI using Phusion polymerase (Finnzymes OY). Segments of gene 5 were amplified using primers that introduce a new SacI site at their 5′ ends and re-joined to the original SacI site to create the deletions. All PCR-amplified segments in the plasmids used for this work were verified by DNA sequencing.
Radiolabeling
Radiolabeling was done in M9-dMAM medium, which is M9 medium (Kellenberger and Sechaud, 1957) supplemented with 0.4 % glucose and 1% DIFCO Methionine Assay Medium. Overnight cultures of pNoDelta1 and pNoDelta2 were diluted into M9-dMAM with 50 μg/ml ampicillin and 25 μg/ml chloramphenicol and incubated with no shaking at 30°C. The next day, the cultures were transferred to a 37°C water bath, grown with shaking at 250 rpm until the OD550 reached ~ 0.3–0.4 and induced with 0.4 mM IPTG for 30 min. Rifampicin was added to 200 μg/ml and later, 35S-Met was added to 1 μCi/ml. Samples were taken after 1, 5, and 45 min, TCA precipitated, and processed for SDS gel analysis. The remaining cultures were centrifuged. The pelleted cells were suspended in 50 mM Tris-HCL (pH 8.0), 5 mM EDTA, and lysed at 24°C after adding Triton X100 to 0.1%. MgSO4 was added to 25 mM, followed by DNase I to 20 μg/ml and the lysates were incubated at 24°C for ~5′ to reduce viscosity and centrifuged at 14,000 × g for 10 min. The supernatant was saved and the pellet was suspended in TKG50 buffer (Tris 20 mM Tris-HCl, pH 7.5, 50 mM Potassium Glutamate). Fractions were TCA precipitated and denatured in SDS for gel analysis.
High-level expression experiments
HK97 capsid genes were expressed using the autoinduction methods and media described by Studier (Studier, 2005), which produce high yields of HK97 procapsids from our expression plasmids. The high protein yields allow the use of autoinduction in small scale cultures for analyzing mutants. Overnight cultures of BL21(DE3)pLysS containing HK97 expression plasmids grown in MDG (non-inducing) media with 50 μg/ml ampicillin and 25 μg/ml chloramphenicol were diluted ~300 fold into 1.5 ml of TYM5052 (autoinduction) media and grown at 37°C with aeration (shaking at 300 rpm) in 18 mm diameter tubes for 24 hrs. Cells were collected and suspended in 0.3 ml of 50 mM Tris-HCL (pH 8.0), 8 mM EDTA. Cells were lysed by adding Triton X100 to a final concentration of 0.15%, freezing in dry ice, and thawing at room temperature. MgSO4 was added to 25 mM, followed by DNase I to 15 μg/ml and the lysates were incubated at 28°C for ~5′ to reduce the viscosity. The lysates were centrifuged at 14,000 × g for 10 min. The supernatant was divided into two tubes; one to act as the supernatant fraction, the other to be used for the PEG (polyethylene glycol) precipitation fraction. Pellets were suspended in 0.3 ml of DL buffer (20 mM Tris HCl, pH 7.5, 40 mM NaCl). For the PEG fraction, 1/2 volume of 12% (w/v) PEG 8000 in 1M NaCl was added, the mixture left on ice for ~20 minutes, and centrifuged at 14,000 × g for 10 min. The PEG supernatant was removed and discarded. Residual liquid was removed after a second quick centrifugation, and the PEG pellet was suspended in 0.15 ml Buffer G (20 mM Tris HCl, pH 7.5, 100 mM NaCl). Fractions were stored cold for biochemical analyses.
Purification of proheads
Control and Del1 proheads were purified from cleared lysates made from induced 0.5 L cultures of BL21(DE3)pLysS containing plasmids pVP0-g4H65A or pVP0-g4H65A-Del1. Purification was done as described previously (Dierkes et al., 2009; Li et al., 2005) and included PEG precipitation, pelleting by ultracentrifugation and separation by velocity sedimentation in glycerol gradients. The final samples were concentrated by ultracentrifugation and resuspended in buffer containing 20 mM Tris-HCL and 50 mM potassium glutamate.
Electrophoresis methods
For native agarose gels, samples were diluted into a dye-glycerol mixture in TAMg buffer (40 mM Tris base, 20 mM acetic acid pH 8.1, and 1 mM magnesium sulfate), loaded onto a native 0.9% agarose gel, electrophoresed, and stained for protein using Coomassie Brilliant Blue R250 as described previously (Duda et al., 1995a). For SDS-polyacrylamide gels, samples were precipitated by adding TCA to ~10%, or diluted 25 fold into 10% TCA with 2 mg/ml sodium deoxycholate (DOC) for autoinduction samples. Following incubation on ice for 10 min, the pellets were collected by centrifugation, washed with acetone, dried, suspended in SDS sample buffer (Laemmli, 1970), and denatured at 100°C. Samples were electrophoresed in 12% low-crosslinking SDS-polyacrylamide gels made from a 33.5% (w/v) acrylamide/0.3% (w/v) methylene bisacrylamide stock (Dreyfuss 1984) using the traditional buffers (Laemmli, 1970), and stained with Coomassie Brilliant Blue R250. The gels in Fig. 4 were made from a 30% (w/v) acrylamide/1.6% (w/v) methylene bisacrylamide stock to help separate the portal band from the mcp band.
Complementation Spot Tests
These tests rapidly evaluate whether a gene expressed from a plasmid makes a protein that can substitute for the one missing from cells infected with a phage carrying an amber mutation. Overnight cultures of the BL21(DE3)plysS mutant strains were mixed with soft agar supplemented with ~1% lactose (to induce gene expression) and used to create a bacterial lawn on an LB plate. Serial dilutions (109 pfu/ml - 104 pfu/ml) of WT HK97 phage, phage am1 (amber mutation in gene 5), phage amU4 (amber mutation in gene 4), and phage amC2 (negative control amber mutation) were spotted as 4 μl drops, and the plates were incubated overnight at 37°C. pV0 served as the wild-type control plasmid. Titers were estimated by assigning a score of 10 for each fully clear spot and 3 for each partially cleared spot (not including those with only a few plaques) and multiplying the results (Dierkes et al., 2009). WT usually gives 5 clear spots and one partially cleared spot, so its plating score is 10 × 10 × 10 × 10 × 10 × 3 = 3×105. Relative complementation values (efficiency of plating, EOP) were calculated by dividing the titer obtained for a mutant plasmid by that obtained for the wild-type plasmid.
Transmission Electron Microscopy
Carbon/formvar grids (400 mesh, Ted Pella, Redding, CA) were treated by glow-discharge before use. Samples were applied, rinsed with water, stained with 1% uranyl acetate in water and blotted dry. Digital micrographs were taken at 56000× using a Morgagni 268(D) EM (FEI, Eindhoven, Netherlands).
HK97 major capsid protein is insoluble when its entire delta domain is deleted
Even short deletions in the HK97 delta domain are deleterious
A short N-terminal delta domain deletion abolishes protease incorporation
Acknowledgments
This work was supported by NIH grant GM47795 from the U.S. Public Health Service to RWH and RLD. We thank Dr. Craig Peebles and our network of colleagues for discussions that were essential for the completion of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benevides JM, Bondre P, Duda RL, Hendrix RW, Thomas GJ., Jr Domain structures and roles in bacteriophage HK97 capsid assembly and maturation. Biochemistry. 2004;43:5428–5436. doi: 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hendrix R. Control Mechanisms in dsDNA Bacteriophage Assembly. In: Calendar R, editor. The Bacteriophages. Springer; US: 1988. pp. 15–91. [Google Scholar]
- Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996;258:286–298. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- Choi KH, Morais MC, Anderson DL, Rossmann MG. Determinants of bacteriophage phi29 head morphology. Structure. 2006;14:1723–1727. doi: 10.1016/j.str.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL. Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. Journal of virology. 2009;83:2088–2098. doi: 10.1128/JVI.02000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T. Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995a;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995b;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- Duda RL, Oh B, Hendrix RW. Functional domains of the HK97 capsid maturation protease and the mechanisms of protein encapsidation. J Mol Biol. 2013;425:2765–2781. doi: 10.1016/j.jmb.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Advances in protein chemistry. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- Greene B, King J. In vitro unfolding/refolding of wild type phage P22 scaffolding protein reveals capsid-binding domain. The Journal of biological chemistry. 1999;274:16135–16140. doi: 10.1074/jbc.274.23.16135. [DOI] [PubMed] [Google Scholar]
- Gubin AN, Kincaid RL. A pressure-extrusion method for DNA extraction from agarose gels. Analytical biochemistry. 1998;258:150–152. doi: 10.1006/abio.1998.2581. [DOI] [PubMed] [Google Scholar]
- Hagen EW, Reilly BE, Tosi ME, Anderson DL. Analysis of gene function of bacteriophage phi 29 of Bacillus subtilis: identification of cistrons essential for viral assembly. Journal of virology. 1976;19:501–517. doi: 10.1128/jvi.19.2.501-517.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: a protein ballet. Advances in virus research. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Howatson AF, Kemp CL. The structure of tubular head forms of bacteriophage lambda; relation to the capsid structure of petit lambda and normal lambda heads. Virology. 1975;67:80–84. doi: 10.1016/0042-6822(75)90405-5. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. Control mechanisms in the morphogeneses of bacteriophage heads. Bio Systems. 1980;12:201–223. doi: 10.1016/0303-2647(80)90018-0. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Sechaud J. Electron microscopical studies of phage multiplication. II. Production of phage-related structures during multiplication of phages T2 and T4. Virology. 1957;3:256–274. doi: 10.1016/0042-6822(57)90092-2. [DOI] [PubMed] [Google Scholar]
- Keller B, Dubochet J, Adrian M, Maeder M, Wurtz M, Kellenberger E. Length and shape variants of the bacteriophage T4 head: mutations in the scaffolding core genes 68 and 22. Journal of virology. 1988;62:2960–2969. doi: 10.1128/jvi.62.8.2960-2969.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B, Maeder M, Becker-Laburte C, Kellenberger E, Bickle TA. Amber mutants in gene 67 of phage T4. Effects on formation and shape determination of the head. J Mol Biol. 1986;190:83–95. doi: 10.1016/0022-2836(86)90077-x. [DOI] [PubMed] [Google Scholar]
- Kennard J, Rixon FJ, McDougall IM, Tatman JD, Preston VG. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. The Journal of general virology. 1995;76 ( Pt 7):1611–1621. doi: 10.1099/0022-1317-76-7-1611. [DOI] [PubMed] [Google Scholar]
- King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- King J, Griffin-Shea R, Fuller MT. Scaffolding proteins and the genetic control of virus shell assembly. The Quarterly review of biology. 1980;55:369–393. doi: 10.1086/411981. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Conway JF, Cheng N, Steven AC, Hendrix RW, Duda RL. Control of Virus Assembly: HK97 “Whiffleball” Mutant Capsids Without Pentons. J Mol Biol. 2005;348:167–182. doi: 10.1016/j.jmb.2005.02.045. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nature structural biology. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988;73:227–235. doi: 10.1016/0378-1119(88)90329-0. [DOI] [PubMed] [Google Scholar]
- Olins PO, Rangwala SH. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. The Journal of biological chemistry. 1989;264:16973–16976. [PubMed] [Google Scholar]
- Parker MH, Casjens S, Prevelige PE., Jr Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- Parker MH, Stafford WF, 3rd, Prevelige PE., Jr Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- Prasad BV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Fane BA. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Advances in experimental medicine and biology. 2012;726:325–350. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- Ray P, Murialdo H. The role of gene Nu3 in bacteriophage lambda head morphogenesis. Virology. 1975;64:247–263. doi: 10.1016/0042-6822(75)90096-3. [DOI] [PubMed] [Google Scholar]
- Showe MK, Black LW. Assembly core of bacteriophage T4: an intermediate in head formation. Nature: New biology. 1973;242:70–75. doi: 10.1038/newbio242070a0. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods in enzymology. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tatman JD, Preston VG, Nicholson P, Elliott RM, Rixon FJ. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. The Journal of general virology. 1994;75 ( Pt 5):1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- Thomsen DR, Roof LL, Homa FL. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. Journal of virology. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugita A, Black LW, Showe MK. Protein cleavage during virus assembly: characterization of cleavage in T4 phage. J Mol Biol. 1975;98:217–215. doi: 10.1016/s0022-2836(75)80116-1. [DOI] [PubMed] [Google Scholar]
- Tuma R, Prevelige PE, Jr, Thomas GJ., Jr Structural transitions in the scaffolding and coat proteins of P22 virus during assembly and disassembly. Biochemistry. 1996;35:4619–4627. doi: 10.1021/bi952793l. [DOI] [PubMed] [Google Scholar]
- Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J Mol Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Wood LJ, Baxter MK, Plafker SM, Gibson W. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. Journal of virology. 1997;71:179–190. doi: 10.1128/jvi.71.1.179-190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]