Abstract
West Nile virus (WNV) is a mosquito-transmitted pathogen, which causes significant disease in humans. The innate immune system is a first-line defense against invading microorganism and many flaviviruses, including WNV, have evolved multifunctional proteins, which actively suppress its activation and antiviral actions. The WNV non-structural protein 1 (NS1) inhibits signal transduction originating from Toll-like receptor 3 (TLR3) and also critically contributes to virus genome replication. In this study we developed a novel FACS-based screen to attempt to separate these two functions. The individual amino acid changes P320S and M333V in NS1 restored TLR3 signaling in virus-infected HeLa cells. However, virus replication was also attenuated, suggesting that the two functions are not easily separated and may be contained within overlapping domains. The residues we identified are completely conserved among several mosquito- and tick-borne flaviviruses, indicating that they are of biological importance to the virus.
Keywords: Flavivirus, West Nile virus, TLR3, NS1, Innate Immunity
Introduction
The Flaviviridae family includes several arthropod-borne viruses, which are important human pathogens, such as dengue virus, yellow fever virus, and West Nile virus (WNV). West Nile virus is a mosquito-transmitted virus that can cause febrile illness, severe encephalitic disease, and death in humans and other vertebrates, and is endemic to regions of Africa, Europe, and North America. The positive-sense RNA genome of WNV codes for three structural proteins (C, prM, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), which are expressed as a single large polyprotein that is co- and post-translationally processed into the ten individual proteins (Chambers et al., 1990). The structural proteins arrange in icosahedral geometry to form mature enveloped virions and package the infectious genome, while the non-structural proteins are required for viral RNA replication and interact with a multitude of cellular pathways. Many viruses have evolved multifunctional proteins to overcome limitations in the coding capacity of their often relatively small genomes and several examples of replication proteins that also interfere with the immune response to infection can be found within the Flaviviridae.
For example, the Hepatitis C virus NS3 protein exhibits RNA helicase activity required for replication, and protease activity in conjunction with its cofactor NS4A, required for polyprotein processing (Reed and Rice, 1998). However, protease activity also cleaves the innate immune adaptor proteins TIR-containing, adaptor-inducing IFN-β (TRIF), and IFN-β promoter stimulator-1 (IPS-1), which transmit signals from the pattern-recognition receptors (PRR) Toll-like receptor 3 (TLR3), TLR4, retinoic acid-inducible gene I (RIG-I), and melanoma-differentiation associated gene 5 (MDA-5) (Li et al., 2005; Loo et al., 2006).
The NS5 protein is the RNA-dependent, RNA polymerase and has been shown for several flaviviruses to also inhibit signal transducer and activator of transcription 1 (STAT1) and/or STAT2 signaling originating from the type I interferon (IFN) receptor by different mechanisms depending on the virus species (Ashour et al., 2009; Laurent-Rolle et al., 2010; Lin et al., 2006; Lin et al., 2004; Mazzon et al., 2009; Park et al., 2007; Werme, Wigerius, and Johansson, 2008). The dengue, West Nile, and Kunjin virus NS2A, NS4A, and NS4B proteins, whose functions in RNA replication are not well understood, are also involved in interfering with Janus-activated kinases (JAK)-STAT signaling from the type I IFN receptor (Evans and Seeger, 2007; Liu et al., 2006; Liu et al., 2005; Munoz-Jordan et al., 2005; Munoz-Jordan et al., 2003). Inhibition of these innate immune signaling pathways decreases both type I IFN and cytokine production, and type I IFN responses in infected cells, which normally lead to interferon-stimulated gene (ISG) expression. The collective function of these non-structural proteins blunts the establishment of an antiviral state in the host cell and allows for increased virus dissemination within the host.
NS1 is a glycoprotein that is expressed intracellularly and is also secreted from infected cells. A hydrophobic signal sequence derived from the C-terminus of the E protein promotes NS1 translocation into the lumen of the endoplasmic reticulum (ER) following polyprotein translation and is subsequently cleaved by cellular signal peptidases (Chambers et al., 1990). Most flavivirus NS1 proteins contain two N-linked glycosylation sites and twelve conserved cysteine residues. Viruses within the Japanese Encephalitis Serocomplex, with exception of Japanese Encephalitis virus itself contain three N-linked glycosylation sites that resides at position 175 for the WNV NS1 protein. NS1 forms heat-labile but detergent-resistant dimers and the secreted form has also been reported to bind to naive cells. In the case of endothelial cells, this is achieved through cell surface interactions between NS1 and the glycosaminoglycans, heparan sulfate and chondroitin sulfate E (Alcon-LePoder et al., 2005; Avirutnan et al., 2007). NS1 has also been shown to bind to and become endocytosed into other cell types such as hepatocytes (Alcon-LePoder et al., 2005). While its function during virus replication is not completely understood, NS1 is absolutely required for RNA replication and plays a role in early negative-sense RNA synthesis (Khromykh et al., 1999; Lindenbach and Rice, 1997; Lindenbach and Rice, 1999; Westaway et al., 1997). Recent work by Youn et al. has demonstrated a direct protein:protein interaction between NS1 and NS4B in infected cells, which may facilitate interactions with the replication complex (Youn et al., 2012). NS1 has recently been implicated in various aspects of flavivirus pathogenesis such as binding to and promoting the degradation of complement factors (Avirutnan et al., 2010; Avirutnan et al., 2011; Chung et al., 2006a). Paradoxically, antibodies to NS1 aid the clearance of infected cells by complement-mediated phagocytosis and NS1-specific antibodies have been shown to confer a protective effect when administered prior to viral challenge (Chung et al., 2006b; Chung et al., 2007; Schlesinger et al., 1990).
TLR3 is an evolutionarily-conserved PRR that recognizes dsRNA, a replication intermediate of many RNA viruses and a component of secondary RNA structures found in viral genomes. TLR3 activation in endosomes leads to the induction of pro-inflammatory cytokines and type I IFN expression, and induces an antiviral state in cells (Nishiya et al., 2005; Yamamoto et al., 2002). Other cytokines, such as IL-6, are also important for the acute phase response to infection, can induce fever, and support the growth and maturation of B cells (Rummel et al., 2006) We have previously reported that NS1 expressed by itself in HeLa and HEK293-TLR3 cells, and in the context of WNV infection inhibits TLR3 (Scholle and Mason, 2005; Wilson et al., 2008). Aside from the HCV protease, Pestivirus N(pro), and dengue virus protease, which target IPS-1, interferon regulatory factor 3 (IRF3), and stimulator of interferon genes (STING), respectively, NS1 is so far the only other flavivirus non-structural protein to interfere with PRR signaling (Aguirre et al., 2012; Hilton et al., 2006; Loo et al., 2006; Yu et al., 2012) In this study we aimed to identify amino acids of NS1 that are required for TLR3 inhibition. Traditional approaches, such as progressive truncation of the NS1 protein, neglect the indispensible role of NS1 in genome replication.
Due to the multifunctional nature of NS1, a screening approach based on trans-complementation was developed to allow selection of randomly mutagenized NS1 proteins that are 1) still able to support viral RNA replication while being 2) defective in TLR3 inhibition. We successfully identified two amino acid changes at positions P320 and M333 of NS1 that restore TLR3 signaling and allow virus genome replication and productive infection in HeLa cells, albeit to levels below wt virus. These amino acids lie within the C-terminal half of the NS1 protein, and each of these residues is conserved among mosquito-borne and tick-borne flaviviruses. We determined that HeLa cells, infected to equivalent levels with recombinant WNV containing the individual mutations, are able to signal through TLR3 while wt WNV infected cells are unresponsive to TLR3 stimulation. However, decreased virus production was observed with both mutant viruses suggesting that TLR3 inhibition and replication support might not be completely separable functions of NS1. These findings suggest that each of the identified amino acid residues are of biological importance to the virus and that the two functions we assessed may be conserved in overlapping domains.
Results
Strategy
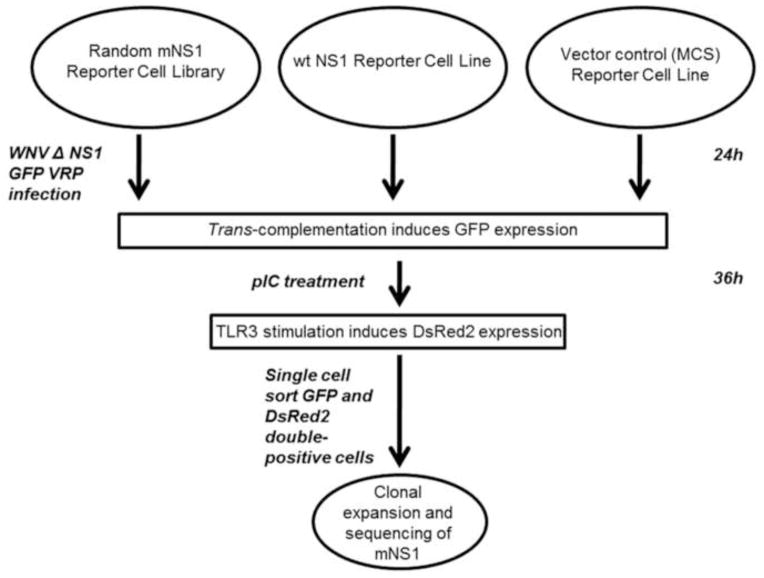
The overall goal of the mutational screen was to identify amino acid changes in the NS1 protein that allow TLR3 signaling and viral replication. The strategy was based on 1) the ability of ectopically expressed wt NS1 protein to rescue replication of a GFP-expressing WNV replicon containing a lethal deletion in NS1, and 2) on the ability of wt NS1 to inhibit TLR3 signaling. Figure 1 outlines the overall strategy. To successfully perform the screen, first a GFP expressing WNV replicon with a lethal deletion in NS1 was created (WNV ΔNS1 GFP rep) and packaged into virus replicon particles (WNV ΔNS1 GFP VRPs). Based on previous studies describing trans-complementation of NS1, this replicon would only be able to replicate and express GFP in the presence of wt NS1 or an NS1 variant that is able to complement the defect (Lindenbach and Rice, 1997). A library of randomly mutagenized NS1 was established in a recombinant lentivirus background to allow expression in a reporter cell line, in which red fluorescent protein DsRed2 is expressed under control of an NFκB-responsive promoter. DsRed2 is only induced by pIC stimulation in cells that express NS1 proteins defective for TLR3 inhibition. During the actual screen, reporter cells expressing mutagenized NS1 were infected with WNV ΔNS1 GFP VRPs and subsequently treated with pIC. The presence of GFP and DsRed2 double-positive cells indicates NS1 proteins of the desired phenotype.
Figure 1. Screen work-flow for the identification of mutagenized NS1 proteins that allow TLR3 signaling and support replication of a WNV NS1-deleted GFP replicon particle.
A HeLa reporter cell library expressing random NS1 mutants (HeLa-NFκB-DsRed2-pLEX-mNS1) was infected at an MOI of 1 with NS1-deleted WNV GFP replicon particles (WNV ΔNS1 GFP VRP) for 24h followed by pIC treatment for an additional 36h. GFP and DsRed2 double-positive cells were sorted by Fluorescence-activated cell sorting (FACS), individual clones expanded and the NS1 sequence of each clone was determined after confirmation of the desired phenotype.
Creation of an NFκB-dependent Red Fluorescent Protein Reporter Cell Line (HeLa-NFκB-DsRed2)
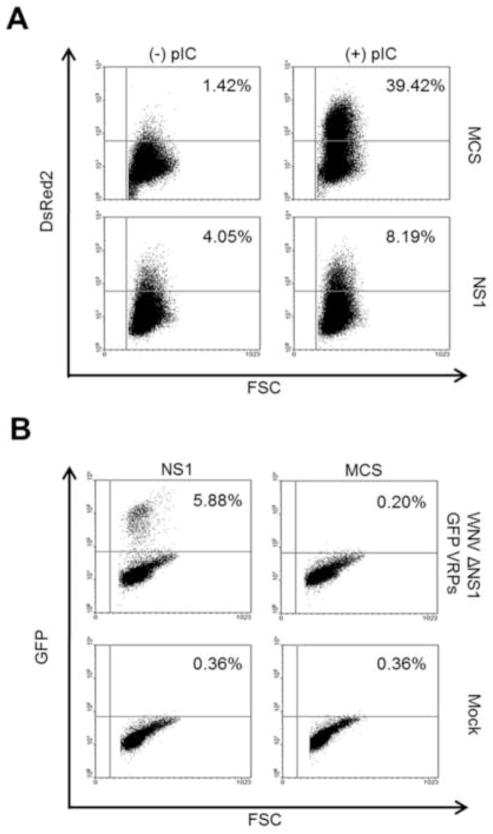
A reporter construct using the canonical NFκB response element and a synthetic ELAM promoter, controlling expression of DsRed2 was assembled and a stable HeLa reporter cell pool (HeLa-NFκB-DsRed2) was established. Background fluorescence was very low and inducible expression of DsRed2 was confirmed by fluorescence microscopy 36h post-treatment with pIC (data not shown). We have previously shown that pIC, when added to the culture medium, will signal through TLR3 alone (Wilson et al., 2008). To create a system for ectopic expression of NS1, WNV NS1 cDNA, including a 6X His epitope tag, was cloned into the pLEX lentivirus vector and recombinant lentiviruses (pLEX-NS1) were produced according to standard procedures. An empty vector control lentivirus was used as a negative control (pLEX-MCS). Stable cell lines were created after transduction of either HeLa-NFkB-DsRed2 cells or standard HeLa cells with either pLEX-NS1 or pLEX-MCS followed by antibiotic selection. HeLa-NFκB-DsRed2-pLEX-MCS and HeLa-NFkB-DsRed2-pLEX-NS1 cells were treated with pIC for 36h and DsRed2 expression was analyzed by flow cytometry. Approximately 40% of pIC treated NFκB-DsRed2-pLEX-MCS cells expressed DsRed2 after 36h (Figure 2A). In agreement with our previously-published work, the number of DsRed2 positive cells was decreased by approximately 80% in wild type NS1-expressing cells after pIC stimulation (Figure 2A) (Scholle and Mason, 2005; Wilson et al., 2008). These results confirm that TLR3 signaling in this cell line results in DsRed2 expression and that wt NS1 can inhibit this process.
Figure 2. Validation of the tools employed by the screen.
(A) HeLa-NFκB-DsRed2 cells stably transduced with either pLEX-NS1 or pLEX-MCS (vector control) lentivirus were treated with pIC for 36h or left untreated and DsRed2 expression was determined by flow cytometry. (B) HeLa-NFκB-DsRed2 cells stably transduced with either pLEX-NS1 or pLEX-MCS (vector control) lentivirus were either infected for 24h with WNV ΔNS1 GFP VRPs or mock infected. GFP expression was determined by flow cytometry. Data are representative of three independent experiments.
Trans-complementation of an NS1-deleted Replicon
A 138nt lethal in-frame deletion within the NS1 coding sequence of a GFP-expressing WNV replicon was introduced to create a replicon (WNV ΔNS1 GFP rep) whose replication was dependent on trans-expressed functional NS1. In vitro transcribed RNA of WNV ΔNS1 GFP rep was replication defective when transfected into naïve HeLa cells or HeLa-pLEX-MCS cells. In contrast, GFP expression from transfected WNV ΔNS1 GFP rep RNA was readily observed in HeLa pLEX-NS1 or HeLa cells expressing NS1 from a CMV promoter (Wilson et al., 2008). GFP positive cells were visible at 24h prost transfection and GFP expression persisted over another 24h period, confirming that trans-expressed wt NS1 was able to sustain WNV ΔNS1 GFP rep replication (supplemental Figure S1).
To facilitate delivery of WNV ΔNS1 GFP rep RNA to reporter cells, VRPs were produced. A BHK packaging cell line (BHK-Pack NS1) was used in which the WNV C, prM, E, and NS1 proteins are expressed from a Venezuelan equine encephalitis (VEE) replicon. HeLa-NFκB-DsRed2-pLEX-MCS and HeLa-NFkB-DsRed2-pLEX-NS1 cells were infected with WNV ΔNS1 GFP VRPs for 24h. Only NS1-expressing cells but not vector control cells, supported replication as evidenced by GFP detection by flow cytometry (Figure 2B) and immunofluorescence (data not shown). Taken together these results demonstrate that the individual reagents and cell lines necessary for the screen functioned as designed.
NS1 Mutagenesis
To produce a library of mutagenized NS1 (mNS1), the coding sequence of NS1, as well as the upstream E signal sequence, were randomly mutated in the pLEX vector using an error-prone PCR approach. Mutagenesis of the E signal sequence was included to address the possibility that signal sequence or signalase cleavage site mutants might alter TLR3 inhibition by the NS1 protein. Reaction conditions were chosen with the objective to introduce 1 or 2nt changes per NS1 copy. Ten independent bacterial clones were sequenced to verify the mutation frequency and to assess gene coverage. All clones were found to harbor at least one nucleotide substitution located throughout the 1146nt NS1 coding sequence (data not shown). As an initial test prior to performing the entire screen, HeLa-NFκB-DsRed2-pLEX-mNS1 cells were plated and either infected with WNV ΔNS1 GFP VRPs for 24h, or treated with pIC for 36h. Fluorescence microscopy confirmed the presence of cells expressing NS1 that supported trans-complementation as well as cells expressing NS1 that allowed TLR3 signaling (data not shown). It is important to note that WNV ΔNS1 GFP VRP infection for up to 72 hours did not induce NFκB activation by itself in wt NS1-expressing cells (Supplemental Figure S2).
Screen
To perform the screen, 2.5 X 106 HeLa-NFκB-DsRed2-pLEX-mNS1 cells were infected with WNV ΔNS1 GFP VRPs at an MOI of 1. GFP fluorescence in some cells was readily observable at 24h post-infection. The cells were subsequently stimulated with pIC for 36h and GFP and DsRed2 double-positive cells were isolated by FACS. The screen was performed twice using independently established HeLa-NFκB-DsRed2-pLEX-mNS1 cell libraries. Approximately 3 X 104 double-positive cells were isolated and plated into 10cm culture plates at low density. 134 clones survived expansion. Phenotypes of clones were confirmed by independently monitoring DsRed2 expression following pIC stimulation and GFP expression post-infection with WNV ΔNS1 GFP VRPs. Ultimately, 18 clones were considered promising candidates and the NS1 sequence was determined, which resulted in four unique sequences [Table 1]. Some sequences were present in more than one of the clones. This can most likely be attributed to allowing the original mutant cell library to expand prior to the sort. Multiple independent selections of the same sequence confirm the reproducibility of our screen.
Table 1.
NS1 nucleotide and amino acid substitutions identified in screen.
| Clone | Nucleotide | Amino Acid | ||
|---|---|---|---|---|
| a Position | Change | b Position | Change | |
| 1 | 530 | C:A | 177 | T: N |
| 883 | G:A | 295 | G:R | |
| 2 | 72 | A:T | 191 | N:I |
| 958 | C:T | 320 | P:S | |
| 3 | 713 | A:T | 238 | E:V |
| 763 | A:G | 255 | N:D | |
| 4 | 950 | C:T | 317 | T:I |
| 997 | A:G | 333 | M:V | |
NS1 is 1046 nucleotides in length
NS1 is 352 amino acids in length
All NS1 sequences identified by the screen had two nucleotide substitutions resulting in point mutations. Notably, no clones were identified that encoded truncated or elongated forms of NS1, and no changes were found in the E signal sequence. Identified amino acid changes were reconstituted both individually, as well as in combination in the pLEX-NS1 sequence, and were packaged into recombinant lentiviruses. These lentivirus particles were used to transduce a HeLa cell line, which contained a luciferase reporter gene driven by the NFκB responsive ELAM promoter, HeLa-NFkB-luc. To verify that the amino acid changes did not adversely affect protein expression, immunoblot analysis was performed in the stably expressing cell lines prior to WNV ΔNS1 GFP VRP infection (Figure 3A). All NS1 mutants, as well as wt NS1 were expressed to similar levels. The mutation at position 177 eliminates one of the NS1 N-linked glycosylation sites at position 175 (see below) explaining the increased mobility of NS1 proteins containing this substitution. In the immunoblot shown in Fig. 3A we had to resort to using an anti-His antibody against the epitope tag, since all NS1-specific antibodies available to us did not detect the mutant proteins efficiently. This was also true for immunofluorescence and flow cytometry.
Figure 3. Efficiency of identified NS1 amino acid substitution proteins to trans-complement WNV ΔNS1 GFP VRPs.
(A) HeLa-NFκB-luc reporter cells were transduced with lentivirus expressing either wt NS1, MCS, or single or double NS1 amino acid changes and selected for puromycin resistance. 5μg of whole cell lysate from each cell line was probed for NS1 expression by immunoblot and normalized to β-actin expression. (B) HeLa-NFκB-luc reporter cells stably transduced with lentivirus expressing either wt NS1, MCS, or single or double NS1 amino acid substitutions were infected at an MOI of 0.15 for 36h with WNV ΔNS1 GFP VRPs or mock infected. Expression of GFP was quantified by flow cytometry by determining the mean fluorescent intensity (MFI). The background MFI of identically infected MCS cells was subtracted from the MFI determined for other infected cells. Data are the average of two independent experiments and error bars represent standard deviation from the mean. (C) GFP expression due to trans-complementation by wild type and NS1 proteins with coding changes was monitored by fluorescence microscopy.
Ability of NS1 Proteins Encoding Amino Acid Changes to Trans-complement
HeLa cell lines expressing NS1 with individual or double amino acid changes were infected at low MOI of 0.15 with WNV ΔNS1 GFP VRPs for 36h and GFP expression was investigated by fluorescence microscopy and by flow cytometry to determine the mean fluorescent intensity (MFI) of the GFP signal (Figure 3B and C).
Trans-complementation by NS1 proteins encoding the individual T177N, N191I, E238V, or G295R changes was similar to wt NS1. In contrast, cells expressing NS1 proteins with the individual changes N255D, T317I, P320S, and M333V trans-complemented to reduced levels compared to wt NS1. All cell lines expressing double amino acid changes in the NS1 protein (T177N/G295R, N191I/P320S, E238V/N255D, and T317I/M333V) also had significantly reduced GFP MFI following infection (Figure 3B and C).
In summary, NS1 protein encoding changes T177N, N191I, E238V, and G295R trans-complemented at levels comparable to wt NS1. The remaining identified amino acid changes in NS1 resulted in less efficient trans-complementation than with wt NS1 protein. Interestingly, the originally identified double mutants showed a greater reduction in trans- complementation ability than the individual single amino acid substitutions.
TLR3 Inhibition by NS1 Proteins Encoding Amino Acid Substitutions
TLR3 signaling in HeLa-NFκB-luc cells expressing NS1 proteins encoding amino acid substitutions was measured by IL-6 ELISA (Figure 4A) and NFκB activation by luciferase reporter assay (Figure 4B) following pIC stimulation. TLR3 signaling was severely inhibited in cells expressing wt NS1, as well as NS1 encoding the T177N, N191I, or E238V substitutions. While the IL-6 response to pIC stimulation was weak in cells expressing G295R NS1 protein, they responded similar to non NS1-expressing cells when NFkB activation was assessed. TLR3 signaling was completely restored in N255D and T317I NS1-expressing cells, and significantly restored in cells expressing P320S and M333V NS1 proteins (Figure 4A and B). Complete responsiveness to pIC was also observed in cells expressing each of the NS1 double amino acid substitutions, with the exception of T177N/G295R NS1.
Figure 4. Efficiency of TLR3 signaling in cells expressing NS1 amino acid substitution proteins.
HeLa-NFκB-luc reporter cells stably transduced with lentivirus expressing either wt NS1, MCS, or single or double NS1 amino acid changes were treated with 20μg/ml pIC for 8h or left untreated. (A) IL-6 secretion in the supernatants of cells was determined by enzyme-linked immunosorbent assay (ELISA) and fold induction was calculated by comparing IL-6 produced by pIC treated cells to that of untreated cells. Data are the average of two independent experiments performed in biological triplicate and error bars represent standard deviation from the mean. Statistical significance was determined by Mann-Whitney U test, where *p≤0.05 NS= not significant. All samples were compared to wt NS1. (B) Fold NFκB induction was determined by luciferase reporter assay. Relative light units were measured and reported as fold induction over untreated cells. Data are the average of two independent experiments and error bars represent standard deviation from the mean. Statistical significance was determined by student’s t-test, where *p≤0.05 NS= not significant. All samples were compared to wt NS1.
The combined results of trans-complementation and TLR3 signaling in cells expressing NS1 variants lead us to conclude that NS1 T177N, N191I, and E238V behave like wt NS1 for the functions we assessed. NS1 G295R and T177N/G295R also functioned most similar to wt NS1, but did allow slightly higher levels of NFκB activation and IL-6 secretion, respectively. Several other substitutions identified by the screen completely restored TLR3 signaling, but significantly impaired trans-complementation (N255D, T317I, N191I/P320S, E238V/N255D, T317I/M333V). Of the remaining NS1 changes identified by the screen, P320S and M333V NS1 supported intermediate levels of trans-complementation and allowed significant TLR3 signaling (Figures 3 and 4). These substitutions were selected for further study in the context of the full length WNV genome.
Identification of Highly-Conserved Amino Acid Residues at the C-terminal Half of the NS1 Protein
The amino acid sequences of 10 mosquito-borne and 6 tick-borne flavivirus NS1 proteins were aligned using ClustalW. The mutations identified by the screen with the most desired phenotype lie in the C-terminal half of the NS1 protein. Residues P320 and M333 are evolutionarily conserved among all flavivirus NS1 proteins that were aligned, suggesting that these positions are of biological importance to the viruses (Figure 5). In contrast, T177N and N191I amino acid residue changes had no effect on the functions we assessed, and were found to have variable residues at those positions (data not shown). E238, while not conserved among all flaviviruses, was completely conserved among mosquito-borne flaviviruses, however, a change at that residue retained a wild type NS1 phenotype. Residues N255 and G295 are completely conserved. A change at position 255 to aspartic acid resulted in a loss of function phenotype, while the change at position 295 to an arginine resulted in a predominantly wild type phenotype. Residue T317 was only partially conserved among mosquito-borne flaviviruses and a change to isoleucine negatively impacted both NS1 functions we assessed.
Figure 5. Shannon entropy (H) and multiple protein alignment of the NS1 proteins of several mosquito- and tick-borne flaviviruses.
The amino acid sequences of 10 mosquito-borne and 6 tick-borne flaviviruses were aligned with ClustalW using the Blosum scoring matrix. The multiple-alignment file was displayed with BOXshade 3.21 where black-shading represents identical residues and gray-shading represents conserved substitutions. Shannon entropy analysis of the multiple protein alignment was performed by the Protein Variability Server (PVS) at the Complutense University of Madrid, and the data generated was displayed in MS Excel. H ≤ 1 are considered highly conserved residues. Mosquito-borne flaviviruses are represented in bolded font and tick-borne flaviviruses in italics.
Not surprisingly, the NS1 proteins of the examined flaviviruses share significant homology as determined by Shannon entropy calculations (average H value = 1.24). Overall, the C-terminal half of the protein was slightly more conserved than the N-terminal half (average H value N-terminal half = 1.36, C-terminal half = 1.11). Taken together, residues 320 and 333 are evolutionarily conserved, and substitutions at these positions showed the most promising phenotype in our screen to separate TLR3 inhibition from NS1’s function in RNA replication.
NS1 Mutations Affect WNV Growth in vitro
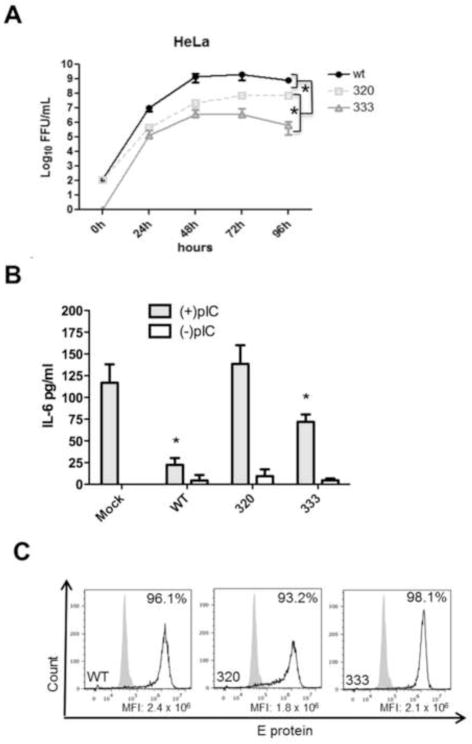
During the screen, NS1 protein was ectopically expressed, independent of other viral proteins. During RNA replication, non-structural proteins need to interact with each other properly to form a functioning replication complex. To investigate the effects of the identified coding changes on genome replication and virus production, the P320S and M333V amino acid substitutions were introduced individually into a WNV infectious clone and recombinant mutant viruses were produced in BHK cells. Multistep growth curves were performed on HeLa cells for up to 96h post-infection. Both WNV mutants were attenuated in HeLa cells with virus titers being lower than wild type virus at all time points examined (Figure 6A). Differences in replication kinetics were also observed between the two mutant viruses with WNV NS1 M333V being significantly more attenuated in HeLa cells than WNV NS1 P320S.
Figure 6. Replication kinetics and TLR3 inhibition of mutant WNV expressing the identified amino acid residue changes in the NS1 protein.
(A) HeLa cells were infected at an MOI of 0.01 with each WNV NS1 mutant and virus containing supernatants were collected every 24h for 96h. Infectious virus was titered on Vero cells by immunofocus assay. Growth curves were performed in biological triplicate and data are representative of two independent experiments. Error bars represent standard deviation from the mean. Statistical significance was determined by student’s t-test, where *p≤0.05. (B) HeLa cells were infected at an MOI of 5 with each WNV NS1 mutant for 1h, washed with PBS, replaced with fresh medium, and allowed to incubate for 20h. Medium was removed and replaced with medium with or without pIC (20μg/ml) for 8h. Cell culture supernatants were collected and IL-6 concentrations were determined by ELISA as pg of IL-6/ml. Data are representative of three independent experiments and error bars represent standard deviation from the mean. Statistical significance was determined by student’s t-test, where *p≤0.05 and NS= not significant. All samples were compared to mock infected cells treated with pIC. (C) HeLa cells infected with identical amounts of wt and mutant viruses were fixed and permeabilized 28h post-infection and infected cells were identified by flow cytometry using an E protein-specific primary monoclonal antibody and were detected with a secondary Alex Fluor 488 antibody. Grey filled histograms represent uninfected control cells and black open histograms represent E protein in WNV infected cell. Data are representative of three independent experiments.
Effect of WNV NS1 Mutants on TLR3 Signaling in HeLa Cells
TLR3 signaling was measured in WNV infected cells in response to pIC. Previous studies with HeLa cells demonstrated that IL-6 secretion in response to pIC treatment is TLR3-dependent and can be abolished in cells transfected with TLR3-specific siRNA (Wilson et al., 2008). HeLa cells were infected for 20h with WNV followed by 8h of pIC treatment. Wild type WNV robustly inhibited IL-6 secretion following pIC treatment compared to mock infected cells. (Figure 6B). Cells infected with WNV NS1 P320S efficiently responded to pIC treatment with IL-6 secretion similar to mock infected cells. WNV NS1 M333V infected cells showed an intermediate phenotype and responded significantly to pIC stimulation compared to wt WNV but still significantly inhibited TLR3 compared to mock infected cells. Given that each mutant virus displayed growth attenuation in HeLa cells, the percentage of infected cells in each case was determined by flow cytometry. All viruses were equally able to infect HeLa cells as determined by expression of the E protein (Figure 6C) and the MFI was similar in all three cases, suggesting that differences in protein expression alone do not account for the observed differences in TLR3 inhibition. Due to the issues of inefficient recognition of the mutant NS1 proteins by anti-NS1 antibodies and the lack of an epitope tag in the context of the full-length virus genome, we were not able to directly compare expression levels of NS1 proteins in these experiments.
Discussion
We have previously demonstrated that the WNV NS1 glycoprotein inhibits TLR3 signaling (Scholle and Mason, 2005; Wilson et al., 2008). In this study, we developed a FACS-based screen to identify randomly-generated amino acid changes within the NS1 protein that restore TLR3 signaling, while allowing the protein to still function in virus genome replication. Initially, the components of the screen were successfully validated and mutagenic coverage was assessed. The mutagenesis was unbiased and nucleotide changes were found throughout the coding sequence of NS1. Following two screens using independently-established mutant NS1 libraries, we identified approximately 3 X 104 double-positive cells of which 134 survived clonal expansion. Of the expanded clones, 18 displayed the desired phenotype. While cell viability due to VRP infection alone was not negatively affected as determined by MTT assay (data not shown), the combination of VRP infection, pIC treatment, the stress of the sorting process, and antibiotic selection likely contributed to a decrease in clonal cell survival. Nevertheless, the identification of several clones with the desired phenotype confirms that our screening strategy was successful.
Overall, four unique double amino acid changes were identified: T177N/G295R, N191I/P320S, E238V/N255D, and T317I/M333V. Analysis of the effect of individual amino acid changes at each of the eight positions demonstrated that changes at residues 177, 191, 238, and 295 did not affect replication in our trans-complementation assay. These same residues also did not seem to be involved in TLR3 inhibition. The notable exception is the change at position 295, which allowed partial activation of NFκB but not IL-6 secretion. The screen only monitored NFκB activation while transcription of IL-6 involves other transcription factors in addition to NFκB, such as AP-1, which may still be inhibited by G295R. The T177N substitution was initially of particular interest because it ablates N-linked glycosylation at position 175, and numerous other studies have described a role for NS1 glycosylation in virulence and pathogenesis of several flaviviruses (Muylaert et al., 1996; Pryor and Wright, 1993; Pryor and Wright, 1994; Tajima, Takasaki, and Kurane, 2008; Whiteman et al., 2010; Whiteman et al., 2011). However, T177N NS1 behaved like wild type protein for trans-complementation and TLR3 inhibition, suggesting that at least this particular glycosylation site is not critical for TLR3 inhibition and/or RNA replication.
The changes at residues 255 and 317 conferred the opposite phenotype, negatively impacting trans-complementation of viral RNA synthesis while allowing TLR3 signaling to occur. These results could either be interpreted that residues 255 and 317 are involved in TLR3 inhibition and that simultaneously, the amino acid substitutions have a negative effect on replication, or that changing these residues render NS1 simply non-functional. In addition to these single amino acid substitutions, the combined residue changes also negatively impacted NS1 protein functions, demonstrating the sensitivity of the NS1 protein to mutation. The ability of our screen to identify these attenuated double-amino acid substitution clones indicates that our FACS-based strategy was highly sensitive. Ultimately the P320S and M333V single substitutions were shown to be the minimal changes necessary to allow TLR3 signal transduction, while still permitting the trans-complementation and rescue of an NS1-deleted VRP. It is important to note, however, that TLR3 signaling was not fully restored and that trans-complementation was inferior to wt NS1. These findings indicate that there is most likely a reciprocal relationship between restoration of TLR3 signal transduction and maintenance of replication competency. Thus, complete separation of the two functions might not be feasible, with a possible explanation being, that the region responsible for TLR3 inhibition is also involved in RNA replication.
Interestingly, a previous clustered charged-amino-acid-to-alanine mutagenesis study of the yellow fever virus NS1 protein identified mutations lethal to virus replication only in the N-terminal half of the protein, while the majority of tolerated mutations mapped to the C-terminal half (Muylaert, Galler, and Rice, 1997). Consistent with these results, none of the amino acid changes we identified in the screen fall into the N-terminal half of the NS1 sequence. One possible explanation for these findings is that the N-terminal half of the protein is critically involved in replication and highly sensitive to mutagenesis. Further, the C-terminal half of the protein also contributes to replication, but in addition, functions in TLR3 inhibition and is more tolerant to mutagenesis. The idea that non-structural proteins may contain overlapping functional domains is not new among the flaviviruses. The Langat NS5 protein is the RdRp and also functions to inhibit JAK/STAT signaling originating from the type I IFN receptor (Park et al., 2007). Fine mapping of the Langat NS5 inhibitory domains indicated that regions and residues necessary for IFN inhibition are located within the highly conserved RdRp domain.
In our case, ClustalW alignment of the C-terminal half of 10 mosquito-borne and 6 tick-borne flavivirus NS1 proteins revealed high sequence similarity in this region of the protein, and residues P320 and M333 are fully conserved, suggesting that their functions in both replication and TLR3 inhibition could be evolutionarily and biologically important.
In a recent study, Baronti et al, were unable to find a role for the NS1 proteins of several flaviviruses in inhibiting TLR3 signaling (Baronti et al., 2010). In that study, the investigators relied on the induction of IFN-β transcripts as a primary readout for TLR3 signaling within HEK293-TLR3 cells and in HeLa cells. We determined previously that HeLa cells fail to produce significant levels of IFN-β transcript and protein following pIC stimulation alone, which may explain these conflicting results (Wilson et al., 2008). Further, the authors of that study had to resort to overexpressing both TLR3 and IRF3 in the HeLa cell lines in order to induce IFN-β transcription following pIC stimulation. As the mechanism of NS1-mediated TLR3 inhibition is currently unknown, IRF3 overexpression in these cells may have bypassed the step(s) in the TLR3 signaling pathway at which NS1 blocks signal transduction. Our current results confirm not only that wt NS1 inhibits TLR3 signaling as previously observed using multiple read-outs, but also that NS1 can be mutated at specific residues to restore NFκB activation and IL-6 transcription and secretion, in response to TLR3 stimulation, while other mutations yield a wt NS1 phenotype.
In addition to ectopic expression studies of the NS1 proteins, mutant WNV growth and TLR3 signaling studies were undertaken in HeLa cells. Both P320S and M333V NS1 WNV were attenuated for growth in HeLa cells and allowed TLR3 signaling in response to pIC. These findings demonstrate that the identified mutations in the context of a WNV infectious clone faithfully recapitulated the phenotypes observed during overexpression of the NS1 protein alone. This is an important finding, as a previous mutagenesis study with the IFN antagonistic WNV NS4B protein was unable to reproduce phenotypes observed with mutant replicons/over expressed protein in the context of an infectious WNV mutant (Evans and Seeger, 2007).
Ultimately, interference with TLR3 signaling is expected to alter the host response to benefit the virus. Future studies will investigate pathogenesis and immune responses to our mutant viruses. P320S NS1 WNV replicated more efficiently and allowed increased TLR3 signaling to occur compared with M333V NS1 WNV, and may represent an important tool to investigate the role of the TLR3 inhibitory function of NS1 during WNV infection. Additionally, the identification of P320 as a residue important for TLR3 inhibition is likely to prove valuable for determining the mechanism by which NS1 antagonizes TLR3 signal transduction.
Materials and Methods
Construction of VSV-G Psudotyped Lentivirus Particles Expressing NS1 (pLEX-NS1)
Full length WNV NS1, including the last 30 amino acids of the E protein and a C-terminal 6X Histidine tag was cloned into the pLEX vector (ThermoScientific/OpenBiosystems) and pLEX-NS1 was packaged into VSV-G pseudotyped, replication-defective, lentivirus particles through co-transfection of HEK 293T cells with vector, packaging (pREV and pMDLg), and envelope (pVSV-G) plasmids. Empty vector (pLEX-MCS) was packaged identically. Lentivirus-containing supernatants were harvested and clarified 40h post-transfection, and stored at −80°C until further use.
Construction of VSV-G Psudotyped Lentivirus Particles Expressing Mutant NS1 (pLEX-mNS1)
NS1 mutants (mNS1) were generated by error-prone PCR of the pLEX-NS1 plasmid using a Gene MorphII EZ clone kit (Agilent Technology/Stratagene), according to the manufacturer’s instructions, with the following primers; mutFw 5′ AAT TCG CCC TTG AAT CGG ATC C 3′ and mutRev 5′ GGT GAT GGT GAT GAT GAC CGG T 3′. The input amount of template DNA was selected to achieve a mutation frequency between 1 nucleotide (nt) and 2nt change(s)/kilobase(kb). The E signal sequence and NS1 are 1146nt in length. Total transformed bacteria were expanded in 250ml of 2X YT broth (Fisher) containing 100μg/ml ampicillin (Sigma), plasmid DNA was purified using a Maxi prep kit (Qiagen), and packaged as lentivirus particles (pLEX-mNS1) as described previously.
Construction of HeLa NFκB-ELAM DsRed2 Reporter Cell Line (HeLa-NFκB-DsRed2)
The CMV promoter of the pDsRed2-C1 plasmid (Clontech) was removed by digestion with PciI and AgeI restriction endonucleases. The 5X NFκB endothelial-leukocyte adhesion molecule-1 (ELAM) promoter of the pNiFtY-SEAP plasmid (Invivogen) was PCR amplified with the following primers; NFκB5XELAM Fw (PciI) 5′ ATT ATA CAT GTA TCG CTG AAT TCT GGG GAC T 3′ and NFκB5XELAM Rev (AgeI) 5′ ATT ATA CCG GTG GCT CTG TCT CAG GTC AGT A 3′, to produce a 278 base pair (bp) amplicon, that was subsequently digested with PciI and AgeI enzymes, and ligated into the pDsRed2-C1 plasmid (NFκB-DsRed2). HeLa cells were transfected (Mirus IT LT-1) with 500ng of NFκB-DsRed2 plasmid. A polyclonal cell population (HeLa-NFκB-DsRed2) was selected with 500μg/ml G418 (Lonza) and response to treatment with 20μg/ml Polyinosinic:polycytidylic acid (pIC) (Calbiochem) was confirmed by detection of DsRed2 with fluorescence microscopy and flow cytometry 36hr later.
Construction of HeLa NFκB-ELAM Luciferase Reporter Cell Line (HeLa-NFκB-luc)
The NFκB 5X ELAM promoter of the pNiFty-SEAP plasmid (Invivogen) was PCR amplified using the following primers: NFκB5XELAM Fw (KpnI): 5′ ATT ATG GTA CCA TCG CTG AAT TCT GGG GAC T 3′ and NFκB5XELAM Rev (HindIII): 5′ ATT ATA AGC TTG GCT CTG TCT CAG GTC AGT A 3′. The amplicon was digested with KpnI and HindIII restriction endonucleases and ligated into KpnI and HindIII sites of the pGL4.15 vector (Promega) upstream of the luciferase reporter gene. HeLa cells were transfected (Mirus, IT LT-1) with 500ng of the reporter construct and a stable, polyclonal cell line was selected by 400μg/ml hygromycin (Cellgro). Responsiveness of the HeLa-NFκB-luc cell line was confirmed by luciferase assay following 8h treatment with 20μg/ml pIC.
VSV-G Pseudotyped Lentivirus Particle Transduction and Titration
For creation of a random mutant NS1 reporter cell library (HeLa-NFκB-DsRed2-pLEX-mNS1), HeLa-NFκB-DsRed2 cells were transduced at an MOI of 0.02 with pLEX-mNS1 particles to ensure fewer than 1 integration event per cell. HeLa-NFκB-DsRed2 cells were plated in 6-well plates and overlaid with 2ml of 1+1+1 DMEM (1X DMEM (CellGro) with 1% Pen/Strep (CellGro), 1% HEPES buffer (CellGro), 1% FBS (Gemini)), containing 8μg/ml polybrene (Sigma), and lentivirus-containing supernatant. Medium was changed 24h post-transduction to 1X DMEM with 10% FBS and cells were placed under puromycin selection (2μg/ml) (Gemini) 48h later. HeLa-NFκB-DsRed2 cells or standard HeLa cells were also transduced with either pLEX-NS1 or pLEX-MCS lentivirus particles to serve as controls.
Lentivirus titers were determined by counting crystal violet-stained, puromycin-resistant HeLa cell colonies following transduction with serial dilutions of lentivirus-containing supernatants.
Construction, Packaging, and Titration of ΔNS1 GFP VRPs
A GFP-expressing WNV replicon (WNV GFP rep) (a gift from P.W. Mason), based on WNR C-NS1-5 with an insertion of GFP and the FMDV 2A autoprotease coding sequences between the capsid and the NS1 signal sequence (Bourne et al., 2007) served as a backbone for introduction of a lethal 138bp (46 amino acid) in-frame deletion in NS1. Initially, the WNV GFP rep plasmid served as template for PCR to amplify the final 60bp of NS1, (incorporating a BstEII restriction site at the 5′ end the amplicon) along with full-length NS2A (which contains a unique XmaI restriction site), using the following primers; delNS1 BstEII Fw 5′ ACG TGG TCA CCA TGG AGA TCA GAC CAC AGA GAC ATG 3′ and delNS1 NS2A Rev 5′ TCT GGC ACA TCG TAT GGG TAG CGT TTA CGG TTG GGA TCA CAT GC 3′. The 790bp amplicon was subsequently digested with BstEII and XmaI restriction endonucleases and ligated into the BstEII and XmaI restriction sites of WNV GFP rep DNA.
WNV ΔNS1 GFP rep DNA was linearized by XbaI digestion followed by phenol:chloroform extraction and RNA was transcribed in vitro using a transcription reaction kit (Ambion) incorporating a m7GpppAmpN2 cap analogue (NEB). BHK packaging cells expressing WNV C, PrM, E, and NS1 (BHK-Pack NS1, a gift from P.W. Mason) from a non-cytopathic Venezuelan equine encephalitis alphavirus (VEE) replicon, were electroporated with WNV ΔNS1 GFP rep RNA. WNV ΔNS1 GFP VRP containing supernatants were collected every 24h in 1+1+1 MEM medium, clarified, and stored at −80°C until further use. For titration, wt NS1 expressing HeLa cells were infected for 24h with WNV ΔNS1 GFP VRPs and GFP positive cells were quantified by flow cytometry.
Screen Conditions
2.5 X 106 HeLa-NFκB-DsRed2-pLEX-mNS1 cells were plated in a T-75 flask and allowed to grow for 24h. Medium was removed and cells were infected at an MOI of 1 with WNV ΔNS1 GFP VRPs in 1+1+1 DMEM for 24h. Infected cells were treated with 20μg/ml pIC in 1+1+1 DMEM for an additional 36h. Cells were removed from the flask by trypsinization, washed, and double-positive cells (GFP and DsRed2) were sorted by Fluorescence-activated Cell Sorting (FACS, Cytomation). Sorted cells were plated at low density in 10cm cell culture dishes and maintained in medium containing 2μg/ml puromycin and 500μg/ml G418. Surviving colonies were isolated and expanded. The phenotypes of the resulting clonal cell lines were confirmed by infecting with an MOI of 1 with WNV ΔNS1 GFP VRPs or treating with 20ug/ml pIC and monitoring GFP or DsRed2 expression by fluorescence microscopy.
Genomic DNA was purified from the clonal cell lines using a QIAamp DNA mini kit (Qiagen). Integrated mNS1 was PCR amplified using the following primers; CMVIEFw 5′ ACT TAC GGT AAA TGG CCC GC 3′, IRESRev and 5′ AAA GGA AAA CCA CGT CCC CG 3′ and purified amplicons were sequenced.
Site-Directed Mutagenesis
NS1 point mutations identified in the clonal cell lines were reconstituted individually or as pairs in the pLEX-NS1 vector, using the PCR based Quick Change II XL kit (Agilent Technologies/Stratagene). Site-directed primer pairs were designed using the Quick Change primer design software. Each primer pair was designed to incorporate a single nucleotide change resulting in a single point mutation. Fidelity of the mutagenesis was confirmed by standard DNA sequencing. Reconstituted mutants were packaged as lentivirus particles and used to produce stable, polyclonal mutant NS1-expressing HeLa-NFκB-luc cells as described previously.
NFκB-luciferase Reporter Assays
HeLa-NFκB-luc cells expressing the reconstituted NS1 mutants were plated at 4 X 104 cells total per well in a 48-well plate. Triplicate wells of each cell line were either treated with 20μg/ml pIC for 8h or left untreated. Supernatants were stored at −80°C for IL-6 ELISA (described below). Cells were washed once with PBS and lysed for 30min in lysis buffer (0.3M NaCl, 0.05M TrisHCL, 0.1%TritonX-100) on ice. Cell debris was removed by centrifugation and supernatants were incubated with luciferase reagent (Promega) and light emission was measured on a luminometer (Berthold) as Relative Light Units/ml (RLU/ml). The RLU/ml of each sample was then normalized to total protein in the whole cell lysate (RLU/ml/μg protein), as determined by a commercial protein assay kit (BioRad). Data were expressed as fold NFκB induction over unstimulated cells.
ELISA
Cell culture supernatants were collected as described and assayed for human IL-6 using a commercial ELISA kit (Ebioscience). IL-6 secretion was then calculated as fold secretion over unstimulated cells or as pg/ml where indicated.
Immunoblotting
Whole cell lysates were prepared in lysis buffer as described. Equivalent amounts of protein (5μg) were mixed with LDS buffer (Invitrogen) containing 2.5mM DTT, heated at 90°C for 5min, and loaded into 4–12% Bis-Tris NuPAGE gels (Invitrogen). Proteins were electrophoretically separated, transferred to a PVDF membrane (Immobilon Millipore), and probed with one of the following primary antibodies: 1:4000 dilution of mouse anti-tetra-His monoclonal antibody (34679; Qiagen) to detect NS1 or 1:10000 dilution of mouse anti-β-actin monoclonal antibody (A1978; Sigma). Bound antibodies were detected with a 1:2000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG (H+L) secondary antibody (474-1806; KPL and an enhanced chemiluminescence detection system (Santa Cruz Biotechnology).
Flow Cytometry
Detection of DsRed2 and GFP
Following the indicated treatments, HeLa-NFκB-DsRed2-pLEX-NS1 or HeLa-NFκB-DsRed2-pLEX-MCS cells were trypsinized, resuspended in 1X DMEM medium with 10% FBS, washed with PBS, and 10,000 events per treatment group were recorded on a BD FACS Calibur flow cytometer using CellQuest Pro (BD) software. Data were analyzed and displayed with WinMDI 2.8 software (The Scripps Institute).
Detection of WNV-infected cells
HeLa cells were infected with WNV or mutant WNV using the indicated conditions, trypsinized, resuspended in 1X DMEM medium with 10% FBS, and washed once with flow staining buffer (1X PBS, 1% BSA, 1% Sodium Azide). All other steps were performed in the dark on ice. Cells were fixed and permeabilized for 20min with cytofix/cytoperm solution (BD), washed, and incubated with a mouse anti-dengue virus E protein monoclonal antibody (D14G2) in cytofix/cytoperm wash buffer. Following several washes, cells were incubated with secondary goat anti-mouse IgG (H+L) Alexa Fluor 488-conjugated antibody (Invitrogen). 5,000 events per treatment group were recorded on an AccuriC6 flow cytometer using CFlow software (BD Accuri) and data were analyzed and displayed with FlowJo version 10 (Tree Star).
Multiple Protein Alignment and Shannon Entropy
The amino acid sequences of 10 mosquito-borne and 6 tick-borne flavivirus NS1 proteins were aligned using ClustalW. The Blosum scoring matrix was employed with an opening gap penalty of 10 and an extending gap penalty of 0.05 (Larkin et al., 2007). The alignment file was graphically displayed with BOXSHADE 3.2, where black shading represents identical residues and grey shading represents conserved substitutions. Shannon entropy was also calculated from the multiple protein alignment based upon the consensus reference sequence, using the Protein Variability Server (PVS) at the Complutense University of Madrid (Garcia-Boronat et al., 2008).
Reconstitution of NS1 Mutations into a West Nile virus Infectious Clone
PCR-mediated, site-directed mutagenesis, as described earlier, was used to clone the identified changes at NS1 amino acid positions 320 and 333, respectively, into an infectious cDNA clone of West Nile virus of a human 2002 isolate from Texas (Rossi et al., 2005). Fidelity of the mutagenesis was confirmed by standard DNA sequencing of the NS1 coding sequence in the cDNA clones, using the primer set listed below. Plasmid DNA was linearized by XbaI digestion followed by phenol:chloroform extraction and RNA was transcribed in vitro. Wild type and mutant WNV RNAs were electroporated into BHK cells, and viruses were captured in the cell culture supernatants every 24h for 120h post-infection. Virus containing supernatants were clarified, aliquoted, and stored at −80°C prior to use (see below).
The NS1 coding sequence of each WNV mutant and wt WNV was assessed following virus production in BHK cells to determine the stability of the mutations. Virus RNA was extracted from each passage 0 (P0) virus stock, reverse transcribed and a 1198bp fragment containing full-length NS1 was amplified using the following primers: WNVmNS1SEQFw 5′ TGC TCG TGA CAG GTC CAT AGC TCT 3′ and WNVmNS1SEQRev 5′ CTG GGT GGC CAA GAA CAC GAC C 3′. PCR products were purified by standard salt:ethanol precipitation and sequenced with the primers listed above.
Titration of WNV
WNV was titered on monolayers of Vero cells in 48-well plates. Virus was allowed to attach to the cells for 1h with rocking every 15min, and cells were overlaid with 1X MEM/1X Tragacanth Gum (MP Biomedicals) and allowed to incubate for 48h. Cells were fixed with a 1:1 ratio of acetone and methanol. The fixed cells were blocked with 1% Normal Horse Serum (NHS) (Gemini Bioproducts) in PBS (1%NHS/PBS). E protein in infected cells was detected with the D14G2 monoclonal antibody, and horseradish peroxidase-conjugated goat anti-mouse IgG (H+L) Immunofoci were visualized using a colorimetric HRP detection kit according to the manufacturer’s instructions (Vector Labs). Virus titer was expressed as focus forming units/ml (FFU/ml).
WNV Growth Kinetics
Multi-step virus growth curves were performed on HeLa cell monolayers in 24-well plates by infecting at an MOI of 0.01 with wt WNV or each WNV NS1 mutant. Following 1h of virus attachment, cells were washed and overlaid with 1ml of 1+1+1 MEM medium. Virus containing supernatants were collected every 24h for 96h and stored at −80°C prior to titration on Vero cells.
TLR3 Signaling in WNV-Infected HeLa cells
TLR3 signaling in WNV-infected HeLa cells was assessed following pIC stimulation. HeLa cells were infected for 1h at an MOI of 5 with wt WNV or each WNV NS1 mutant. Virus inoculum was removed, cells were washed with PBS, and medium was replaced with 1X DMEM medium containing 10% FBS. Twenty hours post-infection medium was removed and replaced with 1X DMEM containing 10% FBS with or without 20μg/ml pIC and allowed to incubate for an additional 8h. IL-6 was quantified by ELISA. The proportion of infected cells was assessed 28h post-infection by flow cytometry to detect the E protein (described above).
Statistical Analyses
Distribution of each data set was determined by Kolmogorov-Smirnov test (KS). Student’s t test was employed to determine significance when data were normally distributed and Mann-Whitney U test was performed when data were not normally distributed. Each test was completed using the statistical software package, GraphPad Prism 4.
Supplementary Material
Highlights.
WNV NS1 was mutated to separate its functions in replication and TLR3 inhibition
Discrete mutations in the C-terminal half of NS1 were identified
Recombinant viruses are slightly attenuated and do not inhibit TLR3
Mutated amino acids are conserved among the flaviviridae
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIH U54 AI 057157-07(SERCEB), project SE-RP-012. We are thankful to Kristen Crook and Jason Wilson for helpful discussions and insights.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8(10):e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcon-LePoder S, Drouet MT, Roux P, Frenkiel MP, Arborio M, Durand-Schneider AM, Maurice M, Le Blanc I, Gruenberg J, Flamand M. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J Virol. 2005;79(17):11403–11. doi: 10.1128/JVI.79.17.11403-11411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83(11):5408–18. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, Atkinson JP. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010;207(4):793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Hauhart RE, Somnuke P, Blom AM, Diamond MS, Atkinson JP. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol. 2011;187(1):424–33. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, Malasit P, Atkinson JP, Diamond MS. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007;3(11):e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronti C, Sire J, de Lamballerie X, Querat G. Nonstructural NS1 proteins of several mosquito-borne Flavivirus do not inhibit TLR3 signaling. Virology. 2010;404(2):319–30. doi: 10.1016/j.virol.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Bourne N, Scholle F, Silva MC, Rossi SL, Dewsbury N, Judy B, De Aguiar JB, Leon MA, Estes DM, Fayzulin R, Mason PW. Early production of type I interferon during West Nile virus infection: role for lymphoid tissues in IRF3-independent interferon production. J Virol. 2007;81(17):9100–8. doi: 10.1128/JVI.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–88. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A. 2006a;103(50):19111–6. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J Virol. 2006b;80(3):1340–51. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Thompson BS, Fremont DH, Diamond MS. Antibody Recognition of Cell Surface-Associated NS1 Triggers Fc-{gamma} Receptor-Mediated Phagocytosis and Clearance of West Nile Virus-Infected Cells. J Virol. 2007;81(17):9551–5. doi: 10.1128/JVI.00879-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Seeger C. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J Virol. 2007;81(21):11809–16. doi: 10.1128/JVI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008;36(Web Server issue):W35–41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. 2006;80(23):11723–32. doi: 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. Journal of Virology. 1999;73(12):10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol. 2010 doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102(8):2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80(12):5908–18. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Liao CL, Lin E, Lin YL. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J Virol. 2004;78(17):9285–94. doi: 10.1128/JVI.78.17.9285-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71(12):9608–17. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73(6):4611–21. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol. 2006;80(5):2396–404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of Interferon Signaling by the New York 99 Strain and Kunjin Subtype of West Nile Virus Involves Blockage of STAT1 and STAT2 Activation by Nonstructural Proteins. J Virol. 2005;79(3):1934–42. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103(15):6001–6. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200(8):1261–70. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79(13):8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muylaert IR, Chambers TJ, Galler R, Rice CM. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology. 1996;222(1):159–68. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- Muylaert IR, Galler R, Rice CM. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71(1):291–8. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya T, Kajita E, Miwa S, Defranco AL. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J Biol Chem. 2005;280(44):37107–17. doi: 10.1074/jbc.M504951200. [DOI] [PubMed] [Google Scholar]
- Park GS, Morris KL, Hallett RG, Bloom ME, Best SM. Identification of residues critical for the interferon antagonist function of Langat virus NS5 reveals a role for the RNA-dependent RNA polymerase domain. J Virol. 2007;81(13):6936–46. doi: 10.1128/JVI.02830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor MJ, Wright PJ. The effects of site-directed mutagenesis on the dimerization and secretion of the NS1 protein specified by dengue virus. Virology. 1993;194(2):769–80. doi: 10.1006/viro.1993.1318. [DOI] [PubMed] [Google Scholar]
- Pryor MJ, Wright PJ. Glycosylation mutants of dengue virus NS1 protein. J Gen Virol. 1994;75 (Pt 5):1183–7. doi: 10.1099/0022-1317-75-5-1183. [DOI] [PubMed] [Google Scholar]
- Reed KE, Rice CM. Molecular characterization of hepatitis C virus. Curr Stud Hematol Blood Transfus. 1998;62:1–37. doi: 10.1159/000060472. [DOI] [PubMed] [Google Scholar]
- Rossi SL, Zhao Q, O’Donnell VK, Mason PW. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology. 2005;331(2):457–70. doi: 10.1016/j.virol.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Rummel C, Sachot C, Poole S, Luheshi GN. Circulating interleukin-6 induces fever through a STAT3-linked activation of COX-2 in the brain. Am J Physiol Regul Integr Comp Physiol. 2006;291(5):R1316–26. doi: 10.1152/ajpregu.00301.2006. [DOI] [PubMed] [Google Scholar]
- Schlesinger JJ, Brandriss MW, Putnak JR, Walsh EE. Cell surface expression of yellow fever virus non-structural glycoprotein NS1: consequences of interaction with antibody. J Gen Virol. 1990;71 (Pt 3):593–9. doi: 10.1099/0022-1317-71-3-593. [DOI] [PubMed] [Google Scholar]
- Scholle F, Mason PW. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology. 2005;342(1):77–87. doi: 10.1016/j.virol.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Tajima S, Takasaki T, Kurane I. Characterization of Asn130-to-Ala mutant of dengue type 1 virus NS1 protein. Virus Genes. 2008;36(2):323–9. doi: 10.1007/s11262-008-0211-7. [DOI] [PubMed] [Google Scholar]
- Werme K, Wigerius M, Johansson M. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol. 2008;10(3):696–712. doi: 10.1111/j.1462-5822.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71(9):6650–61. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman MC, Li L, Wicker JA, Kinney RM, Huang C, Beasley DW, Chung KM, Diamond MS, Solomon T, Barrett AD. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine. 2010;28(4):1075–83. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- Whiteman MC, Wicker JA, Kinney RM, Huang CY, Solomon T, Barrett AD. Multiple amino acid changes at the first glycosylation motif in NS1 protein of West Nile virus are necessary for complete attenuation for mouse neuroinvasiveness. Vaccine. 2011;29(52):9702–10. doi: 10.1016/j.vaccine.2011.09.036. [DOI] [PubMed] [Google Scholar]
- Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82(17):8262–71. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169(12):6668–72. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. Evidence for a Genetic and Physical Interaction between Nonstructural Proteins NS1 and NS4B That Modulates Replication of West Nile Virus. J Virol. 2012;86(13):7360–71. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8(6):e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.