Abstract
Abnormalities in phosphoinositide metabolism are an emerging theme in human neurodegenerative disease. Myotubular myopathy is a prototypical disorder of phosphoinositide dysregulation that is characterized by profound muscle pathology and weakness and that is caused by mutations in MTM1, which encodes a phosphatase that targets 3-position phosphoinositides, including phosphatidylinositol 3-phosphate. Although the association between MTM1 and muscle disease has become increasingly clarified, the normal role(s) of phosphatidylinositol 3-phosphate metabolism in muscle development and homeostasis remain poorly understood. To begin to address the function of phosphatidylinositol 3-phosphate in skeletal muscle, we focused on the primary kinase responsible for its production, and created a muscle-specific conditional knockout of the class III phosphatidylinositol 3-kinase, Pik3c3. Muscle-specific deletion of Pik3c3 did not disturb embryogenesis or early postnatal development, but resulted in progressive disease characterized by reduced activity and death by 2 months of age. Histopathological analysis demonstrated changes consistent with a murine muscular dystrophy. Examination for cellular mechanism(s) responsible for the dystrophic phenotype revealed significant alterations in the autophagolysosomal pathway with mislocation of known dystrophy proteins to the lysosomal compartment. In all, we present the first analysis of Pik3c3 in skeletal muscle, and report a novel association between deletion of Pik3c3 and muscular dystrophy.
Phosphatidylinositols (PIs) comprise a group of low-abundance lipids with hydroxylated inositol head groups that are capable of receiving phosphates at any of the three outer positions. Dynamic phosphorylation of the inositol ring influences many cellular and metabolic processes, including endocytosis, endosomal trafficking, and autophagy.1–5 The quantity and localization of different PIs are regulated by a group of phosphoinositide kinases and phosphatases that function as key regulatory enzymes and that have been implicated in a number of human diseases, including especially oncologic and neurodegenerative diseases.6–9
Myotubular myopathy (MTM) is a severe childhood-onset disease of skeletal muscle caused by mutations in the phosphoinositide phosphatase myotubularin gene (MTM1).10 MTM is characterized by profound weakness and hypotonia at birth, persistent life-long disabilities including wheelchair and ventilator dependence, and early mortality.11 As predicted by the fact that MTM1 has been shown in vitro to be responsible for dephosphorylation of PI(3)P, levels of PI(3)P are significantly elevated in animal models of MTM.12–14 Despite growing knowledge of disease pathogenesis, there are currently no available treatments for MTM. One significant barrier toward therapy development for MTM is the fact that the normal function(s) of PI(3)P in skeletal muscle are unknown.
PI(3)P is created through phosphorylation of PI at the D3 position by PI3 kinases,15,16 or through dephosphorylation of PI(3,5)P2 by the phosphatase FIG4.17 There are three classes of PI3 kinases that produce PI(3)P in mammals, with varying tissue expression and substrate specificity.18,19 In skeletal muscle, the primary sources of PI(3)P are hypothesized (based on gene expression) to be the class III kinase, Pik3c3 (hVPS34), and the class II kinase Pik3c2β, with PIK3C3 considered the major enzymatic regulator of its production.15,16,20 Previous studies of PIK3C3 have identified it as a regulator of several intracellular processes, including endosome-to-Golgi membrane traffic,7 endocytosis,21,22 mTOR-S6K1 signaling,23,24 and autophagy. Perhaps its best-studied function is in autophagy, where PIK3C3 and its regulatory subunit PIK3R4 (Vps15) form multiple complexes with other autophagy gene products to regulate several steps of autophagosome formation and maturation.25–28
The goal of the present study was to begin understanding the role of PI(3)P in skeletal muscle by evaluating the function of PIK3C3. As previously reported, whole-animal gene knockout of Pik3c3 in the mouse results in early embryonic lethality.29,30 Therefore, to study PIK3C3 specifically in muscle, we have used the Cre-lox system. Cre-lox–mediated knockout of Pik3c3 has been performed in kidney,31 liver, and heart32; sensory, cortical, and hippocampal neurons30,33; and T cells,34 but Pik3c3 has yet to be examined in skeletal muscle. We generated mice with conditional knockout of Pik3c3 in skeletal muscle by combining floxed Pik3c330 and Cre recombinase under the muscle creatine kinase promoter: Tg(Ckmm-Cre).35 The resulting mice had normal embryonic and early postnatal development, but died by 2 months of age, presumably from severe cardiomyopathy.32 Somewhat surprisingly, examination of skeletal muscle in knockout animals revealed a murine muscular dystrophy. We present the comprehensive characterization of this dystrophic phenotype. In total, we report for the first time a requirement for Pik3c3 in skeletal muscle homeostasis, and further identify loss of Pik3c3 as a cause of muscular dystrophy in the mouse.
Materials and Methods
Care and Treatment of Animals
All care and treatment of animals was implemented through protocols carefully monitored by the University Committee on Use and Care of Animals. The University of Michigan's Unit for Laboratory Animal Medicine carefully monitored the health of the rodent colonies. The Unit for Laboratory Animal Medicine maintained proper environmental regulation, including temperature and light cycles, unlimited access to water, appropriate food supply, and clean enclosures. Pups were weaned from their mothers according to standard protocols, and tails were clipped for genotyping, as described.
Generation of Mutant Mouse Strains
Floxed Pik3c3 mice, a kind gift from Dr. Fan Wang (Duke University, Trinity, NC), were described in Zhou et al.30 Cre mice are of the strain: B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J, obtained from the Jackson Laboratory (Bar Harbor, ME). Mice were crossed to make Pik3c3fl/+;Cre+ heterozygous mice, and in-crossed to produce the double-floxed allele Pik3c3fl/fl, resulting in the muscle-specific conditional knockout animals.
Genotyping of Mutant Mouse Strains
Protocols and primers for genotyping mice were described previously.30
Quantitative PCR
Total mRNA was isolated from muscle homogenates using QIAGEN RNeasy kits (Qiagen, Valencia, CA), and quantitative PCR (qPCR) was performed using cDNA reverse-transcribed by iScript (BioRad Laboratories, Hercules, CA) on ABI equipment (Applied Biosystems; Life Technologies, Carlsbad, CA) using SYBR Green reagents (Life Technologies). Analysis was done by using the ΔΔCt method. Primers for transcripts analyzed were previously described.36
Western Blot Analysis
Protein was isolated from tissue from KO and littermate mice by mincing and homogenizing in T-PER buffer (Thermo Fisher Scientific, Waltham, MA) with proteinase inhibitors (Roche, Basel, Switzerland), with brief sonication on ice. Loading buffer was added to protein and run on 10% to 15% acrylamide gels, followed by transfer to polyvinylidene difluoride blots. Primary antibodies were applied overnight at 4°C at 1:1000 dilution in 3% bovine serum albumin, 0.5% Triton X-100, NaF.
Indirect analysis of protein expression was performed using standard methodology and the ImageJ program version 1.45k (NIH, Bethesda, MD).
Histology
Animals were sacrificed by anesthetic injection followed by cervical dislocation, and tissues were isolated using sterile surgical methods without the use of laminar flow hoods. Muscle tissue from quadriceps and tibialis anterior was dissected and mounted onto small balsawood pieces that were previously frozen with drops of optimal cutting temperature compound (Tissue-Tek; Thermo Fisher Scientific), and then semithawed with light friction. The mounted muscle tissue was immediately submerged in a −55°C isopentane bath cooled by liquid nitrogen for flash freezing.
Muscles were cut in 12-μm cross sections and mounted on Superfrost Plus slides (Thermo Fisher Scientific) using a Leica cryostat (Leica Microsystems, Wetzlar, Germany) at −20°C, and dried at room temperature before storage at −80°C.
Micrographs were captured with an Infinity1 camera (Lumenera Corporation, Ottawa, ON, Canada) with eponymous software visualized through an Olympus BX43 light microscope (Olympus, Tokyo, Japan). Images were cropped in Adobe Photoshop and arranged for figures with Adobe Illustrator (both programs from Creative Suite 6; Adobe Systems, San Jose, CA).
Standard techniques for indirect fluorescent immunohistology were used. Snap-frozen tissues were cryosectioned and collected on Superfrost slides. After allowing sections to dry, they were washed with PBS to remove residual OCT. Slides were blocked for more than 30 minutes at room temperature in a solution of 0.25 mol/L KCl, 0.20 mmol/L HEPES with 1% bovine serum albumin, 0.1% gelatin, 0.02% NaN3, and 0.1% Triton X-100 (pH 7.9). Slides were incubated overnight at 4°C with Progen guinea pig anti-p62 antibody (PROGEN Biotechnik, Heidelberg, Germany), diluted 1:100 in blocking solution. After three washes in PBS, secondary antibodies (Alexa Fluor 488 conjugated with goat anti-guinea pig IgG; Life Technologies) were applied for 1 hour at room temperature, diluted 1:1000 in blocking solution. Slides were washed several times to remove excess secondary antibody and then mounted with ProLong Gold (Life Technologies).
Bright-Field Imaging and Fiber Size Analysis
Slides were stained with Mayer's H&E following standard protocols, and then mounted with Permount (Thermo Fisher Scientific).
Postcapture analysis, including tests of statistical significance, was performed using Microsoft Excel 2008 (Microsoft, Redmond, WA) and GraphPad Prism version 5 for MacOSX (GraphPad Software, La Jolla, CA).
Transmission Electron Microscopy
Immediately following dissection, tips of quadriceps were carefully cut into approximately 1 × 2-mm fragments, and incubated in Karnovsky's fixative overnight at 4°C.
The fixed tissue was brought to the Microscopy and Image Analysis Laboratory (MIL) core facility at the University of Michigan for processing. Ultrathin sections were analyzed for orientation, and grids were prepared for use on the Phillips CM-100 transmission electron microscope (Philips, Eindhoven, the Netherlands).
Measurements of Serum Creatine Phosphokinase
Serum was collected from the saphenous vein of restrained animals and stored at −80°C. Creatine kinase activity was measured in duplicate using CK NADP Reagent (Cliniqa, San Marcos, CA).
Evans Blue Dye Uptake Assay
Evans Blue dye (EBD) was injected i.p. and allowed to infiltrate tissues over the course of 24 hours before mice were sacrificed and tissues sectioned as described above.
Echocardiographic Analysis
Echocardiograms were taken of the mice as described previously.21
PI(3)P Enzyme-Linked Immunosorbent Assay
Lipid analysis was performed by indirect analysis of whole-muscle lysates using Echelon's PI(3)P Mass ELISA kit (K-3300) (Echelon Biosciences, Salt Lake City, UT), as previously described.14
Results
Generation and Verification of Muscle-Specific Pik3c3 Knockout Mice
We generated muscle-specific Pik3c3 knockout mice by breeding mice harboring a conditional null mutant allele, in which exons 17 and 18 of Pik3c3 are flanked by LoxP sites (Pik3c3fl/+),30 with previously characterized transgenic mice expressing Cre recombinase under the muscle creatine kinase (Ckmm) promoter Tg(Ckmm-Cre).35 Heterozygous animals (Pik3c3fl/+;TgCkmm-Cre+) were intercrossed to produce Pik3c3 muscle-specific knockout mice (TgCkmm-Cre+;Pik3c3fl/fl), referred to as Pik3c3-KO, or simply KO, and sibling mice, referred to as littermate controls or wild type (WT). An example of PCR-based genotyping, using previously published primers,30 from a representative cross is presented in Figure 1A.
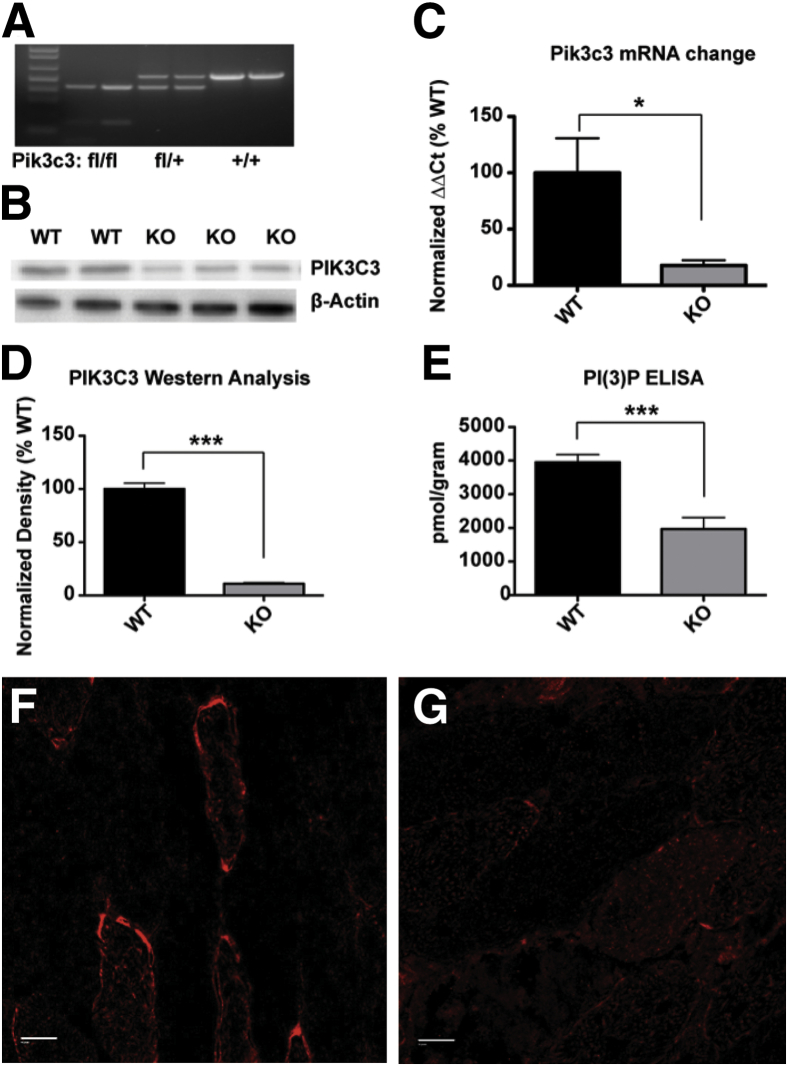
Figure 1.
Muscle-specific Pik3c3 knockout causes a significant decrease in PIK3C3 and PI(3)P levels. Muscle-specific knockout of Pik3c3 was confirmed through direct (DNA, mRNA, protein) and indirect [PI(3)P level] measures. Samples were obtained from quadriceps muscle tissue isolated from 6- to 8-week-old mice, with age-matched littermate controls, unless otherwise noted. A: Genotyping of mice harboring homozygous floxed (fl/fl), heterozygous (fl/+), or WT (+/+) alleles. B: As measured by qPCR, a significant decrease in Pik3c3 transcript levels (17.7 ± 4.7%, n = 4) was detected in skeletal muscle from knockout (KO) animals. C: Values were normalized to Gapdh transcript levels, with WT average value set at 100% (±31%, n = 4). Western blot analysis confirmed reduction of PIK3C3 levels in skeletal muscle from KO animals. D: Quantitation of PIK3C3 protein levels as determined by densitometric analysis, using β-actin protein levels as a loading control, showed a decrease from WT levels (100 ± 5.4%) to 11.0 ± 1.0%. E: PI(3)P levels are reduced in KO animals, as determined by using a PI(3)P ELISA kit [purified lipid (pmol)/mass (g) of muscle tissue] (WT: 3967 ± 225, n = 9; KO: 1982 ± 334, n = 8). Error bars are presented as SEM. Immunostaining with anti-Pik3c3 antibody of sections of quadriceps from WT littermate (F) and KO (G). Staining in WT was along the plasma membrane and in puncta in the sarcoplasm. Staining in KO animals was dramatically reduced, but sarcoplasmic puncta can still be appreciated. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗∗P < 0.001. Scale bars: 20 μm (F and G).
To verify that we successfully achieved knockdown of Pik3c3 expression, we compared both transcript and protein levels from skeletal muscle of knockout mice and age-matched littermates. qPCR revealed an average reduction in skeletal muscle of (mRNA) Pik3c3 levels of 82 ± 31% (SEM) in knockout mice compared to WT (n = 4, P = 0.038) (Figure 1C). Western blot analysis of extracts from quadriceps muscle demonstrated an average reduction of PIK3C3 protein levels to 11 ± 4% (SEM) of WT (n = 3, P < 0.001) (Figure 1, B and D). Lastly, we examined PIK3C3 expression in muscle by immunostaining. In WT quadriceps, we observed a patchy pattern of expression that was both along the plasma membrane and within the sarcoplasm (Figure 1F). This expression was qualitatively and uniformly diminished in the muscle from knockout animals (Figure 1G). Faint expression can be appreciated, and in one sample, we observed a very small number of more brightly staining fibers. These data may indicate that the 11% remaining PIK3C3 observed on Western blot analysis could be from residual protein within the myofiber. Alternatively (or in addition), it may be from other cell types such as fibroblasts and blood vessels.
PIK3C3 is hypothesized to be the primary kinase that generates PI(3)P. To determine the consequence of reduced PIK3C3 levels on PI(3)P levels, we next examined PI(3)P levels from quadriceps muscle of knockout animals and wild type littermates. We did this using an established PI(3)P enzyme-linked immunosorbent assay (ELISA).14 ELISA-based analysis revealed a reduction of PI(3)P levels in skeletal muscle of knockout animals to 50% of controls (n = 9) (Figure 1E). This level of reduction is consistent, though somewhat less than the reduction of the amount of PIK3C3 protein, and supports the previous assumption that PIK3C3 is a primary (but not sole) kinase responsible for PI(3)P production in skeletal muscle. Of note, we cannot exclude that some or even most of the remaining PI(3)P is due either to residual PIK3C3 expression in myofibers or else to PIK3C3 expression in the nonmuscle cell types that are present in total muscle extracts.
Muscle-Specific Knockdown of Pik3c3 Results in Premature Lethality
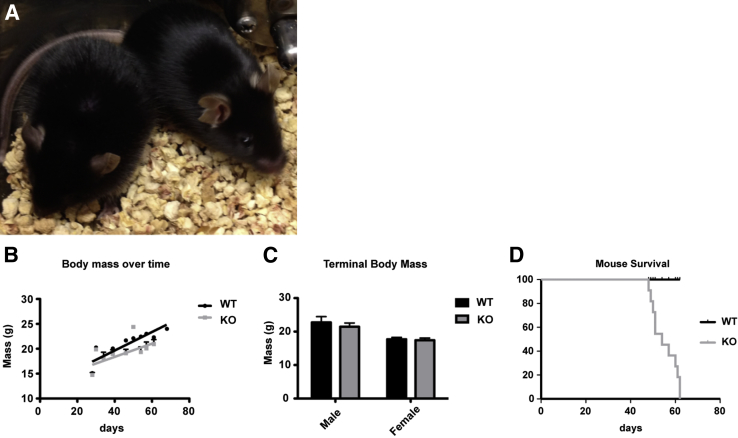
KO animals appeared normal at birth, and are qualitatively similar to littermates through the first month of extra-uterine life (Figure 2, A and B). This was revealed subjectively through animal appearance and activity, and objectively by analysis of body weight of sex-matched animals, because comparisons did not show significant differences (Figure 2, A–C). In addition, skeletal muscle tissue taken from animals sacrificed within the first month appeared histologically normal (Supplemental Figure S1).
Figure 2.
Muscle-specific Pik3c3-cKO animals have a reduced life span. The disease phenotype of knockout (KO) mice develops after 1 month, and affected mice die rapidly after onset of symptoms. A: Despite abnormal levels of activity, KO mice are difficult to distinguish by overall appearance from WT littermates, even at 6 weeks of age (KO mouse is on the left). B: There is no significant difference in the body mass of KO animals (slope, P = 0.21) than that of WT, though weights tend to be smaller than age-matched WT littermates toward the end of KO disease progression (lines represent linear regression; WT, n = 3; KO n = 4). C: Terminal body mass of WT (22.7 ± 0.57 g for males and 17.7 ± 0.24 g for females) and KO animals (21.4 ± 0.38 g for males and 17.4 ± 0.29 g for females) are not significantly different (unpaired t-tests for males, P = 0.075, n = 9; for females, P = 0.45, n = 5). D: KO mice die between 6 and 8 weeks of age, with median survival of 54 days (KO, n = 11, with an equal number of WT littermates were sacrificed to match age and sex of KO mice). Data are expressed as means ± SEM (C).
However, between 40 and 60 days of age, KO animals began to exhibit an overt phenotype consisting mainly of reduced voluntary activity (Supplemental Video S1) and an abnormal suspension test (Supplemental Video S2). The youngest animal observed in our cohort to demonstrate overt phenotypic changes did so at age 42 days (n = 27, KO animals). Shortly after first signs of disease, the KO animals displayed progressively reduced mobility as well as early mortality. The youngest age of death was at 42 days of age, whereas the oldest living knockout animal survived only to 62 days (Figure 2D). The exact cause of death in KO animals was not precisely known, though we strongly suspect it is related to cardiac disease.
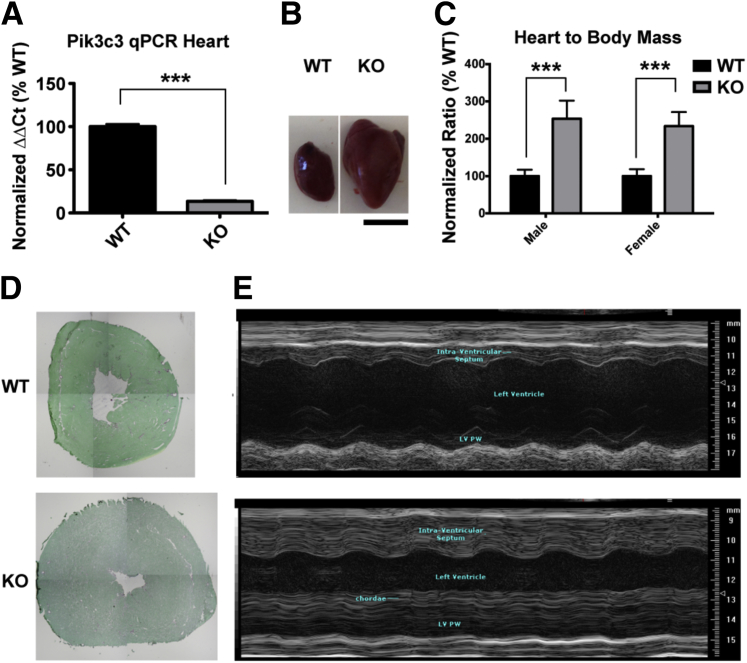
Pik3c3 Knockout Causes a Severe Dilated Cardiomyopathy
Because the Ckmm promoter expresses in cardiac and skeletal muscle, we next examined the hearts of KO animals. We first analyzed levels of Pik3c3 transcript from heart tissue by qPCR (Figure 3A), and showed an average reduction of 87 ± 3%. We next examined the gross anatomical phenotype of the hearts, and observed a dramatic increase in heart size, with the average knockout heart-to-body-mass ratio measuring 250% of age-matched controls (Figure 3, B and C). Low-magnification histopathological analysis corroborated this anatomical observation, revealing qualitative enlargement and thickening of both atrium and ventricle (n = 3) (Figure 3D). In addition, echocardiography showed evidence of significant cardiac hypertrophy and severe cardiac dysfunction (Figure 3E and Supplemental Table S1). Although the B-mode measurements of chamber dimensions were largely unchanged, all measurements of septal and posterior left ventricular wall thickness by M-mode were significantly increased in KO mice (Figure 3E), with an estimated ventricular mass consistent with the morphological findings in isolated hearts. Indices of systolic function, including ejection fraction and stroke volume, and cardiac output were all decreased in KO, whereas the resting heart rate was unchanged (Supplemental Table S1). Diastolic function was also impaired as indicated by an increase in isovolumic relaxation time and ratio of mitral valve E blood velocity to E′ tissue Doppler velocity at the septal annulus. In all, our analysis supports the conclusion that Pik3c3 deletion in the heart causes a severe cardiomyopathy. Of note, our results are consistent with a previously published study using cre-lox knockout of Pik3c3.32
Figure 3.
KO animals display cardiac abnormalities. A: As measured by qPCR, Ckmm-driven cre expression in the heart leads to 87% reduction in levels of Pik3c3 transcript in KO (13.4 ± 1.2%) as compared to WT controls (100 ± 2.9%). B: Obvious cardiomegaly is observed in KO animals . C: Hearts in KO animals are two to three times the size of WT littermates. Increases in heart mass to body mass ratios are consistent in affected males (254 ± 16.1%) and females (234 ± 16.9%). D: Cross-sectional histology of hearts with Sirius Red stain demonstrates increased thickness of walls with no observable fibrosis (aged 45 days). E: Representative M-mode echocardiograms from the left ventricle of KO mice show marked left ventricular wall hypertrophy compared to age-matched WT littermate mice. Data are expressed as means ± SEM. n = 2 (A and C). ∗∗∗P < 0.001. Scale bar = 50 mm (B).
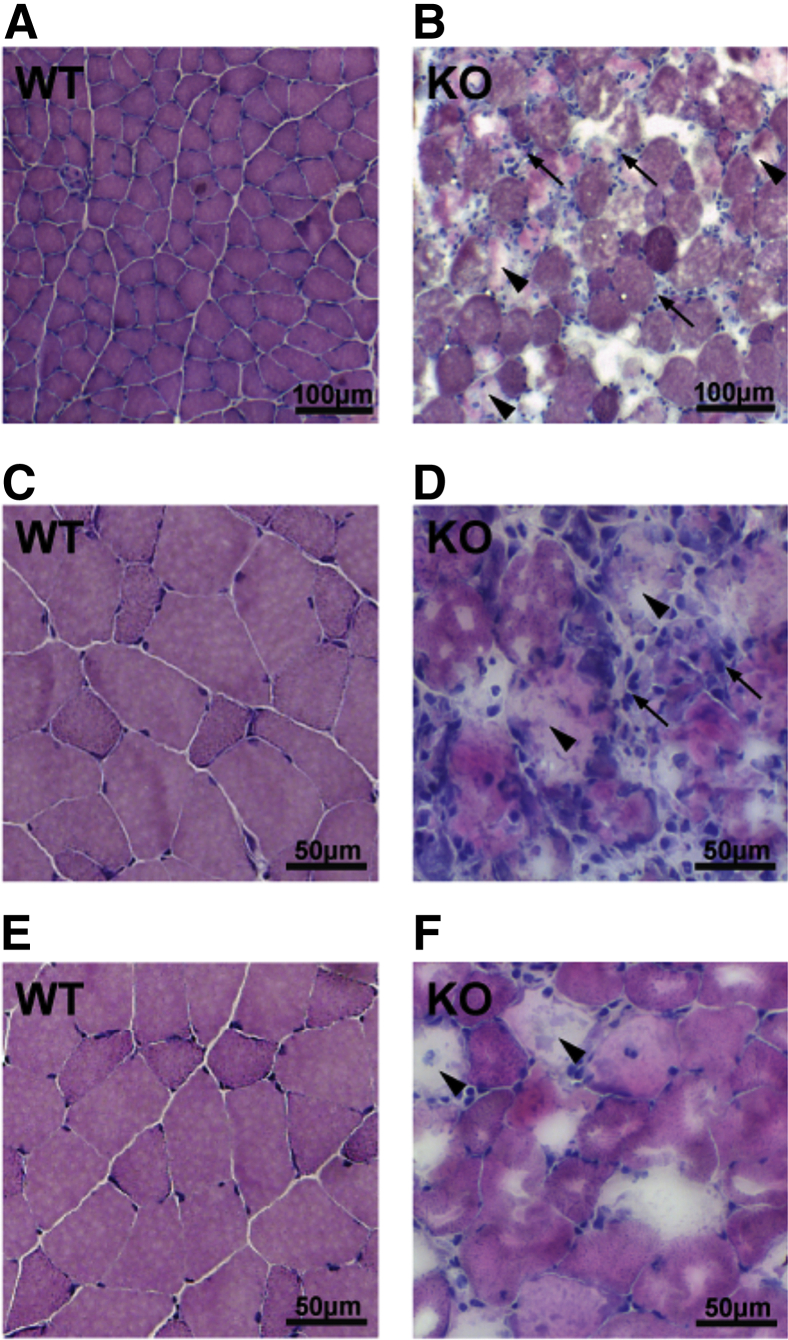
Pik3c3 Knockout Results in a Murine Muscular Dystrophy
Because our primary interest in creating conditional knockouts of Pik3c3 was to examine its requirement for skeletal muscle development and homeostasis, we focused all subsequent studies on skeletal muscle. We began our analysis by performing routine histopathology studies from quadriceps, gastrocnemius, and tibialis anterior muscles. H&E staining of fresh-frozen, cryosectioned muscle from 42- to 60-day-old animals revealed clearly abnormal areas with features that included inflammatory infiltrates (small blue cells), and degenerating and regenerating fibers (Figure 4). Within these areas of abnormal muscle, there was a significant increase in myofibers with internalized nuclei (Supplemental Figure S2A), consistent with muscle regeneration (Figure 4, A and B). In total, the histopathological changes (inflammatory cells, increased internal nuclei, and degenerating fibers) were consistent with a murine muscular dystrophy. These changes were seen in all muscle groups evaluated and in multiple different animals (n = 5).
Figure 4.
KO animals have aberrant histology. Histological analysis of KO skeletal muscle shows signs of muscular dystrophy. Sections were taken from gastrocnemius muscle of 60-day-old WT and KO animals, and stained with H&E. A and B: Low-magnification (20×) images reveal inflammatory infiltrate (arrows) and changes consistent with degeneration and regeneration (arrowheads). C–F: Higher magnification (40×) micrographs show changes as in (A) and (B) but also better reveal abundant internal nuclei.
Of note, there were as well areas of relatively unaffected muscle next to areas with dystrophic changes. In addition, muscles harvested from older knockout animals had more areas of aberrant fibers, whereas (as mentioned above) muscle analyzed before 1 month of age appeared normal (Supplemental Figure S1). Also, the average fiber size was not significantly different between knockouts and their littermates (Supplemental Figure S2B). Lastly, we did not observe an increase in fibrosis in dystrophic muscle areas of the knockout animals (Supplemental Figure S3).
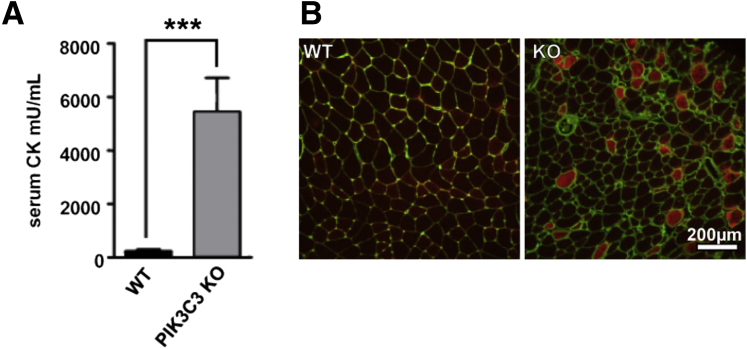
To corroborate the histopathological observation of dystrophic muscle in Pik3c3 knockout mice, we used two additional strategies (Figure 5). The first was to measure serum creatine phosphokinase (CPK) levels. We obtained blood samples from the saphenous vein of animals that were not previously exposed to strenuous exercise or diet modifications. CPK levels were significantly and highly elevated (P = 0.0062). The average level in 45-day-old mice was almost 20-fold higher in conditional knockout animals (5450 ± 1260, n = 4) as compared to their littermate controls (230 ± 66, n = 4) (Figure 5A).
Figure 5.
Pik3c3-cKO mice exhibit dystrophic muscle by independent measures. A: CPK measurements of serum obtained from the distal saphenous vein show increases of nearly 50-fold in excess when KO (mean = 5452 ± 1263 mU/mL) are compared to WT littermates (mean = 233.3 ± 65.51 mU/mL). B: EBD is taken up by dystrophic fibers after 24 hours of incubation following peritoneal injection. EBD is shown in red and counterstained against the extracellular membrane protein Laminin in green. Data are expressed as means ± SEM. n = 4 KO and 4 WT mice. ∗∗∗P < 0.001.
The second measure of muscular dystrophy was assessment of EBD uptake in skeletal muscle (Figure 5B). After systemic injection of EBD, damaged/dystrophic fibers demonstrate dye uptake, whereas healthy fibers do not. As compared to wild types, there was a visible increase in EBD-positive fibers, indicating damaged muscle in the knockouts. Importantly, there was only a slight increase in EBD in the hearts of knockout animals (Supplemental Figure S4, A–D), suggesting that the elevated CPK levels found in the knockouts are most likely from skeletal (and not cardiac) muscle breakdown. In all, based on histopathology, serum CPK levels, and EBD studies, we concluded that reduction of Pik3c3 expression resulted in a murine muscular dystrophy.
Onset of Muscular Dystrophy Correlates with Relative Pik3c3 Expression
To better understand the timing of disease in the Pik3c3-cKO animals, we examined relative RNA expression during muscle development using RT-qPCR. We detected relatively little expression in newborn and 1-month-old muscle, and increasing levels of RNA at both 2 and 3 months (Supplemental Figure S5A). This increase in expression correlates with the timing of dystrophic changes. Another consideration related to both the timing and severity of dystrophy is potential compensation by the class II PI3 kinases. We thus examined expression of Pik3c2a, Pik3c2b, and Pik3c2g in skeletal muscle during muscle development from WT and knockout mice. This analysis revealed two observations. The first is that Pik3c2b, but not Pik3c2a or Pik3c2g, is abundantly expressed in 2- and 3-month-old WT muscle. The second is that there was no statistical change in RNA levels of any class II kinase in knockout samples as compared to WT littermates (Supplemental Figure S5B).
Ultrastructural Analysis Reveals Aberrant Membrane Structures
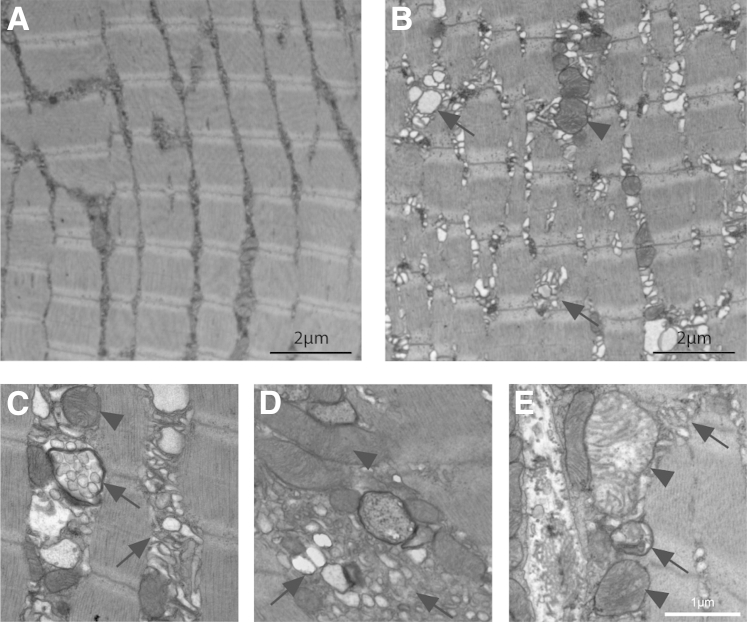
We performed analysis of skeletal muscle ultrastructure to look for clues related to muscle disease pathogenesis. Despite elevated CPK levels and abnormal EBD update, no obvious membrane discontinuities or tears were observed in KO muscle. However, there were widespread and striking abnormalities in several subcellular structures (Figure 6). Specifically, among the various abnormalities we observed, we detected frequent swellings associated with internal sarcolemmal membranes, as well as aberrantly appearing mitochondria (Figure 6B). We also observed degradation compartments surrounded by double membranes, a finding consistent with aberrant autophagy (Figure 6, C and E). Overall, these changes were consistent with a defect in membrane turnover,37 and resemble changes seen in skeletal muscle with defective autophagy.38
Figure 6.
Ultrastructural analyses by transmission electron microscopy show aberrant features in KO mice. Tissue from tibialis anterior muscle of 6-week-old mice was analyzed by electron microscopy. Evaluation of transmission electron micrographs of WT littermate controls (A) and KO (B–E) reveals numerous defects in muscle ultrastructure, including sarcoplasmic swellings in addition to aberrant vacuoles (arrows) and mitochondria (arrowheads). Scale bars: 2 μm (A and B); 1 μm (E).
Autophagy Genes Have Aberrant Expression and Localization in KO Animals
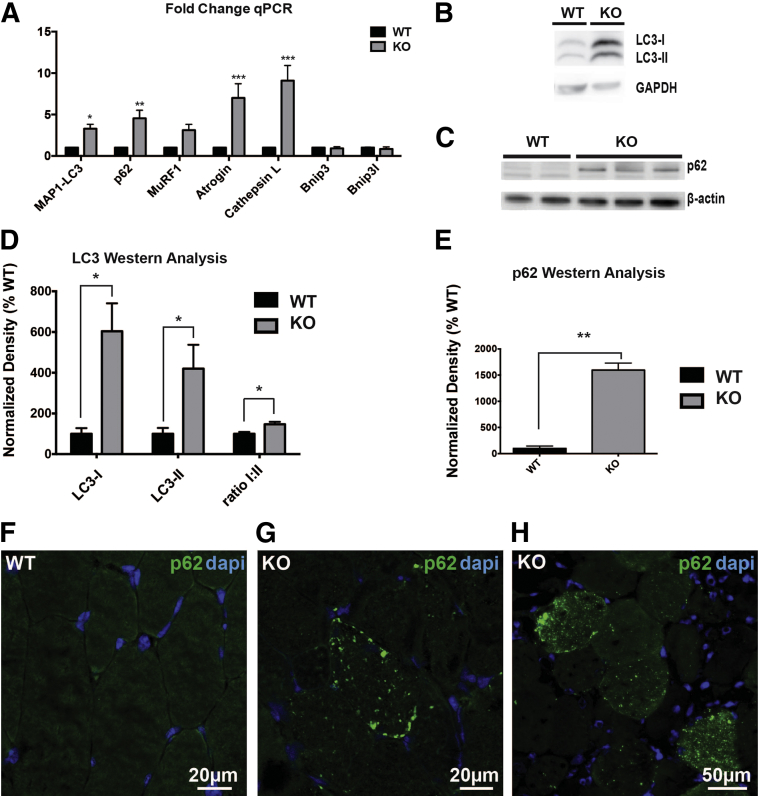
One of the functions of PIK3C3 in vitro is regulation of the initiation of autophagy. Given that the ultrastructural changes were reminiscent of defective autophagy, we examined KO muscle for evidence of aberrant autophagy. We examined autophagy using three approaches: i) measurement of autophagy-related gene expression changes by qPCR; ii) examination of autophagic markers by Western blot analysis; and iii) study of markers by immunocytochemistry. qPCR revealed significant increases in transcripts associated with autophagy, including up-regulation of p62 (4.5 ± 0.97 fold change) and LC3 (3.3 ± 0.53 fold change) (Figure 7A). We also detected increased expression of gene products associated with muscle atrophy, such as ubiquitin ligases atrogin-1 (7.0 ± 1.7 fold change) and MuRF1 (3.1 ± 0.71 fold change). Similarly, we observed significant increases in levels of the lysosomal proteinase, cathepsin L (9.1 ± 1.8 fold change). Although other reports36 on muscle wasting have shown up-regulation in Bnip3 and Bnip3l, levels of these transcripts were unaffected by the Pik3c3 knockout: Bnip3 had a 0.92 ± 0.2 fold change and Bnip3l gave a 0.86 ± 0.23 fold change when normalized to WT animals. This is consistent with the fact that we did not detect any corresponding alterations in overall muscle size or in the size of individual fibers (Supplemental Figure S2).
Figure 7.
KO mice exhibit aberrant markers of basal autophagy. Transcript levels were measured from mRNA isolated from quadriceps of 2-month-old KO and WT littermates. A: Results using primers for Map1-Lc3 (3.3 ± 0.53 fold change), p62 (4.5 ± 0.97 fold change), Murf1 (3.1 ± 0.71 fold change), Atrogin1 (7.0 ± 1.7 fold change), CathepsinL (9.1 ± 1.8 fold change), Bnip3 (0.92 ± 0.2 fold change), and Bnip3l (0.86 ± 0.23 fold change) were normalized to Gapdh internal controls. The y axis indicates fold increase normalized to littermate controls for ΔΔCt (n = 3). B and C: Protein was isolated from quadriceps of 2-month-old KO and WT littermates for analysis by Western blot with antibodies against MAP1-LC3 and p62. D: Densitometric analysis of LC3-I/II, performed using GAPDH as an internal control (n = 2), shows increases in LC3-I (604 ± 137%, ∗P < 0.05) and LC3-II (420 ± 118%), and a ratio of LC3-I:LC3-II that is slightly increased (147 ± 12%). E: Densiometric analysis of blot probed with p62 antibody shows increase in KO muscle to 1593 ± 137% that of WT controls, using β-actin as internal control (n = 3). F–H: Sectioned gastrocnemius muscle from 60-day-old animals stained using immunochemistry shows accumulation of p62/SQSTM1. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
We next examined autophagy by looking at protein markers of the pathway. Western blot analysis showed significant increases in total protein levels of LC3 and p62: LC3-I increased 604 ± 137% and LC3-II by 420 ± 118%, with the ratio of LC3-I:LC3-II only slightly increased (147 ± 12%) (Figure 7, B–E); p62 antibody showed increases in KO muscle to 1593 ± 137% that of WT controls, using β-actin as an internal control (n = 3). These changes are consistent with defective autophagic flux and with the measurements of transcript levels. We performed immunocytochemical analysis using an antibody to p62, a protein associated with the initiation of autophagy that accumulates when autophagic flux is impaired. We observed significant accumulation of p62 with fibers of KO animals, as compared to essentially absent staining in age-matched control siblings (Figure 7, F–H). In total, KO resulted in up-regulation of markers of autophagy, but rather than showing turnover of key proteins, we witnessed an accumulation of the markers of autophagosome formation, consistent with an overall impairment in the initiation of autophagy.
Loss of PIK3C3 Causes Impairment in Lysosomal Function
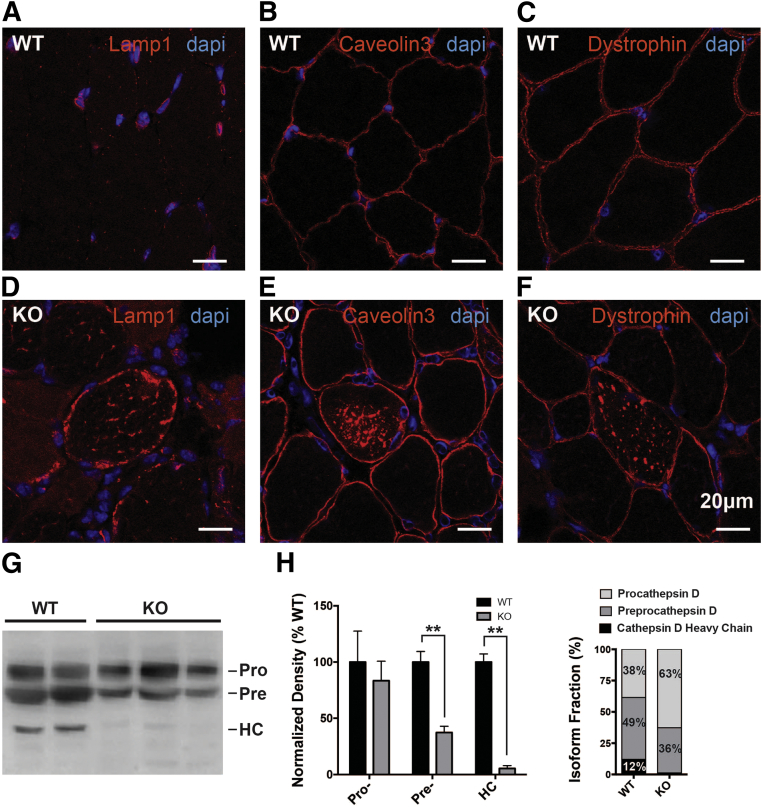
Because aberrant autophagy alone is unlikely to explain the histopathological changes we observed, we sought to uncover additional potential causes for dystrophic muscle pathology in the KO mice. Because elevated CPK levels and increased EBD uptake were suggestive of compromised integrity of the muscle's external membrane, we analyzed expression and localization of integral membrane proteins using immunohistochemistry. We observed striking changes in the localization of both dystrophin and caveolin-3. In numerous muscle fibers from KO (but not WT) animals, these proteins, although still present at the membrane, also accumulated within the myofiber (Figure 8, B, C, E, and F). The mislocalization of these proteins occurred without a change in their overall levels, as determined by Western blot analysis (Supplemental Figure S6).
Figure 8.
Aberrant accumulation of membrane-associated proteins indicates trafficking dysfunction. Sectioned gastrocnemius muscle from 60-day-old animals stained using immunochemistry shows accumulation of LAMP1 (A and D), caveolin-3 (B and E), and dystrophin (C and F) in KO animals compared to littermate controls. G: Western blot analysis of cathepsin D reveals alterations in the processing of the mature form in KO animals from procathepsin D (Pro), preprocathepsin D (Pre), and cathepsin D heavy chain (HC) isoforms. H: Densiometric analysis reveals significant decreases in the levels of Pre and HC isoforms, demonstrated as overall levels normalized to β-actin and as total fraction of cathepsin D. ∗∗P < 0.01.
Internal accumulation of membrane proteins has been previously reported in the setting of abnormal lysosomal function.28 We therefore sought to interrogate lysosomal function in our knockout animals. We examined LAMP1 staining, and found two important changes as compared to WT mice (Figure 8, A and D): accumulation of LAMP1 staining within myofibers as well as increased intensity of LAMP1 staining around the plasma membrane. Both findings have been observed in other models of lysosomal dysfunction.39 To quantitate a possible abnormality in lysosomal function, we looked at cathepsin-D processing by Western blot analysis. Cathepsin-D processing is used as a marker for the normal functioning of the lysosome. We found a significant decrease in the mature forms of cathepsin-D, suggesting a significant abnormality in the ability of the lysosome to cleave cathepsin to its mature, active form (Figure 8, G and H). Taken together with the immunostaining results for LAMP1, dystrophin, and caveolin-3, these results are consistent with impaired lysosomal function and suggest aberrant lysosomal processing as the cause of membrane protein mislocalization.
Discussion
In summary, we have generated muscle-specific knockout mice of Pik3c3 (Vps34) and provided the first phenotypic description of the role of PIK3C3 in skeletal muscle. Knockout mice had reduced levels of Pik3c3 mRNA and protein in striated muscle, and had significantly reduced levels of PI(3)P, the phospholipid generated by PIK3C3. Using both morphological and functional analyses, we identified significant changes in skeletal and cardiac muscle structure and function and in the overall health of the animals, including premature lethality in the second postnatal month. Our results specifically identify PIK3C3 as a gene product required for normal muscle structural integrity and for the regulation of basal autophagy and lysosome function in skeletal muscle.
One of the most significant findings in our study was the appearance of dystrophic muscle pathology in affected mice. This observation was in contrast to our pre-experiment prediction that loss of Pik3c3 would be associated with abnormalities similar to those observed with myotubularin dysfunction, because both overexpression and knockout of Mtm1 cause significant disruption of the sarcotubular network and (at least for Mtm1 knockout) a centronuclear myopathy phenotype. KO mice clearly did not have a centronuclear myopathy, but instead had changes consistent with a muscular dystrophy. The presence of a dystrophy was established based on histopathology, CPK levels, and EBD staining, and our study thus provides a connection between dysfunction in PIP regulation and dystrophic muscle pathology. Of note, a recent study described the muscle-specific knockout of Vps15, which functions as a co-factor of PIK3C3 and is required for many aspects of PIK3C3 function. These mice similarly manifested a murine muscular dystrophy, a finding that corroborates the data from the present paper.28
The appearance of dystrophic pathogenesis gives insight into the involvement of PIK3C3 in pathways other than canonical Beclin-complex–mediated autophagy. On the basis of previous knockout mice studies, it is unlikely that the defects in autophagy we observed in KOs are sufficient to explain the severe muscular dystrophy seen in these animals. Specifically, disruption of the essential components of autophagic machinery Atg7 and Atg5 does not result in dystrophic pathology, but instead a mild myopathy characterized by reduced myofiber size.40,41 Instead, we postulate that abnormalities in lysosomal function are more likely to be the cause of the dystrophic animals. This assertion is supported by two major observations. The first is that mutations in the lysosomal proteins LAMP2 and GAA both result in skeletal muscle disease (Danon disease39 and Pompe disease,42 respectively) with dystrophic features. The second is that lysosomal function (and in particular, lysosomal exocytosis) has been implicated in the regulation of membrane repair,43 and defective membrane repair is a well-established trigger for the development of muscular dystrophy.44 Additional experimentation will obviously be necessary to specifically establish why the loss of PIK3C3 function causes a muscular dystrophy, and to parse out which of the many potential functions of the protein are responsible for regulating membrane integrity in skeletal muscle.
Of note, there are several independent studies of mice with conditionally deleted alleles of Pik3c3, including neurons,30 in T cells,34 in podocytes,31 and in heart and liver.32 Consideration of these studies together reveals several common features of organs lacking PIK3C3 that are seen as well in our muscle-specific deletion. These include aberrant autophagy, disrupted cellular homeostasis, and overall degeneration of tissues. Also, there are some specific differences when comparing studies of different cell types, including controversy over which stages of autophagy are most affected by loss of PIK3C3 function.28,32 It should be noted that there are still many aspects of autophagy initiation and regulation that are not well understood, such as the extent of noncanonical pathways (sans Beclin-1), and cells use multiple processes for sequestering and/or delivering targets to the lysosome.45 Future studies of the various knockout strains of PIK3C3 may help illuminate these issues.
Lastly, it should also be noted that PIK3C3 is likely not the only means for generating PI(3)P in skeletal muscle. This assertion is supported by the fact that we observe a 50% reduction in PI(3)P levels in the muscle of knockouts, despite achieving much higher levels of PIK3C3 reduction. Although the residual expression of PIK3C3 (both in myofibers and perhaps in nonmuscle cell types) in our knockout animals likely accounts for some of the PI(3)P production, we speculate that other PI3 kinases may also participate in its synthesis. One candidate for generating a fraction of PI(3)P in skeletal muscle is PIK3C2B, a class II PI-3 kinase that is expressed in skeletal muscle and whose function is poorly understood. We confirm in this study that Pik3c2b transcript, and not Pik3c2a or 2g, is abundantly detected in skeletal muscle. Experimentation addressing the role of PIK3C2B in muscle, and its interplay with the functions of PIK3C3, will obviously be important to best understand the role of PI(3)P and its regulation in skeletal muscle.
Conclusion
Muscle-specific conditional knockout of PIK3C3 revealed a novel murine dystrophy with aberrant autophagy and lysosomal function. These novel knockout mice provide a useful animal model for the study of degradation pathways and their association to the pathogenesis of muscular dystrophy.
Acknowledgments
We thank Eva Feldman and the Taubman Institute at the University of Michigan, the Physiology Phenotyping Core in partnership with the Frankel Cardiovascular Center Animal Phenotyping Core for animal phenotyping services, including ultrasound imaging and rodent treadmills, and Dr. Fan Wang for providing the Pik3c3 conditional KO mice.
Footnotes
Supported by NIH grants 1K08AR054835 (J.J.D.), HL080388 (D.E.M.), and OD016502 (D.E.M.) and the Muscular Dystrophy Association (J.J.D. and D.E.M.).
Disclosures: None declared.
Supplemental Data
Muscle of young Pik3c3-cKO animals appears normal by histological analysis. Tibialis anterior muscle from animals harvested at 1 month of age appears normal. Left panels are stained with H&E. Right panels are stained with succinic dehydrogenase. Scale bars: 50 μm.
Cross-sectional muscle analysis is typical of a dystrophic phenotype. A: Concentrated areas of aberrant fibers contain elevated numbers of internalized nuclei in knockout (KO) animals (49 ± 11) compared to control littermates (WT) (1 ± 1). B: Muscle fiber area distribution in knockout animals shows a similar pattern to littermates. Data are expressed as means ± SEM (A). n = 4 (A); n = 3 (B).
Fibrosis is not increased in KO muscle. Representative level 3 tibialis anterior muscle of 48-day-old mice—control littermate (A), knockout (B)—stained with Gomori Trichrome as a test for fibrosis. There are no apparent differences between the controls and knockout animals.
Hearts have limited loss of muscle integrity, as measured by EBD uptake. The 48-day-old animals were injected with EBD and sacrificed 24 hours later. Hearts were cryoprotected and sectioned at level 3. Pik3c3 knockout (KO) animals show sparse fibers containing EBD, counterstained with anti-laminin for visualizing individual fibers.
A: RT-qPCR analysis of PI3 kinase expression. RT-qPCR analysis of Pik3c3, Pikc2b, Pik3c2a, and Pik3c2g during four stages of muscle development. RNA was isolated from quadriceps of newborn (0 month), 1-, 2-, and 3-month-old WT mice. Levels of both Pik3c3 and Pik3c2b were low at birth but significantly increased at the 2- and 3-month points. Expression of Pik3c2a and Pik3c2g was essentially nondetectable. Values were normalized to expression at 0 months and to levels of actin. Average values for Pik3c3 at 1, 2, and 3 months were 1.5-, 6-, and 4-fold that of the level at age 0 months, respectively, and for Pik3c2b, were 1-, 1.7-, 9-, and 12-fold. B: RT-qPCR analysis of expression of class II kinases in Pik3c3 knockouts (KO) animals versus littermate controls. There was no statistical change in the levels of any class II kinase in the KO animals compared to their littermates. As also reported in Figure 1, Pik3c3 levels were significantly reduced (8% of WT levels in this experiment). *P < 0.05 (A and B). n = 3 for Pik3c3 and for Pik3c2b.
Overall levels of integral membrane proteins are unaffected by Pik3c3 knockout. Integral membrane proteins showed no appreciable differences in overall expression from skeletal muscle tissue samples isolated from 48-day-old mice. Proteins examined by Western blot analysis were dysferlin (A), α-dystroglycan (B), β-dystroglycan (C), and caveolin-3 (D).
Video demonstrates KO mouse (n = 1, in video) has reduced responsiveness to tactile stimulation compared to control littermates (n = 3). Mice in the video were 48 days old at time of filming.
Video demonstrates the tail suspension assay. Control littermate (right) demonstrates normal leg splay as well as robust sideways movements. Knockout (left) cannot display the hindlegs and has restricted trunk and forelimb movements.
References
- 1.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Itoh T., Takenawa T. Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal. 2002;14:733–743. doi: 10.1016/s0898-6568(02)00028-1. [DOI] [PubMed] [Google Scholar]
- 3.Lecompte O., Poch O., Laporte J. PtdIns5P regulation through evolution: roles in membrane trafficking? Trends Biochem Sci. 2008;33:453–460. doi: 10.1016/j.tibs.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Michell R.H., Heath V.L., Lemmon M.A., Dove S.K. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Payrastre B. Phosphoinositides: lipid kinases and phosphatases. Methods Mol Biol. 2004;273:201–212. doi: 10.1385/1-59259-783-1:201. [DOI] [PubMed] [Google Scholar]
- 6.De Camilli P., Emr S.D., McPherson P.S., Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 7.Lindmo K., Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 8.Volpicelli-Daley L., De Camilli P. Phosphoinositides' link to neurodegeneration. Nat Med. 2007;13:784–786. doi: 10.1038/nm0707-784. [DOI] [PubMed] [Google Scholar]
- 9.Nicot A.S., Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laporte J., Bedez F., Bolino A., Mandel J.L. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet. 2003;12(Spec No 2):R285–R292. doi: 10.1093/hmg/ddg273. [DOI] [PubMed] [Google Scholar]
- 11.Das SD, Dowling J, Pierson CR: X-Linked Centronuclear Myopathy. In GeneReviews [Internet]. Copyright University of Washington, Seattle. 1993–2014. Available at http://www.ncbi.nlm.nih.gov/books/NBK1432, last revised October 6, 2011
- 12.Amoasii L., Hnia K., Chicanne G., Brech A., Cowling B.S., Muller M.M., Schwab Y., Koebel P., Ferry A., Payrastre B., Laporte J. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. J Cell Sci. 2013;126(Pt 8):1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 13.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierson C.R., Dulin-Smith A.N., Durban A.N., Marshall M.L., Marshall J.T., Snyder A.D., Naiyer N., Gladman J.T., Chandler D.S., Lawlor M.W., Buj-Bello A., Dowling J.J., Beggs A.H. Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum Mol Genet. 2012;21:811–825. doi: 10.1093/hmg/ddr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backer J.M. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 16.Meijer A.J., Klionsky D.J. Vps34 is a phosphatidylinositol 3-kinase, not a phosphoinositide 3-kinase. Autophagy. 2011;7:563–564. doi: 10.4161/auto.7.6.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E., Goldowitz D., Meisler M.H., Weisman L.S. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Y., Backer J.M. Regulation of class III (Vps34) PI3Ks. Biochem Soc Trans. 2007;35:239–241. doi: 10.1042/BST0350239. [DOI] [PubMed] [Google Scholar]
- 19.Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 20.Herman P.K., Emr S.D. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michele D.E., Kabaeva Z., Davis S.L., Weiss R.M., Campbell K.P. Dystroglycan matrix receptor function in cardiac myocytes is important for limiting activity-induced myocardial damage. Circ Res. 2009;105:984–993. doi: 10.1161/CIRCRESAHA.109.199489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson E.E., Overmeyer J.H., Gunning W.T., Maltese W.A. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J Cell Sci. 2006;119:1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- 23.Nobukuni T., Kozma S.C., Thomas G. hvps34, an ancient player, enters a growing game: mTOR Complex1/S6K1 signaling. Curr Opin Cell Biol. 2007;19:135–141. doi: 10.1016/j.ceb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 25.Funderburk S.F., Wang Q.J., Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris D.P., Vogel P., Wims M., Moberg K., Humphries J., Jhaver K.G., DaCosta C.M., Shadoan M.K., Xu N., Hansen G.M., Balakrishnan S., Domin J., Powell D.R., Oravecz T. Requirement for class II phosphoinositide 3-kinase C2alpha in maintenance of glomerular structure and function. Mol Cell Biol. 2011;31:63–80. doi: 10.1128/MCB.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B.T., Shokat K.M., Williams R.L. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R.C., Guan K.L., Nishino I., Sandri M., Pende M., Panasyuk G. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med. 2013;5:870–890. doi: 10.1002/emmm.201202057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X., Takatoh J., Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6:e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Wang L., Hasegawa H., Amin P., Han B.X., Kaneko S., He Y., Wang F. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechtel W., Helmstadter M., Balica J., Hartleben B., Kiefer B., Hrnjic F., Schell C., Kretz O., Liu S., Geist F., Kerjaschki D., Walz G., Huber T.B. Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J Am Soc Nephrol. 2013;24:727–743. doi: 10.1681/ASN.2012070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaber N., Dou Z., Chen J.S., Catanzaro J., Jiang Y.P., Ballou L.M., Selinger E., Ouyang X., Lin R.Z., Zhang J., Zong W.X. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L., Budolfson K., Wang F. Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience. 2011;172:427–442. doi: 10.1016/j.neuroscience.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willinger T., Flavell R.A. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruning J.C., Michael M.D., Winnay J.N., Hayashi T., Horsch D., Accili D., Goodyear L.J., Kahn C.R. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 36.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., Goldberg A.L., Schiaffino S., Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Millay D.P., Maillet M., Roche J.A., Sargent M.A., McNally E.M., Bloch R.J., Molkentin J.D. Genetic manipulation of dysferlin expression in skeletal muscle: novel insights into muscular dystrophy. Am J Pathol. 2009;175:1817–1823. doi: 10.2353/ajpath.2009.090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino I. Autophagic vacuolar myopathies. Curr Neurol Neurosci Rep. 2003;3:64–69. doi: 10.1007/s11910-003-0040-y. [DOI] [PubMed] [Google Scholar]
- 39.Malicdan M.C., Nishino I. Autophagy in lysosomal myopathies. Brain Pathol. 2012;22:82–88. doi: 10.1111/j.1750-3639.2011.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raben N., Roberts A., Plotz P.H. Role of autophagy in the pathogenesis of Pompe disease. Acta Myol. 2007;26:45–48. [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarti S., Kobayashi K.S., Flavell R.A., Marks C.B., Miyake K., Liston D.R., Fowler K.T., Gorelick F.S., Andrews N.W. Impaired membrane resealing and autoimmune myositis in synaptotagmin VII-deficient mice. J Cell Biol. 2003;162:543–549. doi: 10.1083/jcb.200305131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int J Biochem Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Codogno P., Mehrpour M., Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Muscle of young Pik3c3-cKO animals appears normal by histological analysis. Tibialis anterior muscle from animals harvested at 1 month of age appears normal. Left panels are stained with H&E. Right panels are stained with succinic dehydrogenase. Scale bars: 50 μm.
Cross-sectional muscle analysis is typical of a dystrophic phenotype. A: Concentrated areas of aberrant fibers contain elevated numbers of internalized nuclei in knockout (KO) animals (49 ± 11) compared to control littermates (WT) (1 ± 1). B: Muscle fiber area distribution in knockout animals shows a similar pattern to littermates. Data are expressed as means ± SEM (A). n = 4 (A); n = 3 (B).
Fibrosis is not increased in KO muscle. Representative level 3 tibialis anterior muscle of 48-day-old mice—control littermate (A), knockout (B)—stained with Gomori Trichrome as a test for fibrosis. There are no apparent differences between the controls and knockout animals.
Hearts have limited loss of muscle integrity, as measured by EBD uptake. The 48-day-old animals were injected with EBD and sacrificed 24 hours later. Hearts were cryoprotected and sectioned at level 3. Pik3c3 knockout (KO) animals show sparse fibers containing EBD, counterstained with anti-laminin for visualizing individual fibers.
A: RT-qPCR analysis of PI3 kinase expression. RT-qPCR analysis of Pik3c3, Pikc2b, Pik3c2a, and Pik3c2g during four stages of muscle development. RNA was isolated from quadriceps of newborn (0 month), 1-, 2-, and 3-month-old WT mice. Levels of both Pik3c3 and Pik3c2b were low at birth but significantly increased at the 2- and 3-month points. Expression of Pik3c2a and Pik3c2g was essentially nondetectable. Values were normalized to expression at 0 months and to levels of actin. Average values for Pik3c3 at 1, 2, and 3 months were 1.5-, 6-, and 4-fold that of the level at age 0 months, respectively, and for Pik3c2b, were 1-, 1.7-, 9-, and 12-fold. B: RT-qPCR analysis of expression of class II kinases in Pik3c3 knockouts (KO) animals versus littermate controls. There was no statistical change in the levels of any class II kinase in the KO animals compared to their littermates. As also reported in Figure 1, Pik3c3 levels were significantly reduced (8% of WT levels in this experiment). *P < 0.05 (A and B). n = 3 for Pik3c3 and for Pik3c2b.
Overall levels of integral membrane proteins are unaffected by Pik3c3 knockout. Integral membrane proteins showed no appreciable differences in overall expression from skeletal muscle tissue samples isolated from 48-day-old mice. Proteins examined by Western blot analysis were dysferlin (A), α-dystroglycan (B), β-dystroglycan (C), and caveolin-3 (D).
Video demonstrates KO mouse (n = 1, in video) has reduced responsiveness to tactile stimulation compared to control littermates (n = 3). Mice in the video were 48 days old at time of filming.
Video demonstrates the tail suspension assay. Control littermate (right) demonstrates normal leg splay as well as robust sideways movements. Knockout (left) cannot display the hindlegs and has restricted trunk and forelimb movements.