Abstract
Metakaryotic cells and syncytia with large, hollow, bell shaped nuclei demonstrate symmetrical and asymmetrical amitotic nuclear fissions in microanatomical positions and numbers expected of stem cell lineages in tissues of all three primordial germ layers and their derived tumors. Using fluorescence in situ hybridization mononuclear metakaryotic interphase cells have been found with only 23 centromeric and 23 telomeric staining regions. Syncytial bell shaped nuclei of found in weeks ~5–12 of human gestation display 23 centromeric and either 23 or 46 telomeric staining regions. These images suggest that (a.) homologous chromatids pair at centromeres and telomeres, (b.) all paired telomeres join end-to-end with other paired telomeres in all mononuclear and some syncytial metakaryotic cells and (c.) telomere junctions may open and close during the syncytial phase of development. Twenty-three telomeric joining figures could be accounted by 23 rings of one chromatid pair each, a single pangenomic ring of 23 joined chromatid pairs or any of many possible sets of oligo-chromatid pair rings. As telomeric end-joining may affect peri-telomeric gene expression a programmed sequence of telomeric end-joining associations in metakaryotic stem cells could guide developmental arboration and errors in, or interruptions of, this program could contribute to carcinogenesis.
INTRODUCTION
Non-eukaryotic metakaryotic cells with large, hollow, bell shaped nuclei arise from amitotic divisions of human embryonic stem cells at the time of anlagen appearance (4–7 wks of gestation). The numbers of bell shaped nuclei increase within large tubular syncytia through the 9th–14th week and are then regularly distributed as single cells throughout the fetal meta-organs. The bell shaped nuclei appear to be appended to, rather than enclosed by, a large, balloon shaped, Feulgen fluorescent, mucinous cytoplasm. They are rarely observed scattered as single cells within adult epithelia but are plentiful in preneoplastic lesions (colon) and neoplastic lesions of many organs. Feulgen DNA image cytometry reveals that bell shaped nuclei increase by either of two forms of symmetrical amitoses concomitant with DNA doubling without chromosomal condensation. Furthermore, cells with bell-shaped nuclei undergo asymmetric amitoses to create a diverse set of cells with at least nine distinguishable nuclear morphotypes including those of the fetal and tumor parenchymal cells that subsequently increased in number by mitoses [1, 2, 3].
Because of marked differences between these previously unreported nuclear forms and eukaryotic nuclei in size, shape, DNA synthesis and segregation these cells have been denominated metakaryotes [1, 2]. The cytological evidence of asymmetric nuclear fissions creating specific nuclear morphotypes of cells that subsequently divide by mitosis and comprise the fetal/juvenile tissue or tumor parenchyma supports the hypothesis that cells with bell shaped nuclei comprise a stem cell lineage in organogenesis and carcinogenesis. The metakaryotic stem cell lineage is observed in fetal tissues derived from all three primordial layers of the human embryonic blastula: ectoderm, mesoderm and ectoderm [3]. Nearly identical metakaryotic forms populate fetal mouse and rat tissues as well as the developing plant Arabidopsis suggesting an evolutionary origin prior to the separation of plants and animals [3]. The distribution of the number of point mutant colonies as a function of size in adult human lungs indicates that the stem cell lineage of fetal/juvenile growth and development is mutator or hypermutable in phenotype and that maintenance stem cells of adult tissues have much lower mutation rates [4]. These observations support the long-standing hypothesis that tumor initiation mutations are restricted to stem cells of the fetal/juvenile period [5–8).
Herein we report unexpected findings about the pangenomic organization of chromatids in metakryotic interphase nuclei in syncytia and mononuclear cells in developing human tissues and tumors.
MATERIALS AND METHODS
1. Samples
All samples were obtained under a protocol approved in advance by the MIT Committee on the Use of Humans as Experimental Subjects and the IRBs of each contributing hospital. Anonymous surgical discards were fixed immediately upon surgical removal in Carnoy’s solution and stored under refrigeration in 70% ethanol [1, 3]. Fetal tissues of hindgut (5–7 wks gestation) and of spinal cord (9 –10 wks) were used for the cytometric analyses reported here. Tumor samples were obtained from adults of the organs indicated.
2.Slide preparation
~2 mm3 tissue samples were macerated for spreading on slides in two ways:
Samples were rinsed in distilled water, placed in 1N HCl at 60° C for precisely 8 minutes, rinsed in distilled water, placed in 45% acetic acid at room temperature for 15 minutes and then applied to slides for spreading [1]
Samples were cut into ~1 mm3 pieces, placed in 1 ml tubes filled with collagenase II (10 U/µl), and digested 30 minutes on a shaker at 37°C. The resulting macerated sample was twice spun down and washed in distilled water and then applied to slides for spreading [3].
Samples were spread in 100 µl of 45% acetic acid at room temperature. Slides were frozen on dry ice; cover slips were removed and slides were air dried at room temperature [1].
3. Histochemical staining
Feulgen purple staining of DNA
Dry slides were stained in a Schiff’s reagent for 1 hour, rinsed twice in 2 × SSC, stained in 1% Giemsa solution in Sörenssen buffer for 5 minutes, rinsed in Sörenssen buffer, then in distilled water and air dried. Dry slides were placed in xylene for 3 hours and cover slips glued with DePex mounting media.
4. Fluorescence in Situ Hybridization
Pan-telomeric PNA probe, pan-centromeric and whole chromosome #18 DNA probes were applied as previously described [9].
Telomeric PNA probe
Hybridization mixture (20 µl) containing 70% deionized formamide, 1% blocking reagent (Boehringer, Mannheim, Germany) in 10 mM Tris (pH 7.2) and 6 ng of Cy-3-labeled telomeric PNA probe (Cy-3- (CCCTAA)3 synthesized by Perspective Biosystems, Framingham, MA, USA) was added to each slide and mounted under a 24×50 mm cover slip. DNA was denatured by 3 min at 80° C followed by a hybridization period of 3 h at room temperature. After removal of the cover slips, the slides were washed twice for 15 min with 70% formamide/10 mM Tris (pH 7.2) followed by Tris washing for 5 min with 50 mM Tris (pH 7.50/0.15 M NaCl/0.05% Tween-20). The slides were dehydrated with an ethanol series and air-dried.
Chromosome and pan-centromeric probes
Whole chromosome-specific and centromeric painting probes (Cambio, UK) were applied together (3 µl each). For each slide 3 µl FITC-labeled pan-telomeric probe was also added and mixed with 17 µl hybridization buffer (Cambio, UK). Probes were denaturated separately, whole chromosome-specific 18 probe by incubation at 65° C for 10 min followed by incubation at 37° C for 60 min; pan-centromeric probe by incubation at 85°C for 10 min.The slides were hybridized at 37° C overnight.
For post-hybridization washes slides were preincubated with 2×SSC until cover slips could be removed then washed twice with 65% formamide in 1 × SSC at 37°C, followed by three washes in 1 × SSC at 37°C, 5 min each. Washing buffer (4 × SSC/0.05% Tween-20) was mixed with blocking protein (15%) and used to dilute antibodies as follows: F1 rabbit anti-FITC (1:200), F2 FITC goat anti-rabbit IgG (1:100) (Cambio, UK).
For immunofluorescence detection of FITC-labeled probes, slides were incubated with 100 µl of each of the antibody solutions at 37°C for 25 min, followed by 3 × 5 min washes in buffer and dehydrated through an ethanol series. Slides were embedded with 25 µl Vectashield mounting medium (Vector Laboratories, Burlingame, USA) containing 0.15 µg/ml DAPI counterstain.
5. Microscopy and image analysis
Feulgen DNA image cytometry
The KS 400 program supporting the Axioscop 2 MOT microscope imaging system (Carl Zeiss, Germany) to estimate the DNA content of each individual metakaryotic nucleus relative to the DNA content of the human eukaryotic genome observed on the same slide as mitotic figures and interphase nuclei.
Telomere and centromere counting
Telomeric and centromeric fluorescent signals were counted by combining 2D images of a series of focal planes. 3D images were also observed and analyzed using the Apo Tome capability of the imaging system (Carl Zeiss, Germany) [1].
RESULTS
Genomic organization of the metakaryotic interphase nucleus
Metakaryotic bell shaped nuclei were found to carry a normal human diploid DNA content by quantitative Feulgen cytometry consistent with 46 chromatids and were considered to be cells in “interphase”, the period between nuclear fissions and, for metakaryotes, the concomitant period of DNA doubling [1–3] Applications of FISH probes specific for human centromeric and telomeric sequences to both eukaryotic and metakaryotic nuclei from human fetal organs were expected to display 46 centromeric and 92 telomeric staining foci. In fetal organ eukaryotic cells in mitosis the expected number of centromeres and telomeres were observed in early prophase (Fig. 1a) and metaphase (Fig. 1b,c), the latter revealing 46 centromeric and ~184 telomeric staining foci. In tumors eukaryotic mitoses displayed a variable DNA content, centromeric and telomeric numbers but the numbers of centromeric and telomeric staining regions were greater, not less, than those expected for diploid mitoses.
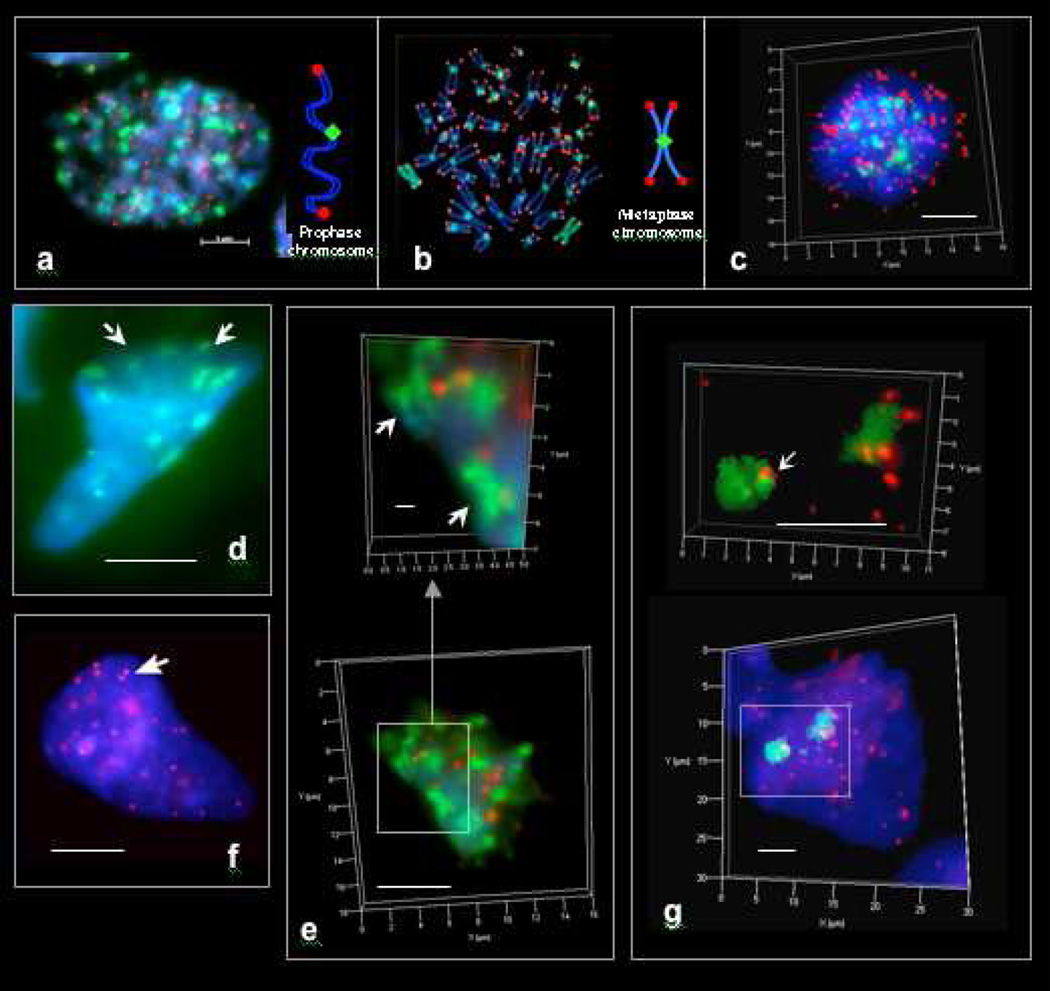
Figure 1. Pan-centromeric (FITC, green) and pan-telomeric (Cy3, red) FISH staining regions in cells and syncytia of early human fetuses and tumors.
a. Fetal mitotic (eukaryotic) nucleus (colon, 12 wks) in early prophase. b. Cultured human mitotic (eukaryotic) lymphoblastoid cell nucleus (TK6) in metaphase. c. Metaphase of fetal colonic mitotic cell as seen in 3D imaging. 46 centromeric and 92 telomeric staining regions appear in eukaryotic prophases. 46 centromeric and ~ 184 telomeric staining regions appear in eukaryotic metaphases. d. Pan-centromeric FISH in an extra-syncytial bell shaped metakaryotic nucleus of human fetal colonic epithelium, 12 weeks. Centromeric staining regions sum to about 23 per nucleus and appear to be paired (arrowed). e. Bell shaped metakaryotic nucleus in human colon adenocarcinoma presented in 3D. Note doublets of centromeres (green) and telomeres (red). f. Pan-telomeric FISH in an extra-syncytial bell shaped metakaryotic nucleus in human colonic epithelium, 12 weeks. Telomeric staining regions summed to about 23 in extra-syncytial nuclei and many appeared to be doublets (arrow). g. Pan-telomeric (red) and whole chromosome #18 (FITC, green) FISH in a syncytial bell shaped nucleus in human spinal cord ganglia, 9 weeks. Note two separate staining masses. Bar scale, 5 µm.
In contrast extra-syncytial bell shaped nuclei of fetal tissues that populate fetal proto-organs from the beginning of the second trimester through maturity [3] displayed only ~23 staining foci using either centromeric or telomeric FISH or antibody probes (Fig. 1d,e). Most bell shaped syncytial nuclei found in first trimester anlagen formation showed about 23 centromeric and telomeric foci but many showed about 46 telomeric foci. Use of a Zeiss 'Apotome' microscope permitted observation of centromeric and telomeric foci in three dimensions facilitating enumeration by reducing error due to coincidence of staining regions (overlap) when viewed in two dimensions. Images using both centromeric and telomeric probes revealed each focus to consist of a doublet as would be expected for chromatids homologously paired at centromeres and telomeres (Fig. 1d,f). However, the finding of only ~23 telomeric foci would require a hypothesis not only of homologous pairing, which would reduce the expected telomeric foci to 46, but a means of physically associating telomeric regions in groups of four to create the observed 23 telomeric foci.
Similar doublet associations of centromeric and telomeric FISH-staining regions were observed in tumor metakaryotic nuclei but with greater clumping of centromeric and telomeric foci (Fig. 1e).
The process of sorting out the three dimensional positions of each maternal and paternal chromatid copy has begun: FISH probes for two chromosomes, #6 (not shown) and #18 (Fig. 1g) in interphase metakaryotic nuclei found two physically separated masses for each chromosome. But whether these are separations of maternal/paternal genomes or of short and long chromatid arms joined at centromeres and telomeres has not yet been determined.
DISCUSSION
Genomic organization of the metakaryotic interphase nucleus
Human-specific pancentromeric and pantelomeric FISH probes hybridized to targets within the bell-shaped nuclei (Fig. 1) of free and syncytial cells of early fetal tissue and tumors. En passant such images demonstrate the important point that these cellular forms are indeed human and not some non-human symbiont peculiar to fetal/tumor tissue as multiple reviewers have suggested. Controls demonstrating that bell-shaped nuclei are not artifacts of fixation, staining maceration or spreading steps have been previously addressed, a principle control being the demonstration of the bell shaped nuclei in tumor samples in snap-frozen thin sections and human adenocarcinomaderived cell culture (HT-29) [3].
The interphase genomic organization of the open-mouthed, amitotic metakaryotic nuclei is distinct from that of the closed, mitotic eukaryotic nuclei (Figure 1a). Extra-syncytial bell shaped nuclei of free mononuclear cells contained 23 human centromeric and 23 telomeric FISH staining foci. The number 23 is consistent with a simple interphase model of homologous chromosomal pairing at centromeres and telomeres with end-to-end telomeric joining within or among chromosomes or both. This level of organization was independently observed at MIT (ANG, EVG) and the University of Leiden (JNF, FD) on multiple samples derived from fetal tissues and tumors. Search of the cytogenetic literature has failed to discover a previous report of any cell type with centromeric and telomeric pairing and all telomeric sequences apparently involved in end-to-end junctions. Taken together with the quantitative cytometric evidence of amitotic fission [2, 3] of the bell shaped nuclei, DNA doubling occurring during and soon after nuclear fission [3] and images of clear asymmetric amitotic fissions creating the mitotic eukaryotic forms of various tissue parenchyma [1–3], these data establish these putative stem cells as “metakaryotic” organisms with modes of genomic organization, DNA segregation and replication distinct from eukaryotic cells.
Homologous chromosomal pairing characteristic of meiosis has also been found in adult insect parenchymal cell interphase eukaryotic nuclei; paired insect chromosomes also demonstrated ring structures suggestive of intra-chromosomal telomeric linkage [10]. Spontaneous and x-ray induced ring chromosomes in pollen of Zea mays were recognized as structures that could undergo changes during plant development and account for clonal variegation suggestive of a change in a stem cell lineage [11]. End-to-end telomeric joinings are common observations in human tumor mitoses both as ring chromosomes and inter-chromosomal telomeric joinings [12]. These have been generally interpreted as metastable products of a random a breakage-fusion-bridge cycle in mitotic cells [13–15]. Ring chromosomes and end-joined chromosomes were used by Boveri as cytogenetic evidence of tumor monoclonality in mitotic metaphases of tumor parenchyma [16]. Insofar as metakaryotic cells appear to be the stem cells of monoclonal preneoplasia, neoplasia and their derived metastases, we suggest that such rings and end-junctions may be unresolved products of normal metakaryotic end-joined intermediates. Of course, chromosomal aberrations of all kinds have been induced in eukaryotic cell lines in vitro by x-rays and radiomimetic chemicals so our suggestion relates specifically to chromosomal variations that are found to be expanded as clones in organogenesis, carcinogenesis and other pathogeneses involving cell growth and differentiation.
There are really two separate sets of observations reported here that deserve reflection. The first are images of metakaryotic nuclei with 23 homologously paired chromatids all of which are involved in telomeric joinings of unknown structure.
The second are images of some metakaryotic interphase nuclei within the tubular syncytia that show 23 centromeric but ~46 telomeric staining areas raises the possibility that the telomeric junctions may open and shut. Between opening and shutting different specific telomeric junctions might form. We note that in the post-syncytial period (>12–14 wks) the mononuclear metakaryotic cells of various organs display genomes with 23 centromeric and 23 telomeric staining areas as if the syncytial period (~5–12 wks) specifically delineates developmental period in which such telomeric junction opening and closings occur. Images of metakaryotic mononuclear cells of tumors are more difficult to analyze but our impression to date is that nearly all, if not all, of their telomeres are involved in end-joining.
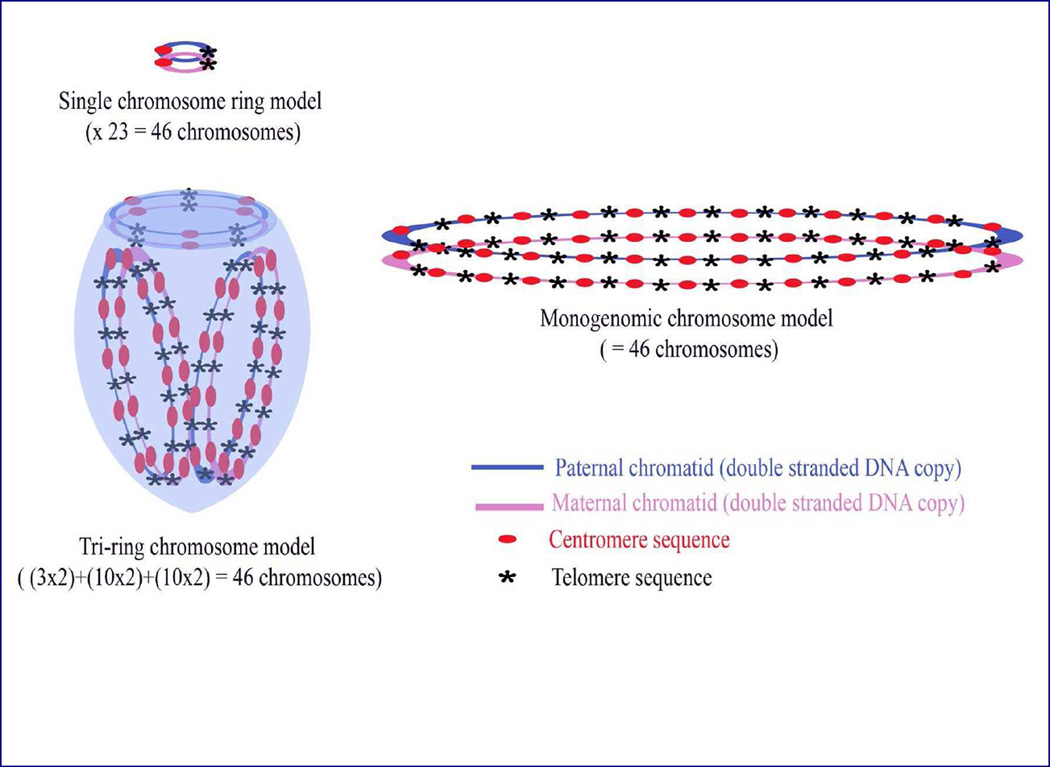
A human genome with 23 pairs of homologously paired chromatids (chromosomes) in which all telomeres are intra- or inter-chromosomally joined may be created by any of numerous possible arrangements. Several of these are illustrated in Figure 2. At the simplest level intrachromosomal telomeric joining of all homologously paired chromosomes could create a set of 23 paired ring chromosomes which would be observed as 23 centromeric and 23 telomeric staining regions.
Figure 2. Hypothetical possibilities of metakaryotic genome organization of homologously paired chromosomes with pangenomic telomere joining that result in the observed 23 doublets of centromeric staining and 23 doublets of telomeric staining.
This could be accomplished simply by a set of 23 single homologously paired chromatid rings (chromosomes) or a single pangenomic ring (chromosome) of homologously paired chromatids. However, a large number of permutations of paired chromatid order and combinations as oligo-chromosomal rings are possible, One such set is illustrated here as a tri-ring or chromosome model containing a ring with three specific homologously paired telomere-joined chromatids and two different rings each containing ten specific other chromatid pairs with joined telomeres, totaling 23 chromatid pairs.
Alternately, the paired chromosomes could be joined as any of a large series of single pangenomic rings and any of which would contain 23 centromeric and 23 telomeric staining regions.
One of us (WGT) has explored the potential of pair-wise telomeric junctions to code for the hypothetical “switches” that could govern the reproducible, non-random, step-wise transitions that are the hall mark of metazoan development and on a more limited scale of carcinogenesis. The potential for telomeric junctions to define a stepwise program lies in the huge number of pair wise joining sets represented by the specific joinings of 46 homologously paired telomeric ends reported herein. Consider the simple case of a genome organized as a single pangenomic ring as in Figure 2. Allowing interchromosomal junctions of any permuted order of homologous 23 chromatid pairs (in which the orientation of each chromatid pair telomeric junction may be p->q or q->p) yields 23! × 223 = 2.2× 1029 unique possibilities. This number is dwarfed by the number of all possible unique possibilities in the set of all possible ring chromosomes formed by all possible modes of pair-wise interchromosomal telomeric joining, i.e. rings of different sizes totaling 23 telomeric junctions among them.
As the adult human body contains about 3 × 1013 or ~245 cells and is created as a first approximation by binary fissions from a fertilized egg, approximately 246 unique cells constitute the cells of the developing and growing human body up to maturity. Were our development programmed in a binary code, any mode of 46 binary switches would be sufficient to create a unique “address” for each and every cell. It is clear that such “switches” could be represented by some small number of paired chromatid telomeric junctions in metakaryotic interphase stem cell nuclei. This is a testable hypothesis.
Pangenomic scans using chromosome-specific FISH can discover the nature and changes, if any, of the telomeric junctions in the metakaryotic syncytial and post-syncytial nuclei periods up to maturity in different organs [17]. Our collaborative group is seeking support for this study. If specific changes occur they can be mapped throughout the developmental arboration of organogenesis and the relationship of specific junctions to coordinate expression of specific genes defining the phenotype of each developmental state. Such changes of telomeric junctions among organs during their development would be consistent while a static unchanging set of telomeric junctions would invalidate the hypothesis that telomeric junctions constitute binary switches of a developmental program.
These observations and derived conjectures may be not unreasonably extended to the area of telomere “shortening” observed in parenchymal cells of aging humans and not in parenchymal cells of human tumors. Aging tissue maintenance stem cells are apparently eukaryotic and expose their telomeric ends with each mitotic cycle of programmed turnover. Metakaryotic stem cells in development and carcinogenesis are, however, continuously amitotic. Their end-joined telomeric regions might not be subject to attrition. If the end-joining were accomplished by continuous double stranded DNA sequences such paired chromosomes would not have “telomeres” at all.
If metakaryotes are the stem cells of fetal/juvenile organs and tumors the phenomenon of telomeric shortening might be restricted to maintenance, eukaryotic, stem cells in a tissue that commences with the last metakaryotic stem cell doubling defining maturation. Aging of tissues in terms of eukaryotic maintenance stem cell doublings could represent telomere attrition in mitotic cells while preneoplastic and neoplastic stem cells
Conclusions
The characteristics peculiar to metakaryotic cells that differentiate them from eukaryotic cells now include large, open-mouthed, bell shaped nuclei, homologous centromeric/telomeric pairing, pangenomic end-to-end telomeric association, DNA increase concomitant with genomic segregation, and symmetrical and asymmetrical amitotic nuclear fissions, the latter of which give rise to eukaryotic parenchymal cells of fetal/juvenile tissues and their derived tumors. The metakaryotic nuclear morphology and processes of genomic segregation and replication are clearly different from those found in mitotic eukaryotic cellular forms of yeast, insect and mammalian cells. The possibility that programmed telomeric end pairing is used in the machinery of stem cell differentiation is mathematically enticing. The positions of specific chromosomes in interphase and amitosis as well as the extent of homologous pairing along chromosomes may now be revealed by an orderly FISH-based investigation. Whether this particular hypothesis is sustained by further study or not, it would appear prudent to more deeply explore these bizarre, metakaryotic, forms of life in order to better understand the processes of human development and carcinogenesis.
ACKNOWLEDGMENTS
This work was supported primarily by personal funds (EVG, WGT) and internal funds of the Leiden University Medical Centre (JNF, FD). ANG was supported principally as a Doctoral Trainee of the National Institute of Environmental Health Sciences. Two of us (EVG, WGT) are recipients of a grant to study metakaryotic phenomena in carcinogenesis from United Therapeutics, Inc., Silver Spring MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gostjeva EV, Zukerberg L, Chung D, Thilly WG. Bell-shaped nuclei dividing by symmetrical and asymmetrical nuclear fission have qualities of stem cells in human colonic embryogenesis and carcinogenesis. Cancer Genet and Cytogenet. 2006;164:16–24. doi: 10.1016/j.cancergencyto.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Gostjeva EV, Thilly WG. Stem cell stages and the origins of colon cancer: a multidisciplinary perspective. Stem Cell Rev. 2005;1:243–251. doi: 10.1385/SCR:1:3:243. [DOI] [PubMed] [Google Scholar]
- 3.Gostjeva EV, Tomita-Mitchell A, Mitchell ME, Varmuza S, Fomina JN, Darroudi F, Thilly WG. Metakaryotic stem cell lineages in organogenesis of humans and other metazoans. Organogen. 2009;5:1–10. doi: 10.4161/org.5.4.9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudo H, Li-Sucholeiki X-C, Marcelino LA, Gruhl AN, Herrero-Jimenez P, Zarbl H, et al. Fetal-juvenile origins of point mutations in the adult human tracheal-bronchial epithelium: Absence of detectable effects of age, gender or smoking status. Mutat Res. 2008;646:25–40. doi: 10.1016/j.mrfmmm.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Thilly WG. Looking ahead: Algebraic thinking about genetics, cell kinetics and cancer. In: Bartsch H, Hemminiki K, O'Neill IK, editors. Methods for Detecting DNA Damaging Agents in Humans: Applications in Cancer Epidemiology and Prevention, IARC Monograph No. 89. IARC Scientif Publ; 1988. pp. 486–492. [PubMed] [Google Scholar]
- 6.Moolgavkar SH. Biologically motivated two-stage model for cancer risk assessment. Toxicol Lett. 1988;43:139–150. doi: 10.1016/0378-4274(88)90025-2. [DOI] [PubMed] [Google Scholar]
- 7.Thilly WG. What actually causes cancer? Technology Review. 1991 Mar-Apr;:48–54. [Google Scholar]
- 8.Meza R, Luebeck EG, Moolgavkar SH. Gestational mutations and carcinogenesis. Math Biosci. 2005;197:188–210. doi: 10.1016/j.mbs.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Fomina J, Darroudi F, Boei JJ, Natarajan AT. Discrimination between complete and incomplete chromosome exchanges in X-irradiated human lymphocytes using FISH with pan-centromeric and chromosome specific DNA probes in combination with telomeric PNA probe. Int J Radiat Biol. 2000;76:807–813. doi: 10.1080/09553000050028968. [DOI] [PubMed] [Google Scholar]
- 10.Diaz G, Lewis KR. Interphase chromosome arrangement in Anopheles atroparvus. Chromosoma. 1975;52:27–35. doi: 10.1007/BF00285786. [DOI] [PubMed] [Google Scholar]
- 11.McClintock B. A Correlation of Ring-Shaped Chromosomes with Variegation in Zea Mays. Proc Natl Acad Sci (USA) 1932;18:677–681. doi: 10.1073/pnas.18.12.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadamori N, Han T, Kakati S, Sandberg AA. Chromosomes and causation of human cancer and leukemia. LI. A hairy cell leukemia case with 14q+ and ring chromosomes: significance of ring chromosomes in blood disorders. Cancer Genet Cytogenet. 1983;10:67–77. doi: 10.1016/0165-4608(83)90107-3. [DOI] [PubMed] [Google Scholar]
- 13.McClintock B. The fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphase. Missouri Univ Agr Exp Sta Bull. 1938;290:1–48. [Google Scholar]
- 14.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–76. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faberge’ AC. Relation between Chromatid-Type and Chromosome-Type Breakage-Fusion-Bridge Cycles in Maize Endosperm. Genetics. 1958;43:737–749. doi: 10.1093/genetics/43.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boveri T. The origin of malignant tumors. 1914 [English translation by M.Boveri Baillière, Tindall& Cox 1929] [Google Scholar]
- 17.Darroudi F, Bergs JW, Bezrookove V, Buist MR, Stalpers LJ, Franken NA. PCC and COBRA-FISH a new tool to characterize primary cervical carcinomas: To assess hallmarks and stage specificity. Cancer Letters. 2010;287:67–74. doi: 10.1016/j.canlet.2009.05.034. [DOI] [PubMed] [Google Scholar]