A glucosyltransferase forms geranyl and neryl glucosides during grape ripening.
Abstract
Terpenoids represent one of the major classes of natural products and serve different biological functions. In grape (Vitis vinifera), a large fraction of these compounds is present as nonvolatile terpene glycosides. We have extracted putative glycosyltransferase (GT) sequences from the grape genome database that show similarity to Arabidopsis (Arabidopsis thaliana) GTs whose encoded proteins glucosylate a diversity of terpenes. Spatial and temporal expression levels of the potential VvGT genes were determined in five different grapevine varieties. Heterologous expression and biochemical assays of candidate genes led to the identification of a UDP-glucose:monoterpenol β-d-glucosyltransferase (VvGT7). The VvGT7 gene was expressed in various tissues in accordance with monoterpenyl glucoside accumulation in grape cultivars. Twelve allelic VvGT7 genes were isolated from five cultivars, and their encoded proteins were biochemically analyzed. They varied in substrate preference and catalytic activity. Three amino acids, which corresponded to none of the determinants previously identified for other plant GTs, were found to be important for enzymatic catalysis. Site-specific mutagenesis along with the analysis of allelic proteins also revealed amino acids that impact catalytic activity and substrate tolerance. These results demonstrate that VvGT7 may contribute to the production of geranyl and neryl glucoside during grape ripening.
Terpenoids, a class of secondary metabolites, are involved in interactions between plants and insect herbivores or pollinators and are implicated in general defense and stress responses (Gershenzon and Dudareva, 2007). The C10 and C15 members of this family were also found to affect the flavor profiles of most fruits and the scent of flowers at varying levels (Lund and Bohlmann, 2006; Schwab et al., 2008). The biosynthetic pathway and molecular genetics of terpenoids have been intensively and widely studied in different species (Loza-Tavera, 1999; Trapp and Croteau, 2001; Mahmoud and Croteau, 2002; Dudareva et al., 2004, Pichersky et al., 2006). Recent analyses of the grapevine (Vitis vinifera) genome (Jaillon et al., 2007; Velasco et al., 2007; Martin et al., 2010) revealed a large family of terpenoid synthases, many of which have been shown to produce fragrant monoterpenols (Lücker et al., 2004; Martin and Bohlmann, 2004; Martin et al., 2010, 2012).
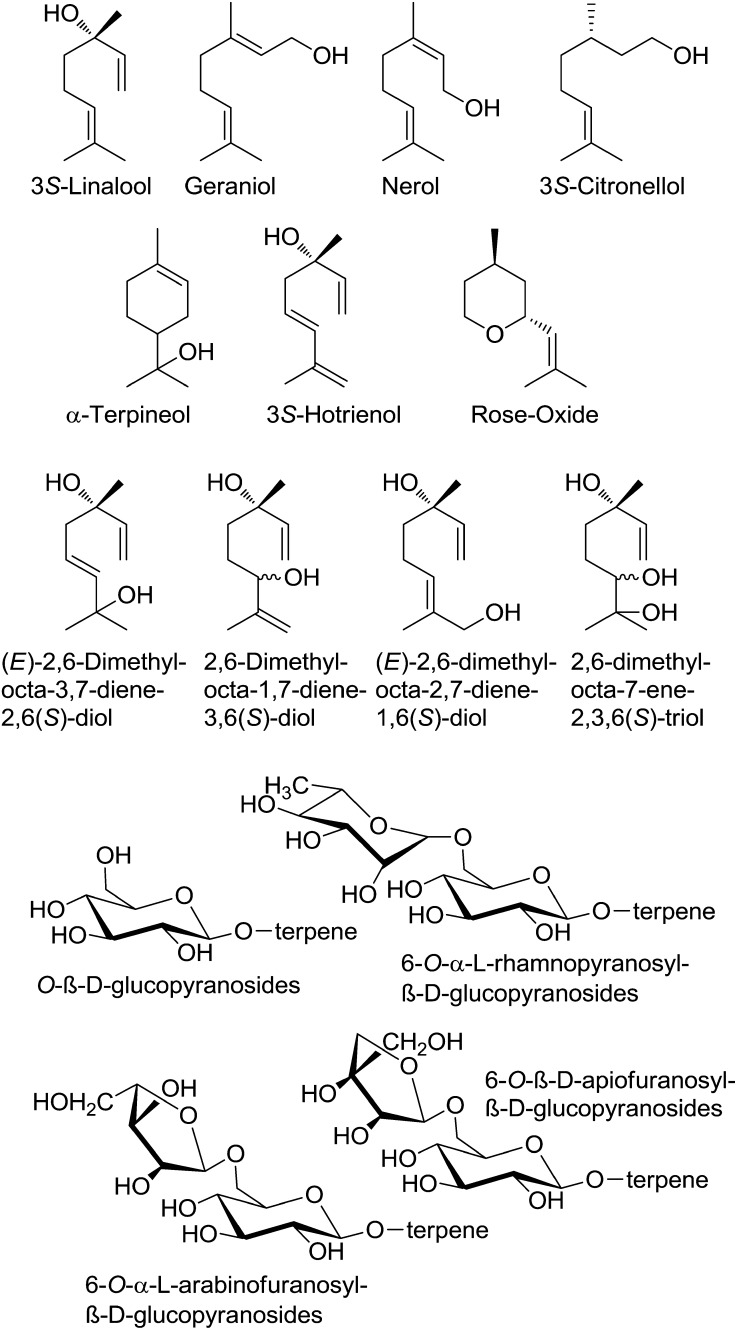
Monoterpenes are one major class of positive aroma compounds in traditional grapevine varieties, as they impart floral and citrus notes to some white wines (Mateo and Jiménez, 2000; Ebeler, 2001). The quality and quantity of terpenes vary strongly among grapevine varieties and even show differences between lines (clones) of the same variety. Berries of grapes such as cv Muscat of Alexandria and cv Gewurztraminer contain various and large amounts of C10 terpenoids, which are essential for the typical cv Muscat-like aroma. The characteristic aroma of cv Muscat, the most terpene-laden of all grapevine varieties, is primarily determined by a combination of just three terpenoid alcohols: geraniol, 3S-linalool, and nerol. Geraniol is considered to be the most crucial (Fig. 1). The same terpenes are also essential to the varietal aroma of cv Riesling, in addition to which α-terpineol, 3S-citronellol, and 3S-hotrienol are deemed equally important (Bayrak, 1994; Luan et al., 2005). Among the odoriferous monoterpenes, the cyclic ether rose oxide is a potent odorant in cv Scheurebe and cv Gewurztraminer (Guth, 1996, 1997). Terpenes are found mainly in the exocarp of grape berries, and the concentration of many terpenes accumulates as the grape ripens (Martin et al., 2012). The absolute levels of terpenes are highly affected by environmental conditions. This makes a systematic search for clones with preferable phenotypes difficult (Rivoal et al., 2010).
Figure 1.
Structural formulae of selected monoterpenes and their glycosylated conjugates found in grapes and wines.
In the early 1980s, it was discovered that most terpenes found in aromatic grapevine varieties are oxidized to polyhydroxylated derivatives or chemically bound to sugars in the form of O-β-d-glucopyranosides, 6-O-α-l-rhamnopyranosyl-β-d-glucopyranosides, 6-O-α-l-arabinofuranosyl-β-d-glucopyranosides, and 6-O-β-d-apiofuranosyl-β-d-glucopyranosides, making them odorless (Fig. 1; Williams et al., 1982). Glycosylation is one of the most widespread modifications of plant secondary metabolites, and merely 17% to 23% of the terpenes found in cv Riesling grapes are present in their free (unglycosylated) form (Gunata et al., 1985b; Razungles et al., 1993; Maicas and Mateo, 2005). The temporal accumulation of monoterpenol glycosides has been investigated in two different grapevine varieties (Luan et al., 2005, 2006a, 2006b). The high proportion of bound monoterpenes is considered as a wine’s “hidden aromatic potential” (Gunata et al., 1985a), since only free terpenes contribute to the aroma of a wine. Some of the bound terpenes are converted to their free form by either acid or enzymatic hydrolysis, usually both, during must fermentation and wine maturation. However, not only positive aroma contributors are released but also musty-smelling phenols are liberated that also occur as glycosides (Rocha et al., 2004).
Glycosides are formed by the action of glycosyltransferases (GTs), which are a ubiquitous group of enzymes that catalyze the transfer of a sugar moiety from an activated sugar donor, usually UDP-Glc, to acceptor molecules (Ross et al., 2001; Bowles et al., 2006). Sugar conjugation results in increased stability and water solubility. It has also been considered to control the compartmentalization of metabolites (Zhao et al., 2011). Over the past few years, the completion of several plant genome sequencing projects has unraveled unsuspected complexity within modifying enzymes such as GTs. These enzymes are encoded by large multigene families comprising several hundred genes. Thousands of GTs (http://www.cazy.org/) have been proposed and classified into 91 families to date (Coutinho et al., 2003). GTs in class 1, which recognize small-molecule scaffolds, are encoded by 120, 165, and 210 genes in Arabidopsis (Arabidopsis thaliana), Medicago truncatula (Gachon et al., 2005), and grapevine, respectively (http://genomes.cribi.unipd.it/grape/). The encoded proteins that contain a conserved plant secondary product GT motif toward the N terminus belong to the GT family 1 of the classification and typically use UDP-α-d-Glc as sugar donor (UGT). Heterologous expression of all Arabidopsis secondary metabolism UGTs (AtUGTs) in Escherichia coli and in vitro testing of the substrate specificity led to the identification of enzymes capable of catalyzing key glycosylation reactions (Lim et al., 2001, 2002; Paquette et al., 2003). It has also been shown that 27 AtUGTs glycosylate a diversity of monoterpenes, sesquiterpenes, and diterpenes such as geraniol, linalool, terpineol, and citronellol (Caputi et al., 2008). Although only linaloyl glucoside has been tentatively identified in Arabidopsis (Aharoni et al., 2003), this plant seems to have the capacity to glycosylate a variety of terpenes, comprising monoterpenols that are key flavor compounds in grape.
Using protein extracts prepared from leaves and berries of grape cultivars, it has been shown that glucosyltransferase activities can be detected against a wide range of substrates (Ford and Høj, 1998). Several classes of phenylpropanoids, including flavonols, anthocyanidins, flavanones, flavones, isoflavones, a stilbene, simple phenols, and monoterpenols, were among the substrates glucosylated. Total soluble leaf proteins separated into fractions with differing glucosyltransferase activities. Furthermore, it was demonstrated that glycosylation proceeds with a clearly detectable enantiodiscrimination for linalool and its hydroxylated derivatives in cv Morio-Muscat and cv Muscat Ottonel using enantioselective gas chromatography (GC)-mass spectrometry (MS; Luan et al., 2004). These findings show that monoterpene glycosylation in grape has a big impact on the resulting wine flavor. However, to date, only an anthocyanidine 3-O-glucosyltransferase (VvGT1; Ford et al., 1998) and 5-O-glucosyltransferase (VvB12H3_5-GT; Jánváry et al., 2009), a resveratrol/hydroxyl cinnamic acid O-glucosyltransferase from Concord grape (Vitis labrusca [VlRSGT]; Hall and DeLuca 2007), three phenolic acid O-glucosyltransferases (Vitis vinifera galloyl glucosyl transferase1 [VvgGT1]–VvgGT3; Khater et al., 2012), a flavonol 3-O-glucuronosyltransferase (VvGT5), and a bifunctional 3-O-glucosyltransferase/galactosyltransferase (VvGT6; Ono et al., 2010) have been cloned from grapes and functionally characterized. The substrate specificities of their encoded proteins are restricted to phenolics. On the other hand, recombinant GT proteins encoded by genes from Arabidopsis, Eucalyptus perriniana, Rauvolfia serpentine, Sorghum bicolor, and M. truncatula are multisubstrate enzymes that glucosylate monoterpenols, although with much lower efficiency than their assumed in planta substrates (Hefner et al., 2002; Hansen et al., 2003; Nagashima et al., 2004; He et al., 2006; Caputi et al., 2008).
Studies on the improvement of grape aroma are mainly focused on the enhancement of monoterpene biosynthesis. However, high levels of terpene glycosides (up to 80% of total terpenes) show that the reduction of terpene metabolism toward aroma-inactive glycosides could be an alternative approach, as higher levels of aroma-active terpenes would be present.
The now publicly available genome database of the grapevine variety cv Pinot Noir provides the opportunity to systematically identify monoterpenol GT genes in Vitis spp. (Jaillon et al., 2007; Velasco et al., 2007). In this study, we used 27 GT sequences of Arabidopsis that encode putative terpene alcohol GTs (Caputi et al., 2008) as a basis for the functional characterization of similar genes in the Vitis spp. genome. Metabolite profiling analyses in combination with gene expression analyses in different varieties and biochemical characterization of recombinant proteins resulted in the identification of a nerol/geraniol glucosyltransferase. Detection of functional and nonfunctional allelic forms of the enzyme followed by site-directed mutagenesis led to the identification of three amino acid residues that affect GT activity.
RESULTS
In Silico Analysis
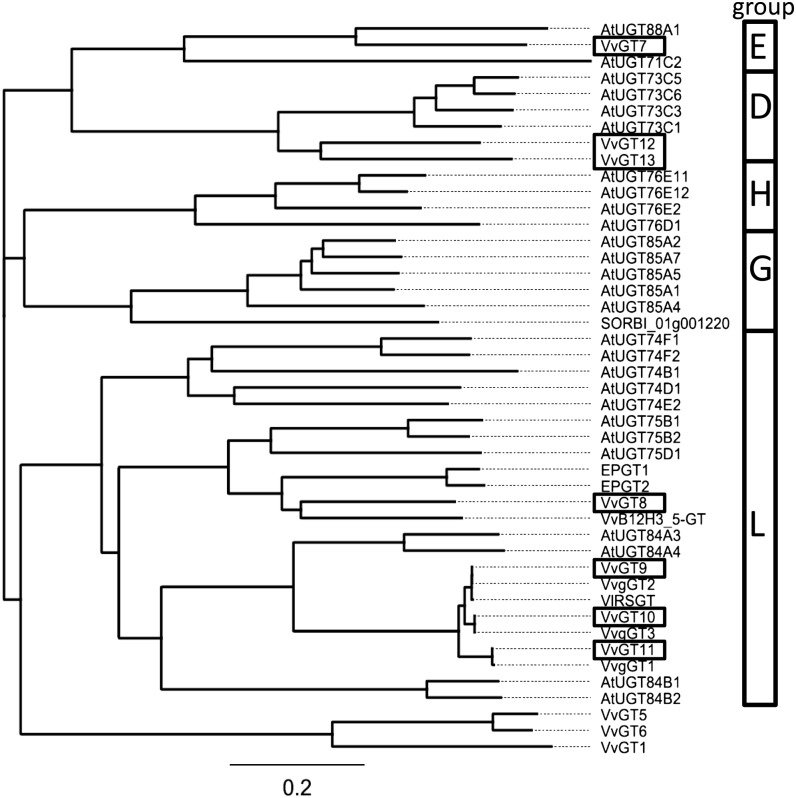
To understand the metabolism of terpene alcohols, we analyzed the GT family of grape for specific terpene GTs. Recently, a platform of 107 AtUGTs, comprising the multigene family of small molecule GTs, was screened against model terpenoid acceptors. Twenty-seven of the recombinant UGTs were shown to glycosylate monoterpene, sesquiterpene, and diterpene alcohols, such as geraniol, α-terpineol, linalool, and citronellol (Caputi et al., 2008). We performed BLASTX searches of the 27 GT sequences from Arabidopsis against the cv Pinot Noir genome database (Jaillon et al., 2007; Velasco et al., 2007). The first six hits of every search were chosen for further examination. Out of 162 candidate genes, 67 unique entries were obtained (Supplemental Table S1). These nucleotide sequences represent putative terpene GT genes of grape (VvGT). The vast numbers of potential candidates correspond with the multitude of terpene glycosides found in different grapevine varieties (Mateo and Jiménez, 2000). A phylogenetic tree was constructed based on the 67 unique sequences and divided the genes into five subgroups (Supplemental Fig. S1). Seven sequences, namely VvGT7 (CAO49526.1.pro; XP_002276546; XM_002276510), VvGT8 (CAN67608.1.pro; XP_002262883; XM_002262847), VvGT9 (CAN78291.1.pro; XP_002285379; XM_002285343), VvGT10 (CAN71972.1.pro; XP_002285412; XM_002285376), VvGT11 (CAN59771.1.pro; XP_002274256; XM_002274220), VvGT12 (CAN83016.1.pro; XP_002265326; XM_002265290), and VvGT13 (CAO62987.1.pro; XP_002265216; XM_002265180), were chosen for further analysis because of their high sequence similarity to terpene GTs (Caputi et al., 2008). A phylogenetic tree constructed from protein sequences with known GT activity for terpenes from Arabidopsis, E. perriniana, and S. bicolor and biochemically characterized proteins from Vitis species showed that the selected sequences sort into groups D, E, and L of the GT classification (Fig. 2). Comparison of the seven VvGT sequences with the physical map of the cv Pinot Noir genotype showed the locations of the genes on four linkage groups (LG3, LG5, LG8, and LG16; Supplemental Fig. S2). VvGT7 and VvGT8 are located together with VvGT7-like and VvGT8-like (96% and 94% nucleotide identity to VvGT7 and VvGT8, respectively) in the same orientation on LG16 and LG5, respectively. They are surrounded by a cluster of further GT-like genes. VvGT9 to VvGT11 can be found on LG3, whereas VvGT12 and VvGT13 occur in a cluster of UDP-GT genes on LG8.
Figure 2.
Phylogenetic tree of GT protein sequences. Protein sequences are from Arabidopsis (AtUGT), E. perriniana (EPGT1 and EPGT2), and S. bicolor (SORBI) with known glucosyltransferase activity toward terpenes and biochemically characterized proteins from Vitis spp. (Vitis vinifera [Vv] and Vitis labrusca [Vl]). VvGT7 to VvGT13 were investigated in this study. For methods used to construct the phylogeny, see “Materials and Methods.” GT subgroup assignments are shown in the boxes.
Expression Analysis
Gene expression analyses of the seven putative GT genes were performed by GeXP profiling in up to five cultivars in different tissues and at different sampling dates to narrow down the number of candidate genes (Supplemental Figs. S3–S6). The method was first validated by comparison of the transcript levels obtained by quantitative real-time PCR and GeXP for VvGT7 in different tissues of cv Gewurztraminer 11-18 Gm. Similar data were provided by both methods except in leaves, which showed slightly lower values, when analyzed by GeXP (Supplemental Fig. S3; Supplemental Table S2). This is in accordance with other studies on the reliability of the GeXP method (Rai et al., 2009; Drew et al., 2011).
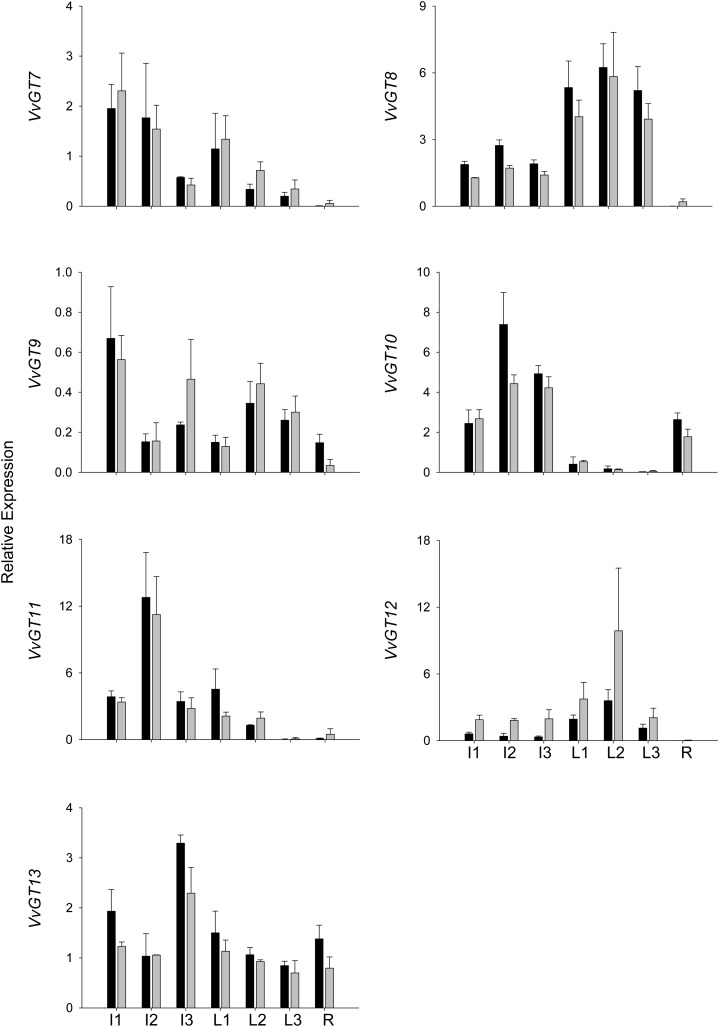
The expression levels of the putative GTs varied strongly between vegetative tissues (for primer selection, see Supplemental Table S3) and sampling dates. Only in the case of VvGT12 did they vary considerably between varieties (Figs. 3 and 4). VvGT8 and VvGT12 showed their highest transcript levels in leaves, whereas VvGT7, VvGT10, VvGT11, and VvGT13 were mainly expressed in inflorescence. GT mRNAs were barely found in roots, except for VvGT10, which showed very low expression levels in leaves of different ages.
Figure 3.
Gene expression analysis of VvGTs by GeXP in nonberry tissues. The relative expression was quantified in cv Gewurztraminer 11-18 Gm (black bars) and cv White Riesling 239-34 Gm (gray bars). Sampled tissues were as follows: inflorescences 4 weeks (I1) and 2 weeks (I2) before flowering and at full bloom (I3); leaves at approximate ages of 1 week (L1), 3 weeks (L2), and 5 weeks (L3); and roots (R). Mean values and se of three independent experiments (biological replicates) are shown.
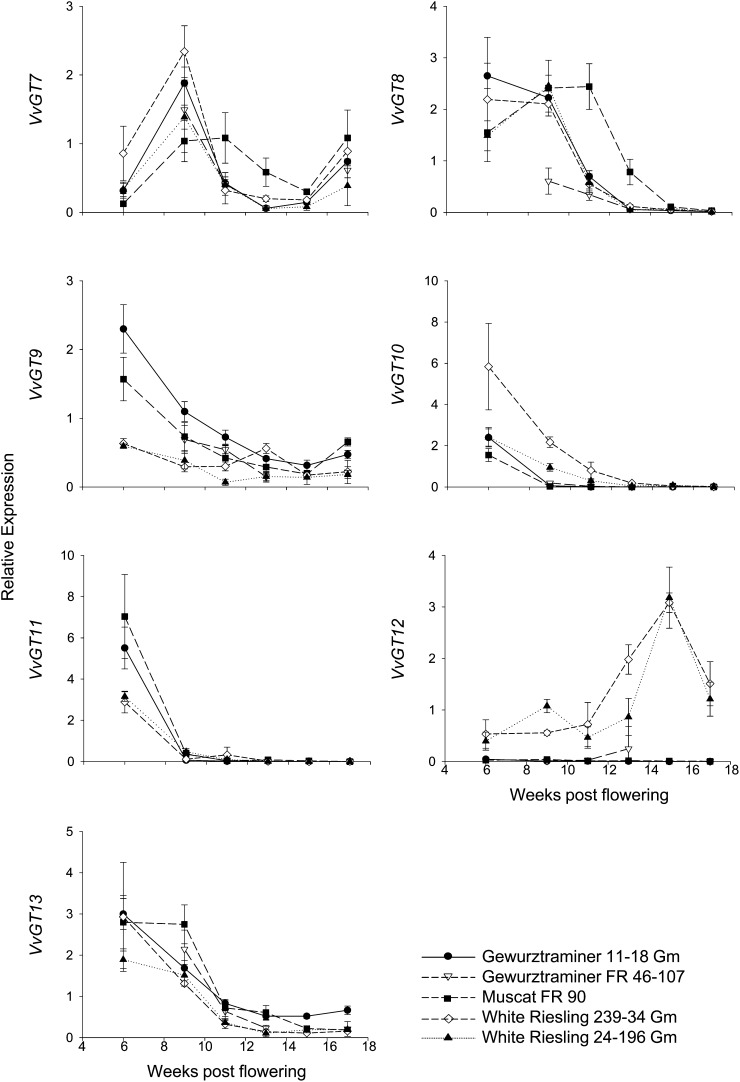
Figure 4.
Gene expression analysis of VvGTs by GeXP. Different stages of berry development are given as weeks after flowering. Expression was determined in berry exocarp of five different varieties and clones. Mean values and se of three independent experiments (biological replicates) are shown. Cultivars are as follows: black circles, cv Gewurztraminer 11-18 Gm; white triangles, cv Gewurztraminer FR 46-107; black squares, cv Muscat FR 90; white diamonds, cv White Riesling 239-34 Gm; and black triangles, cv White Riesling 24-196 Gm.
The putative GT genes showed pronounced changes in mRNA levels during berry development in all five cultivars (Fig. 4). VvGT8, VvGT9, VvGT10, VvGT11, and VvGT13 are highly transcribed in the exocarp of young green berries up to 9 weeks after flowering. In later stages of development, transcript levels declined and stabilized at a low level. Profiles of VvGT7 and VvGT12 deviated from this pattern. VvGT7 showed a two-phase expression pattern, reaching a maximum level of transcripts in berry exocarp at the second sampling date, 9 weeks after flowering, and then declining during the following month to reach a second peak at complete ripeness. VvGT12 transcripts increased substantially in berries of cv Riesling during ripening until the levels collapsed to one-half during the last 2 weeks before harvest time. Although VvGT12 and VvGT13 cluster in the same GT subgroup (Fig. 2), they showed completely different transcription patterns in the tissues analyzed (Figs. 3 and 4). Expression levels of VvGT12 differed considerably in berries of different cultivars. Transcript levels were approximately 100 times higher in berry exocarp of cv Riesling compared with cv Gewurztraminer and cv Muscat a Petits Grains Blancs (Fig. 4). In summary, the expression levels of VvGT8, VvGT9, VvGT10, VvGT11, and VvGT13 gradually decreased during grape berry development, while VvGT7 and VvGT12 transcript levels rose at later ripening stages.
Metabolite Analysis
Metabolite analysis was performed to correlate candidate gene expression with the levels of monoterpenyl glucosides in different tissues of cv White Riesling 239-34 Gm and cv Gewurztraminer 11-18 Gm and during berry development of all five analyzed cultivars. Free monoterpenes as well as glycosylated monoterpenes were isolated from different tissues (inflorescence, leaf, root, and exocarp) and subsequently separated by solid-phase extraction (Gunata et al., 1988; Mateo and Jiménez, 2000). Grape exocarp was chosen because it accumulates high levels of terpene metabolites (Luan et al., 2004). GC-MS was employed for the detection and quantification of volatile monoterpenes. Also, a stable isotope dilution analysis method was developed for the quantitative determination of nonvolatile monoterpenyl glucosides by HPLC-tandem mass spectrometry (MS/MS; Supplemental Fig. S7). For this purpose, isotopically labeled internal standards were chemically synthesized.
The grape tissues differed not only in the amount of total terpenes but also in their terpene profiles at different stages of development (Table I). The free-to-glycosidically bound monoterpene ratio varied remarkably between the different tissues. While leaves generally accumulated similar levels of free and bound monoterpenes, the amount of monoterpenyl glucosides prevailed in inflorescences and roots. In inflorescences of cv White Riesling 239-34 Gm and cv Gewurztraminer 11-18 Gm, linalool concentrations were lower than geraniol concentrations; on the other hand, levels of linaloyl β-d-glucoside were higher than geranyl β-d-glucoside levels. This implicates the expression of a linalool-specific GT in inflorescence.
Table I. Amounts of free monoterpenes and monoterpenyl β-d-glucosides in different grapevine tissues of cv White Riesling 239-34 Gm and cv Gewurztraminer 11-18 Gm.
Plant material was prepared and analyzed as described in “Materials and Methods.” Except for roots, a continuous sampling at fixed time intervals took place. Plant materials are as follows: inflorescence 1, 4 weeks before flowering; inflorescence 2, 2 weeks before flowering; inflorescence 3, week of full bloom; leaf 1, 1 week old; leaf 2, 3 weeks old; and leaf 3, 5 weeks old. n.d., Not detectable. Amounts are listed in mg kg−1 plant material ( n = 2).
| Cultivar | Substrate | Free or Bound | Inflorescence 1 | Inflorescence 2 | Inflorescence 3 | Leaf 1 | Leaf 2 | Leaf 3 | Root |
|---|---|---|---|---|---|---|---|---|---|
| White Riesling 239-34 Gm | Linalool | Free | 0.17 ± 0.06 | n.d. | n.d. | 0.02 ± 0.04 | n.d. | n.d. | n.d. |
| β-d-Glucosides | 4.78 ± 1.64 | 3.18 ± 0.63 | 7.91 ± 4.09 | 1.41 ± 1.10 | 0.45 ± 0.31 | 1.14 ± 0.71 | n.d. | ||
| Nerol | Free | n.d. | n.d. | n.d. | 0.04 ± 0.06 | n.d. | n.d. | n.d. | |
| β-d-Glucosides | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| Geraniol | Free | 2.93 ± 0.28 | 2.07 ± 0.62 | 1.79 ± 0.39 | 3.57 ± 0.92 | 2.42 ± 0.29 | 0.74 ± 0.32 | 0.57 ± 0.19 | |
| β-d-Glucosides | 2.28 ± 0.93 | 2.00 ± 0.56 | 5.32 ± 1.86 | 3.01 ± 1.30 | 1.30 ± 0.11 | 2.42 ± 2.60 | 1.52 ± 0.68 | ||
| Citronellol | Free | 0.39 ± 0.20 | 0.42 ± 0.21 | n.d. | n.d. | n.d. | n.d. | 0.27 ± 0.02 | |
| β-d-Glucosides | 0.06 ± 0.09 | n.d. | n.d. | 0.28 ± 0.20 | n.d. | 0.19 ± 0.10 | n.d. | ||
| Total | Free | 3.49 | 2.49 | 1.79 | 3.63 | 2.42 | 0.74 | 0.84 | |
| β-d-Glucosides | 7.12 | 5.18 | 13.23 | 4.70 | 1.75 | 3.75 | 1.52 | ||
| Gewurztraminer 11-18 Gm | Linalool | Free | 0.32 ± 0.01 | 0.28 ± 0.04 | 0.44 ± 0.03 | 0.28 ± 0.02 | n.d. | n.d. | n.d. |
| β-d-Glucosides | 10.74 ± 1.13 | 10.43 ± 0.04 | 106.25 ± 45.92 | 2.60 ± 1.52 | 0.98 ± 0.13 | 0.67 ± 0.07 | n.d. | ||
| Nerol | Free | 0.66 ± 0.38 | 0.28 ± 0.09 | n.d. | n.d. | n.d. | n.d. | n.d. | |
| β-d-Glucosides | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| Geraniol | Free | 20.63 ± 12.63 | 7.7 ± 2.4 | 9.33 ± 0.57 | 8.07 ± 0.18 | 3.12 ± 0.55 | 2.06 ± 0.16 | 1.20 ± 0.19 | |
| β-d-Glucosides | 7.90 ± 1.52 | 5.46 ± 0.90 | 94.22 ± 16.03 | 1.95 ± 1.23 | 2.67 ± 2.60 | 0.47 ± 0.10 | 8.26 ± 3.62 | ||
| Citronellol | Free | 1.46 ± 0.85 | 1.32 ± 0.41 | n.d. | n.d. | n.d. | 0.08 ± 0.13 | 0.18 ± 0.01 | |
| β-d-Glucosides | 1.00 ± 0.20 | 0.75 ± 0.1 | 1.27 ± 1.79 | 0.17 ± 0.08 | n.d. | n.d. | 0.96 ± 1.36 | ||
| Total | Free | 23.07 | 9.58 | 9.77 | 8.35 | 3.12 | 2.14 | 1.38 | |
| β-d-Glucosides | 19.64 | 16.64 | 201.74 | 4.72 | 3.65 | 1.14 | 9.22 |
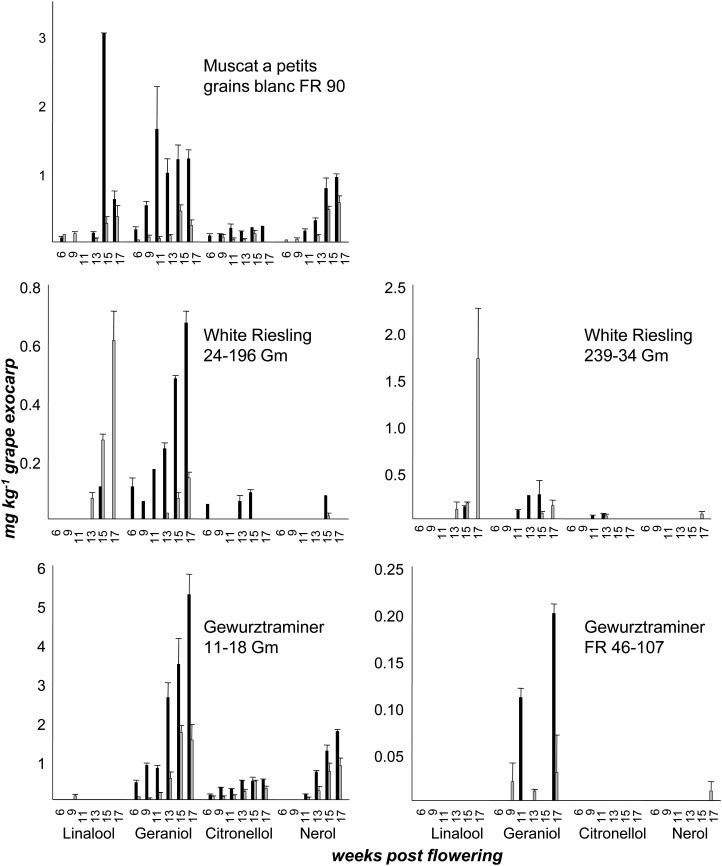
Berry exocarp was analyzed during grape development of five cultivars (Fig. 5; Supplemental Fig. S6; Supplemental Table S4). In exocarp of cv Gewurztraminer FR 46-107, probably a clone with impaired monoterpene biosynthesis, free and bound monoterpenes were hardly detected (less than 0.25 mg kg−1 berry exocarp). The cv Muscat a Petits Grains Blancs FR 90 and cv Gewurztraminer 11-18 Gm exocarp (Fig. 5) showed a diversified terpene spectrum at every stage of ripening, with high levels of geraniol, citronellol, and nerol derivatives (up to 5.5 mg kg−1 berry exocarp), while in both cv Riesling clones, small amounts of the metabolites were mainly observed at the end of the ripening process. Levels of free and bound monoterpenes increased during grape ripening in berry exocarp of all cultivars investigated. The cv Gewurztraminer 11-18 Gm accumulated the highest amounts of free and bound monoterpenes; however, levels of linalool and linaloyl β-d-glucoside rarely exceeded the limit of detection (0.1 mg kg−1 grape exocarp; Fig. 5). Geraniol and geranyl β-d-glucoside were the predominant terpene metabolites during the ending phase of ripening of all varieties except for cv White Riesling. Different ratios of free to bound forms of the individual monoterpenes were observed (Fig. 5). This observation points to differential preferences of the involved GTs for their monoterpene substrates.
Figure 5.
Amounts of free monoterpenes and monoterpenyl β-d-glucosides in grape exocarp of different cultivars during ripening. Note that graphs have different scales. Grape exocarp was peeled and extracted. Free (black bars) and glycosidically bound (gray bars) monoterpenes were isolated by solid-phase extraction. Free monoterpenes were measured by GC-MS and monoterpenyl β-d-glucosides by HPLC-MS/MS (n = 2).
Gene Sequencing and Allelic Variation
VvGT7 and VvGT12 were selected for further analysis due to their high transcript levels in grape berries at the end of the ripening process (Fig. 4). Nucleotide sequences were amplified from five cultivars (cv White Riesling 239-34 Gm and 24-196 Gm, cv Gewurztraminer FR 46-107 and 11-18 Gm, and cv Muscat a Petits Grains Blancs FR 90) using gene-specific primers, designed on the basis of predicted UGT mRNAs from the reference genome of PN40024 (Jaillon et al., 2007; accession nos. XM_002276510 and XM_002265290, respectively; Supplemental Table S5). PCR products were cloned, and a minimum of six plasmids were sequenced for each gene and cultivar. Six alleles of VvGT12 were obtained. Although we cannot exclude the sequencing of paralogous genes, we use the term “alleles” throughout this article for simplification reasons (see “Discussion”). The three deduced amino acid sequences differed in residue substitutions at two sites and contained a four-amino acid insertion compared with the reference protein PN40024. The 12 alleles of VvGT7 translated into 10 protein sequences of 477 to 481 amino acids with a calculated molecular mass of 52 to 53 kD (Supplemental Fig. S8). The differences resulted from missense mutations at 19 sites and insertions of amino acids at four sites. From all cultivars except cv Gewurztraminer FR 46-107, more than two alleles of VvGT7 were isolated and translated into three protein sequences.
Heterologous Expression of VvGT7 Alleles and Enzymatic Activity
The 10 alleles of VvGT7 (a–j) and three VvGT12 alleles were cloned in the expression vector pGEX-4T-1. The recombinant enzymes were expressed with an N-terminal glutathione S-transferase (GST) tag, affinity purified, and verified by SDS-PAGE and western blot using GST-specific antibody (Supplemental Fig. S9). Enzyme activity studies were performed with UDP-[14C]Glc and various putative substrates (terpenols, flavonoids, and different monoalcohols). The alcohols were selected because they have been identified as aglycones of glycosides in grape (Sefton et al., 1994; Baek and Cadwallader, 1999; Mateo and Jiménez, 2000; Wirth et al., 2001). The VvGT12 proteins exhibited no activity toward any of the substrates and were not further analyzed. Alternative UDP sugars were not tested. In contrast, eight VvGT7 alleles (a–h) encoded catalytically active GT proteins that displayed distinct substrate specificities (Table II; Supplemental Fig. S10). The active VvGT7 proteins preferred nerol as the main substrate but also efficiently converted citronellol, geraniol, eugenol, and benzyl alcohol. Additionally, they were also able to glucosylate the flavonol quercetin, several short-chain monoalcohols (hexanol, octanol, cis-3-hexenol and trans-2-hexenol, 3-methyl-3-butenol, and 3-methyl-2-butenol), and 8-hydroxylinalool as well as terpineol, farnesol, and mandelonitrile, albeit with lower efficiency (Table II). No activity was found with linalool and anthocyanidins (pelargonidin and cyanidin). The formation of terpenyl glucosides was confirmed by HPLC-MS analysis in comparison with chemically synthesized reference material (Fig. 6). The retention times and fragmentation pattern of the reference material and VvGT7 products were identical and in accordance with the proposed fragmentation mechanism (Supplemental Fig. S7). Additionally, the glucosides of nerol, citronellol, and geraniol were visualized by radioactivity-thin-layer chromatography (radio-TLC; Supplemental Fig. S11). The extracted radioactivity of the enzyme assays consisted exclusively of the monoterpenyl β-d-glucosides. Diglucosides were not detected by radio-TLC analysis. Except for a very low activity of VvGT7i and VvGT7j toward eugenol, nerol, citronellol, benzyl alcohol, farnesol, and 3-methyl-2-butenol, the two allelic enzymes VvGT7i and VvGT7j were unable to glucosylate the tested substrates (Table II).
Table II. Relative enzymatic activities of 10 allelic VvGT7 proteins from grape toward putative substrates as determined by radiochemical analysis with UDP-[14C]Glc.
The relative activities (%) refer to the highest level of extractable radioactivity that was measured for VvGT7g with the substrate nerol (100%). Values greater than 5% are shown in different fonts: 5%–10%, italic; 11%–50%, bold; 51%–80%, bold italic; and 81%–100%, underscored). For se, see Supplemental Figure S10.
| Substrate | VvGT7a | VvGT7b | VvGT7c | VvGT7d | VvGT7e | VvGT7f | VvGT7g | VvGT7h | VvGT7i | VvGT7j | Empty Vector |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Citronellol | 48 | 67 | 73 | 71 | 71 | 33 | 45 | 10 | 2 | 0 | 0 |
| Geraniol | 27 | 51 | 60 | 37 | 32 | 19 | 32 | 6 | 0 | 0 | 0 |
| 8-Hydroxylinalool | 1 | 5 | 4 | 3 | 1 | 2 | 1 | 0 | 0 | 0 | 0 |
| Linalool | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nerol | 70 | 99 | 91 | 90 | 76 | 72 | 100 | 54 | 4 | 0 | 0 |
| Terpineol | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 0 |
| Cyanidin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Kaempferol | 1 | 4 | 3 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 1 |
| Pelargonidin | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Quercetin | 4 | 17 | 17 | 11 | 6 | 14 | 9 | 1 | 0 | 0 | 0 |
| Benzyl alcohol | 15 | 21 | 23 | 11 | 22 | 9 | 19 | 2 | 1 | 1 | 0 |
| Phenylethanol | 4 | 5 | 2 | 5 | 5 | 3 | 20 | 1 | 0 | 0 | 0 |
| Eugenol | 16 | 38 | 46 | 43 | 40 | 26 | 40 | 23 | 10 | 0 | 0 |
| Farnesol | 2 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 1 | 0 |
| Hexanol | 5 | 2 | 1 | 5 | 4 | 4 | 6 | 3 | 0 | 0 | 0 |
| Octanol | 1 | 6 | 5 | 4 | 5 | 3 | 6 | 1 | 0 | 0 | 0 |
| Mandelonitrile | 3 | 2 | 0 | 3 | 2 | 3 | 4 | 1 | 0 | 0 | 0 |
| 3-Methyl-2-butenol | 3 | 5 | 4 | 4 | 6 | 4 | 6 | 1 | 1 | 1 | 0 |
| 3-Methyl-3-butenol | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | 0 | 0 |
| Cis-3-hexenol | 4 | 5 | 4 | 4 | 5 | 3 | 8 | 1 | 0 | 0 | 0 |
| Trans-2-hexenol | 8 | 8 | 8 | 7 | 8 | 5 | 11 | 4 | 0 | 0 | 0 |
Figure 6.
Detection of monoterpenyl β-d-glucosides as products of VvGT7 by HPLC-electrospray ionization-MS/MS. A, HPLC-MS/MS analysis of linaloyl-β-d-glucoside (peak 1), neryl-β-d-glucoside (peak 2), geranyl-β-d-glucoside (peak 3), and citronellyl-β-d-glucoside (peak 4) of 0.1 mg mL−1. B to D, Neryl-β-d-glucoside (B), citronellyl-β-d-glucoside (C), and geranyl-β-d-glucoside (D) produced by VvGT7. Traces display the total ion current of the characteristic transitions (see “Materials and Methods”). Gaussian smoothing was applied.
Biochemical Characterization of VvGT7a to VvGT7c
To determine the kinetic constants, the assay conditions were optimized for the conversion of nerol. The highest activity for the three alleles VvGT7a, VvGT7b, and VvGT7c was found in Tris-HCl buffer between pH 8 and 8.5 at 30°C to 37°C, and product formation was linear for at least 2 to 4 h, depending on the allelic form (Supplemental Fig. S12). Km and turnover number (kcat) values were obtained for nerol, citronellol, and geraniol with a constant UDP-Glc level (502 µm) and for UDP-Glc with a fixed nerol concentration (200 or 400 µm). The kinetic parameters were determined from hyperbolic Michaelis-Menten saturation curves and corroborated the preference of the three enzymes for the acceptor nerol. Whereas VvGT7b and VvGT7c showed almost identical Km and kcat values for the substrates nerol, citronellol, geraniol, and UDP-Glc (Table III), the kinetic constants of VvGT7a were significantly different compared with VvGT7b and VvGT7c and confirmed the lower activity of this allelic form (Table II).
Table III. Kinetic constants of VvGT7a, VvGT7b, and VvGT7c determined for the substrates nerol, citronellol, geraniol, and UDP-Glc.
| Allele | Substrate | Km | kcat | kcat/Km |
|---|---|---|---|---|
| mm | 10−3 s−1 | 10−3 s−1 mm−1 | ||
| VvGT7a | Nerol | 0.417 ± 0.004 | 0.4 | 1.0 |
| Citronellol | 0.433 ± 0.062 | 0.2 | 0.5 | |
| Geraniol | 0.464 ± 0.056 | 0.1 | 0.2 | |
| UDP-Glc | 0.052 ± 0.009 | 0.3 | 5.8 | |
| VvGT7b | Nerol | 0.204 ± 0.027 | 2.0 | 9.8 |
| Citronellol | 0.306 ± 0.075 | 0.2 | 0.7 | |
| Geraniol | 0.396 ± 0.012 | 0.2 | 0.5 | |
| UDP-Glc | 0.048 ± 0.001 | 1.2 | 25.0 | |
| VvGT7c | Nerol | 0.211 ± 0.016 | 1.4 | 6.6 |
| Citronellol | 0.445 ± 0.021 | 0.3 | 0.7 | |
| Geraniol | 0.321 ± 0.002 | 0.3 | 0.9 | |
| UDP-Glc | 0.056 ± 0.003 | 1.4 | 25.0 |
Site-Directed Mutagenesis of VvGT7i and VvGT7j
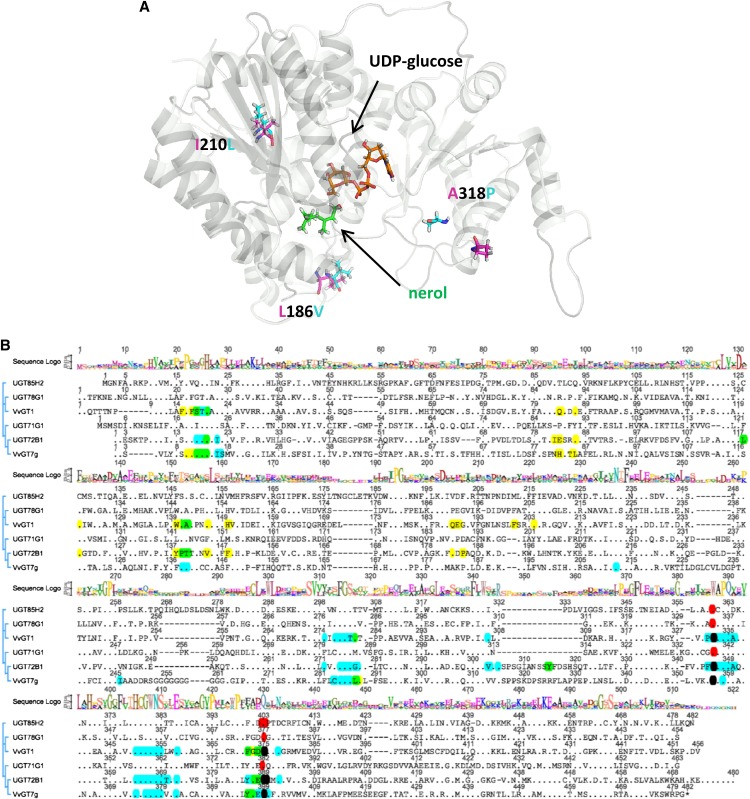
The protein sequence alignment of the 10 allelic forms of VvGT7 showed pairwise identities of more than 98%. Interestingly, the active allozymes (a–h) consistently differed from the inactive ones (i and j) in only three amino acids (L186V, I210L, and P318A; Supplemental Fig. S8). The structures of the five proteins were modeled by YASARA (Geoffrey Behrens, University of Greifswald; http://www.yasara.org/) to get an impression of the localization of the three amino acids (Fig. 7). The modeling was performed by using the protein data bank files (www.rcsb.org) of 2PQ6 (structure of UGT85H2), 2ACV (UGT71G1), 2ACW (UGT71G1 with UDP-Glc), and 3HBF (UGT78G1) as templates. The three-dimensional (3D) structure revealed that the three amino acids were not located near the active site of the enzymes. To restore the catalytic activity of VvGT7i and VvGT7j and to verify the importance of the three amino acids for the enzymatic activity, we performed site-directed mutagenesis. Initially, the amino acid Ala-318 of VvGT7i and VvGT7j was mutated to the corresponding amino acid Pro found in the active alleles. This residue is located at the entrance of the cleft, which leads to the active site of the enzyme (Wang, 2009). Due to the known effect of Pro on the protein structure, it was supposed that the mutation A318P could affect the activity of VvGT7i and VvGT7j. However, the replacement of the amino acid in VvGT7i and VvGT7j did not cause a significant change in the activity of the two allelic proteins (Table IV). Only a second mutation (V186L or L210I) yielded active mutants (VvGT7i-V186L-A318P, VvGT7i-L210I-A318P, VvGT7j-V186L-A318P, and VvGT7i-L210I-A318P; Table IV). Both double mutants of VvGT7j converted only nerol, while the mutants VvGT7i-V186L-A318P and VvGT7i-L210I-A318P glucosylated nerol, citronellol, geraniol, eugenol, and benzyl alcohol (Table IV). In the following step, the two triple mutants of VvGT7i (V186L, L210I, and A318P) and VvGT7j (V186L, L210I, and A318P) were created and the activity of the corresponding proteins was tested. Both triple mutants showed similar substrate tolerance (in this context, tolerance describes the property of the enzyme much better than specificity, as specificity relates to a very limited number of substrates) as the eight active alleles VvGT7a to VvGT7h, although they did not act upon the full range of substrates. The mutant VvGT7i-V186L-L210I-A318P converted nerol, citronellol, and geraniol as well as eugenol, benzyl alcohol, quercetin, hexanol, octanol, cis-3-hexenol, trans-2-hexenol, and terpineol (Table IV). No activity toward 8-hydroxylinalool, linalool, cyanidin, kaempferol, pelargonidin, phenylethanol, farnesol, mandelonitrile, 3-methyl-2-butenol, and 3-methyl-3-butenol was detected. The activity of VvGT7j-V186L-L210I-A318P was lower compared with VvGT7i-V186L-L210I-A317P. This triple mutant was active toward nerol, citronellol, kaempferol, quercetin, benzyl alcohol, eugenol, farnesol, and 3-methyl-2-butenol. Remarkably, geraniol was not accepted, and kaempferol was a better substrate than quercetin, which was not observed on the other VvGT7 proteins.
Figure 7.
Ribbon diagram of the calculated 3D structure of VvGT7b with UDP-Glc (orange) and nerol (green; A) and amino acid sequence alignment of VvGT7g and five plant GTs that have been crystallized and their 3D structures solved (B). UDP-Glc and nerol are shown as sticks and marked with arrows. β-Strands and α-helices are shown in gray. The residues at positions 186, 210, and 318, which differentiated the active from the inactive forms, are shown as sticks and are denominated. The alignment was performed using ClustalX. The plant GTs include M. truncatula UGT71G1 (protein data bank identification 2ACW), UGT78G1 (3HBF), and UGT85H2 (2PQ6), grape VvGT1 (2C1Z), and Arabidopsis UGT72B1 (2VCE). A consensus sequence logo is shown in the top traces, conserved amino acids are displayed as dots, and amino acids within 5 Å to the acceptor and sugar donor molecules (UDP-Glc) are marked in yellow and blue, respectively. Overlaps are shown in green (blue + yellow) and black (blue + red). The plant secondary glucosyltransferase box is confined by the amino acids marked in red.
Table IV. Relative activities of the generated mutants of VvGT7i and VvGT7j toward the tested substrates compared with VvGT7b, which showed the highest activity toward nerol (set to 100%).
The substrate screening was performed as described in “Materials and Methods.” Values greater than 5% are shown in different fonts: 5%–10%, italic; 11%–50%, bold; 51%–80%, bold italic; and 81%–100%, underscored).
| Substrate | VvGT7b | VvGT7i | VvGT7i-A318P | VvGT7i-L210I-A318P | VvGT7i-V186L-A318P | VvGT7i-V186L-L210I-A318P | VvGT7j | VvGT7j-A318P | VvGT7j-L210I-A318P | VvGT7j-V186L-A318P | VvGT7j-V186L-L210I-A318P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Citronellol | 67 | 2 | 1 | 14 | 13 | 49 | 0 | 0 | 0 | 0 | 20 |
| Geraniol | 51 | 0 | 0 | 4 | 4 | 20 | 0 | 0 | 0 | 0 | 0 |
| 8-Hydroxylinalool | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Linalool | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nerol | 100 | 4 | 0 | 23 | 22 | 79 | 0 | 0 | 5 | 6 | 28 |
| Terpineol | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Cyanidin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Kaempferol | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
| Pelargonidin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Quercetin | 18 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 8 |
| Benzyl alcohol | 21 | 1 | 4 | 4 | 4 | 10 | 0 | 6 | 0 | 0 | 9 |
| Phenylethanol | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eugenol | 38 | 10 | 6 | 10 | 12 | 13 | 0 | 0 | 0 | 0 | 13 |
| Farnesol | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 3 |
| Hexanol | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| Octanol | 6 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| Mandelonitrile | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3-Methyl-2-butenol | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 5 |
| 3-Methyl-3-butenol | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| Cis-3-Hexenol | 5 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 |
| Trans-2-Hexenol | 8 | 0 | 0 | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
Enantioselectivity of VvGT7
Grape berries accumulate S-citronellol in their exocarp, although in lower levels than nerol and geraniol (Fig. 5; Luan et al., 2005). To investigate the enantiomeric preference of VvGT7, racemic R,S-citronellol was used as substrate for VvGT7a, VvGT7b, and VvGT7c. Chiral phase GC-MS analysis of residual citronellol demonstrated the consumption of R-citronellol (Fig. 8B). Enantiomerically pure R-citronellol was released when citronellyl β-d-glucoside formed by VvGT7a, VvGT7b, and VvGT7c was hydrolyzed by acid treatment (Fig. 8C). In two control experiments, enantiomerically pure R- or S-citronellol was utilized with UDP-[14C]Glc as substrate. Only R-citronellol was converted to radiolabeled β-d-glucoside. Thus, VvGT7a, VvGT7b, and VvGT7c showed high chiral discrimination ability for racemic citronellol, as only the R-enantiomer was glucosylated.
Figure 8.
Enantioselectivity of VvGT7a determined by chiral phase GC-MS analysis of citronellol. A, A racemic mixture of R,S-citronellol was used as a substrate for VvGT7a. B, Residual citronellol is depleted in R-citronellol. C, R-Citronellol is released by acid-catalyzed hydrolysis from citronellyl β-d-glucoside. Signals labeled with x are by-products of the hydrolysis. D, Enantiomerically pure R-citronellol was used as reference material. Traces in A, C, and D were smoothed.
DISCUSSION
Putative Monoterpenyl GTs in Grape
The possible functions of monoterpenyl glucosides and the glycosylation of monoterpenes in plants are described in Supplemental Figure S13. In this study, seven putative GT genes (VvGT7–VvGT13) were selected from grape because of their high similarity with monoterpenol GT genes from Arabidopsis (Caputi et al., 2008, 2010). VvGT9, VvGT10, and VvGT11 were identical to VvgGT2, VvgGT3, and VvgGT1, respectively. They were recently characterized as phenolic acid O-glucosyltransferases (Fig. 2; Khater et al., 2012). Furthermore, VvGT9 has been described as VlRSGT of V. labrusca (Fig. 2; Hall and Deluca, 2007). Heterologous expression of VvGT9 to VvGT11 genes followed by enzyme activity assays confirmed the published data (data not shown). Since only VvGT7 and VvGT12 showed a ripening-related expression pattern in accordance with the accumulation of monoterpenyl glycosides in mature grape berries (Figs. 4 and 5), they were subjected to further analysis. Three allelic VvGT12 proteins were expressed in E. coli but showed no activity toward the target substrates. Eight out of 10 allelic VvGT7 proteins (12 different nucleotide sequences) displayed a broad substrate tolerance and catalyzed the glucosylation of terpenols, phenols, and short-chain alcohols (Table II). VvGT7 belongs to group E of the GT nomenclature (Fig. 2; Ross et al., 2001), which has expanded considerably during the evolution of higher plants (Caputi et al., 2012). The promiscuous monoterpenol GT proteins from E. perriniana and S. bicolor (Hansen et al., 2003; Nagashima et al., 2004) are members of groups L and G, respectively. Thus, comparative genomics can successfully be applied to identify putative related genes in diverse genomes. The results also reveal that monoterpenyl GTs can originate from different subgroups of the GT gene family in various genomes, as has been shown for flavonoids (Lim et al., 2004).
Monoterpenyl Glucoside Pattern and Expression Pattern of VvGT7 in Grape Cultivars
Monoterpenyl glucosides were quantified in different tissues of grape by a highly accurate stable isotope dilution analysis. Significant qualitative and quantitative differences of the terpene glucosides between the cultivars analyzed were observed in the different tissues (Table I; Fig. 5). As far as grape berry exocarp is concerned, the cultivars can be grouped into three classes. The first group, including cv Gewurztraminer 11-18 Gm and cv Muscat a Petits Grains Blancs FR 90, accumulated appreciable levels of terpenes in all ripening stages, whereas in both cv Riesling clones, the metabolites were mainly detected at the end of ripening (second group). This is in agreement with previously published data and reflects the maturation characteristics of the investigated cultivars (Williams et al., 1982; Wilson et al., 1984; Gunata et al., 1985b; Martin et al., 2012). The VvGT7 expression patterns (Fig. 4) were similar in all varieties analyzed and showed maxima at 9 to 11 weeks after flowering and at complete ripeness (17 weeks after flowering). It appears that VvGT7 has different functions during berry development. In berry exocarp of cv Gewurztraminer FR 46-107, probably a clone with impaired monoterpene biosynthesis, free and bound monoterpenes were scarcely detected (third group).
Bound and free monoterpenes were also analyzed in roots and inflorescences of the cv Gewurztraminer 11-18 Gm and cv White Riesling 239-34 Gm (Table I). The cv Gewurztraminer 11-18 Gm accumulated larger amounts of bound terpenes in roots than cv White Riesling clone 239-34 Gm. Roots are important organs for plant fitness and are responsible for the absorption of water and the storage of nutrients and other metabolites. However, in soil, the root is exposed to a multitude of herbivores and microbes. Monoterpenes might provide protection against these pathogens. The concentration of monoterpenes and phenols, like geraniol and eugenol, increased when roots were infested by phylloxera (Lawo et al., 2011). In the case of eugenol, the level in infested tissue was more than 10-fold higher than in uninfested root tips. However, VvGT7 appears to be not involved in the production of monoterpenyl glucosides in roots, as its expression level is low in this tissue.
Inflorescences produced high amounts of linaloyl and geranyl β-d-glucoside, with a sudden increase toward full bloom. However, free linalool was hardly detectable. Linalool is frequently encountered in floral scents, where it is formed by linalool synthase (Dudareva et al., 1996, 1999). In petals of grape, linalool appears to be efficiently glucosylated and metabolized to other metabolites, such as linalool oxides (Raguso and Pichersky, 1999). The expression of VvGT7 did not increase toward full bloom. In accordance with previous studies (Gunata et al., 1986), during leaf growth the amounts of free and bound monoterpenes decreased. The VvGT7 expression pattern followed the decrease of monoterpenyl glucoside levels in leaves.
VvGT7 seems not to be induced by free monoterpenes, as both cv Gewurztraminer clones share similar VvGT7 transcript profiles, although they differ completely in their monoterpene levels. The cv Muscat a Petits Grains Blancs FR 90 contains the two inactive VvGT7 allozymes (VvGT7i and VvGT7j) as well as one active allozyme (VvGT7a). Thus, one catalytically active VvGT7 allozyme is sufficient to account for the production of monoterpenyl glucosides, or additional GTs contribute to their formation.
Polymorphism
More than two alleles of VvGT7 were isolated from several cultivars of the diploid grape. In cv Muscat a Petits Grains Blancs FR 90, four VvGT7 alleles were amplified by proofreading polymerase. The nucleotide sequences translated into three different proteins, two of them (VvGT7i and VvGT7j) being almost inactive. The two inactive allozymes differed only by the insertion of one amino acid (Arg-443). The third allozyme, VvGT7a, showed a single amino acid exchange (G11A) to the cv Gewurztraminer-specific allozyme VvGT7g (Supplemental Table S6). Cloning and sequencing errors can be excluded as a source for the high polymorphism of VvGT7, as sequences were obtained from several cultivars or they contained single-nucleotide polymorphisms and insertions/deletions also present in other sequences. On the contrary, sequencing errors and nucleotide changes introduced during PCR are expected to be randomly distributed.
All sequenced alleles of VvGT7 align to a single locus on chromosome 16 in the genome of grape when applying BLASTN on the 12-genome assembly (http://genomes.cribi.unipd.it), which normally signifies that all alleles belong to a single gene. However, segmental duplications are very frequent in plant genomes, and recently tandem duplicated genes may be similar enough to be misinterpreted as one locus (Claros et al., 2012). The highly similar genes may be merged into the same sequence during genome assembly, even if Sanger technology with its long read was used. That is especially true for the polyploidized genome of grape, where three ancestral genomes contribute to the haploid content (Jaillon et al., 2007; Velasco et al., 2007; Giannuzzi et al., 2011). The estimated number of grape genes is about 30,000, but EST records showed that a minimum of about 40,000 genes are present in Vitis spp. (Grimplet et al., 2009). Thus, it is probable that the VvGT7 sequences belong to at least two genes.
An alternative explanation for the occurrence of more than two alleles of a single gene in a diploid species is the presence of intraorganismal genetic heterogeneity. Cell layers of different genotypes can coexist in grapevine, because somatic mutations can be fixed and transmitted to new individuals by vegetative propagation, resulting in periclinal chimeras. For microsatellites, more than two alleles for one locus have been frequently observed in grape as a result of the occurrence of periclinal chimerism (Franks et al., 2002; Riaz et al., 2002; Hocquigny et al., 2004; Bertsch et al., 2005).
Biochemical Analysis of VvGT7
VvGT7 is presumably a cytosolic protein due to the absence of a signal peptide, as predicted by SignalP 4.1 (Nielsen et al., 1997; Petersen et al., 2011; Caputi et al., 2012). The pH optimum for VvGT7 is 8 to 8.5, a value that is slightly higher than that observed in the cytosol of plant cells. The pH optimum observed for a partially purified l-menthol GT from Mentha × piperita is slightly lower (7.3–7.7; Martinkus and Croteau, 1981). Eight out of 10 identified VvGT7 allozymes (a–h) displayed a similar substrate spectrum, whereas two (i and j) were almost inactive. The allelic forms a to h preferably converted nerol followed by citronellol, geraniol, eugenol, benzyl alcohol, and quercetin (Table II). However, they can be distinguished by their activities and substrate preferences. While the allozyme VvGT7g shows highest selectivity and overall activity for nerol, it also displays a broad substrate tolerance with relatively high activity toward phenylethanol as well as trans-2-hexenol when compared with the other allozymes. That way, each allozyme forms its characteristic product spectrum. The kinetic data of VvGT7a to VvGT7c were determined for the acceptors nerol, geraniol, and citronellol and for UDP-Glc. The Km values of VvGT7 for these acceptors are similar to those of GTs from strawberry (Fragaria × ananassa; Lunkenbein et al., 2006; Landmann et al., 2007) and grape (Khater et al., 2012) acting on (hydroxyl)cinnamic acids and those of a promiscuous GT from S. bicolor (Hansen et al., 2003) for nerol, geraniol, and citronellol. Noteworthy, the Km value of a purified GT from M. piperita for l-menthol (monoterpenol) is five times lower and that for UDP-d-Glc is five times higher than the corresponding values of VvGT7 (Martinkus and Croteau, 1981). The kcat values for the conversion of nerol and UDP-Glc are in the same range as those of a sterol (a triterpene) converting GT from Withania somnifera (Madina et al., 2007) as well as the UGT71G1 (an isoflavonoid GT) from M. truncatula (He et al., 2006). However, the kcat values of VvGT7 for nerol, geraniol, and citronellol are 5- to 971-fold smaller than those of UGT85B1, a cyanohydrin glucosyltransferase with monoterpenol promiscuous activity (Hansen et al., 2003). The kcat/Km value (9.8 × 10−3 s−1 mm−1) is highest for nerol and comparable to the data (16 × 10−3 s−1 mm−1) obtained for 24-methylenecholesterol (a triterpene) for a sterol GT (Madina et al., 2007). This indicates that nerol and to a lesser extent geraniol and citronellol could serve as substrates for VvGT7 in planta.
The protein most similar to VvGT7 in Arabidopsis is UGT88A1. This protein reacts with citronellol, geraniol, and farnesol (Fig. 2; Caputi et al., 2008). It also glucosylated the flavonol quercetin at positions 3, 3′, 4′, and 7 (Lim et al., 2004), the coumarin esculetin at position 6, and scopoletin (Lim et al., 2003). However, quercetin was a minor substrate for VvGT7 (Table II), while esculetin and scopoletin have not been tested.
Comparative genomics can identify highly similar nucleotide sequences in a number of genomes. However, their encoded proteins can differ greatly in their substrate preference and biological function, as the results show. Thus, heterologous expression and biochemical testing of putative enzymes are still essential to elucidate the in planta roles of genes.
Structural Analysis of VvGT7 and Identification of Amino Acids Affecting Substrate Preference and Activity
Plant GTs show a horseshoe-like structure with N- and C-terminal domains connected directly via an interdomain linker. Additionally, two α-helices at the C terminus stabilize the conformation of the GTs (Jánváry et al., 2009; Wang, 2009). Two highly conserved amino acids (His and Asp) are observed in the active site of most plant GTs. They are assumed to be involved in the catalysis and are located at positions 14 and 116 in VvGT7 (Fig. 7; Supplemental Fig. S8). Although the eight catalytically active allelic VvGT7 proteins share high pairwise sequence identity of 98.4%, they can be clearly discriminated by their characteristic substrate spectrum (Table II). Thus, we generated the 3D structures of VvGT7a to VvGT7c by homology modeling (Fig. 7) and identified residues that account for the substrate selectivity and activity of the GT by pairwise comparison of the amino acid sequences (Supplemental Table S6).
The allelic VvGT7a and VvGT7g showed similar substrate preference except for nerol, eugenol, and phenylethanol, which were more efficiently transformed by VvGT7g. The comparison of the two protein sequences identified a single amino acid exchange, Gly to Ala, at position 11. This residue is close to His-14, which participates in the catalysis in GTs (Hans et al., 2004; Brazier-Hicks et al., 2007) and is part of the acceptor-binding pocket (Fig. 7B). Substitution of Ala-11 by Gly significantly enhanced the selectivity for nerol, eugenol, and phenylethanol, whereas the activity toward the other substrates remained almost untouched.
VvGT7d converted the alcohols with similar activities compared with VvGT7b and VvGT7c, except for geraniol (Table II). The sequence alignment showed that the allelic protein VvGT7d differed from VvGT7b in a single amino acid exchange (G2E), while in VvGT7c, 11 amino acids were changed. Glu-2 appears to selectively promote the catalysis of geraniol and is located at the N-terminal domain of VvGT7b and VvGT7c. This domain mainly binds the acceptor (Wang, 2009).
Similarly, VvGT7e and VvGT7c differed in a single amino acid (R28H), which resulted in altered enzymatic activities toward distinct substrates (geraniol, nerol, and quercetin), while the specificities for the other alcohols remained unchanged (Table II). Again, this residue is located in the N terminus involved in acceptor binding (Fig. 7B).
Interestingly, VvGT7h displayed significantly lower enzymatic activity and deviating substrate preference in comparison with the other catalytically active allelic proteins under the same screening conditions. VvGT7h was active toward nerol, citronellol, geraniol, and eugenol and showed weak activity with terpineol, quercetin, benzyl alcohol, phenylethanol, hexanol, octanol, mandelonitrile, 3-methyl-2-butenol, cis-3-hexenol, and trans-2-hexenol. The alignment of all 10 wild-type protein sequences showed that VvGT7h contains Gly at position 190, while the other nine alleles have Asp-190 (Supplemental Fig. S8). The mutation of G190D of VvGT7h would result in the sequence of VvGT7b. Sequence alignment of VvGT7g and crystallized GT proteins revealed that position 190 in VvGT7g falls into a region of amino acids (Fig. 7B, marked in yellow for UGT72B1 and VvGT1) that are in the vicinity (less than 5 Å) of the acceptor molecules.
Thus, residues in positions 2, 11, and 28 correlated with the activity for certain substrates and affected the substrate tolerance of VvGT7, whereas amino acids in position 190 impact the catalytic power of the entire protein.
Site-Directed Mutagenesis Identified Important Residues for Activity
We also identified two allelic forms, VvGT7i and VvGT7j, which were almost inactive. Only eugenol was glucosylated by VvGT7i to a minor extent. Comparison of all VvGT7 sequences revealed three amino acids that separated the active from the inactive forms (Supplemental Fig. S8). These residues are not part of the active site, as was illustrated by the calculated 3D structures of VvGT7i and VvGT7j (Fig. 7A). The residue Ala-318 of VvGT7i and VvGT7j is located at the entrance of the cleft leading to the active site and is replaced by Pro in the enzymatically active proteins. Sequence comparison with structurally characterized GTs showed that residue 318 is contained in an extra loop region that has also been identified in UGT72B1 (Fig. 7B). In the structure of UGT72B1, a GT that accepted phenols, this loop is partially disordered but extended to the top of the acceptor-binding pocket. Mutations (D312N and F315Y) in this loop installed high levels of N-GT activity in addition to O-GT activity (Brazier-Hicks et al., 2007). However, A318P exchange in VvGT7i and VvGT7j did not influence the activity of the enzymes (Table IV). In contrast, the double mutants VvGT7i-V186L-A318P and VvGT7i-L210I-A318P showed significant and similar activities for nerol, citronellol, and eugenol, whereas VvGT7j-V186L-A318P and VvGT7j-L210I-A318P converted only nerol to a minor extent. The exchange of the third amino acid in the triple mutants VvGT7i-PIL (for proline, isoleucine, leucine) and VvGT7j-PIL led to a further increase of the activity. Position 186 is located close to Asp-190, which has been identified as an essential amino acid during the comparison of the VvGT7 alleles, whereas position 210 is in the neighborhood of Asn-212, which is conserved in all GT proteins analyzed and found in the vicinity of the acceptor in the calculated 3D structure (Fig. 7B). VvGT7i-PIL showed similar activity for the tested substrates to VvGT7a and converted nerol, citronellol, geraniol, and eugenol. Although VvGT7j-PIL catalyzed the glucosylation of its substrates less efficiently than VvGT7i-PIL, it displayed a high preference for kaempferol, which was not observed for any of the other allelic forms. Comparison of the amino acid sequences of the triple mutants showed that VvGT7j-PIL has an insertion of an additional Arg at position 443. This residue is part of the two C-terminal α-helices that are situated at the opposite site of the cleft. It was assumed that the two helices help to stabilize the spatial structure of the cleft by fixing the N- and C-terminal domains (Jánváry et al., 2009). Since the active site is located within the cleft, the insertion of Arg-443 modifies the spatial arrangement of the enzyme’s catalytic center and thus its substrate preference.
The site-directed mutagenesis of VvGT7i and VvGT7j demonstrated that three residues that are not directly part of the active site have a remarkable effect on the reaction catalyzed by VvGT7, as they probably affect the overall shape of the protein and thus influence activity and selectivity. Furthermore, we observed a synergistic effect of the stepwise exchange of the three amino acids, which indicated that these residues (Leu-186, Ile-210, and Pro-318) are essential for the conversion of nerol, geraniol, citronellol, and eugenol.
Overall, we observed that amino acid exchanges in the N terminus (positions 2, 11, and 28) mainly shifted the substrate preference and tolerance of VvGT7, whereas exchanges in the C terminus (positions 186, 190, 210, 318, and 443) affected the total activity. This is in accordance with the fact that the nucleotide sugar donor mainly interacts with the C-terminal domain of the GT proteins, whereas the acceptor primarily binds to the N-terminal domain (Wang, 2009). It is noteworthy that the substrate preferences (Table II) and kinetics (Table III) of VvGT7b and VvGT7c were almost identical, although their sequences differed by 10 amino acids. This demonstrates some plasticity of the enzyme activity, as these replacements did not affect the performance of the catalytic protein.
In Planta Substrates of VvGT7
The catalytically active VvGT7 proteins glucosylate geraniol, nerol, and R-citronellol in vitro (Table II). Neryl β-d-glucoside was primarily detected in exocarp of mature berries and was not observed in nonberry tissue. In contrast, geranyl β-d-glucoside was present in all surveyed tissues at all developmental stages. Similarly, citronellyl β-d-glucoside was isolated from leaf, inflorescence, and berry exocarp. At the same time, all tissues expressed VvGT7 transcripts at varying levels, indicating that VvGT7 can play a role in the glucosylation of monoterpenols in vivo. However, free and glycosylated citronellol is usually S-configured in grape berries and showed high enantiomeric purities, with an enantiomeric excess greater than 90% (Luan et al., 2005). Since VvGT7 preferred R- over S-citronellol as substrate, it is obvious that VvGT7 is not involved in the production of S-citronellyl glucoside.
A correlation between the pattern of monoterpenyl glucoside accumulation and VvGT7 transcript levels is not obvious. In berry exocarp (Fig. 5), the amount of geranyl and neryl β-d-glucoside increased almost constantly from week 6 to week 17, whereas mRNA levels of VvGT7 reached two peaks during berry development (Fig. 4). This can be explained by the fact that GTs represent a large enzyme family frequently displaying broad substrate tolerance and overlapping enzymatic activities. Arabidopsis contains at least 27 GTs that each glucosylated more than one terpenoid in vitro (Caputi et al., 2008). Similarly, the VvGT7 enzymes glucosylated a variety of substrates. VvGT7 may have different functions at various stages of berry development as well as in other vegetative tissues. VvGT7 is probably only one of many terpene GTs in grapevine, as has been proposed earlier (Ford and Høj, 1998). Additional GTs may also contribute to the biosynthesis of neryl and geranyl β-d-glucoside. Moreover, monoterpenyl glucosides are only intermediates of a glycosylation pathway, as neryl and geranyl glucosides are decorated with additional sugars, thus forming diglycosides and triglycosides (Fig. 1; Williams et al., 1982). These were not quantified in this study, but their formation rate may affect the amount of the monoglucosides, independently of the VvGT7 activity. Nerol and geraniol availability can influence β-d-glucoside accumulation. Despite similar VvGT7 expression levels in grape berries of different cultivars (Fig. 4), the levels of free and bound geraniol and nerol varied significantly (Fig. 5). Possibly, VvGT7 may also be involved in the metabolism of eugenol, as the level of glycosidically bound eugenol increased during berry development (Fenoll et al., 2009). In addition to the above, translational control and protein half-life may explain the lack of direct correlation between monoterpenyl glucoside profiles and VvGT7 transcript levels.
CONCLUSION
The genome sequence of grape contains a number of putative GT genes. Those with the highest similarity to GTs of Arabidopsis, whose encoded proteins catalyze the formation of terpene glucosides in vitro, were selected as candidate genes. Expression analysis, metabolite profiling, and biochemical studies led to the identification of the first terpene GT of grape. The encoded enzyme glucosylates a range of alcohols, including the monoterpenols nerol, geraniol, and citronellol. Site-specific mutagenesis identified three amino acids that are located outside the active site but are essential for enzymatic activity. These results can be useful for breeding programs for the selection of genotypes with low terpene GT activities and presumably higher levels of aroma-active, free terpenols. They might also be a basis for the development of new biotechnological processes.
MATERIALS AND METHODS
Plant Material
Grape (Vitis vinifera) cv Gewurztraminer 11-18 Gm, cv Gewurztraminer FR 46-107, cv White Riesling 239-34 Gm, cv White Riesling 24-196 Gm, and cv Muscat a Petits Grains Blancs FR 90 were grown at the Institute of Grapevine Breeding of Geisenheim University during 2011 and 2012. Sampling of berries during development from pea size to complete ripeness (6 and 17 weeks after flowering) was conducted at six dates in 2011 (Supplemental Fig. S5). For terpenoid analysis, 250 g of berries was stored at −20°C. Of an additional subset of berries, exocarp had been peeled off and immediately frozen in liquid nitrogen for subsequent RNA extraction. Roots were obtained from ungrafted plants of cv White Riesling 239-34 Gm and cv Gewurztraminer 11-18 Gm grown in the greenhouse. From the same cultivars in the field, leaves were sampled at approximate ages of 1, 3, and 5 weeks (Supplemental Fig. S3). In addition, inflorescences were collected 4 and 2 weeks before flowering and at full bloom (Supplemental Fig. S4). All samples were stored at −20°C
Chemicals
Except where stated otherwise, all chemicals, solvents, and reference compounds were purchased from Sigma-Aldrich, Fluka, and Roth. UDP-[14C]Glc (300 mCi mmol−1, 0.1 mCi mL−1) was obtained from American Radiolabeled Compounds. (R,S)-3,7-Dimethyl-1,6-octadien-3-ol (linalool) and (E)-3,7-dimethyl-2,6-octadien-1-ol (geraniol) were obtained from Roth. (R,S)-3,7-Dimethyl-6-octen-1-ol (citronellol) and pure (R)-(+)-β-citronellol were purchased from Sigma-Aldrich. (Z)-3,7-Dimethyl-2,6-octadien-1-ol (nerol) was purchased from Alfa Aesar. (3R,3S)-3,7-Dimethyl-6-octenyl β-d-glucopyranoside (citronellyl β-d-glucoside), (Z)-3,7-dimethyl-2,6-octadienyl β-d-glucopyranoside (neryl β-d-glucoside), and (E)-3,7-dimethyl-2,6-octadienyl β-d-glucopyranoside (geranyl β-d-glucoside) were synthesized according to the Koenigs-Knorr procedure (Paulsen et al., 1985). (R,S)-3,7-Dimethyl-1,6-octadienyl β-d-glucopyranoside (linaloyl β-d-glucoside), a less reactive tertiary alcohol, was synthesized according to a modified Koenigs-Knorr procedure using another catalyst (Hattori et al., 2004). Deuterium-labeled 1,1-[2H2]citronellyl β-d-glucoside was prepared as described (Hill et al., 1994; Wüst et al., 1998). The spectral data of the synthesized compounds were in accordance with the given data (Paulsen et al., 1985; Salles et al., 1991; Konda et al., 1997).
Nucleic Acid Extraction
For total RNA extraction, 1 g of plant material was ground to a fine powder in liquid nitrogen using mortar and pestle. The extraction was carried out in triplicate for each sample using the cetyl-trimethyl-ammonium bromide method following an established protocol (Zeng and Yang, 2002) adapted by Reid et al. (2006) to meet the requirements of different grapevine tissues. Remaining genomic DNA was digested by DNase I and cleaned up with the High Pure RNA Isolation kit (Roche). DNA was extracted using the innuPREP Plant DNA kit (AJ Innuscreen) following the manufacturer’s instructions. Nucleic acid integrity was confirmed by agarose gel electrophoresis. Concentrations and 260:280-nm ratios were determined using the Nanodrop 1000 (Thermo Scientific).
Transcription Analysis
The gene expression patterns of seven VvGT genes were analyzed together with five reference genes (VvACTIN, VviAP47 [for Vitis vinifera aspartic protease47], VvPP2A [for Vitis vinifera protein phosphatase 2A], VvSAND, and VvTIP41 [for Vitis vinifera TAP42-interacting protein]) using the Genome Lab GeXP Genetic Analysis System (Beckman Coulter) for multiplex, quantitative gene expression analysis (for details, see Supplemental Fig. S14).
Comparative Sequencing
The reference genome of PN40024 (Jaillon et al., 2007) was used to design gene-specific primers in the untranslated regions of the seven putative VvGT genes using the tool Primer-BLAST (Ye et al., 2012). Primers were purchased from Eurofins MWG Operon (Supplemental Table S7). PCR was performed with Phusion DNA Polymerase (Thermo Fisher Scientific) using high-fidelity buffer. Thermal cycling conditions were 98°C for 30 s, followed by 32 cycles consisting of 98°C for 5 s, 60°C for 5 s, and 72°C for 30 s, and a final elongation step of 72°C for 1 min. Genomic DNA extracted from leaves was used as a template at a final concentration of 1 ng µL−1. PCR products were gel purified with the Wizard SV Gel and PCR Clean-Up System (Promega). A-tailing of purified PCR products was performed with Taq DNA Polymerase (Thermo Fisher Scientific). A-tailed PCR products were ligated into pGEM-T Easy vector (Promega) and cloned in One Shot TOP10 Chemically Competent Escherichia coli (Life Technologies). Plasmids were isolated with the PureYield Plasmid Miniprep System (Promega) and sequenced with the vector-specific primers M13 uni (−21) and M13 rev (−29) by an ABI 3730 capillary sequencer (StarSEQ). Primer walking was performed when necessary. Raw data were edited with the FinchTV software (Geospiza). Sequences were assembled with SeqMan and aligned with MegAlign (DNASTAR).
Metabolite Analysis: Sample Preparation
Grape berries were peeled and 10 g (fresh weight) of the exocarp was used for each analysis. In case of root, leaf, and inflorescence, 4 g of plant material was used. The material was ground in liquid nitrogen and extracted with a mixture of phosphate buffer (0.1 m, pH 7) and 13% (v/v) ethanol for 24 h under nitrogen with exclusion of light (Jesús Ibarz et al., 2006). 2-Octanol was used as an internal standard for the determination of free monoterpenes. For the determination of monoterpenyl β-d-glucosides, stable isotope dilution analysis was applied using [2H2]citronellyl β-d-glucoside as a labeled internal standard. The concentration of the internal standards was adapted for each variety, each type of tissue, and each stage of development. 2-Octanol was added in a range of 0.3 to 6.8 mg kg−1 plant material and [2H2]citronellyl β-d-glucoside in a range of 0.1 to 3.5 mg kg−1. To purify the sample, Carrez reagents (Merck) were added (1 mL each), and the sample was then centrifuged at 14,500 rpm for 20 min at 5°C. The supernatant was taken for subsequent solid-phase extraction to isolate and separate free monoterpenes from glycosidically bound monoterpenes. Therefore, a 200-mg Lichrolut EN column (Merck) was conditioned as described (Piñeiro et al., 2004). Free monoterpenols were eluted with dichloromethane and glycosidically bound monoterpenols with methanol. For GC-MS detection, the dichloromethane fractions were dried with Na2SO4, concentrated using nitrogen to 200 µL, and analyzed. For HPLC-MS/MS detection, the methanolic fractions were concentrated under reduced pressure and the residues were dissolved in water:acetonitrile (7:3, v/v). The samples were analyzed by liquid chromatography-MS/MS.
Enantioselective Analysis of Citronellol
To determine the enantioselectivity of VvGT7, the enantiomeric composition of residual citronellol and glucosidically bound citronellol was determined by an enantioselective GC column. Citronellol that was not converted by VvGT7 was isolated by solid-phase microextraction (SPME) for 10 min at 60°C using a fiber coated with an 85-µm film of polyacrylate (Supelco). After extraction, the SPME fiber was desorbed for 10 min at 250°C in the injector port of the GC-MS system. Subsequently, residual citronellol was completely removed by extraction with 5 mL of dichloromethane. Citronellyl β-d-glucoside that remained in the aqueous phase was hydrolyzed by HCl (5 mL, 0.1 m, pH 1) for 1 h at 100°C to release citronellol (Skouroumounis and Sefton, 2000). After hydrolysis, citronellol was analyzed by SPME as described above.
Construction of Expression Plasmids
The full-length open reading frames were subcloned to the pGEM-T Easy vector (Promega). All VvGT genes were amplified with primers introducing BamHI and NotI restriction sites. Subsequently, the genes were cloned in frame with the N-terminal GST-tag into the pGEX-4T-1 expression vector (Amersham Bioscience). Gene identity was confirmed by sequencing (Microsynth; Eurofins MWG Operon).
Site-Directed Mutagenesis
The site-directed mutagenesis of VvGT7i (V186L, L210I, A317P), VvGT7j (V186L, L210I, A317P), and VvGT7h (G190D) was performed according to the QuikChange Protocol (Novagen). The primers were designed as described in the manual (Supplemental Table S7). Mutations were verified by sequencing (Microsynth; Eurofins MWG Operon).
Heterologous Protein Expression
Recombinant protein was expressed in E. coli BL21 (DE3) pLysS (Novagen). Precultures were grown overnight at 37°C in Luria-Bertani medium containing 100 µg mL−1 ampicillin and 23 µg mL−1 chloramphenicol. The following day, the cultures with a final volume of 1 L of Luria-Bertani medium containing the appropriate antibiotics were inoculated with 50 mL of the preculture. The culture was grown at 37°C at 160 rpm until optical density at 600 nm reached 0.4 to 0.6. After cooling the culture to 16°C, 1 mm isopropyl-β-d-thiogalactopyranoside was added to induce the protein expression. Cultures were incubated overnight at 16°C to 18°C at 160 rpm. Cells were harvested by centrifugation and stored at −80°C. Negative controls were carried out with E. coli BL21 (DE3) pLysS cells containing the empty expression vector pGEX-4T-1.
Cell Lysis and Purification
Recombinant GST fusion proteins were purified by GST Bind resin (Novagen) following the manufacturer’s instructions. Briefly, the cells were resuspended in the binding buffer containing 10 mm 2-mercaptoethanol. Cells were disrupted by sonication (Branson). The crude protein extract was incubated for 2 h with the GST Bind resin to bind GST fusion protein. The recombinant protein was eluted with GST elution buffer containing reduced glutathione and quantified by Bradford solution (Sigma-Aldrich). The presence of the expressed proteins was confirmed by SDS-PAGE and western blot using anti-GST antibody and goat anti-mouse IgG fused to alkaline phosphatase. Bands were visualized by colorimetric detection of alkaline phosphatase using 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium.
Activity Assay and Kinetics
In the initial screening, each reaction mixture (200 µL in total) contained Tris-HCl buffer (100 mm, pH 7.5, and 10 mm 2-mercaptoethanol), 37 pmol of UDP-[14C]Glc (0.01 µCi), substrate (50 µL of a 1 mg mL−1 stock solution), and purified protein (0.5–0.8 µg µL−1). The reaction mixture was incubated at 30°C overnight. The assays were stopped by adding 1 µL of 24% (v/v) TCA and extracted with 500 µL of water-saturated 1-butanol. The organic phase was mixed with 2 mL of Pro Flow P+ cocktail (Meridian Biotechnologies), and radioactivity was determined by liquid scintillation counting (Tri-Carb 2800TR; Perkin Elmer). Additionally, negative controls without substrate and with heat-inactivated protein were performed. The kinetic data were determined with increasing concentrations of nerol, geraniol, and citronellol from 1 to 500 µm and a fixed UDP-Glc concentration of 502.5 µm (500 µm unlabeled UDP-Glc and 2.5 µm UDP-[14C]Glc). The total volume was 40 µL, and 15 µg of purified protein was used. The measurements were performed under the following conditions: the assays of VvGT7a and VvGT7c were carried out at 30°C and 37°C, respectively, using a Tris-HCl buffer (100 mm) with 10 mm 2-mercaptoethanol, pH 8, and 2 h of incubation. The assay of VvGT7b was performed at 37°C using a Tris-HCl buffer (100 mm) with 10 mm 2-mercaptoethanol, pH 8.5, and 1 h of incubation. The kinetic analyses of citronellol and geraniol were performed with a doubled incubation time. The reaction was stopped by adding 1 µL of 24% (v/v) TCA, and glucosides were extracted with 100 µL of water-saturated 1-butanol. Radioactivity was determined by liquid scintillation counting. To determine the kinetic data of UDP-Glc, the value of nerol was fixed according to the measured Km value of nerol (400 or 200 µm), and UDP-[14C]Glc was mixed with nonradiolabeled UDP-Glc to obtain concentrations ranging from 5 to 300 µm. The Km and Vmax values were calculated from Lineweaver-Burk plots, Hanes-Woolf plots, and nonlinear fitting of the experimental data.
HPLC-MS Analysis
Liquid chromatography-MS was performed to verify the glucosylated products. The reaction mixtures (200 µL in total) contained Tris-HCl buffer (100 mm, pH 8.0 or 8.5, and 10 mm 2-mercaptoethanol), 12.5 mm UDP-Glc, substrate (50 µL of a 1 mg mL−1 stock solution), and purified protein (0.5–0.8 µg µL−1) and were incubated overnight at 30°C or 37°C. The reaction was stopped by adding 1 µL of 24% (v/v) TCA and extracted with 500 µL of ethyl acetate. The organic solvent was vaporized and analyzed by HPLC-MS. For HPLC-MS/MS analysis of monoterpenyl-β-d-glucosides, an HP 1050 HPLC system coupled to an API 2000 (Applied Biosystems, AB Sciex) triple-quadrupole mass spectrometer was used. Data acquisition was performed using Analyst software version 1.4.2. (Applied Biosystems, AB Sciex). The column (Phenomenex Gemini-NX 5u C18, 250 × 3 mm) was eluted by water:acetonitrile (7:3, v/v) containing 0.2% (v/v) ammonia until 12 min and then by a linear gradient to water:acetonitrile (4:6, v/v; 0.2% [v/v] ammonia) until 18 min. The column temperature was maintained at 40°C. The mass spectrometer was operated in electrospray ionization-multiple reaction-monitoring negative ion mode. Nitrogen was used as curtain (setting 20), nebulizing, and collision gas (collision energy was −20 eV). Monoterpenyl β-d-glucosides were identified by the following characteristic multiple reaction monitoring transitions: linaloyl β-d-glucoside, mass-to-charge ratio (m/z) 315→161(Glu), 315→113(Glu); neryl β-d-glucoside, m/z 315→119(Glu), 315→113(Glu); geranyl β-d-glucoside, m/z 315→119(Glu), 315→113(Glu); citronellyl β-d-glucoside, m/z 317→101(Glu), 317→161(Glu); [2H2]citronellyl β-d-glucoside, m/z 319→101(Glu), 319→161(Glu) (Domon and Costello, 1988; Cole et al., 1989; Salles et al., 1991).
Radio-TLC Analysis
The reaction mixture for thin-layer chromatography analysis was composed as described above except that 37 pmol of UDP-[14C]Glc was used. The extraction was carried out with 500 µL of ethyl acetate. The organic solvent was vaporized, and the pellet was resuspended in 10 µL of methanol and applied on Silica Gel 60 F254 plates (Merck). The dried plates were developed in a solvent system consisting of chloroform:acetic acid:water (50:45:5, v/v/v). Plates were dried and analyzed using a digital autoradiograph (EG&G Berthold).
GC-MS System
GC-MS analysis was performed with a Varian Saturn 3900 gas chromatograph coupled to a Varian Saturn 2100T-MS ion trap. Separation was performed on a Phenomenex Zebron ZB-WAXplus column (30 m × 0.25 mm × 0.25 µm). Helium flow rate was 1 mL min−1. The analysis was carried out in split mode with 220°C injector temperature. Electron impact ionization-MS spectra were recorded from m/z 40 to 300 (ionization energy, 70 eV; trap temperature, 170°C). The oven temperature program started at 60°C for 3 min, followed by an increase rate of 10°C min−1, up to a final temperature of 250°C for a duration of 5 min. Enantioselective GC-MS analysis was performed with a Varian GC-450 coupled to a Varian MS-240 ion trap. The column was a DiAcß [heptakis-(2,3-di-O-acetyl-6-O-tert.-butyldimethylsilyl)-β-cyclodextrin], 26 m × 0.32 mm i.d. with a 0.1-µm film. Helium was used as the carrier gas with a flow rate of 1 mL min−1, and injector temperature was 250°C with a split ratio of 1:100 (liquid injections) or splitless (SPME measurement). The oven temperature program was 70°C (for 3 min), ramped by 0.5°C min−1 to 130°C, and then increased by 20°C min−1 up to 200°C (for 3 min). GC-MS measurements were recorded in full-scan mode. Selected ion monitoring mode was used for quantification.
In Silico Analysis
Alignments of protein and nucleotide sequences were created using MUSCLE of the GENEIOUS Pro 5.5.6 program with its default parameters (Biomatters; http://www.geneious.com/). The phylogenetic tree was constructed utilizing the neighbor-joining method with no outgroup by Geneious tree builder embedded in GENEIOUS Pro. Protein 3D structures were calculated by homology modeling using YASARA (http://www.yasara.org) and were visualized by PyMOL 1.2r1 (Delano Scientific; http://www.pymol.org/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers VvGT7 (CAO49526.1.pro; XP_002276546; XM_002276510), VvGT8 (CAN67608.1.pro; XP_002262883; XM_002262847), VvGT9 (CAN78291.1.pro; XP_002285379; XM_002285343), VvGT10 (CAN71972.1.pro; XP_002285412; XM_002285376), VvGT11 (CAN59771.1.pro; XP_002274256; XM_002274220), VvGT12 (CAN83016.1.pro; XP_002265326; XM_002265290), and VvGT13 (CAO62987.1.pro; XP_002265216; XM_002265180).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of 67 putative VvGTs sequences.
Supplemental Figure S2. Location of the GT genes placed in the map of the reference genome PN40024.
Supplemental Figure S3. Validation of GeXP analysis.
Supplemental Figure S4. Largest and smallest leaves of Gewurztraminer 11-18 Gm and White Riesling 239-34 Gm used for RNA extraction and terpenoid analysis.
Supplemental Figure S5. Inflorescences of Gewurztraminer 11-18 Gm and White Riesling 239-34 Gm used for RNA extraction and terpenoid analysis.
Supplemental Figure S6. Grape berries of Gewurztraminer 11-18 Gm and White Riesling 239-34 Gm used for RNA extraction and terpenoid analysis.
Supplemental Figure S7. Fragmentation mechanism of monoterpenyl β-d-glucosides.
Supplemental Figure S8. Multiple protein sequence alignment of the 10 alleles of VvGT7.
Supplemental Figure S9. SDS-PAGE and western blot analysis.
Supplemental Figure S10. Relative enzymatic activity (%) of 10 allelic VvGT7 proteins from V. vinifera.
Supplemental Figure S11. Radio-TLC analysis of products formed by VvGT7a from different monoterpenes.
Supplemental Figure S12. Time course experiments showing product formation over time.
Supplemental Figure S13. Functions of monoterpenyl glucosides and the glycosylation of monoterpenes in plants.
Supplemental Figure S14. Gene expression analysis by Genome Lab GeXP Genetic Analysis System.
Supplemental Table S1. Putative VvGTs.
Supplemental Table S2. Primers used for quantitative real-time PCR.
Supplemental Table S3. Gene-specific primers used for GeXP.
Supplemental Table S4. Amounts of free monoterpenes and monoterpene-β-d-glucosides in grape exocarp during grape ripening.
Supplemental Table S5. Primers used in PCR for subsequent cloning and sequencing.
Supplemental Table S6. Pairwise comparison of the amino acid sequences of 18 tested VvGT7 alleles and mutants.
Supplemental Table S7. Primers used for site-directed mutagenesis as indicated in the primer name.
Supplementary Material
Acknowledgments
We thank Frank Manty for acting as native speaker, correcting the article, and suggesting stylistic improvements.
Glossary
- GC
gas chromatography
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- GST
glutathione S-transferase
- radio-TLC
radioactivity-thin-layer chromatography
- 3D
three-dimensional
- SPME
solid-phase microextraction
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SCHW634/17–1).
The online version of this article contains Web-only data.
References
- Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FW, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ. (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek HH, Cadwallader KR. (1999) Contribution of free and glycosidically bound volatile compounds to the aroma of Muscadine grape juice. J Food Sci 64: 441–444 [Google Scholar]
- Bayrak A. (1994) Volatile oil composition of Turkish rose (Rosa damascena). J Agric Food Chem 64: 441–448 [Google Scholar]
- Bertsch C, Kieffer F, Maillot P, Farine S, Butterlin G, Merdinoglu D, Walter B. (2005) Genetic chimerism of Vitis vinifera cv. Chardonnay 96 is maintained through organogenesis but not somatic embryogenesis. BMC Plant Biol 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Lim EK, Poppenberger B, Vaistij FE. (2006) Glycosyltransferases of lipophilic small molecules. Annu Rev Plant Biol 57: 567–597 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, Bowles DJ, Davies GJ, Edwards R. (2007) Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA 104: 20238–20243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L, Lim EK, Bowles DJ. (2008) Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry 14: 6656–6662 [DOI] [PubMed] [Google Scholar]
- Caputi L, Lim EK, Bowles DJ, inventors June 10, 2010. Monoterpenoid modifying enzymes. US Patent Application No. US2010/0143975 [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. (2012) A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J 69: 1030–1042 [DOI] [PubMed] [Google Scholar]
- Claros MG, Bautista R, Guerrero-Fernández D, Benzerki H, Seoane P, Fernández-Pozo N. (2012) Why assembling plant genome sequences is so challenging. Biology 1: 439–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RB, Tabet JC, Salles C, Crouzet J. (1989) Structural “memory effects” influencing decompositions of glucose alkoxide anions produced from monoterpene glycoside isomers in tandem mass spectrometry. Rapid Commun Mass Spectrom 3: 59–61 [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Domon B, Costello CE. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate 5: 397–409 [Google Scholar]
- Drew JE, Mayer CD, Farquharson AJ, Young P, Barrera LN. (2011) Custom design of a GeXP multiplexed assay used to assess expression profiles of inflammatory gene targets in normal colon, polyp, and tumor tissue. J Mol Diagn 13: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J. (2004) Biochemistry of plant volatiles. Plant Physiol 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Piechulla B, Pichersky E. (1999) Biogenesis of floral scent. Hortic Rev (Am Soc Hortic Sci) 24: 31–54 [Google Scholar]
- Ebeler SE. (2001) Analytical chemistry: unlocking the secrets of wine flavor. Food Rev Int 17: 45–64 [Google Scholar]
- Fenoll J, Manso A, Hellín P, Ruiz L, Flores P. (2009) Changes in the aromatic composition of the Vitis vinifera grape Muscat Hamburg during ripening. Food Chem 114: 420–428 [Google Scholar]
- Ford CM, Boss PK, Høj PB. (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273: 9224–9233 [DOI] [PubMed] [Google Scholar]
- Ford CM, Høj PB. (1998) Multiple glucosyltransferase activities in the grapevine Vitis vinifera. Aust J Grape Wine Res 4: 48–58 [Google Scholar]
- Franks T, Botta R, Thomas MR, Franks J. (2002) Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet 104: 192–199 [DOI] [PubMed] [Google Scholar]
- Gachon CM, Langlois-Meurinne M, Saindrenan P. (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10: 542–549 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Giannuzzi G, D’Addabbo P, Gasparro M, Martinelli M, Carelli FN, Antonacci D, Ventura M. (2011) Analysis of high-identity segmental duplications in the grapevine genome. BMC Genomics 12: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Cramer GR, Dickerson JA, Mathiason K, Van Hemert J, Fennell AY. (2009) VitisNet: “omics” integration through grapevine molecular networks. PLoS ONE 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunata YZ, Bayonove C, Baumes R, Cordonnier R. (1985a) The aroma of grapes. I. Extraction and determination of free and glycosidically bound fractions of some grape aroma components. J Chromatogr A 331: 83–90 [Google Scholar]
- Gunata YZ, Bayonove CL, Baumes RL, Cordonnier RE. (1986) Changes in free and bound fractions of aromatic components in vine leaves during development of Muscat grapes. Phytochemistry 25: 943–946 [Google Scholar]
- Gunata YZ, Bayonove CL, Baumes RL, Cordonnier RE. (1985b) The aroma of grapes: localisation and evolution of free and bound fractions of some grape aroma components cv Muscat during first development and maturation. J Sci Food Agric 36: 857–862 [Google Scholar]
- Gunata Z, Bitteur S, Brillouet JM, Bayonove C, Cordonnier R. (1988) Sequential enzymic hydrolysis of potentially aromatic glycosides from grape. Carbohydr Res 184: 139–149 [Google Scholar]
- Guth H. (1996) Determination of the configuration of wine lactone. Helv Chim Acta 79: 1559–1571 [Google Scholar]
- Guth H. (1997) Identification of character impact odorants of different white wine varieties. J Agric Food Chem 45: 3022–3026 [Google Scholar]
- Hall D, De Luca V. (2007) Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J 49: 579–591 [DOI] [PubMed] [Google Scholar]
- Hans J, Brandt W, Vogt T. (2004) Site-directed mutagenesis and protein 3D-homology modelling suggest a catalytic mechanism for UDP-glucose-dependent betanidin 5-O-glucosyltransferase from Dorotheanthus bellidiformis. Plant J 39: 319–333 [DOI] [PubMed] [Google Scholar]
- Hansen KS, Kristensen C, Tattersall DB, Jones PR, Olsen CE, Bak S, Møller BL. (2003) The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry 64: 143–151 [DOI] [PubMed] [Google Scholar]
- Hattori S, Kawaharada C, Tazaki H, Fujimori T, Kimura K, Ohnishi M, Nabeta K. (2004) Formation mechanism of 2,6-dimethyl-2,6-octadienes from thermal decomposition of linalyl β-D-glucopyranoside. Biosci Biotechnol Biochem 68: 2656–2659 [DOI] [PubMed] [Google Scholar]
- He XZ, Wang X, Dixon RA. (2006) Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J Biol Chem 281: 34441–34447 [DOI] [PubMed] [Google Scholar]
- Hefner T, Arend J, Warzecha H, Siems K, Stöckigt J. (2002) Arbutin synthase, a novel member of the NRD1beta glycosyltransferase family, is a unique multifunctional enzyme converting various natural products and xenobiotics. Bioorg Med Chem 10: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hill RK, Abächerli C, Hagishita S. (1994) Synthesis of (2S,4S)- and (2S,4R)-[5,5,5-2H3]leucine from (R)-pulegone. Can J Chem 72: 110–113 [Google Scholar]
- Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir MC, Merdinoglu D. (2004) Diversification within grapevine cultivars goes through chimeric states. Genome 47: 579–589 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jánváry L, Hoffmann T, Pfeiffer J, Hausmann L, Töpfer R, Fischer TC, Schwab W. (2009) A double mutation in the anthocyanin 5-O-glucosyltransferase gene disrupts enzymatic activity in Vitis vinifera L. J Agric Food Chem 57: 3512–3518 [DOI] [PubMed] [Google Scholar]
- Jesús Ibarz M, Ferreira V, Hernández-Orte P, Loscos N, Cacho J. (2006) Optimization and evaluation of a procedure for the gas chromatographic-mass spectrometric analysis of the aromas generated by fast acid hydrolysis of flavor precursors extracted from grapes. J Chromatogr A 1116: 217–229 [DOI] [PubMed] [Google Scholar]
- Khater F, Fournand D, Vialet S, Meudec E, Cheynier V, Terrier N. (2012) Identification and functional characterization of cDNAs coding for hydroxybenzoate/hydroxycinnamate glucosyltransferases co-expressed with genes related to proanthocyanidin biosynthesis. J Exp Bot 63: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konda Y, Toida T, Kaji E, Takeda K, Harigaya Y. (1997) First total synthesis of two new diglycosides, neohancosides A and B, from Cynanchum hancockianum. Carbohydr Res 301: 124–143 [Google Scholar]
- Landmann C, Fink B, Schwab W. (2007) FaGT2: a multifunctional enzyme from strawberry (Fragaria × ananassa) fruits involved in the metabolism of natural and xenobiotic compounds. Planta 226: 417–428 [DOI] [PubMed] [Google Scholar]
- Lawo NC, Weingart GJF, Schuhmacher R, Forneck A. (2011) The volatile metabolome of grapevine roots: first insights into the metabolic response upon phylloxera attack. Plant Physiol Biochem 49: 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng 87: 623–631 [DOI] [PubMed] [Google Scholar]
- Lim EK, Baldauf S, Li Y, Elias L, Worrall D, Spencer SP, Jackson RG, Taguchi G, Ross J, Bowles DJ. (2003) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13: 139–145 [DOI] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Elias L, Worral D, Li Y, Ro J, Bowles DJ. (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Lim EK, Li Y, Parr A, Jackson RG, Ashford DA, Bowles DJ. (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Loza-Tavera H. (1999) Monoterpenes in essential oils: biosynthesis and properties. Adv Exp Med Biol 464: 49–62 [DOI] [PubMed] [Google Scholar]
- Luan F, Hampel D, Mosandl A, Wüst M. (2004) Enantioselective analysis of free and glycosidically bound monoterpene polyols in Vitis vinifera L. cvs. Morio Muscat and Muscat Ottonel: evidence for an oxidative monoterpene metabolism in grapes. J Agric Food Chem 52: 2036–2041 [DOI] [PubMed] [Google Scholar]
- Luan F, Mosandl A, Degenhardt A, Gubesch M, Wüst M. (2006a) Metabolism of linalool and substrate analogs in grape berry mesocarp of Vitis vinifera L cv Morio Muscat: demonstration of stereoselective oxygenation and glycosylation. Anal Chim Acta 563: 353–364 [Google Scholar]
- Luan F, Mosandl A, Gubesch M, Wüst M. (2006b) Enantioselective analysis of monoterpenes in different grape varieties during berry ripening using stir bar sorptive extraction- and solid phase extraction-enantioselective-multidimensional gas chromatography-mass spectrometry. J Chromatogr A 1112: 369–374 [DOI] [PubMed] [Google Scholar]
- Luan F, Mosandl A, Münch A, Wüst M. (2005) Metabolism of geraniol in grape berry mesocarp of Vitis vinifera L. cv. Scheurebe: demonstration of stereoselective reduction, E/Z-isomerization, oxidation and glycosylation. Phytochemistry 66: 295–303 [DOI] [PubMed] [Google Scholar]
- Lücker J, Bowen P, Bohlmann J. (2004) Vitis vinifera terpenoid cyclases: functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (-)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65: 2649–2659 [DOI] [PubMed] [Google Scholar]
- Lund ST, Bohlmann J. (2006) The molecular basis for wine grape quality: a volatile subject. Science 311: 804–805 [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Bellido M, Aharoni A, Salentijn EM, Kaldenhoff R, Coiner HA, Muñoz-Blanco J, Schwab W. (2006) Cinnamate metabolism in ripening fruit: characterization of a UDP-glucose:cinnamate glucosyltransferase from strawberry. Plant Physiol 140: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madina BR, Sharma LK, Chaturvedi P, Sangwan RS, Tuli R. (2007) Purification and physico-kinetic characterization of 3beta-hydroxy specific sterol glucosyltransferase from Withania somnifera (L) and its stress response. Biochim Biophys Acta 1774: 392–402 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB. (2002) Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci 7: 366–373 [DOI] [PubMed] [Google Scholar]
- Maicas S, Mateo JJ. (2005) Hydrolysis of terpenyl glycosides in grape juice and other fruit juices: a review. Appl Microbiol Biotechnol 67: 322–335 [DOI] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Bohlmann J. (2004) Identification of Vitis vinifera (−)-alpha-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry 65: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Martin DM, Chiang A, Lund ST, Bohlmann J. (2012) Biosynthesis of wine aroma: transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta 236: 919–929 [DOI] [PubMed] [Google Scholar]
- Martinkus C, Croteau R. (1981) Metabolism of monoterpenes: evidence for compartmentation of l-menthone metabolism in peppermint (Mentha piperita) leaves. Plant Physiol 68: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo JJ, Jiménez M. (2000) Monoterpenes in grape juice and wines. J Chromatogr A 881: 557–567 [DOI] [PubMed] [Google Scholar]
- Nagashima S, Tomo S, Orihara Y, Yoshikawa T. (2004) Cloning and characterization of glucosyltransferase cDNA from Eucalyptus perriniana cultured cells. Plant Biotechnol 21: 343–348 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8: 581–599 [DOI] [PubMed] [Google Scholar]
- Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T. (2010) Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22: 2856–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S. (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Paulsen H, Le-Nguyén B, Sinnwell V, Heemann V, Seehofer F. (1985) Synthesis of glycosides from mono-, sesqui- and diterpene alcohols. EurJOC 8: 1513–1536 [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. (2006) Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro Z, Palma M, Barroso CG. (2004) Determination of terpenoids in wines by solid phase extraction and gas chromatography. Anal Chim Acta 513: 209–214 [Google Scholar]
- Raguso RA, Pichersky E. (1999) New perspectives in pollination biology: floral fragrances. A day in the life of a linalool molecule: chemical communication in a plant-pollinator system. Part 1. Linalool biosynthesis in flowering plants. Plant Species Biol 14: 95–120 [Google Scholar]
- Rai AJ, Kamath RM, Gerald W, Fleisher M. (2009) Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal Bioanal Chem 393: 1505–1511 [DOI] [PubMed] [Google Scholar]
- Razungles A, Gunata Z, Pinatel S, Baumes RY, Rayonove C. (1993) Quantitative studies on terpenes, norisoprenoids and their precursors in several varieties of grapes. Sci Aliments 13: 59–72 [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz S, Garrison KE, Dangl GS, Boursiquot JM, Meredith CP. (2002) Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J Am Soc Hortic Sci 127: 508–514 [Google Scholar]
- Rivoal A, Fernandez C, Lavoir AV, Olivier R, Lecareux C, Greff S, Roche P, Vila B. (2010) Environmental control of terpene emissions from Cistus monspeliensis L. in natural Mediterranean shrublands. Chemosphere 78: 942–949 [DOI] [PubMed] [Google Scholar]
- Rocha SM, Coutinho P, Delgadillo I, Dias Cardoso A, Coimbra MA. (2004) Effect of enzymatic aroma release on the volatile compounds of white wines presenting different aroma potentials. J Sci Food Agric 85: 199–205 [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ. (2001) Higher plant glycosyltransferases. Genome Biol 2: S3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles C, Jallageas JC, Beziat Y, Cristau HJ. (1991) Synthesis of 4- and 10-deuterated neryl and geranyl-β-D-glucosides and their use in corroboration of a mechanism proposed for the fragmentation of heterosides in tandem mass spectrometry. J Labelled Comp Radiopharm 31: 11–22 [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E. (2008) Biosynthesis of plant-derived flavor compounds. Plant J 54: 712–732 [DOI] [PubMed] [Google Scholar]
- Sefton MA, Francis IL, Williams PJ. (1994) Free and bound volatile secondary metabolites of Vitis vinifera grape cv Sauvignon Blanc. J Food Sci 59: 142–147 [Google Scholar]
- Skouroumounis GK, Sefton MA. (2000) Acid-catalyzed hydrolysis of alcohols and their β-D-glucopyranosides. J Agric Food Chem 48: 2033–2039 [DOI] [PubMed] [Google Scholar]
- Trapp SC, Croteau RB. (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al. (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2009) Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett 583: 3303–3309 [DOI] [PubMed] [Google Scholar]
- Williams PI, Strauss CR, Wilson B, Massy-Westropp RA. (1982) Studies on the hydrolysis of Vitis vinifera monoterpene precursor compounds and model monoterpene β-D-glucosides rationalizing the monoterpene composition of grapes. J Agric Food Chem 30: 1219–1223 [Google Scholar]
- Wilson B, Strauss CR, Williams PJ. (1984) Changes in free and glycosidically bound monoterpenes in developing Muscat grapes. J Agric Food Chem 32: 919–924 [Google Scholar]
- Wirth J, Guo W, Baumes R, Günata Z. (2001) Volatile compounds released by enzymatic hydrolysis of glycoconjugates of leaves and grape berries from Vitis vinifera Muscat of Alexandria and Shiraz cultivars. J Agric Food Chem 49: 2917–2923 [DOI] [PubMed] [Google Scholar]
- Wüst M, Rexroth A, Beck T, Mosandl A. (1998) Mechanistic aspects of the biogenesis of rose oxide in Pelargonium graveolens L’Héritier. Chirality 10: 229–237 [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yang T. (2002) RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep 20: 417 [Google Scholar]
- Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, Dixon RA. (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23: 1536–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.