An acyl-CoA-binding protein changes the acyl-CoA and oil composition in seeds of transgenic Arabidopsis.
Abstract
Low-molecular mass (10 kD) cytosolic acyl-coenzyme A-binding protein (ACBP) has a substantial influence over fatty acid (FA) composition in oilseeds, possibly via an effect on the partitioning of acyl groups between elongation and desaturation pathways. Previously, we demonstrated that the expression of a Brassica napus ACBP (BnACBP) complementary DNA in the developing seeds of Arabidopsis (Arabidopsis thaliana) resulted in increased levels of polyunsaturated FAs at the expense of eicosenoic acid (20:1cisΔ11) and saturated FAs in seed oil. In this study, we investigated whether alterations in the FA composition of seed oil at maturity were correlated with changes in the acyl-coenzyme A (CoA) pool in developing seeds of transgenic Arabidopsis expressing BnACBP. Our results indicated that both the acyl-CoA pool and seed oil of transgenic Arabidopsis lines expressing cytosolic BnACBP exhibited relative increases in linoleic acid (18:2cisΔ9,12; 17.9%–44.4% and 7%–13.2%, respectively) and decreases in 20:1cisΔ11 (38.7%–60.7% and 13.8%–16.3%, respectively). However, alterations in the FA composition of the acyl-CoA pool did not always correlate with those seen in the seed oil. In addition, we found that targeting of BnACBP to the endoplasmic reticulum resulted in FA compositional changes that were similar to those seen in lines expressing cytosolic BnACBP, with the most prominent exception being a relative reduction in α-linolenic acid (18:3cisΔ9,12,15) in both the acyl-CoA pool and seed oil of the former (48.4%–48.9% and 5.3%–10.4%, respectively). Overall, these data support the role of ACBP in acyl trafficking in developing seeds and validate its use as a biotechnological tool for modifying the FA composition of seed oil.
Cytosolic low-molecular mass (approximately 10 kD) acyl-coenzyme A-binding protein (ACBP) consists of a four-α-helix domain capable of binding acyl-CoAs with high affinity in a wide range of eukaryotic organisms (Faergeman et al., 2007). It is believed to serve a housekeeping function of maintaining free acyl-CoA concentrations at low nanomolar levels and, thus, prevents micelle formation and the partitioning of acyl-CoA into membranes (Knudsen et al., 1999). This protein is also considered to contribute to another facet of acyl-CoA pool maintenance via its role in the intracellular transport of acyl-CoAs in the aqueous environment of the cytosol (Rasmussen et al., 1994). Moreover, it has also been shown to exhibit more specialized functions in metabolic processes in which acyl-CoA is actively involved, depending on the tissue and physiological state (Guerrero et al., 2006; Xiao and Chye 2011; Yurchenko and Weselake, 2011).
In the developing seeds of oleaginous plants, fatty acids (FAs) are synthesized de novo in plastids and are activated to acyl-CoAs upon their transfer to the cytosol, after which time they can undergo additional modifications (e.g. elongation and desaturation) on the membranes of the endoplasmic reticulum (ER; for review, see Rawsthorne, 2002). While FA elongation is performed on the acyl-CoA substrate, the introduction of the second and third double bonds requires the acyl group to be esterified to phosphatidylcholine (PC; Jaworski, 1987). The composition of the acyl-CoA pool, therefore, is highly dynamic and represents a net result of both de novo synthesis and acyl-editing processes (Bates et al., 2009).
The acyl-CoA pool provides substrate for acyltransferases involved in the biosynthesis of triacylglycerol (TAG), which is a major component of seed oil (Weselake et al., 2009). More specifically, TAG synthesis typically occurs via a series of acyl-CoA-dependent acylations of a glycerol backbone derived from sn-glycerol-3-phosphate in a pathway known as the sn-glycerol-3-phosphate or Kennedy pathway (for review, see Snyder et al., 2009; Weselake et al., 2009), although acyl-CoA-independent reactions can also be involved in the production of TAG (Stobart et al., 1997; Banaś et al., 2000; Dahlqvist et al., 2000) and thus contribute to its final composition. Low-molecular mass ACBPs have been demonstrated to modulate the activities of Kennedy pathway acyltransferases in a manner dependent upon the ratio of ACBP to acyl-CoA, stimulating TAG biosynthesis under conditions of acyl-CoA excess and inhibiting acyltransferase activities when relative amounts of acyl-CoA are low compared with ACBP, thus regulating the size of the acyl-CoA pool (for review, see Yurchenko and Weselake, 2011).
The acyl-CoA pool in seeds is also influenced through a distinct route involving lysophosphatidylcholine acyltransferase (LPCAT), which catalyzes the acyl-CoA-dependent acylation of lysophosphatidylcholine at the sn-2 position to form PC (Ichihara et al., 1995). Acyl groups esterified to PC become substrates for FA desaturation and other modifications (Miquel and Browse, 1992; Broun et al., 1998) and can then be returned back to the acyl-CoA pool or channeled into TAG through acyl-CoA-independent mechanisms (Stymne and Stobart, 1984; Weselake, 2005; Lager et al., 2013). The efficiency of this acyl group channeling to and from PC is an important determinant of the overall composition of FAs in the acyl-CoA pool and, subsequently, in seed oil.
Previously, we demonstrated that the expression of the Brassica napus low-molecular mass ACBP (hereafter referred to as BnACBP) in the presence of Arabidopsis (Arabidopsis thaliana) LPCAT isoforms in an in vitro system enhanced the incorporation of oleic acid (18:1cisΔ9; hereafter referred to as 18:1) into PC and the release of linoleic acid (18:2cisΔ9,12; hereafter referred to as 18:2) from PC into acyl-CoA (Yurchenko et al., 2009). In line with these results, the expression of BnACBP complementary DNA (cDNA) in Arabidopsis developing seeds was also shown to result in elevated levels of the polyunsaturated fatty acids (PUFAs) 18:2 and α-linolenic acid (18:3cisΔ9,12,15; hereafter referred to as 18:3) in seed oil, mainly at the expense of eicosenoic acid (20:1cisΔ11; hereafter referred to as 20:1) and saturated fatty acids (SFAs; Yurchenko et al., 2009). Based on these findings, BnACBP was proposed to be involved in acyl exchange between acyl-CoA and PC pools, which may affect the rate of FA modifications and, ultimately, the FA composition of seed oil (Yurchenko et al., 2009).
In this study, we endeavored to provide further evidence that low-molecular mass ACBP functions in acyl trafficking by investigating whether changes in the FA composition of TAG in Arabidopsis seeds expressing BnACBP were correlated with modifications in the composition of the acyl-CoA pool. In addition, since FA modifications such as elongation and desaturation as well as TAG synthesis occur on ER membranes, we also examined the effect of changing the subcellular localization of BnACBP (from the cytosol to the ER) on the acyl composition of TAG and the acyl-CoA pool in transgenic Arabidopsis. Consequently, we generated localized pools of acyl-CoAs that could be readily accessed by acyltransferases involved in seed oil biosynthesis. Taken together, our findings provide insight into the role of low-molecular mass ACBP in seed oil metabolism and suggest that ACBP (either in its native cytosolic form or as an ER-targeted fusion protein) may serve as a useful tool in biotechnological modifications of FA composition in oil crops.
RESULTS
Molecular Genetic Constructs for Subcellular Targeting of BnACBP
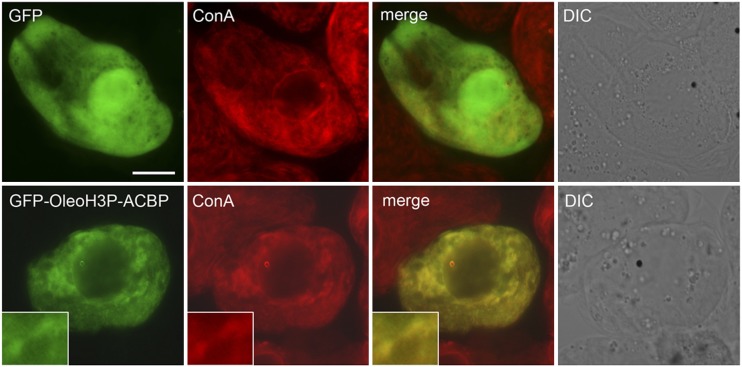
Two experimental constructs were designed for the seed-specific production of BnACBP targeted to different subcellular locations in transgenic Arabidopsis (Fig. 1). The first construct comprised a cDNA encoding BnACBP inserted between the seed-specific β-phaseolin promoter and transcriptional terminator and is referred to as ACBP. The second construct, referred to as OleoH3P-ACBP, was identical in structure to the ACBP construct except that it included an in-frame fusion of BnACBP with a modified version of the Arabidopsis 18-kD OLEOSIN1 gene (OLEO1). Unmodified OLEO1 encodes a protein that is cotranslationally synthesized on the ER and subsequently targeted to oil bodies, where it plays a major role in developing seeds (Qu and Huang, 1990; Beaudoin et al., 1999). In the modified version utilized in this study, the OLEO1 cDNA was altered to encode a protein lacking two segments (20 and 17 amino acids in length, respectively) normally present within its hydrophobic domain, thereby potentially reducing its targeting efficiency to oil bodies. Consistent with this premise, microscopy-based localization studies with OleoH3P-ACBP fused to GFP (GFP-OleoH3P-ACBP) revealed that the fusion protein localized exclusively to the ER in transiently transformed tobacco (Nicotiana tabacum) cells (Fig. 2).
Figure 1.
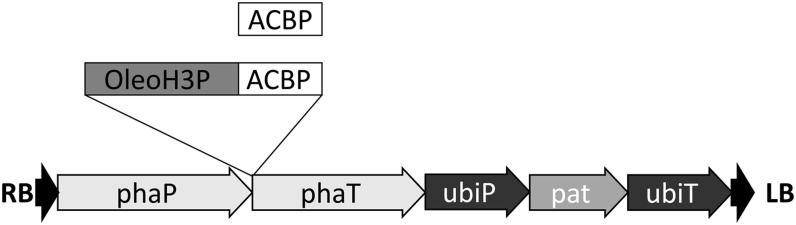
BnACBP-bearing transgenic constructs designed for seed-specific expression in Arabidopsis. Rectangles indicate BnACBP- and OLEO1-derived coding sequences, while arrows indicate the orientation of sequences within the pSBS binary background vector. RB, Right transfer DNA border; phaP, seed-specific β-phaseolin promoter; phaT, β-phaseolin transcriptional terminator; ubiP, ubiquitin promoter; pat, phosphinothricin acetyltransferase herbicide resistance gene; ubiT, ubiquitin transcriptional terminator; LB, left transfer DNA border.
Figure 2.
Subcellular localization of the OleoH3P-BnACBP fusion protein in tobacco suspension-cultured cells. Tobacco cv Bright Yellow-2 cells were transiently transformed (via biolistic bombardment) with either the GFP control (top row) or GFP-OleoH3P-ACBP (bottom row), along with LEC2 in order to transactivate the β-phaseolin promoter in nonseed tissue, and were subsequently visualized using epifluorescence microscopy. Images depict GFP fluorescence (green), ER-localized concanavalin A (ConA) fluorescence (red), and the corresponding merged and differential interference contrast (DIC) images. While GFP was localized throughout the cytosol and nucleus, as expected, GFP-OleoH3P-ACBP colocalized with concanavalin A-stained ER, as evidenced by the yellow color in the merged images. Shown in the insets is a portion of the GFP-OleoH3P-ACBP-transformed cell shown at higher magnification. All micrographs are representative images obtained from at least three replicate biolistic experiments. Bar = 10 μm.
Expression of BnACBP in Transgenic Arabidopsis Seeds
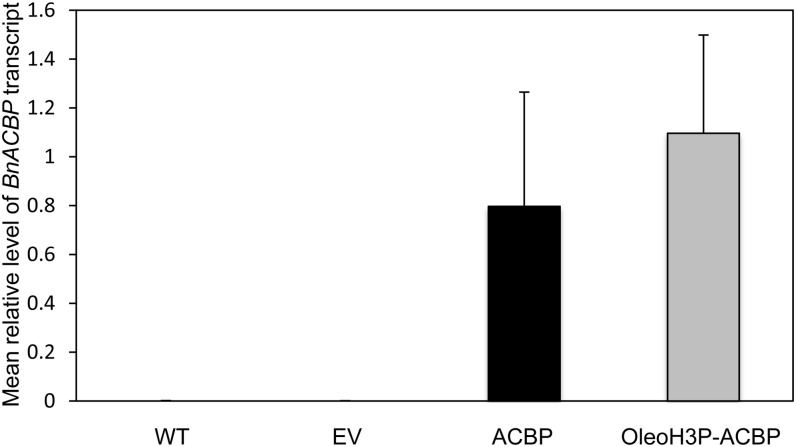
In order to ascertain that both the ACBP and OleoH3P-ACBP constructs resulted in the expression of BnACBP transcripts in the seeds of transgenic Arabidopsis, quantitative real-time reverse transcription (RT)-PCR assays were carried out using total RNA isolated from developing T2 seeds (16 d after flowering [DAF]) of four independent lines bearing each experimental construct as well as empty vector and untransformed lines (wild-type) as negative controls (Fig. 3). As expected, neither wild-type plants nor lines bearing empty vector generated BnACBP transcript at detectable levels in any case. Conversely, lines containing both ACBP and OleoH3P-ACBP constructs resulted in the production of BnACBP transcript at levels that were not significantly different from one another, although the levels were rather variable from line to line.
Figure 3.
Quantitative real-time RT-PCR analysis of BnACBP expression in developing seeds (16 DAF) from four independent transgenic Arabidopsis lines bearing empty vector (EV), ACBP, and OleoH3P-ACBP constructs along with untransformed wild-type controls (WT). Each bar represents the mean level of BnACBP transcript relative to the constitutively expressed internal control, AtPP2AA3. Error bars denote sd.
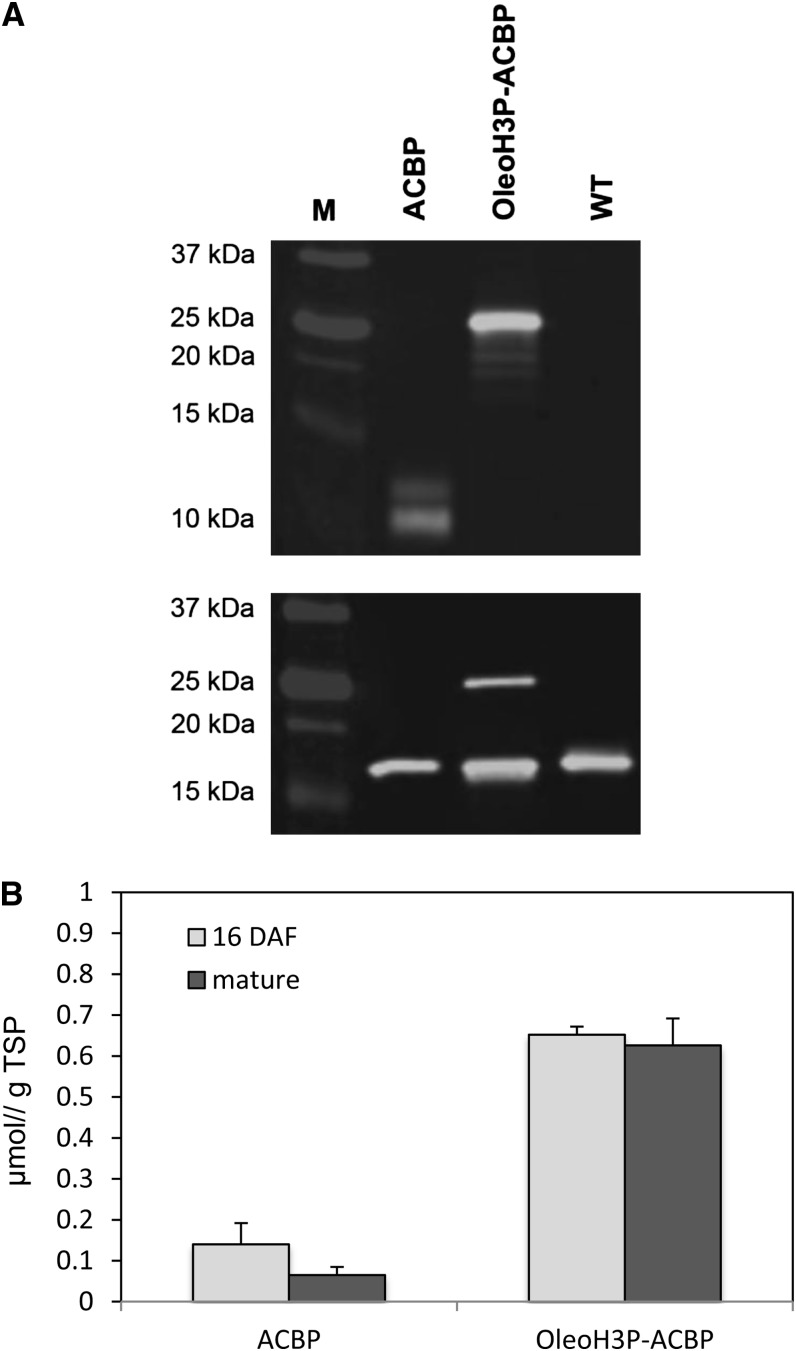
To provide further confirmation of BnACBP expression in transgenic Arabidopsis seeds, the levels of BnACBP protein produced in the various transgenic lines were compared in both mature and developing (16-DAF) T3 seeds using western-blot analysis. Equivalent amounts of total seed protein from four lines bearing each construct as well as an untransformed control were separately probed with anti-BnACBP antibodies and anti-Arabidopsis 18-kD oleosin D9 antibodies, which were used as an internal standard to normalize for equal sample loading (Fig. 4). While no BnACBP protein was detected in wild-type lines, both ACBP and OleoH3P-ACBP lines produced BnACBP protein at detectable levels (Fig. 4A). A more in-depth analysis of the levels of BnACBP protein present in T3 seeds indicated that significantly higher levels of BnACBP were observed in lines containing the OleoH3P-ACBP fusion construct (0.65 ± 0.02 μmol g−1 total seed protein at 16 DAF and 0.63 ± 0.07 μmol g−1 total seed protein in mature seeds) than in ACBP lines (0.14 ± 0.05 μmol g−1 total seed protein at 16 DAF and 0.07 ± 0.02 μmol g−1 total seed protein in mature seeds). Differences between BnACBP levels in developing seeds were not significantly different from those observed in corresponding mature seeds in either case (Fig. 4B).
Figure 4.
Levels of BnACBP protein in Arabidopsis seeds bearing ACBP and OleoH3P-ACBP constructs. A, Representative western blots of total protein from mature T3 seeds. Blots incubated with anti-BnACBP antibodies (top) and anti-OLEO1 antibodies (bottom), which were included to normalize for sample loading, are shown. M, Protein marker (Precision Plus Protein Standards, Bio-Rad); WT, wild-type Arabidopsis. B, Levels of BnACBP in developing (16 DAF) and mature T3 seeds from four independent transgenic lines bearing ACBP and OleoH3P-ACBP constructs. TSP, Total seed protein. Bars represent mean values, and error bars indicate sd.
Arabidopsis T2 and T3 Seeds Expressing BnACBP Displayed Changes in the FA Composition of Their Oil
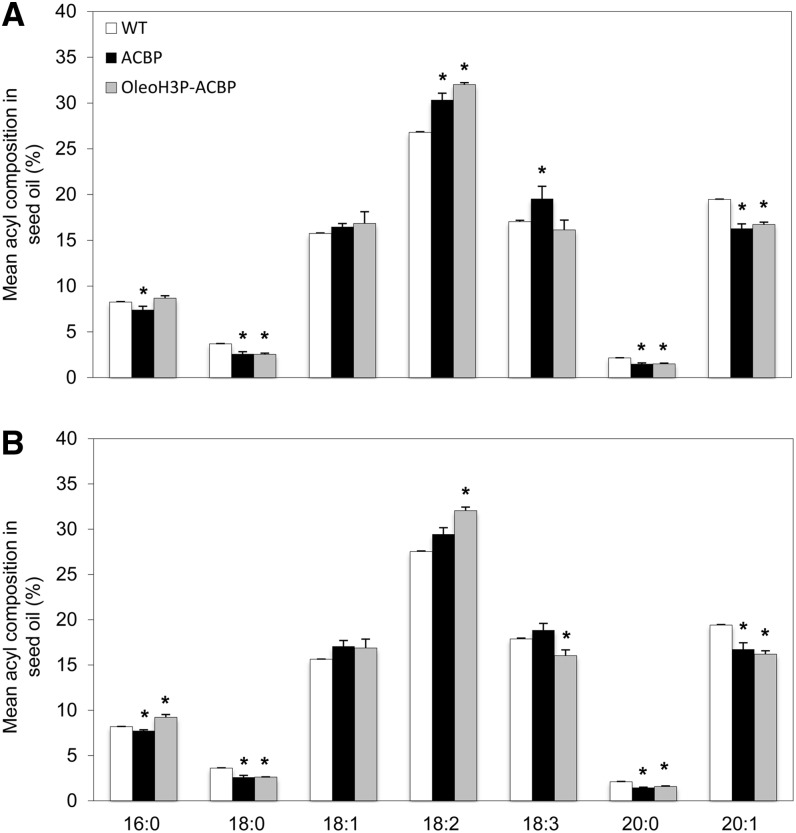
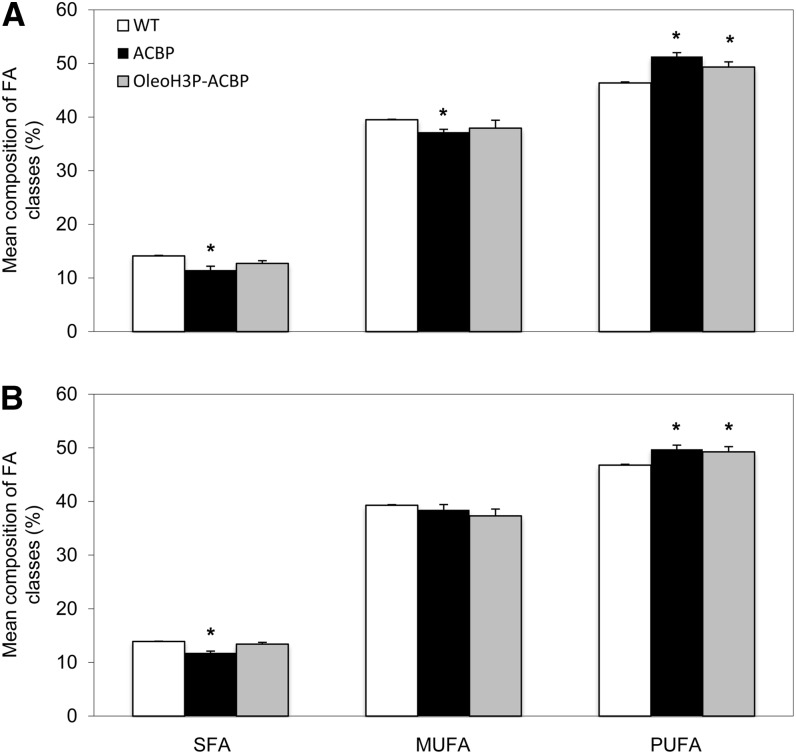
Examination of the composition of oil from mature Arabidopsis T2 seeds bearing both the ACBP and OleoH3P-ACBP constructs revealed a significant increase in 18:2 along with a significant decrease in stearic acid (18:0), eicosanoic acid (20:0), and 20:1 in seed TAG compared with wild-type controls (Fig. 5). Lines transformed with the ACBP construct also exhibited significantly elevated levels of 18:3 as well as decreased palmitic acid (16:0) content in seed oil compared with wild-type plants. No significant changes were observed in oleic acid (18:1) levels in lines bearing either transgenic construct compared with wild-type plants. Overall, T2 lines transformed with both BnACBP-bearing constructs showed a significant increase in PUFAs compared with wild-type plants (Fig. 6), which occurred at the expense of monounsaturated FAs and SFAs. The majority of T2 lines analyzed displayed no changes in seed oil content compared with wild-type controls (data not shown). Indeed, of all the BnACBP-bearing T2 lines initially analyzed (10 ACBP lines and seven OleoH3P-ACBP lines), only two showed a slight but statistically significant increase in seed oil content, one line exhibited a significant decrease in oil content, and the remainder had unaltered seed oil content compared with wild-type seeds (data not shown).
Figure 5.
Fatty acid composition of oil from Arabidopsis wild-type and transgenic T2 (A) and T3 (B) mature seeds. Each bar indicates the mean molar percentage of the particular fatty acid within the seeds of four independent lines bearing each construct as well as four untransformed wild-type control plants (WT). Error bars represent sd, and asterisks denote means that are significantly different from wild-type controls at P ≤ 0.05.
Figure 6.
Composition of fatty acid classes in the oil of Arabidopsis wild-type and transgenic T2 (A) and T3 (B) mature seeds. Each bar indicates the mean molar percentage of the particular fatty acid group within the seeds of four independent lines bearing each construct as well as four untransformed wild-type control plants (WT). Error bars represent sd, and asterisks denote means that are significantly different from wild-type controls at P ≤ 0.05. MUFA, Monounsaturated FAs.
In order to further substantiate these results, an examination of the FA composition from the seed oil of T3 lines was also carried out. The four ACBP and OleoH3P-ACBP lines displaying the highest PUFA content were utilized to produce the next generation of seeds. Analysis of the oil from T3 seeds derived from homozygous lines containing the two BnACBP constructs confirmed our previous findings in T2 seeds, whereby significant increases in PUFA content were evident compared with wild-type controls, mainly due to an increase in 18:2 and a decrease in 18:0, 20:0, and 20:1 (Figs. 5 and 6). T3 seeds containing the ACBP construct also displayed an increase in the proportion of 18:3 (although this was not significant) and a decrease in 16:0 content, while those bearing the OleoH3P-ACBP construct exhibited a decrease in 18:3 and an increase in 16:0. Although these FA composition changes in the oil of T3 seeds were more subtle than those associated with T2 seeds, the results from both data sets point out the significant effect of BnACBP expression on the FA composition of seed oil, which appears to occur mainly through an increase in the proportion of 18:2 (and 18:3 for construct ACBP) and a decrease in the proportion of 18:0, 20:0, and 20:1.
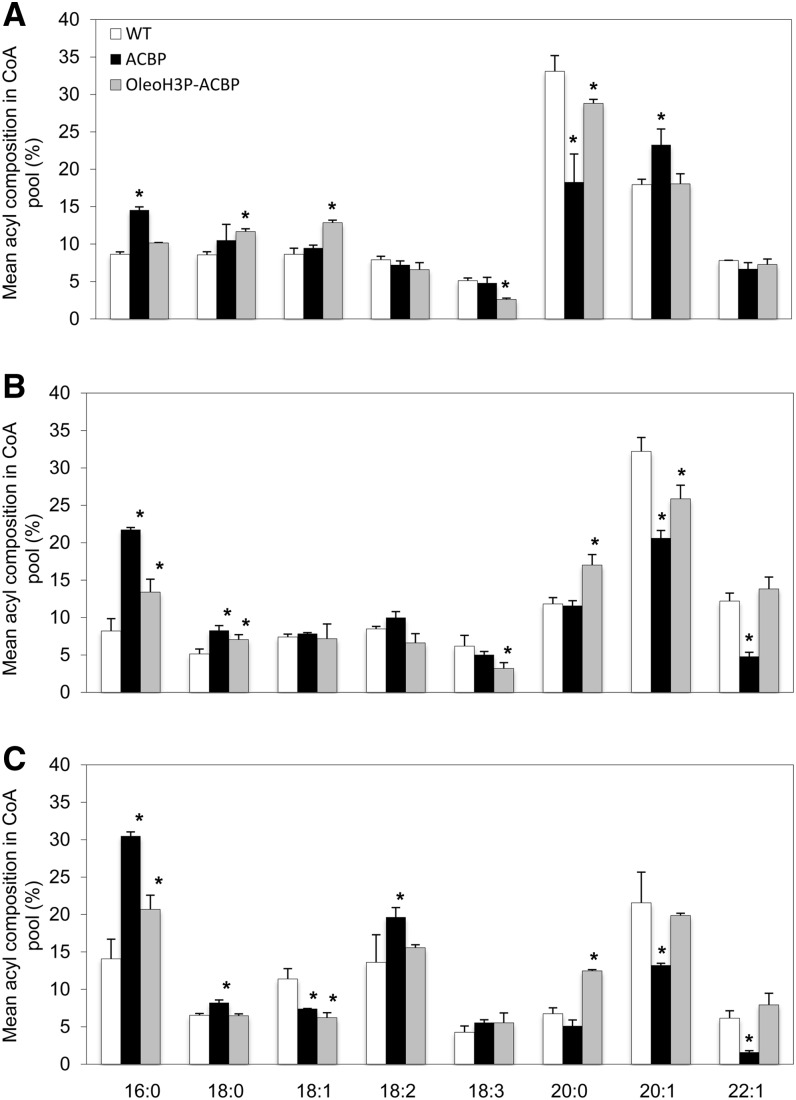
Composition of the Acyl-CoA Pool in Developing T3 Seeds
Developing T3 seeds were harvested at different developmental stages (10, 15, and 20 DAF) from four homozygous lines bearing each construct as well as an untransformed control, and the compositions of the acyl-CoA pools were compared between the different stages of development (Supplemental Fig. S1). In both transgenic and wild-type plants, saturated acyl groups (16:0, 18:0, and 20:0) represented the largest portion of the acyl-CoA pool at 10 DAF. While both 18:0-CoA and 20:0-CoA levels decreased by later stages of development, 16:0-CoA content increased during the course of seed development. In the case of monounsaturated acyl groups, the proportion of 18:1-CoA did not appear to differ dramatically in wild-type seeds over the range of developmental stages tested but decreased slightly in transgenic plants. In wild-type and OleoH3P-ACBP lines, the proportion of 20:1-CoA increased from 10 to 15 DAF but then decreased by 20 DAF, while lines transformed with construct ACBP exhibited a decrease in the proportion of 20:1-CoA in the acyl-CoA pool as seed development progressed. A similar pattern was observed for erucoyl (22:1cisΔ13)-CoA. Finally, although the proportions of polyunsaturated acyl groups (18:2 and 18:3) in the acyl-CoA pool were relatively low at early developmental stages (10–15 DAF), the proportion of 18:2-CoA increased substantially from 15 to 20 DAF in both wild-type and transgenic seeds.
Comparisons were also made between the acyl-CoA pool compositions of transgenic and wild-type seeds at each developmental stage (Fig. 7). At 10 DAF, transgenic seeds bearing the ACBP construct exhibited significantly higher proportions of 16:0-CoA compared with wild-type controls, while the same was true of both ACBP and OleoH3P-ACBP lines at 15 and 20 DAF. The content of 18:0-CoA also tended to be higher in transgenic seeds compared with wild-type seeds, especially during the early stages of seed development (10–15 DAF). Furthermore, OleoH3P-ACBP seeds had higher proportions of 18:1-CoA at 10 DAF compared with those from wild-type plants, with both transgenic lines exhibiting significant reductions in this acyl moiety by 20 DAF. The proportion of 18:2-CoA in transgenic seeds was similar to that in wild-type seeds at 10 and 15 DAF but increased by 20 DAF in lines transformed with the ACBP construct. Interestingly, OleoH3P-ACBP lines, but not those lines transformed with the ACBP construct, had decreased levels of 18:3-CoA at 10 and 15 DAF compared with wild-type plants. While the major acyl group at 10 DAF in wild-type seeds was 20:0, this contributed a smaller proportion of the acyl-CoA pool in all transgenic seeds at this stage of development. However, by 15 to 20 DAF, this acyl group was significantly increased compared with wild-type seeds in OleoH3P-ACBP, but not ACBP, lines. Furthermore, at later stages of seed development (15 and 20 DAF), 20:1-CoA tended to be proportionally lower in transgenic lines than in wild-type seeds, while lines bearing the ACBP construct exhibited decreased 22:1-CoA levels compared with wild-type controls. Taken together, the main changes in acyl-CoA pool composition in developing seeds of ACBP transgenic lines were an increase in saturated fatty acyl-CoAs (16:0 and 18:0) and polyunsaturated 18:2-CoA and a decrease in monounsaturated acyl-CoAs (18:1, 20:1, and 22:1). Lines transformed with the OleoH3P-ACBP construct also displayed an increase in saturated acyl-CoAs (16:0 and 18:0 at 10–15 DAF and 20:0 at 15–20 DAF) and a decrease in 18:1-CoA at 20 DAF. No significant differences in polyunsaturated acyl-CoAs or 20:1-CoA were observed at 20 DAF in OleoH3P-ACBP lines. Thus, seeds from lines bearing the ACBP and OleoH3P-ACBP constructs exhibited different changes in their acyl-CoA compositions compared with wild-type seeds, including increased levels of 18:2-CoA and decreases in 20:1- and 22:1-CoA during later stages of seed development in the case of ACBP lines and decreased 18:3-CoA during early stages of seed development in the case of OleoH3P-ACBP lines.
Figure 7.
Acyl composition of the total acyl-CoA pool from Arabidopsis wild-type and transgenic T3 developing seeds 10 DAF (A), 15 DAF (B), and 20 DAF (C). Each bar denotes the mean molar percentage of the particular acyl group within the seeds of four independent lines bearing each construct as well as four untransformed wild-type control plants (WT). Error bars represent sd, and asterisks indicate means that are significantly different from wild-type controls at P ≤ 0.05.
DISCUSSION
BnACBP is a small protein consisting of 92 amino acid residues and is, to the best of our knowledge, the only ACBP identified to date in this species. As it is expressed at high levels in developing embryos (Hills et al., 1994), it has been proposed that ACBP may play an important role in TAG accumulation in developing seeds. This hypothesis was later supported by the fact that recombinant BnACBP affects acyltransferase activities in vitro (Brown et al., 1998; Yurchenko et al., 2009) and that overexpression of ACBP in developing seeds affects the FA composition of seed oil (Enikeev and Mishutina, 2005; Yurchenko et al., 2009).
In this study, we designed two separate constructs for the seed-specific expression of BnACBP in transgenic Arabidopsis, with the first encoding an unmodified version of cytosolic BnACBP (construct ACBP) and the second encoding a modified OLEO1 fused to BnACBP (construct OleoH3P-ACBP; Fig. 1). More specifically, this modified OleoH3P protein, which lacks a portion of its hydrophobic domain and thus disrupts the ability of the protein to target from the ER to oil bodies, resulted in the localization of the OleoH3P-ACBP fusion protein to the ER (Fig. 2). As such, we were able to differentially increase the concentration of BnACBP protein at two distinct subcellular locations (the cytosol and ER) and examine the effects of ACBP on the acyl composition of the acyl-CoA pool in developing seeds and oil at seed maturity.
Both the ACBP and OleoH3P-ACBP constructs resulted in the production of BnACBP transcript at levels that were not significantly different from one another in developing seeds (16 DAF; Fig. 3). In contrast, the amount of BnACBP protein in developing and mature seeds appeared to be affected by the fusion partner, with OleoH3P-ACBP lines exhibiting higher levels of BnACBP than ACBP lines (Fig. 4). Whether the stability of the OleoH3P-ACBP fusion protein is a virtue of its fusion partner (i.e. OleoH3P), retention at the ER, and/or proximity to an acyl-rich environment is currently unknown. However, the elevation in the quantity of ER-targeted OleoH3P-ACBP products could be explained by the fact that oleosin normally accumulates in developing seeds and remains stable even upon desiccation (Murphy et al., 1989), while the relatively low levels of BnACBP protein might be a result of natural protein turnover in the cell.
The heterologous expression of BnACBP in Arabidopsis developing seeds resulted in increased amounts of PUFAs in the seed oil (Figs. 5 and 6). In transgenic seeds bearing the ACBP construct, this increase was due to an elevation in the proportions of 18:2 and 18:3 at the expense of 16:0, 18:0, 20:0, and 20:1. Conversely, transgenic lines bearing the OleoH3P-ACBP construct resulted in an increased proportion of 18:2 and a decrease in 18:3 in seed oil compared with wild-type controls. Furthermore, rather than a decrease in 16:0 levels, OleoH3P-ACBP lines displayed a slight increase in the levels of this FA. Decreased levels of 18:0, 20:0, and 20:1 in the seed oil from lines transformed with the OleoH3P-ACBP construct were similar to those observed in seeds transformed with the ACBP construct. Similarly, modifications in the acyl-CoA pools within the seeds of transgenic lines were observed. For instance, lines transformed with either BnACBP-containing construct tended to show an increase in the proportion of saturated acyl groups, such as 16:0 and 18:0, at the expense of some monounsaturated groups (18:1 and 20:1) compared with wild-type plants (Fig. 7). During later stages of seed development (15–20 DAF), ACBP lines, but not OleoH3P-ACBP lines, also displayed significant reductions in 20:1-CoA and 22:1-CoA as well as a significant increase in 18:2 acyl groups compared with wild-type plants. In contrast, seeds bearing OleoH3P-ACBP exhibited significantly lower proportions of 18:3 in the acyl-CoA pool at 10 and 15 DAF compared with those from wild-type controls, while the levels of this acyl group remained unchanged in seeds transformed with the ACBP construct. Taken together, these data support our previous findings that ACBP levels in developing seeds might play an important role in the partitioning of acyl-CoA between FA elongation and desaturation (Yurchenko et al., 2009). Also, our results provide evidence that the modified OLEO1 sequence used in this study in an attempt to target BnACBP to the ER also affected the changes observed in FA composition within the acyl-CoA pool and seed oil in transgenic lines. The mechanism(s) underlying this phenomenon, however, remains to be determined.
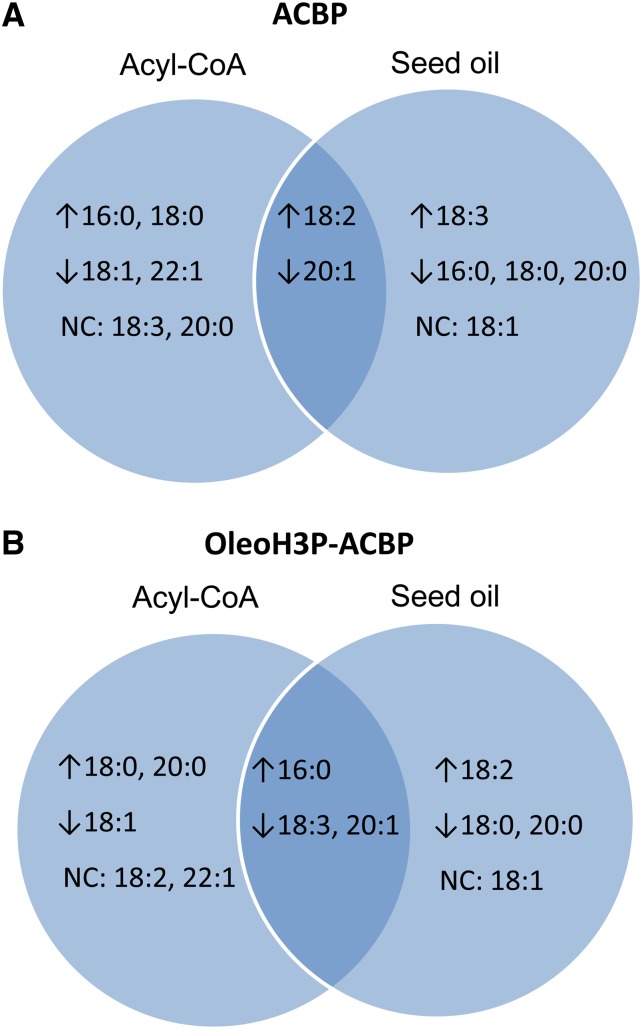
The changes in FA composition of seed oil in lines transformed with either BnACBP-bearing construct (Figs. 5 and 6) can be attributed, at least in part, to modifications in the makeup of the acyl-CoA pool (Figs. 7 and 8). In the case of ACBP lines, the increase in 18:2-CoA observed during late stages of seed development (20 DAF) could be explained by more efficient acyl editing of membrane phospholipids in the presence of higher ACBP levels (Huang et al., 2005; Yurchenko et al., 2009). It is also reasonable to speculate that enhanced channeling of 18:2 into TAG in these lines occurred, at least to some extent, via acyl-CoA-dependent pathways. On the other hand, despite the unchanged levels of 18:3 in the acyl-CoA pool of developing seeds in ACBP lines, relative amounts of this FA were increased in the seed oil. From this observation, we can conclude that acyl-CoA-independent pathways may be responsible for the increased levels of 18:3 in the seed oil of ACBP transgenic seeds. Conversely, the decrease in 20:1-CoA levels at later stages of seed development (15–20 DAF) correlated well with the changes observed in the seed oil of ACBP transgenic lines. Since FA elongation takes place on acyl-CoA substrates, a decrease in 20:1 and 22:1 in the acyl-CoA pool of transgenic ACBP seeds could be explained by lower levels of 18:1-CoA compared with wild-type controls. Finally, even though the relative levels of 16:0 and 18:0 increased significantly in the acyl-CoA pool of ACBP developing seeds, the opposite was observed for these FAs in TAG.
Figure 8.
Comparison of the overall changes in fatty acid composition in the acyl-CoA pool of developing seeds (15–20 DAF) and oil from mature seeds of transgenic Arabidopsis lines bearing ACBP (A) or OleoH3P-ACBP (B) constructs compared with wild-type controls. Changes in the relative amounts of each fatty acid in transgenic seeds are denoted by ↑ (increase compared with wild-type plants), ↓ (decrease compared with wild-type plants), or NC (no change compared with wild-type plants). Overlapping sections indicate fatty acid contents that were similarly modified in both the acyl-CoA pool and seed oil in each case. [See online article for color version of this figure.]
Changes in the FA compositions of the acyl-CoA pool and TAG in transgenic seeds harboring the OleoH3P-ACBP construct were similar to those occurring in ACBP transgenic lines, with only a few exceptions. The most prominent difference between lines bearing the two constructs was an apparent decrease in 18:3 in both the acyl-CoA pool and TAG of T3 seeds in OleoH3P-ACBP lines (Figs. 5, 7, and 8). It is likely that 18:2 produced on PC in developing seeds transformed with an ER-localized construct (OleoH3P-ACBP) would be readily channeled to TAG, resulting in reduced production/liberation of 18:3 and unchanged levels of 18:2 in the acyl-CoA pool compared with wild-type seeds. These differences in the changes observed in the acyl compositions of acyl-CoA pools and TAG between ACBP and OleoH3P-ACBP transgenic seeds likely resulted from a number of contributing factors, including subcellular localization, stability, mobility, total and local concentrations of the recombinant ACBP protein, and perhaps their ability to interact with other proteins and/or lipids.
The fact that overproduction of acyl-CoA-binding sites affected the composition of the acyl-CoA pool (Fig. 7) is not surprising and was observed previously in both Saccharomyces cerevisiae and animal systems (Mandrup et al., 1993; Knudsen et al., 1994; Gaigg et al., 2001; Huang et al., 2005; Oikari et al., 2008). Indeed, the increase in the contribution of saturated acyl-CoAs to the total acyl-CoA pool observed in this study is similar to results reported in yeast and mice overexpressing ACBP (Mandrup et al., 1993; Huang et al., 2005). In a plant system, this could be explained by the premature release of acyl groups from the plastidial fatty acid synthase complex and conversion to acyl-CoA catalyzed by acyl-CoA synthetase, which is enhanced due to product removal by ACBP (Mandrup et al., 1993). Our finding that increases in 16:0 and 18:0 in the acyl-CoA pool of transgenic lines were not reflected in changes observed in TAG indicates that the levels of SFA in TAG may be tightly regulated through specificity and selectivity characteristics of the acyltransferases involved in oil biosynthesis.
Based on the data obtained here, as well as results acquired in a previous study (Yurchenko et al., 2009), we propose that the increase in the prevalence of acyl-CoA-binding sites in developing seeds affected the partitioning of 18:1-CoA between desaturation and elongation, thus resulting in an increase in the proportion of 18:2 in the seed oil, which occurred at the expense of 20:1. The mechanism of ACBP changing the ratio of 18:1-CoA channeling toward elongation and desaturation is not fully understood but could be explained to some extent by enhancing the activity of LPCAT (in both forward and reverse reactions) in the presence of higher levels of BnACBP (Yurchenko et al., 2009). This, in turn, could affect the overall turnover rate of acyl groups (acyl editing) on phospholipids (mainly on PC), enhancing channeling of the substrate (18:1) to the site of desaturation and the products (18:2 and 18:3) back to the acyl-CoA pool or directly to neutral lipids.
CONCLUSION
In this study, we present new findings regarding the effect of low-molecular mass ACBP on seed oil biosynthesis. We found that, in addition to changes in the FA composition of seed oil in transgenic Arabidopsis expressing BnACBP (Yurchenko et al., 2009), the acyl group composition of the acyl-CoA pool in their developing seeds was also significantly altered compared with wild-type control plants. Some changes in the composition of the acyl-CoA pool (such as an increase in 18:2-CoA and a decrease in 20:1-CoA at 20 DAF) confirmed our hypothesis that cytosolic ACBP is involved in regulating the partitioning of 18:1 between elongation and desaturation on the ER, possibly by stimulating the acyl editing of PC. Other changes in the composition of the acyl-CoA pool (such as increases in 16:0-CoA and 18:0-CoA) differed from those observed in the seed oil of transgenic Arabidopsis (decreases in 16:0 and 18:0) and are thus more difficult to interpret. However, these differences indicate that additional mechanisms are involved in the regulation of FA composition in TAG, such as the selectivity of acyltransferases.
In addition to Arabidopsis lines bearing a BnACBP construct that produces a cytosolic protein, we also tested a novel construct in which BnACBP was fused in frame with an OLEO1 variant (OleoH3P) that was modified to enhance protein retention on the ER. Lines bearing the OleoH3P-ACBP fusion construct exhibited changes in the FA composition of the acyl-CoA pool and seed oil that were similar to those observed in lines transformed with unmodified BnACBP, with some exceptions. Taken together, our findings provide insight into the role of low-molecular mass ACBP in seed oil metabolism. While additional studies are required to fully understand the role of ACBP in TAG composition, our findings suggest that this protein (either in its native cytosolic form or as an ER-targeted fusion) may be useful in the biotechnological modification of FA composition in oil crops.
MATERIALS AND METHODS
Preparation of Genetic Constructs for BnACBP Expression in Developing Seeds
Diagrammatic representations of the vectors utilized in this study for Arabidopsis (Arabidopsis thaliana) transformation can be found in Figure 1. Full-length cDNA encoding Brassica napus cytosolic low-molecular mass ACBP (BnACBP) was obtained from Picoscript based on the cDNA sequence available in the GenBank database (accession no. X77134). Restriction endonuclease sites were added to the 5′ and 3′ ends (NcoI and XhoI at the 5′ end and HindIII at the 3′ end) to facilitate molecular cloning. In the case of the ACBP construct, the BnACBP cDNA was inserted into the pSBS4006 binary vector (SemBioSys) between the Phaseolus vulgaris β-phaseolin promoter and transcriptional terminator to allow seed-specific expression. Construct OleoH3P-ACBP included an in-frame fusion between a modified Arabidopsis OLEO1 (GenBank accession no. X62353) fragment with a shortened hydrophobic domain and the full-length BnACBP cDNA. To generate the modified OleoH3P fragment, primers NTD2 (5′-TATTCTCGAGCCATGGCGGATACTGCTAGAGG-3′), which contains XhoI and NcoI restriction sites (in boldface), and NTR (5′-CAGTGGCGCCTTTAGCAATCTGTCTAGAC-3′), which contains an NarI restriction site (in boldface), were utilized to amplify a 136-nucleotide fragment of the OLEO1 cDNA beginning at the start codon. A second 102-nucleotide fragment was amplified from the same source using primers HN3D (5′-CCTTAGCGGCGCCGGAACTGTCATAGCTTTG-3′), which contains an NarI restriction site (in boldface), and HC3R (5′-AGAGTTAAAAATACCGGTGATGAGGAGT-3′), which contains an MseI restriction site (in boldface). This second fragment comprised nucleotides 137 through 297 of the OLEO1 cDNA, where nucleotide 1 is the first nucleotide of the start codon. Both PCR fragments were digested with NarI and subsequently ligated. This fragment was then amplified with primers NTD2 and HC3R using the ligation mixture as a template, while a third 165-bp fragment encoding the C-terminal portion of OLEO1 was amplified from cDNA using primers CTD (5′-TGGATTTTTAAGTACGCAACGGGAGAGC-3′), which contains an MseI site (in boldface), and CTR (5′-AGCCATACTAGTAGTGTGTTGACCACCACGAG-3′), which contains an SpeI site in place of the stop codon (in boldface). PCR fragments were digested with MseI and ligated to generate the OleoH3P sequence. This modified OLEO1 fragment bore a deletion of nucleotides 136 through 195 (corresponding to amino acids 46–65), which were replaced by an NarI site (generating a Gly and an Ala), as well as nucleotides 301 through 351 (corresponding to amino acids 101–117). The OleoH3P fragment was subsequently inserted between the β-phaseolin promoter and terminator in a pSBS background (SemBioSys), and the BnACBP cDNA was then fused in frame to the C terminus of the OleoH3P sequence to generate construct OleoH3P-ACBP.
In order to confirm that the OleoH3P-ACBP cassette did, indeed, target BnACBP to the ER, we also produced an additional vector bearing an in-frame fusion of OleoH3P-ACBP to GFP for localization studies (construct GFP-OleoH3P-ACBP). This was achieved by initially amplifying the mGFP-4 sequence from the pSK-GFP vector with primers OY32 (5′-TTTCATGACGCACAATCCCACTATCC-3′) and OY33 (5′-TTTCATGAGTTTGTATAGTTCATCCATGCC-3′), which have added BspHI sites (in boldface). The resulting mGFP-4 fragment was then inserted into the NcoI site (which has compatible cohesive ends with BspHI-digested fragments) downstream of the β-phaseolin promoter and upstream of the OleoH3P fragment in the OleoH3P-ACBP construct. A control vector including a β-phaseolin promoter::mGFP-4::β-phaseolin transcriptional terminator was also produced by replacing the BnACBP fragment from the ACBP construct with the mGFP-4 sequence, which had been amplified with primers mGFPF1SmaI (5′-GACCCGGGACAATGAGTAAAGGAGAAGAAC-3′) and mGFPR1HindIII (5′-AGGAAGCTTATTTGTATAGTTCATCCATGC-3′).
All binary vectors also contained an antibiotic resistance marker cassette (spectinomycin) for bacterial selection and an herbicide resistance cassette (PHOSPHINOTHRICIN ACETYLTRANSFERASE) inserted between the parsley (Petroselinum crispum) polyubiquitin promoter and transcriptional terminator for plant selection. Conventional procedures were used for molecular cloning and bacterial transformation (Sambrook et al., 1989). All molecular constructs were sequenced prior to plant transformation.
Plant Transformation and Growth Conditions
Arabidopsis transformation and growth conditions were conducted according to Nykiforuk and Boothe (2012). In brief, Arabidopsis plants (C-24) were grown in Sunshine LA4 soil mix (SunGro) in a growth chamber with a 16-h light period and 350 μE of light intensity at a constant temperature of 20°C. ACBP and OleoH3P-ACBP vectors were introduced into Agrobacterium tumefaciens strain EHA101 via electroporation, with subsequent A. tumefaciens-mediated transformation of Arabidopsis performed using the floral dip method (Clough and Bent, 1998). Treated plants were placed under a dome for 16 to 24 h to maintain high humidity and then grown normally until seeds reached maturity. T1 seeds were germinated on selective medium containing 80 μm dl-phosphinothricin, and herbicide-resistant transgenic T1 seedlings were transferred to soil 7 d after germination. Plants were grown individually in a growth chamber to produce mature T2 seeds for seed oil analysis. T2 seeds included a mixture of heterozygous and homozygous genotypes for the presence of the transgene, while approximately 25% were null segregants with a wild-type genotype. T2 seeds from four lines bearing each construct were selected based on their ability to generate the highest levels of PUFAs in their seed oil for producing the next generation of transgenic seeds. Ten T2 plants were grown from the seeds of each line to produce homozygous T3 seeds, which were harvested separately and used for further analyses. Sixty T3 seeds from each plant were utilized for segregation analyses to determine the zygosity of the parent T2 plant in each case.
Quantitative Real-Time RT-PCR Analyses
Total RNA was extracted from developing siliques (16 DAF) from four independent transgenic lines bearing ACBP, OleoH3P-ACBP, and empty vector constructs as well as untransformed controls using the Qiagen Plant RNeasy kit according to the manufacturer’s instructions. Contaminating genomic DNA was removed using the TURBO DNA-free system (Ambion, Life Technologies). First-strand cDNA synthesis was performed using the SuperScript III first-strand cDNA synthesis kit (Invitrogen, Life Technologies) with approximately 100 ng of total RNA as template and an oligo(dT) primer in a final volume of 10 μL. Subsequent quantitative real-time PCR assays were performed in triplicate with 1 μL of a 1:5 dilution of each cDNA as template along with SYBR Green PCR master mix in a final volume of 10 μL according to the manufacturer’s recommendations (Applied Biosystems, Life Technologies). Reactions were carried out using the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). BnACBP-specific primers BnACBPF2 (5′-TGACCACCAGTCGTCCTGGGA-3′) and BnACBPR2 (5′-CACCATCAAGCTGAAGCGGAG-3′) were designed to amplify a 162-nucleotide fragment of the BnACBP transcript without concomitant amplification of Arabidopsis ACBP sequences. In addition, a 146-nucleotide fragment of a constitutively expressed internal standard transcript encoding protein phosphatase type 2A subunit A3 (AtPP2AA3; GenBank accession no. BT002601; Dekkers et al., 2012) was amplified using primers AtPP2AAF1 (5′-TCAATCCGTGAAGCTGCTGCAAAC-3′) and AtPP2AAR1 (5′-ACTGCACGAAGAATCGTCATCCGA-3′). Reactions lacking template cDNA and those without reverse transcriptase were included in each case as negative controls. Thermal parameters for amplification were 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Dissociation curves were generated to ascertain that only a single product was produced in each case. Relative levels of gene expression were obtained through the use of standard curves produced via a set of serial dilutions. All BnACBP expression data represent mean values normalized to those of AtPP2AA3.
Western-Blot Analyses
Developing embryos from six siliques collected 16 DAF and 40 mature homozygous T3 Arabidopsis seeds (approximately 25 mg) obtained from four independent transgenic lines were ground in 0.5 mL of extraction buffer (0.4 m Suc, 0.5 m NaCl, and 50 mm Tris-HCl, pH 8.0). Total seed proteins were solubilized through the addition of 10% (w/v) SDS (to a final concentration of 2% [w/v] SDS) followed by boiling for 10 min. Total protein content was determined using the bicinchoninic acid protein assay (Pierce). Approximately 2 μg of total seed protein per sample was then analyzed by 15% (w/v) SDS-PAGE using standard protocols (Sambrook et al., 1989) and stained with Coomassie Brilliant Blue R 250 or blotted for western analysis. Blotted samples were probed with polyclonal antibody directed against BnACBP (Brown et al., 1998) or monoclonal antibody directed against Arabidopsis OLEO1 (raised by SemBioSys). BnACBP was detected using secondary donkey anti-rabbit IRDye 800 CW (Li-Cor Biosciences), while both the full-length and modified oleosin were detected using secondary goat anti-mouse IRDye 800 CW (Li-Cor Biosciences). Both were subsequently analyzed using the Odyssey Infrared Imaging System (Li-Cor Biosciences). The OLEO1 protein was used as an internal standard to normalize for equal sample loading, while a range of known amounts of BnACBP produced in Escherichia coli were used to quantify the levels of BnACBP in total seed protein samples (Nykiforuk et al., 2006; Yurchenko et al., 2009).
Analysis of the Fatty Acid Composition of Oil from Mature Seeds
Seed oil was extracted from 10 mg of mature T2 and T3 seeds using the hexane-isopropanol method (Hara and Radin, 1978). In the case of T2 seeds, 10 ACBP and seven OleoH3P-ACBP independent transgenic lines were analyzed initially, and the four lines displaying the most prominent alterations in PUFA content in seed TAG were used for statistical analyses and the production of T3 seeds. For all experiments, three technical replicates were carried out per line. Methanolic HCl was used as a methylation agent for preparing FA methyl esters for subsequent separation by gas chromatography. Internal standard (tripentadecanoylglycerol) was added to each sample before seed oil extraction to estimate the total FA content by gas chromatography. FA methyl esters were analyzed on an Agilent 6890N Gas Chromatograph with the 5975 Inert XL Mass Selective Detector equipped with an autosampler (Agilent) and were separated using a capillary DB-23 (30-m) column (0.25 mm × 0.25 µm × 30 m) with a constant helium flow of 1.2 mL min−1 and a temperature program of 90°C to 180°C at 10°C min−1, followed by a hold at 180°C for 5 min, and finally 180°C to 230°C at 5°C min−1. Integration events were detected and identified between 9 and 20 min and compared with a NuChek 463 gas-liquid chromatography standard.
Analysis of the Acyl-CoA Pool in Developing Seeds
Derivatization and analysis of the acyl-CoA pool in Arabidopsis developing seeds were conducted according to the technique described by Larson and Graham (2001). Briefly, developing homozygous T3 seeds derived from four independent transgenic lines were obtained from five to six frozen siliques (at 10, 15, and 20 DAF). Seeds were placed in a centrifuge tube with a 3-mm glass bead (Fisher Scientific) and homogenized on a bead beater (Biospec Products) for 1 min. Two hundred microliters of freshly prepared extraction buffer (50% [v/v] isopropanol, 25 mm KH2PO4, pH 7.2, 1.25% [v/v] glacial acetic acid, and 1 mg mL−1 essentially FA-free bovine serum albumin) and 400 µL of petroleum ether saturated with isopropanol:water (1:1, v/v) were added to each tube. Samples were vortexed and centrifuged for 1 min at 200g, and the upper phase was discarded. The lower phase was washed with 400 µL of petroleum ether saturated with isopropanol:water (1:1, v/v) a further three to four times. Finally, 5 µL of saturated ammonium sulfate and 600 µL of methanol:chloroform (2:1, v/v) were added to the washed lower phase. Samples were mixed, incubated at room temperature for 20 min, and centrifuged at 12,000g for 2 min. The supernatant was transferred to 2-mL HPLC vials and dried under a vacuum (Savant Automatic Environmental SpeedVac with VaporNet AES 2000; Fisher Scientific). When dry, 100 µL of the derivatizing agent (0.5 m chloroacetaldehyde in 0.15 m trisodium citrate/citric acid, pH 4.0, and 0.5% [w/v] SDS) was added to each vial and incubated for 20 min at 85°C. Subsequently, the reaction mixture was filtered through a 0.45-µm nylon filter in a 2-mL tube and transferred back to an HPLC vial with a spring insert. Samples were analyzed on an Agilent Technologies 1200 HPLC system with a LUNA-18(2) (150 mm × 2 mm × 5 µm) column equipped with an in-line fluorescence detector using the method described by Larson and Graham (2001).
Analysis of Subcellular Localization
Tobacco (Nicotiana tabacum ‘Bright Yellow-2’) suspension-cultured cells were biolistically cobombarded with plasmid DNA encoding the GFP-OleoH3P-ACBP or GFP (control) construct along with the 35S::LEAFY COTYLEDON2 vector (Petrie et al., 2010) to allow expression from the seed-specific β-phaseolin promoter in nonseed tissue. Transformations were carried out using a biolistic particle delivery system (1000/HE; Bio-Rad Laboratories). Following bombardment, cells were incubated for 20 h to allow for expression and sorting of the introduced gene product(s) and were then fixed in formaldehyde and incubated with 0.01% (w/v) pectolyase Y-23 (Kyowa Chemical Products) followed by permeabilization with Triton X-100. Procedures for cv Bright Yellow-2 cell maintenance, biolistic bombardment, and processing for microscopy are described elsewhere (Lingard et al., 2008). Concanavalin A conjugated to Alexa 594 (Molecular Probes) was added to cells at a final concentration of 5 μg mL−1 and incubated for 20 min, as described elsewhere (Shockey et al., 2006).
Epifluorescent images of cells were acquired using an Axioscope 2 MOT epifluorescence microscope (Carl Zeiss) with a 63× Plan Apochromat oil-immersion objective. Images were captured using a Retiga 1300 CCD camera (Qimaging) and Openlab software (Improvision). Figures were composed using Adobe Photoshop CS (Adobe Systems).
Statistical Analyses
SAS 9.1 software (SAS Institute) was used for performing statistical procedures. FA profile, defined here as the composition of FA classes (saturated, monounsaturated, and polyunsaturated), and FA composition of seed oil or the acyl-CoA pool from seeds bearing each construct were compared with those from wild-type controls using ANOVA (General Linear Model procedure) with Dunnett’s test. In the case of quantitative real-time RT-PCR data, two-tailed paired Student’s t tests were carried out to determine the significance of differences in BnACBP expression levels between ACBP and OleoH3P-ACBP lines. Four biological replicates (consisting of independent transgenic lines representing distinct integration events) per construct were used in the analyses. In every case, differences were considered significant at P ≤ 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Acyl composition of the total acyl-CoA pool from Arabidopsis seeds at various developmental stages.
Supplementary Material
Acknowledgments
We thank Dr. Matthew Hills (John Innes Centre) for the primary antibodies against B. napus ACBP, Dr. Rodrigo Siloto (University of Alberta) for assistance with the generation of transgenic constructs, and Crystal Snyder (University of Alberta) for technical support in the biochemical analysis of seed oil and acyl-CoAs.
Glossary
- FA
fatty acid
- ER
endoplasmic reticulum
- PC
phosphatidylcholine
- TAG
triacylglycerol
- LPCAT
lysophosphatidylcholine acyltransferase
- cDNA
complementary DNA
- PUFA
polyunsaturated fatty acid
- SFA
saturated fatty acid
- DAF
days after flowering
- RT
reverse transcription
Footnotes
This work was supported by the Alberta Crop Industry Development Fund Student Development Program (to O.Y.), the Natural Sciences and Engineering Research Council of Canada (Discovery Grants to R.J.W. and R.T.M.), the University of Guelph Research Chairs Program (to R.T.M.), and the Canada Research Chairs Program (to R.J.W.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Banaś A, Dahlqvist A, Ståhl U, Lenman M, Stymne S. (2000) The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem Soc Trans 28: 703–705 [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Lacey DJ, Napier JA. (1999) The biogenesis of the plant seed oil body: oleosin protein is synthesised by ER-bound ribosomes. Plant Physiol Biochem 37: 481–490 [Google Scholar]
- Broun P, Boddupalli S, Somerville C. (1998) A bifunctional oleate 12-hydroxylase: desaturase from Lesquerella fendleri. Plant J 13: 201–210 [DOI] [PubMed] [Google Scholar]
- Brown AP, Johnson P, Rawsthorne S, Hills MJ. (1998) Expression and properties of acyl-CoA binding protein from Brassica napus. Plant Physiol Biochem 36: 629–635 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Ståhl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97: 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HW, Bentsink L. (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53: 28–37 [DOI] [PubMed] [Google Scholar]
- Enikeev AG, Mishutina UO. (2005) Physiological effects of rapeseed transformation with the acb gene as affected by the genetic vector structure. Russian Journal of Plant Plysiology 52: 668–671 [Google Scholar]
- Faergeman NJ, Wadum M, Feddersen S, Burton M, Kragelund BB, Knudsen J. (2007) Acyl-CoA binding proteins: structural and functional conservation over 2000 MYA. Mol Cell Biochem 299: 55–65 [DOI] [PubMed] [Google Scholar]
- Gaigg B, Neergaard TB, Schneiter R, Hansen JK, Faergeman NJ, Jensen NA, Andersen JR, Friis J, Sandhoff R, Schrøder HD, et al. (2001) Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol Biol Cell 12: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C, Martín-Rufián M, Reina JJ, Heredia A. (2006) Isolation and characterization of a cDNA encoding a membrane bound acyl-CoA binding protein from Agave americana L. epidermis. Plant Physiol Biochem 44: 85–90 [DOI] [PubMed] [Google Scholar]
- Hara A, Radin NS. (1978) Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem 90: 420–426 [DOI] [PubMed] [Google Scholar]
- Hills MJ, Dann R, Lydiate D, Sharpe A. (1994) Molecular cloning of a cDNA from Brassica napus L. for a homologue of acyl-CoA-binding protein. Plant Mol Biol 25: 917–920 [DOI] [PubMed] [Google Scholar]
- Huang H, Atshaves BP, Frolov A, Kier AB, Schroeder F. (2005) Acyl-coenzyme A binding protein expression alters liver fatty acyl-coenzyme A metabolism. Biochemistry 44: 10282–10297 [DOI] [PubMed] [Google Scholar]
- Ichihara K, Mae K, Sano Y, Tanaka K. (1995) 1-Acylglycerophosphocholine O-acyltransferase in maturing safflower seeds. Planta 196: 551–557 [Google Scholar]
- Jaworski JG (1987) Biosynthesis of monoenoic and polyenoic fatty acids. In PK Stumpf, ed, The Biochemistry of Plants, Vol 9. Academic Press, New York, pp 159–173 [Google Scholar]
- Knudsen J, Faergeman NJ, Skøtt H, Hummel R, Børsting C, Rose TM, Andersen JS, Højrup P, Roepstorff P, Kristiansen K. (1994) Yeast acyl-CoA-binding protein: acyl-CoA-binding affinity and effect on intracellular acyl-CoA pool size. Biochem J 302: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen J, Jensen MV, Hansen JK, Faergeman NJ, Neergaard TB, Gaigg B. (1999) Role of acylCoA binding protein in acylCoA transport, metabolism and cell signaling. Mol Cell Biochem 192: 95–103 [DOI] [PubMed] [Google Scholar]
- Lager I, Yilmaz JL, Zhou XR, Jasieniecka K, Kazachkov M, Wang P, Zou J, Weselake R, Smith MA, Bayon S, et al. (2013) Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J Biol Chem 288: 36902–36914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TR, Graham IA. (2001) A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. (2008) Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrup S, Jepsen R, Skøtt H, Rosendal J, Højrup P, Kristiansen K, Knudsen J. (1993) Effect of heterologous expression of acyl-CoA-binding protein on acyl-CoA level and composition in yeast. Biochem J 290: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Browse J. (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Murphy DJ, Cummins I, Kang AS. (1989) Synthesis of the major oil-body membrane protein in developing rapeseed (Brassica napus) embryos: integration with storage-lipid and storage-protein synthesis and implications for the mechanism of oil-body formation. Biochem J 258: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykiforuk CL, Boothe JG. (2012) Transgenic expression of therapeutic proteins in Arabidopsis thaliana seed. Methods Mol Biol 899: 239–264 [DOI] [PubMed] [Google Scholar]
- Nykiforuk CL, Boothe JG, Murray EW, Keon RG, Goren HJ, Markley NA, Moloney MM. (2006) Transgenic expression and recovery of biologically active recombinant human insulin from Arabidopsis thaliana seeds. Plant Biotechnol J 4: 77–85 [DOI] [PubMed] [Google Scholar]
- Oikari S, Ahtialansaari T, Heinonen MV, Mauriala T, Auriola S, Kiehne K, Fölsch UR, Jänne J, Alhonen L, Herzig KH. (2008) Downregulation of PPARs and SREBP by acyl-CoA-binding protein overexpression in transgenic rats. Pflugers Arch 456: 369–377 [DOI] [PubMed] [Google Scholar]
- Petrie JR, Shrestha P, Liu Q, Mansour MP, Wood CC, Zhou XR, Nichols PD, Green AG, Singh SP. (2010) Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu RD, Huang AH. (1990) Oleosin KD 18 on the surface of oil bodies in maize: genomic and cDNA sequences and the deduced protein structure. J Biol Chem 265: 2238–2243 [PubMed] [Google Scholar]
- Rasmussen JT, Faergeman NJ, Kristiansen K, Knudsen J. (1994) Acyl-CoA-binding protein (ACBP) can mediate intermembrane acyl-CoA transport and donate acyl-CoA for beta-oxidation and glycerolipid synthesis. Biochem J 299: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S. (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ. (2009) Acyltransferase action in the modification of seed oil biosynthesis. New Biotechnol 26: 11–16 [DOI] [PubMed] [Google Scholar]
- Stobart K, Mancha M, Lenman M, Dahlqvist A, Stymne S. (1997) Triacylglycerols are synthesised and utilized by transacylation reactions in microsomal preparations of developing safflower (Carthamus tinctorius L.) seeds. Planta 203: 58–66 [Google Scholar]
- Stymne S, Stobart AK. (1984) Evidence for the reversibility of the acyl-CoA:lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons and rat liver. Biochem J 223: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ (2005) Storage production metabolism in microspore-derived cultures of Brassicaceae. In T Nagata, H Lörz, JM Widholm, eds, Biotechnology in Agriculture and Forestry: 56. Haploids in Crop Improvement II. Springer-Verlag, Berlin, pp 97–122 [Google Scholar]
- Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PB, Harwood JL. (2009) Increasing the flow of carbon into seed oil. Biotechnol Adv 27: 866–878 [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. (2011) New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res 50: 141–151 [DOI] [PubMed] [Google Scholar]
- Yurchenko OP, Nykiforuk CL, Moloney MM, Ståhl U, Banaś A, Stymne S, Weselake RJ. (2009) A 10-kDa acyl-CoA-binding protein (ACBP) from Brassica napus enhances acyl exchange between acyl-CoA and phosphatidylcholine. Plant Biotechnol J 7: 602–610 [DOI] [PubMed] [Google Scholar]
- Yurchenko OP, Weselake RJ. (2011) Involvement of low molecular mass soluble acyl-CoA-binding protein in seed oil biosynthesis. New Biotechnol 28: 97–109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.