Change in light intensity, rather than light/dark transition, regulates the activity of pyruvate orthophosphate dikinase in maize via reversible phosphorylation at Thr-527.
Abstract
In C4 plants, pyruvate orthophosphate dikinase (PPDK) activity is tightly dark/light regulated by reversible phosphorylation of an active-site threonine (Thr) residue; this process is catalyzed by PPDK regulatory protein (PDRP). Phosphorylation and dephosphorylation of PPDK lead to its inactivation and activation, respectively. Here, we show that light intensity rather than the light/dark transition regulates PPDK activity by modulating the reversible phosphorylation at Thr-527 (previously termed Thr-456) of PPDK in maize (Zea mays). The amount of PPDK (unphosphorylated) involved in C4 photosynthesis is indeed strictly controlled by light intensity, despite the high levels of PPDK protein that accumulate in mesophyll chloroplasts. In addition, we identified a transit peptide cleavage site, uncovered partial amino-terminal acetylation, and detected phosphorylation at four serine (Ser)/Thr residues, two of which were previously unknown in maize. In vitro experiments indicated that Thr-527 and Ser-528, but not Thr-309 and Ser-506, are targets of PDRP. Modeling suggests that the two hydrogen bonds between the highly conserved residues Ser-528 and glycine-525 are required for PDRP-mediated phosphorylation of the active-site Thr-527 of PPDK. Taken together, our results suggest that the regulation of maize plastid PPDK isoform (C4PPDK) activity is much more complex than previously reported. These diverse regulatory pathways may work alone or in combination to fine-tune C4PPDK activity in response to changes in lighting.
Pyruvate orthophosphate dikinase (PPDK) is an abundant mesophyll-chloroplast enzyme involved in C4 photosynthesis. It plays an essential role in regenerating phosphoenolpyruvate (PEP), the primary cellular CO2 acceptor molecule. PPDK activity strongly correlates (r = 0.96) with the photosynthetic rate (Edwards et al., 1985). Therefore, PPDK may limit the rate of CO2 assimilation in the C4 cycle (Hatch, 1987). PPDK regulatory protein (PDRP), a unique bifunctional enzyme, catalyzes this light-dependent regulation by reversible phosphorylation of an active-site Thr in PPDK (Thr-527 in maize [Zea mays] in full amino acid sequence [http://www.maizegdb.org]; previously termed Thr-456; Ashton and Hatch, 1983; Burnell and Hatch, 1985; Roeske and Chollet, 1987; Ashton et al., 1990; Burnell, 1990; Chastain et al., 2000, 2011). PDRP is an unusual regulatory protein for three reasons (Chastain et al., 1997, 2008; Burnell and Chastain, 2006; Astley et al., 2011): (1) it is bifunctional, catalyzing both PPDK activation/dephosphorylation and PPDK inactivation/phosphorylation; (2) it uses ADP instead of ATP as the phosphoryl donor; and (3) it employs an inorganic phosphate-dependent, inorganic pyrophosphate-forming dephosphorylation mechanism as opposed to the simple hydrolysis mechanism common to most protein phosphatases.
The functional properties of PDRP have been examined by selective substitutions at His-458 and active-site Thr-456 in the maize plastid PPDK isoform (C4PPDK; Ashton and Hatch, 1983; Burnell and Hatch, 1984, 1985). These studies confirmed that PDRP is a Ser/Thr kinase that requires a phosphorylated His in the target enzyme (Burnell and Hatch, 1986). This regulatory threonyl phosphorylation of PPDK is a monocyclic cascade (Stadtman and Chock, 1977) in which the covalent modification system is assumed to be a continuous process that allows the extent of PPDK activation to be attuned to the metabolic needs (Roeske and Chollet, 1989). Therefore, PDRP can alter the activation state of its target enzyme, PPDK, according to the concentrations of metabolites (e.g. ADP, inorganic phosphate, pyruvate, and PEP) involved in the regulatory cycle. In addition, PPDK activity also can be modulated by Mg2+ and temperature (Hatch and Slack, 1968; Wang et al., 2008).
In all plants, PPDK is located in both cytoplasmic and plastid compartments (Chastain and Chollet, 2003). Regulation of the bidirectional activities of C4PPDK has been proposed to be the consequence of light/dark-mediated changes in the stromal ADP level via its action as a potent competitive inhibitor of the PDRP phospho-PPDK dephosphorylation function (Burnell and Hatch, 1985; Chastain et al., 2011). However, GDP can serve as a substrate for the regulatory phosphorylation of the cytoplasmic PPDK isoform (Chastain et al., 2011). Two genes that encode chloroplastic (RP1) and cytosolic (RP2) isoforms of PDRP have been identified in the C3 plant Arabidopsis (Arabidopsis thaliana). Both of them have kinase and phosphotransferase activities, although RP2 catalyzes PPDK dephosphorylation at a slower rate than does RP1 (Chastain et al., 2008; Astley et al., 2011). Bacterial genomic databases show that PDRP homologs, referred to as Domain of Unknown Function299 (DUF299) genes, are present in all PPDK-containing bacteria (Burnell, 2010). In Escherichia coli, which lacks PPDK, DUF299 regulates the on/off activity of phosphoenolpyruvate synthetase (PEPS) via reversible phosphorylation of the PEPS active-site Thr (Burnell, 2010). This specific target Thr residue for PDRP in C4PPDK is highly conserved in all dikinases from C3 angiosperms and prokaryotes that have been examined (Rosche et al., 1994; Fisslthaler et al., 1995; Agarie et al., 1997; Imaizumi et al., 1997; Wei et al., 2000). Taken together, these results suggest that this regulatory threonyl phosphorylation of the PPDK is a very ancient mechanism. This notion implies a common evolutionary pathway for C4 photosynthesis facilitated by the preexistence of homologs of C4 enzymes in C3 plants (Edwards et al., 2001; Hibberd and Quick, 2002; Wang et al., 2009). The most significant adaptation for the enzyme to be utilized in C4 photosynthesis may have already occurred well before the emergence of the pathway in modern angiosperms (Chastain et al., 2011).
A previous empirical study showed that PPDK activity is insensitive to variations in PPDK level when a cold-tolerant ppdk is inserted into the genome of maize (Ohta et al., 2006). Enzyme activity measurements were performed on 48 strains, each with a different PPDK expression level, showing that there was only about a 20% change in the PEP formation rate despite a 5.7-fold variation in PPDK level. A similar phenomenon was also observed in transgenic rice (Oryza sativa) leaves, in which maize PPDKs accumulated at very high levels but failed to activate fully, even after 14 h of illumination and complete inactivation in darkness (Taniguchi et al., 2008). These findings suggest that the mechanism for regulating PPDK is far more complicated than previously thought. It is not known whether all or only part of the PPDK that accumulates in mesophyll chloroplasts is required for C4 photosynthesis, because the protein level does not affect its enzyme activity. If it is only a select portion of PPDK that is required, then it is also unknown how light regulates the amount of PPDK involved in C4 photosynthesis.
To address these issues, we created a comprehensive profile of PPDK posttranslational modifications. We identified the cleavage site of the transit peptide, its N-terminal acetylated form, and four phosphorylated residues. We found that it is not the light/dark transition per se but rather a change in light intensity that regulates PPDK activity by modulating reversible phosphorylation at Thr-527. Importantly, we also partially determined the catalytic mechanism of PDRP. Taken together, these results suggest that the mechanisms via which PPDK is regulated are more complex than previously described (Ashton and Hatch, 1983; Chastain et al., 2000) and provide a foundation for studies on the molecular mechanism of PPDK regulation in the C4 pathway.
RESULTS
Phosphorylation at Thr-527 of PPDK Is Independent of the Light/Dark Transition
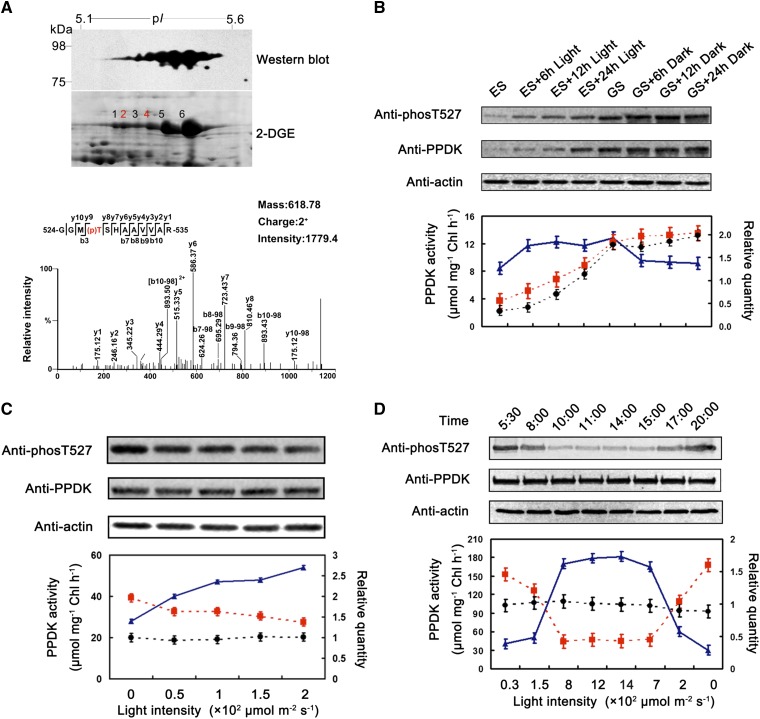
We performed two-dimensional gel electrophoresis (2DGE) coupled with immunoblotting to characterize the posttranslational modification of PPDK in young maize leaves. Six spots displayed a pearls on a string pattern on the western blot, and the corresponding spots on the 2DGE map had molecular masses of approximately 95 kD with pI values of 5.1 to 5.6 (Fig. 1A). After in-gel digestion, these spots were analyzed using ultra-high-performance liquid chromatography (UHPLC) coupled with tandem mass spectrometry (MS/MS). As anticipated, all six spots were identified as maize PPDK, and a doubly charged mass peak ([M + 2H]2+) at mass-to-charge ratio (m/z) 618.78 was detected for each spot. This ion corresponds to a 12-residue peptide (GGMTSHAAVVAR; residues 524–535) of PPDK with phosphorylated Thr-527 (Fig. 1A).
Figure 1.
Light intensity regulated the reversible phosphorylation of maize chloroplast PPDK at Thr-527. A, Identification of phosphorylated Thr-527 of PPDK in illuminated maize seedlings. The top gel shows a western blot of maize PPDK. The protein was extracted from 6-d-old green maize seedlings that had been illuminated for 12 h. The specific antibody raised against phosphorylated Thr-527 of PPDK detected six spots, which were visible in a pearls on a string pattern on the PVDF membrane. The corresponding 2DGE map (bottom gel) highlights the six PPDK isoforms (75–98 kD, pI 5.1–5.6). The graph at bottom displays the phospho-MS/MS spectra observed in all six spots (m/z 618.78, charge 2+). Ions corresponding to y and b fragments of the GGMTSHAAVVAR sequence are labeled and shown in the corresponding sequence insets. The assignment of the phosphorylated Thr-527 site was deduced from the mass of b and y ions. B to D, The gels at top show variations at Thr-527 and the abundance of PPDK in maize seedlings exposed to different light regimens (varied intensity of light); the graphs at bottom illustrate the correlations between PPDK phosphorylation levels, abundance, and activity. The triangles, squares, and circles represent PPDK activity, phosphorylation levels, and abundance, respectively. Chl, Chlorophyll.
With high-resolution mass spectrometry (MS; Waters Synapt HD-MS), the relative abundance of unphosphorylated and phosphorylated peptide isoforms can be estimated by the number of MS/MS spectral counts. For example, the spectral numbers of the [M + 2H]2+ at m/z 618.78 corresponding to phosphopeptide GGM(p)TSHAAVVAR were 29 and 36 for spots 2 and 4, respectively, which were considerably greater than the single spectrum observed for the unphosphorylated peptide GG(ox)MTSHAAVVAR ([M + 2H]2+ at m/z 586.79; here, the oxidation state of Met-526 is sulfoxide [15.9994 D]; Supplemental Fig. S1). This result suggests that the amount of the phosphorylated isoform PPDK at Thr-527 must be higher than that of the unphosphorylated isoform. In other words, PPDK must be highly phosphorylated at Thr-527. Surprisingly, the ion corresponding to the peptide containing phosphorylated Thr-527 could be detected in each PPDK isoform on the 2DGE gel of 6-d-old seedlings that had been illuminated for 12 h immediately before harvesting. This result is inconsistent with previous reports that maize chloroplast PPDK is only phosphorylated in darkness and dephosphorylated upon exposure to light (Ashton and Hatch, 1983; Chastain et al., 2000). We suspected that this inconsistency might be the result of differences in light intensity used to illuminate maize seedlings. In our experiment, maize seedlings were illuminated with light intensity of 200 μmol m–2 s–1, in comparison with one previous study (Chastain et al., 2000), where maize seedlings were illuminated from dawn until 12 noon under full sunlight (peak illumination at noon was 1,500 μmol m–2 s–1), and no light intensity was mentioned in the earlier study (Ashton and Hatch, 1983).
We first tested whether light/dark transitions strictly regulated phosphorylation at the active-site Thr-527 of maize PPDK. Western blotting was performed on protein extracted from young maize seedlings treated with different illumination regimens: a 24-h-light/0-h-dark cycle yielding green seedlings (GS; under a light intensity of 200 μmol m–2 s–1); GS grown in darkness for 6 h (GS + 6 h), 12 h (GS + 12 h), or 24 h (GS + 24 h); a 0-h-light/24-h-dark cycle yielding etiolated seedlings (ES); and ES grown in light (200 μmol m–2 s–1) for 6 h (ES + 6 h), 12 h (ES + 12 h), or 24 h (ES + 24 h; Supplemental Fig. S2). To accurately detect variations in the phosphorylation levels at Thr-527 of PPDK, a polyclonal antibody was generated using a synthetic 11-residue phosphopeptide as antigen in which Thr-527 was phosphorylated (see “Materials and Methods”). This antibody was highly specific to the phosphorylated form of maize PPDK because there was no cross-reaction with the synthetic unphosphorylated peptide, with unphosphorylated PPDK, or with other phosphoproteins in the soluble leaf extracts (Supplemental Fig. S3). As a control, an anti-PPDK antibody was used to detect changes in PPDK abundance in maize leaves treated with different light regimens. As shown in Figure 1B, (1) PPDK was strongly phosphorylated at Thr-527 in GS leaves that had not been exposed to the dark condition; (2) the phosphorylation level at Thr-527 gradually increased with increasing PPDK abundance in ES exposed to light; and (3) PPDK activity increased in the ES + 6 h group relative to the ES group and remained at similar elevated levels in the ES + 12 h and ES + 24 h groups. In contrast, PPDK activity decreased in the GS + 6 h group compared with the GS group and remained at similar decreased levels in the GS + 12 h and GS +24 h groups. Taken together, these results suggested that the light/dark transition had no influence on the phosphorylation at Thr-527 of maize PPDK and that PPDK activity was insensitive to the abundance of PPDK.
Phosphorylation at Thr-527 of Maize PPDK Is Strictly Regulated by Light Intensity
We next tested whether light intensity has an influence on phosphorylation at Thr-527. Western blotting was performed on the GS + 12 h dark samples illuminated at different levels of light intensity (50, 100, 150, and 200 μmol m−2 s−1) for 30 min. As shown in Figure 1C, the phosphorylation levels on Thr-527 gradually decreased with increasing light intensity, while the PPDK activity levels gradually increased with increasing light intensity. In the control, no change was evident in the abundance of PPDK. These results suggested that light intensity positively regulated the PPDK activity by modulating the reversible phosphorylation at Thr-527 of PPDK.
To confirm that the degree of phosphorylation at Thr-527 of PPDK is strictly regulated by light intensity, we examined the variation in Thr-527 phosphorylation and PPDK activity in leaves of maize seedlings illuminated with natural sunlight at different times of day (varied light intensity). The fourth leaves of the maize seedlings were harvested at eight time points (see “Materials and Methods”). As expected, the phosphorylation at Thr-527 decreased dramatically with the increasing light intensity in the morning and increased with the decreasing light intensity in the evening (Fig. 1D). The pattern of PPDK activity was completely opposite to that of Thr-527 phosphorylation at these levels of light intensity. The phosphorylation levels of Thr-527 reached the minimum at the light intensity of 800 μmol m−2 s−1, where PPDK activity reached its maximum. Once light intensity exceeded this level, the phosphorylation of Thr-527 and PPDK activity did not appear to be further affected and remained stable. We also noted that even at peak PPDK activity levels, a part of PPDK was still phosphorylated at Thr-527 (Fig. 1D). To verify this finding, the relative abundance of unphosphorylated and phosphorylated Thr-527 isoforms was estimated by measuring the number of MS/MS spectral counts. Although not accurate, this approach provided a qualitative estimate of the degree of phosphorylation at Thr-527. 2DGE was performed on the maize seedlings harvested at 2 pm (1,400 μmol m–2 s–1) and 8 pm (0 μmol m–2 s–1). The protein spots representing PPDK on each 2DGE gel (Fig. 1A; Supplemental Fig. S4) were excised and then subjected to MS analysis after trypsin digestion. As expected, 36 peptides containing phosphorylated Thr-527 and 60 peptides containing unphosphorylated Thr-527 were identified in the maize seedling harvested at 2 pm (1,400 μmol m–2 s–1). In comparison, 107 peptides containing phosphorylated Thr-527 and only five peptides containing unphosphorylated Thr-527 were identified in the maize seedling harvested at 8 pm (0 μmol m–2 s–1). These results suggested that approximately 30% and 90% of the total PPDK were phosphorylated at Thr-527 in the maize seedlings when harvested at 2 and 8 pm, respectively (Supplemental Fig. S4); this finding was highly consistent with our western-blot results (Fig. 1D). Additionally, we also assessed the net photosynthesis rate, which showed a good correlation with PPDK activity (Supplemental Fig. S5). To test whether PPDK activity is also regulated by the circadian clock, the maize seedlings were treated with 200 and 800 μmol m–2 s–1 illuminations constantly and respectively, 24 h for each. Samples were taken from the seedlings under each level of illumination every 4 h during the 24-h treatment, and both PPDK activity and the phosphorylation levels in all samples were assayed. There were no evident changes in either the phosphorylation levels or PPDK activities in maize seedlings illuminated with identical light intensity; the enzyme activities and phosphorylation levels exhibited good correlation in a maize seedling illuminated at two levels of light intensity (Supplemental Fig. S6). This result indicated that the PPDK activity is independent of circadian regulation. To summarize, these results suggested that the amount of PPDK in mesophyll chloroplast was in excess of what is required for C4 photosynthesis, and light intensity strictly regulated PPDK activity via the regulation of reversible phosphorylation at Thr-527 of PPDK.
C4ppdk Is Specifically Expressed in Maize Leaves
There are two loci for PPDK genes in maize chromosomes. One locus can transcribe two overlapping genes (C4 chloroplast PPDK [C4ppdk] and cytosolic PPDK [CyppdkZm1]) that are divergent at their 5′ ends owing to different transcription initiation sites (Sheen, 1991). To confirm whether only C4ppdk, but not the two Cyppdk genes, was expressed in maize leaves, we quantified transcript levels of the three genes encoding C4PPDK, CyPPDKZm1, and CyPPDKZm2 using quantitative reverse transcription-PCR. Specific primers were designed from the 5′ untranslated region (UTR) and the first exon to distinguish the three different genes, particularly C4ppdk (relative to the transcription site from the 5′ UTR position, –15 bp to the first exon’s position +67 bp) and CyppdkZm1 (from the 5′ UTR, –384 bp to –224 bp). Only C4ppdk was detected in maize leaves under the different illumination regimens, indicating that C4ppdk was specifically expressed in maize leaves (Supplemental Fig. S7). Transcription of C4ppdk was also partially regulated by light; C4ppdk transcription dramatically increased after ES were exposed to light for 6 h, then markedly decreased at 12 h, and remained low until 24 h. In contrast, C4ppdk transcription started to decrease at 12 h during the transition from light to dark.
N-Terminal Amino Acid Sequence Analysis and MS/MS
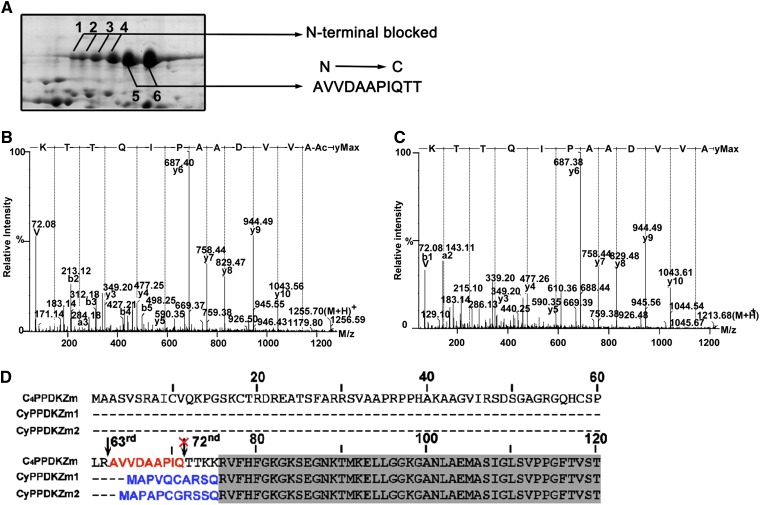
To verify that the identified PPDKs were all C4PPDKs, we analyzed the N-terminal sequences of the six PPDK spots. The three PPDKs were almost identical in sequence, differing only at a few residues in the N terminus (Sheen, 1991). The mature C4PPDK contains the specific N-terminal sequence TTKK, CyPPDKZm1 contains the 11-residue sequence MAPAPCGRSSQ, and CyPPDKZm2 contains the 10-residue sequence MAPVQCARSQ (Matsuoka et al., 1988; Sheen, 1991). Therefore, we performed N-terminal sequence analysis of these proteins to distinguish the CyPPDKs from C4PPDK.
Edman degradation performed on the PPDK spots on 2DGE gels of seedlings exposed to different illumination regimens identified an 11-residue peptide (AVVDAAPIQTT) in spots 5 and 6, corresponding to residues 63 to 74 of the deduced full-length PPDK sequence. No sequence information was obtained for spots 1 to 4 (Fig. 2A), suggesting that the N-terminal residue of these proteins was modified. We performed UHPLC-MS/MS on the in-gel-digested peptides obtained for spots 1 to 6 to identify which modifications occurred at the N terminus of the PPDKs in these spots. We also performed a database search with N-terminal acetylation selected as a variable modification in addition to the usual parameters. The [M + 2H]2+ ion at m/z 628.32 (1,254.68 D) was detected in spots 1 to 4, which corresponded to the N-terminally acetylated peptide acetyl-AVVDAAPIQTTK (residues 63–74) of maize chloroplast PPDK (Fig. 2B). The [M + 2H]2+ ion at m/z 607.33 (1,212.67 D) was detected in spots 5 and 6, which corresponded to residues 63 to 74 (AVVDAAPIQTTK) of maize chloroplast PPDK (Fig. 2C). The two peptides differed by 42 D, which confirmed that the identified peptide in spots 1 to 4 was N-terminally acetylated, consistent with the sequence analysis. N-terminal acetylation of nucleus-encoded chloroplast proteins in the cytosol is required for proper chloroplast characteristics, such as the stability and/or import competence of organellar precursor proteins (Pesaresi et al., 2003). Interestingly, both N-terminally acetylated and unacetylated peptides were identified with high confidence in spot 3, indicating that it was a mixture of at least two chemically distinct PPDK isoforms. This result confirmed that the chloroplast PPDK underwent differential posttranslational modifications. These data indicated that spots 1 to 4 shared an identical N-terminal sequence with spots 5 and 6, except that they were acetylated. Thus, the N-terminal sequence information of the six spots confirmed that they contained C4PPDK and not CyPPDK (Fig. 2D). The N terminus of the mature protein is at residue 63 of PPDK rather than residue 72, as reported previously (Matsuoka et al., 1988). This difference may reflect potential proteolysis at the N terminus during protein purification. Indeed, rapid proteolytic cleavage during the isolation of other key C4 pathway enzymes, namely PEP carboxykinase and PEP carboxylase, provides a precedent for this possibility (McNaughton et al., 1989; Walker and Leegood, 1995; Walker et al., 1997). Using 2DGE and MS, we also identified the N termini of CyPPDKZm1 and CyPPDKZm2 in 25-d-old maize endosperm where the two proteins were highly expressed (Supplemental Fig. S8). Mass spectra demonstrated that the N terminus of mature CyPPDKZm1 was APVQCAR (residues 2–8; Met-1 was removed to yield the mature protein), whereas that of CyPPDKZm2 was APAPCGR (residues 2–8). These results confirmed the prediction of Sheen (1991) regarding the sequences inferred from the respective mRNAs. These results indicated that C4PPDK, but not CyPPDKs, is specifically expressed in maize leaves.
Figure 2.
Determination of the N-terminal amino acid sequences of different PPDK isoforms using Edman degradation and UHPLC-MS/MS. A, The first 11 N-terminal residues identified in spots 5 and 6 were determined using Edman degradation. UHPLC-MS/MS showed that the other four spots contained N-terminally acetylated protein. B and C, Collision-induced dissociation mass spectra are shown for the N-terminal acetylated peptides from spot 3 (B) and N-terminal unacetylated peptides from spot 5 (C). D, Alignment of N-terminal sequences of three maize PPDKs encoded by individual genes. The arrow shows the N terminus of mature C4PPDK at Ala-63. [See online article for color version of this figure.]
Identification of Multiple Phosphorylation Sites on C4PPDK
Protein spots appearing in a pearls on a string pattern on a 2DGE gel usually indicate multiple phosphorylation events. To determine whether the six spots arose from protein phosphorylation, the in vivo phosphorylation status of PPDK was analyzed using 2DGE and MS. Each of the six spots was excised from 2DGE gels of 6-d-old seedlings and digested with trypsin. The candidate phosphopeptides were analyzed with UHPLC-MS/MS after enrichment using a titanium dioxide (TiO2) microcolumn, and four phosphorylation sites were identified (Supplemental Fig. S9). In addition to the phosphorylated Thr-527 found in every spot, phosphorylated Thr-309, Ser-506, and Ser-528 were also identified (Table I). Among the four identified phosphorylation sites, Thr-527 and Ser-528 have been identified in maize (Casati et al., 2005; Bi et al., 2011) and Arabidopsis (Reiland et al., 2009). Phosphorylation at the other two sites has not been reported previously. Phosphopeptides and their corresponding unphosphorylated peptides were all identified in the same spot. For example, spot 4 contained phosphorylated Ser-506, Thr-527, and Ser-528 and their corresponding unphosphorylated peptides (Table I). Three peptides with an identical sequence but containing different phosphorylated sites, Thr-527 and/or Ser-528, were identified in spot 5, indicating that one PPDK spot contained different forms of PPDK resulting from different posttranslational modifications.
Table I. Peptides with different posttranslational modifications and their unmodified counterparts identified in the protein spots corresponding to maize C4PPDK.
ox and p, Oxidation and phosphorylation of amino acids, respectively; acetyl, N-terminal acetylation of a peptide.
| Spot No. | Peptide Sequence | Posttranslational Modification | Calculated Mr | m/z | Protein Sequence Coverage | Score |
|---|---|---|---|---|---|---|
| % | ||||||
| 1 | 63AVVDAAPIQTTK74 | Acetyl | 1,254.68 | 628.32 (+2) | 72 | 51 |
| 524GGoxMpTSHAAVVAR535 | Thr-527 | 1,251.54 | 626.78 (+2) | 90 | ||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,749.26 | 917.43 (+3) | 101 | |||
| 501VRAETSPEDVGGMHAAVGIL520 | 2,024.00 | 1,013.02 (+2) | 126 | |||
| 2 | 63AVVDAAPIQTTKKRVF78 | Acetyl | 1,785.01 | 893.52 (+2) | 88 | 98 |
| 524GGoxMpTSHAAVVAR535 | Thr-527 | 1,251.54 | 626.78 (+2) | 94 | ||
| 524GGoxMTSHAAVVAR535 | 1,172.58 | 586.79 (+2) | 113 | |||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,749.26 | 917.43 (+2) | 116 | |||
| 501VRAETSPEDVGGMHAAVGIL520 | 2,024.00 | 675.67 (+2) | 81 | |||
| 3 | 63AVVDAAPIQTTK74 | Acetyl | 1,254.68 | 628.35 (+2) | 92 | 72 |
| 63AVVDAAPIQTTK74 | 1,212.67 | 607.34 (+2) | 57 | |||
| 524GGMpTSHAAVVAR535 | Thr-527 | 1,235.55 | 618.79 (+2) | 94 | ||
| 308GpTAVNVQCMVFGNMGNTSGTGVLFTR333 | Thr-309 | 2,740.22 | 914.42 (+3) | 62 | ||
| 503AETSPEDVGGMHAAVGILTER523 | 2,155.02 | 719.34 (+3) | 107 | |||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,749.26 | 1,375.65 (+2) | 157 | |||
| 4 | 503AETSPEDVGGMHAAVGILTER523 | 1,785.01 | 893.52 (+2) | 94 | 93 | |
| 63AVVDAAPIQTTKKRVF78 | Acetyl | 2,234.99 | 1,118.51 (+2) | 83 | ||
| 503AETpSPEDVGGMHAAVGILTER523 | Ser-506 | 1,235.55 | 618.78 (+2) | 102 | ||
| 503AETSPEDVGGMHAAVGILTER523 | 2,139.03 | 714.01 (+3) | 118 | |||
| 524GGoxMpTSHAAVVAR535 | Thr-527 | 1,172.58 | 586.79 (+2) | 98 | ||
| 524GGoxMTSHAAVVAR535 | 1,251.54 | 626.78 (+2) | 49 | |||
| 524GGMTpSHAAVVAR535 | Ser-528 | 2,733.27 | 1,367.64 (+2) | 151 | ||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,155.02 | 719.32 (+3) | 146 | |||
| 5 | 63AVVDAAPIQTTK74 | 1,212.67 | 607.33 (+2) | 97 | 60 | |
| 503AETpSPEDVGGMHAAVGILTER523 | Ser-506′ | 2,234.99 | 1,118.51 (+2) | 77 | ||
| 503AETSPEDVGGMHAAVGILTER523 | 2,139.03 | 714.01 (+3) | 121 | |||
| 524GGMpTSHAAVVAR535 | Thr-527 | 1,235.55 | 618.78 (+2) | 100 | ||
| 524GGoxMTpSHAAVVAR535 | Ser-528 | 1,251.54 | 626.78 (+2) | 83 | ||
| 524GGMpTpSHAAVVAR535 | Thr-527, Ser-528 | 1,373.52 | 687.73 (+2) | 43 | ||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,733.27 | 912.09 (+3) | 57 | |||
| 6 | 63AVVDAAPIQTTK74 | Thr-527 | 1,212.67 | 607.34 (+2) | 98 | 60 |
| 524GGoxMpTSHAAVVAR535 | 1235.55 | 618.78 (+2) | 99 | |||
| 503AETSPEDVGGMHAAVGILTER523 | 2,155.02 | 719.34 (+3) | 123 | |||
| 308GTAVNVQCMVFGNMGNTSGTGVLFTR333 | 2,749.26 | 917.43 (+3) | 113 |
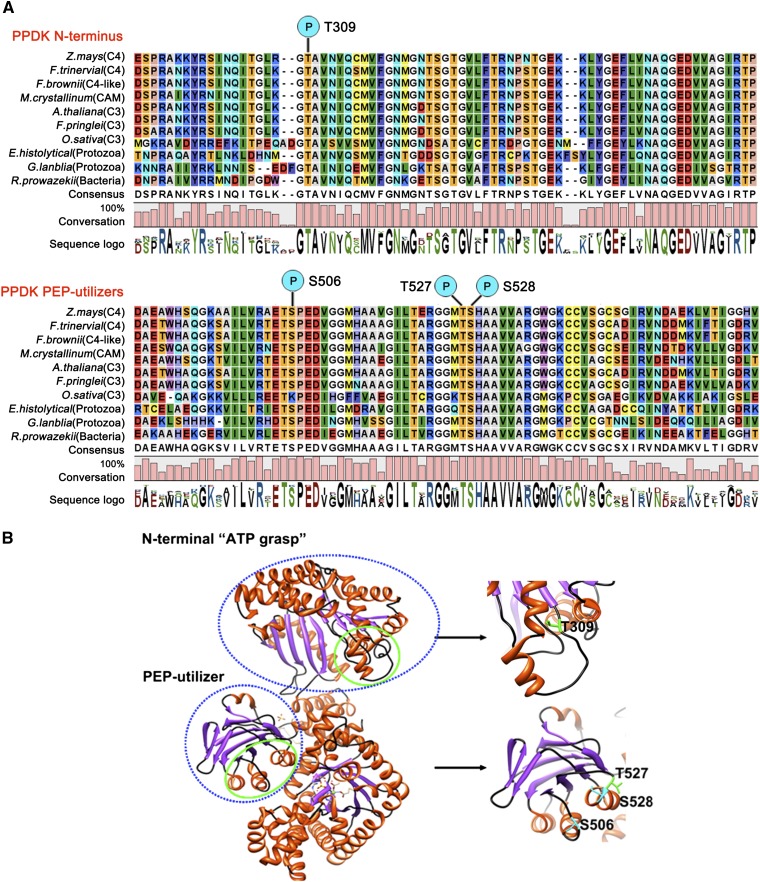
Sequence alignment of different C3, Crassulacean acid metabolism, and C4 plant species indicated that the identified phosphoresidues were highly conserved among these species (Fig. 3A). The x-ray structure of maize PPDK reveals a three-domain structure consisting of a consecutive N-terminal ATP-grasp region, a central domain containing His-455 (His-529 in the full sequence of maize PPDK), and a C-terminal domain (Herzberg et al., 1996; McGuire et al., 1998). The identified phosphorylation sites at (1) Thr-309 and (2) Ser-506, Thr-527, and Ser-528 were located in the N-terminal ATP-grasp domain or the central His-529 PEP-utilizer domain, respectively (Fig. 3B). The two domains function in the first two catalytic steps of the interconversion of ATP, inorganic phosphate, and pyruvate to AMP, inorganic pyrophosphate, and PEP, respectively, for Clostridium symbiosum PPDK (Lin et al., 2006). The phosphorylation of these residues may play an important role in the regulation of PPDK function, particularly for Thr-527 and Ser-528, which are adjacent to the key residue His-529 in this domain.
Figure 3.
Locations of the four phosphorylation residues in the primary sequence and the three-dimensional structure of C4PPDK. A, Alignment of the PPDK N terminus and PEP-utilizer domain sequences using the CLC Main Workbench 5.1 software (http://www.clcbio.com). Individual residues are color coded based on software default parameters. A high degree of conservation is evident in the regions corresponding to these two motifs of PPDK. Among the Ser and Thr residues, Thr-309 (N-terminal domain), Ser-506, Thr-527, and Ser-528 (PEP-utilizer domain) were the most conserved in green plants, protozoa, and bacteria. The sequence logo under each column represents the degree of amino acid conservation (0%–100%). CAM, Crassulacean acid metabolism. B, Three-dimensional cartoon depicting the backbone structure of C4PPDK (PDB identifier 1VBG). The blue dashed lines mark the N-terminal ATP-grasp domain and PEP-utilizer fold structures. The green solid lines mark the four phosphorylation sites.
Ser-528 Is Another Target of PDRP
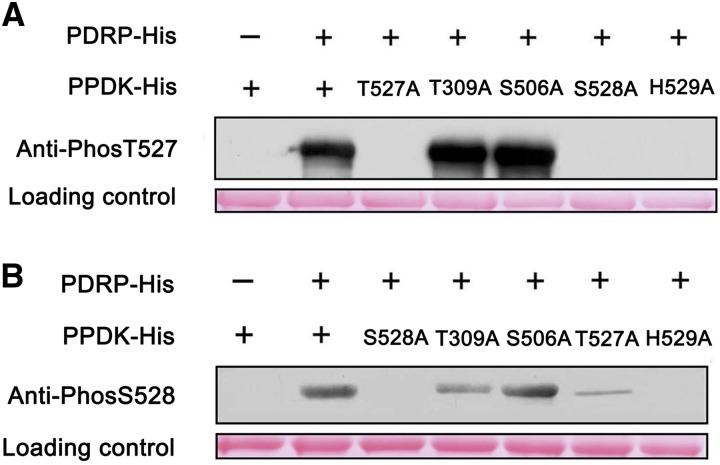
The reversible phosphorylation at Thr-527 of PPDK is catalyzed by PDRP in maize (Burnell and Hatch, 1986), and His-529 phosphorylation is required as a catalytic intermediate during this process (Burnell and Hatch, 1984). To determine whether phosphorylation at Thr-309, Ser-506, and Ser-528 is also catalyzed by PDRP, we used recombinant His-tagged maize C4PPDK for directed mutagenesis of these phosphoresidues. Ala was selected as a suitable substitute amino acid because of its inability to be phosphorylated. Recombinant His-C4PPDK, His-PDRP, and four mutant His-C4PPDKs were expressed in E. coli and purified. Purified His-PDRP was incubated with its candidate substrate in the presence of ADP (phosphoryl donor) and then separated using SDS-PAGE. Immunoblotting was performed on the recombinant PPDK using antibodies specific to phosphorylated Thr-309, Ser-506, Thr-527, and Ser-528. No phosphorylation was detected with antibodies that recognized phosphorylated Thr-309 or Ser-506, suggesting that phosphorylation at these two sites was catalyzed by a kinase(s) different from PDRP. Using an antibody specific to phosphorylated Thr-527, phosphorylation was detected in His-C4PPDK, His-C4PPDK-T309A, and His-C4PPDK-S506A but not in His-C4PPDK-T527A, His-C4PPDK-S528A, or His-C4PPDK-H529A (Fig. 4A). These results suggested that Ser-528 may be important for the phosphorylation at Thr-527. Surprisingly, the antibody against phosphorylated Ser-528 showed high (His-C4PPDK, His-C4PPDK-T309A, and His-C4PPDK-S506A), low (His-C4PPDK-T527A), or no (His-C4PPDK-S528A and His-C4PPDK-H529A) phosphorylation signals (Fig. 4B). These results suggested that PDRP may catalyze phosphorylation at Ser-528 in the presence of phosphorylated His-529.
Figure 4.
Site-directed mutagenesis of maize C4PPDK reveals that Ser-528 is another target of PDRP. After incubation with PDRP, the phosphorylation status of wild-type maize C4PPDK and point mutants was identified using specific antibodies against Thr-527 (A) and Ser-528 (B), as described in “Results.” His-C4PPDKs (2 μg per lane) stained with Ponceau Q served as a loading control. [See online article for color version of this figure.]
To test whether Ser-528 phosphorylation can be regulated by light intensity because Ser-528 is also a target of PDRP, we performed western-blot analysis of the proteins extracted from maize leaves illuminated with 0, 100, 200, and 800 μmol m−2 s−1 using antibodies specific to phosphorylated Ser-528. Very low signals were detected on the proteins extracted from the leaves illuminated with 0, 100, and 200 μmol m−2 s−1, but no signal was detected in the proteins extracted from the leaves illuminated with the light intensity of 800 μmol m−2 s−1 (Supplemental Fig. S10A). This finding suggested that phosphorylation at Ser-528 might also be regulated by light intensity. At the same time, we suspect that the level of C4PPDK phosphorylated at Ser-528 is likely to be much lower than that at Thr-527 in maize leaves, meaning that phosphorylation at Ser-528 is not as important as at Thr-527 for the regulation of PPDK activity, because the signal was very weak in all protein samples. To verify this assumption, we first assayed the target amino acid preference of PDRP when it is phosphorylating PPDK in vitro. Western-blot analysis showed that the phosphorylation signal could be detected in 1 min using the antibody specific to Thr-527, and this signal dramatically increased to a saturation level in 5 min or less. In contrast, the phosphorylation signal of Ser-528 could be detected in at least 20 min, and this signal was much weaker than that of Thr-527 (Supplemental Fig. S10B). These results suggested that the efficiency of PDRP’s phosphorylation of Thr-527 is much higher than that of Ser-528. We also estimated the relative in vivo abundance of phosphorylated Thr-527 and phosphorylated Ser-528 isoforms by measuring the number of MS/MS spectral counts. 2DGE was performed on the proteins extracted from dark-treated maize seedlings and from the seedlings illuminated with the light intensity of 800 μmol m−2 s−1. The phosphopeptides were enriched in the protein spots representing PPDK; these spots were excised from each 2DGE gel and then analyzed using MS. A total of 953 peptides containing phosphorylated Thr-527 but only eight peptides containing phosphorylated Ser-528 were identified in dark-treated seedlings (Supplemental Fig. S10C). In contrast, 127 peptides containing phosphorylated Thr-527 but no phosphorylated Ser-528 were identified in the seedlings illuminated with the light intensity of 800 μmol m−2 s−1. This result indicated that the abundance of PPDK isoforms with phosphorylated Ser-528 is much lower in vivo compared with phosphorylated Thr-527.
To uncover the reason why Thr-527 is more amenable to phosphorylation by PDRP than Ser-528, we carefully analyzed the structure of PPDK (Protein Data Bank [PDB] identifier 1VBG). We found that the side chain of Thr-527 is more exposed to solvent than that of Ser-528 (Supplemental Fig. S10D). In line with this observation, the solvent-accessible surface area of Thr-527 is 114.55 Å2, whereas that of Ser-528 is only 23.94 Å2, as calculated by the Proteins, Interfaces, Structures, and Assemblies server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). These findings suggested that Thr-527 has a much better chance than Ser-528 to be phosphorylated by PDRP, even though both of them are targets of PDRP. This state of affairs must be the reason why the amount of phosphorylated Thr-527 PPDK is much greater than the amount of phosphorylated Ser-528 PPDK in maize leaves.
Ser-528 Plays an Essential Role in PDRP Regulation of PPDK Phosphorylation at Thr-527
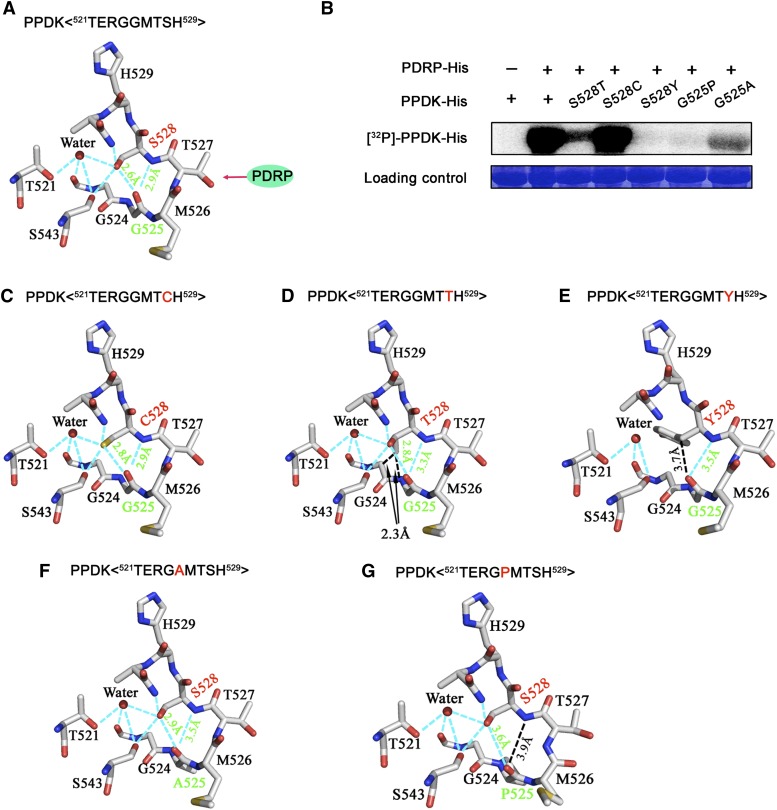
To find out why Ser-528 affected the phosphorylation state at Thr-527, we examined the relationship of these residues in the available three-dimensional crystal structure of maize C4PPDK (Nakanishi et al., 2005) using UCSF Chimera (Pettersen et al., 2004). With unphosphorylated Thr-527, the hydroxyl hydrogen and backbone amide hydrogen of Ser-528 could each form a hydrogen bond (H-bond; 2.6 and 2.9 Å) with the carboxyl oxygen of Gly-525, according to the FindHBond function of the program (Fig. 5A). We suspected that the two H-bonds might play an important role in the phosphorylation of Thr-527 in PPDK by PDRP. Of the 20 common amino acids, only Ser, Thr, and Cys have the potential to form two H-bonds with Gly, because they have a hydroxyl or sulfhydryl group on their side chain. To test these predictions experimentally with PPDK, we substituted Ser-528 with Thr, Cys, or Tyr and we substituted Gly-525 with Ala, which is structurally similar to Gly, or with Pro, which is not expected to form an H-bond because of a steric hindrance.
Figure 5.
The two H-bonds between Ser-528 and Gly-525 play an essential role in the regulation of phosphorylation at Thr-527 by PDRP. A, The atomic-level environment of H-bond formation between Gly-525 and Ser-528. The ribbon diagram was obtained from the coordinates of the C4PPDK (PDB identifier 1VBG) corresponding sequence 521TERGGMTSH529. Atoms are colored as follows: oxygen, red; carbon, white; nitrogen, blue; sulfur, yellow. Cyan dashed lines represent intramolecular H-bonds. B, Autoradiographic analysis of the phosphorylation of wild-type and mutant (S528T, S528C, S528Y, G525P, and G525A) recombinant C4PPDK by means of [γ-32P]ADP and His-tagged recombinant PDRP. The loading control occupies the position at approximately 95 kD in PPDK, as revealed by Coomassie blue staining of the same SDS polyacrylamide gel. C to G, Mutated structures were constructed and optimized using Coot software (Emsley et al., 2010), which revealed the detailed atomic-level environment of H-bond formation between Gly-525 and Cys-528 (C), Gly-525 and Thr-528 (D), Gly-525 and Tyr-528 (E), Ala-525 and Ser-528 (F), and Pro-525 and Ser-528 (G). Atoms are colored as in A. The cyan dashed lines show intramolecular H-bonds. The red letters in the sequence indicate mutant amino acid residues.
The recombinant proteins were incubated with His-PDRP, [β-32P]ADP (which was converted from [γ-32P]ATP), and AMP. To accurately measure phosphorylation rates, we first established the time course of PPDK phosphorylation by PDRP. Maximal phosphorylation of PPDK was achieved within 40 min, but 50% of maximal phosphorylation was achieved within 2 min (Supplemental Fig. S11). Therefore, 2 min was chosen as the time to detect the phosphorylation efficiency of these recombinant proteins. Strong phosphorylation was detected for the wild-type PPDK and His-C4PPDK-S528C; however, His-C4PPDK-S528T and His-C4PPDK-G525A exhibited weak signals, and no signals were detected on His-C4PPDK-S528Y or His-C4PPDK-G525P (Fig. 5B). All the mutated structures were constructed and optimized using the Coot program (Emsley et al., 2010). As expected, the His-C4PPDK-S528C mutant maintained the same H-bonds as the wild type, with similar H-bond lengths (2.8 and 2.9 Å; Fig. 5, A and C). The introduced methyl group of the side chain in the His-C4PPDK-S528T mutant was slightly repulsed by the main chain of Gly-524 and thus weakened the two H-bonds (2.8 and 3.3 Å; Fig. 5D). Moreover, the larger side chain of the His-C4PPDK-S528Y mutant came into contact with Gly-524 and Ala-531 and thus disrupted one of the two H-bonds (Fig. 5E). The His-C4PPDK-G525A mutant slightly changed the main-chain conformation near this residue and thus weakened the two H-bonds (2.9 and 3.5 Å; Fig. 5F). In contrast, the rigid ring of the G525P mutant disrupted one of the two H-bonds (Fig. 5G). These experimental data and structural analyses indicated that the two H-bonds between Ser-528 and Gly-525 resulted in an optimal conformation for PDRP to phosphorylate Thr-527 of PPDK. In other words, PDRP catalyzes the reversible phosphorylation of PPDK rapidly and efficiently, but a subtle conformational change around PPDK’s active site Thr-527 will affect its catalytic efficiency. We also measured the enzymatic activity of these mutant proteins. As expected, the activity of His-C4PPDK-S528C was approaching that of PPDK, whereas the activity of other mutants markedly decreased (Supplemental Fig. S12). These data suggested that disruption of either of the two H-bonds or a change in the H-bond length between Gly-525 and Ser-528 had an impact on the enzymatic activity of PPDK.
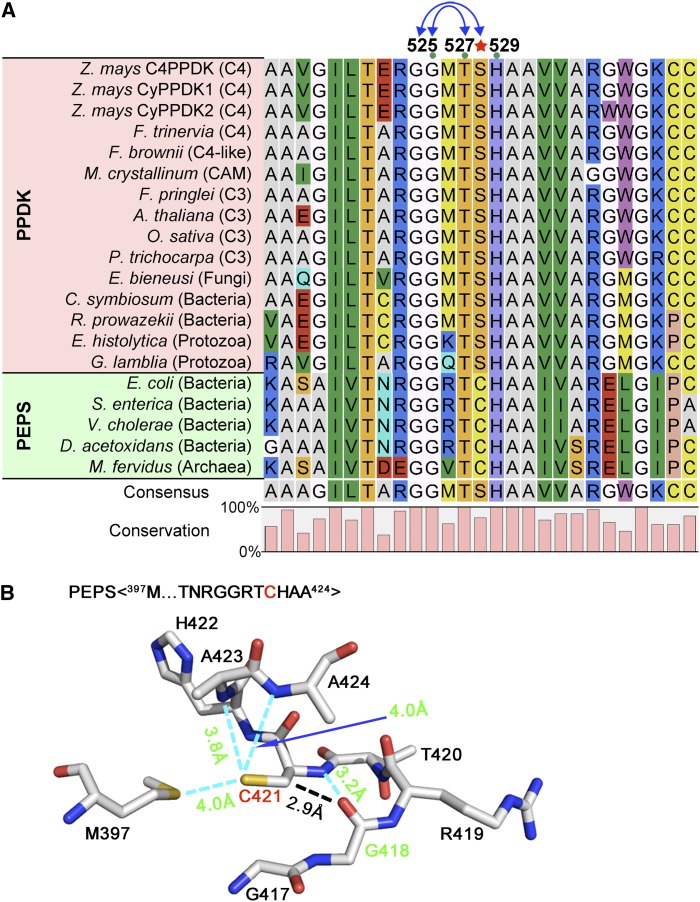
To see if this catalytic mechanism exists in PPDKs of other species of plants, amino acid sequences were aligned with PPDK and its homolog PEPS from different species. Gly-524, Gly-525, Thr-527, and His-529 were all highly conserved among the species examined, whereas Ser-528, which is highly conserved in PPDKs of plants and fungi, was replaced by Cys (Cys-421) in most bacterial PEPS (Fig. 6A). These results suggested that Cys at this position was present earlier in evolutionary history and was later substituted with Ser because the latter can form stronger double bonds with Gly-525. Although Cys-421 only formed one H-bond with Gly-418 (corresponding to Gly-525 in maize; PDB identifier 2OLS), it also had a van der Waals interaction with this Gly in PEPS (Fig. 6B). The H-bond plus a van der Waals bond would have a similar effect to the two H-bonds in PPDK, which would allow the Thr-420 (corresponding to Thr-527 in maize) to be easily phosphorylated by DUF299.
Figure 6.
Mechanism of convergent evolution of H-bond formation in the PEP-utilizer domain of C4PPDK and PEPS. A, Alignment of primary sequences of C4PPDK and its homolog PEPS among different species. The H-bond-forming residues Gly-525 and Ser-528 are highly conserved, and Ser-528 (red star) is an evolutionarily conserved substitution of Cys by Ser. CAM, Crassulacean acid metabolism. B, Atomic structure of the PEPS sequence 397M…GGRTCHAA424 (PDB identifier 2OLS). Cyan dashed lines highlight the intermolecular H-bonds between Cys-421 and Gly-418 in PEPS, which were identified with the FindHBond extension in UCSF Chimera.
DISCUSSION
PPDK is the key enzyme of the C4 pathway, and its activity may limit the photosynthesis rate in maize leaves (Usuda et al., 1984, 1985). Previous studies have shown that PPDK activity is strictly and reversibly regulated by light. This light/dark modulation is mediated by reversible phosphorylation of a conserved Thr residue in the active-site domain by PDRP (Burnell and Hatch, 1985; Roeske and Chollet, 1987; Burnell and Chastain, 2006; Chastain et al., 2011). Nonetheless, if phosphorylation of PPDK at the active-site Thr serves only as an on/off switch tightly regulated by light/dark, then full activation of PPDK would occur at very low light intensity and would be much lower than that required for maximum rates of photosynthesis.
In this study, we found that phosphorylation of maize C4PPDK at Thr-527 is regulated by a change in the light intensity and not necessarily by the light/dark transition (Fig. 1, B–D). At different levels of light intensity, the fluctuation in PPDK activity levels was contradictory to that for the phosphorylation of PPDK at Thr-527 but highly consistent with the net photosynthetic rates (Fig. 1, B–D; Supplemental Fig. S5). These results suggest that light intensity positively regulates PPDK activity through reversible phosphorylation at Thr-527, and PPDK activity precisely limits the photosynthetic rate. Light intensity in plant growth environments is constantly changing. Various levels of phosphorylation of PPDK at Thr-527 could provide more flexibility in controlling the C4 photosynthetic rate in response to variations in light intensity. Therefore, our findings provide a better understanding of the regulatory mechanism of PPDK activity.
Notably, light-responsive regulation of chloroplast PPDK activity appears to be a precise and robust regulatory mechanism in C4 plants. First, chloroplast PPDKs accumulate in excess, constituting approximately 7% to 10% of the protein content of mesophyll cells (Baer and Schrader, 1985; Edwards et al., 1985), ensuring that a sufficient supply of PPDKs is available for C4 photosynthesis, as required. Our results demonstrate that even when PPDK activity reaches its maximum level (when the phosphorylation level at Thr-527 reaches its minimum) at a light intensity of 800 μmol m–2 s–1, approximately one-third of the total of PPDK is still phosphorylated (Fig. 1D; Supplemental Fig. S4). This finding suggests that the amount of PPDK accumulated in the mesophyll chloroplasts is in excess of what is required for photosynthesis, even at peak PPDK activity levels. Second, the levels of PPDK activation can be rapidly and precisely tuned to adapt to changes in metabolic needs arising from altered light intensity (Fig. 1, C and D), without any need for the initiation of gene transcription and protein expression. The precision of this regulatory mechanism might be a good reason for the transition of PPDK from serving as only an ancillary glycolytic enzyme in C3 plants (Chastain et al., 2011), where it is in low abundance, to a key enzyme that limits the rate of photosynthesis in C4 plants.
This regulatory mechanism provides a reasonable explanation for why PPDK activity is insensitive to variations in PPDK level (Ohta et al., 2006; Taniguchi et al., 2008). Some signal representing the rate of photosynthesis at different levels of light intensity, probably a concentration of some metabolite, controls the number of PPDK molecules involved in photosynthesis; therefore, PPDK not involved in photosynthesis is phosphorylated. Merely increasing PPDK expression could not increase enzyme activity. Therefore, these findings have profound implications for the efforts to introduce C4 photosynthesis in C3 crop plants to increase crop yield.
In this study, we found that Ser-528 is another target of PDRP (Fig. 4B). Structurally, the side chain of Thr-527 is more exposed to the solvent than that of Ser-528 (Fig. 5A; Supplemental Fig. S10D). Thus, Ser-528 is far less likely to be phosphorylated than Thr-527, so that the amount of PPDK with phosphorylated Ser-528 is much lower compared with Thr-527 in vivo (Supplemental Fig. S10, A and C). Therefore, it is likely not as important as phosphorylation at Thr-527 for the regulation of PPDK activity in response to a change in light intensity. Nonetheless, Ser-528 plays an essential role in the PDRP-mediated phosphorylation of maize C4PPDK at the active-site Thr-527. The two H-bonds between Ser-528 and Gly-525 might form an optimal conformation near the position of the active-site Thr-527, which promotes the optimal phosphorylation of PPDK by PDRP (Fig. 5A). Disruption of either of the two H-bonds, or a change in the H-bond length, should seriously affect regulatory efficiency (Fig. 5, B and D–G). This finding has broad implications for studies of the mechanism of the reversible phosphorylation of PPDK by PDRP.
In fact, this regulatory threonyl phosphorylation of PPDK was already in place before the emergence of the C4 pathway in modern angiosperms (Chastain et al., 2011). DUF299, a PDRP homolog in E. coli, can regulate the activity of its PEPS (homolog of PPDK) via reversible phosphorylation of the active-site Thr (Burnell, 2010). Sequence alignment shows that most residues are highly conserved in the catalytic domain between PPDKs of fungal and plant species and PEPS of bacteria, except that Cys in bacteria is replaced by Ser in PPDKs (Fig. 6A). The crystal structure of PEPS indicates that an H-bond and a van der Waals interaction between Cys-421 (Ser-528 in maize) and Gly-418 (Gly-525 in maize; Fig. 6B) made the Thr-420 (Thr-527 in maize) an easy target for phosphorylation by DUF299. The substitution of Cys for Ser at this position may have important evolutionary significance for PPDK. First, the substitution for Ser helps PPDK exempted from the possible redox regulation, which is a common mechanism in plant cells. Second, the Cys sulfur atom has a weaker polarization (lower electronegativity) than the oxygen atom does in the side chain of Ser, thus enabling the formation of two H-bonds (with Ser) that perhaps make the PPDK structure more stable than the one containing an H-bond and a van der Waals bond (with Cys). Third, substitution with Ser promotes the selection of an alternative target of PDRP because Ser, and not Cys, can be phosphorylated.
In addition to Thr-527 and Ser-528, Thr-309 and Ser-506 were found to be phosphorylated in maize seedlings in this work. Thr-309 is located in the ATP-binding domain and Ser-506 is located near His-529 in the three-dimensional structure (Fig. 3B). Phosphorylation of these two residues may play an important role in fine-tuning of the C4PPDK activity, perhaps via repulsion of the two negatively charged moieties or via a substrate hindrance. Additionally, C4PPDK is partially acetylated at the N terminus (Fig. 2B). N-terminal acetylation is a common posttranslational modification in chloroplast proteins of several plant species (Wang et al., 2006; Kleffmann et al., 2007). As predicted by Zybailov et al. (2008), maize C4PPDK is acetylated at the N-terminal Ala, and both the second and third residues are Val. Although the function of the N-terminal acetylation is unclear, this finding may lead to an interesting line of research in further studies of the function of PPDK.
Taken as a whole, our results indicate that multiple posttranslational modifications might be involved in the regulation of C4PPDK activity. These diverse regulatory pathways may work alone or in combination to fine-tune C4PPDK activity in response to changes in light intensity.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of maize (Zea mays) ecotype B73 were used in all experiments. For the light/dark treatments, seeds were separated into two groups: one group was grown in the dark, and the other was grown in the light (200 μmol m–2 s–1). Dry mature maize seeds were soaked in water at 28°C for 24 h before transfer to soil. The seeds in soil were then incubated at 28°C with constant light for 6 d to obtain GS or in darkness at the same temperature to obtain ES. After 6 d, some ES were exposed to light for 6 h (ES + 6 h), 12 h (ES + 12 h), or 24 h (ES + 24 h). Conversely, GS were transferred to darkness for 6 h (GS + 6 h), 12 h (GS + 12 h), or 24 h (GS + 24 h). For experiments in which low light intensity was varied, the GS + 12 h and ES seedlings were illuminated with 50, 100, 150, or 200 μmol m–2 s–1 for 30 min each. Leaves from the seedlings were harvested for protein extraction and enzyme activity analysis at different time points. The fourth leaves of 30-d-old seedlings that had been illuminated by full sunlight were harvested in Beijing, at a temperature of approximately 30°C ± 3°C, at eight time points: 5:30 am (30 μmol m–2 s–1), 8 am (150 μmol m–2 s–1), 10 am (800 μmol m–2 s–1), 12 noon (1,200 μmol m–2 s–1), 2 pm (1,400 μmol m–2 s–1), 3 pm (700 μmol m–2 s–1), 5 pm (200 μmol m–2 s–1), and 8 pm (0 μmol m–2 s–1). To examine whether the circadian clock influences PPDK activity, maize seedlings were illuminated with sulfur plasma lamps with light intensities of 200 and 800 μmol m–2 s–1.
2DGE and Western Blotting of Plant Proteins
Total protein extracts were prepared for 2DGE as described (Wang et al., 2006). Briefly, plant materials were ground in liquid nitrogen, then dissolved in 10% (w/v) TCA in acetone (at –20°C) containing 0.07% (v/v) β-mercaptoethanol, and stored overnight at –20°C. After centrifugation at 40,000g for 1 h, the supernatant was discarded and the pellet was rinsed with cold acetone (–20°C) containing 0.07% (v/v) β-mercaptoethanol. The pellet was vacuum dried and solubilized in 3 mL of a solution of 7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 40 mm dithiothreitol (DTT), 1% (v/v) protease inhibitor mixture (Sigma), 0.2 mm Na2VO3, and 1 mm NaF at room temperature for 1 to 2 h. Insoluble material was removed by centrifugation at 100,000g at 4°C for 1 h. Protein concentration was determined using a 2-D Quant kit (GE Healthcare).
Using a rehydration loading method (Wang et al., 2006), 1 mg of protein was applied to a 24-cm Immobiline DryStrip with a linear pH 4 to 7 gradient (GE Healthcare) and subjected to isoelectric focusing for 28 h using the EttanIPGphor 3 IEF system (GE Healthcare). For the second-dimension electrophoresis, SDS-PAGE gels (12.5% [w/v] acrylamide) were used in the Ettan DALTsix 230 electrophoresis unit at 220 V (GE Healthcare). Gels were stained with PhastGel Blue R-350 (PhastGel Staining Kit; GE Healthcare). Western blotting was performed using PPDK-specific antibodies (see below). Supersignal West Dura Extended Duration Substrate (Pierce) was used to detect horseradish peroxidase-conjugated secondary antibodies.
Immunoblotting Using Phosphopeptide-Specific Antibodies
Rabbit polyclonal antibodies, raised against synthetic phosphopeptide conjugates corresponding to the four phosphorylation sites (Thr-309, Ser-506, Thr-527, and Ser-528) of maize C4PPDK, were used to detect phosphorylated PPDK. The peptide sequences were as follows: GLRG(p)TAVNVQC (residues 305–315), VRAET(p)SPEDVG (residues 501–511), ERGGM(p)TSHAAV (residues 522–532), and RGGMT(p)SHAAVV (residues 523–533). In addition, the antibody raised against recombinant His-C4PPDK was used as a probe for total PPDK protein. After production and affinity purification of these antibodies, soluble protein was extracted from seedling leaves that had been subjected to the various treatments described earlier. Fresh maize leaves (0.5 g wet weight) were homogenized with a mortar and pestle in 1 mL of ice-cold extraction buffer (50 mm Tris-HCl [pH 8.0], 0.1 mm EDTA, 20 mm ammonium sulfate, 25% [v/v] glycerol, 2 mm DTT, 0.2 mm phenylmethanesulfonyl fluoride, and 1% [v/v] Protease Inhibitor Cocktail [Sigma]). The homogenate was clarified by centrifugation at 14,000g for 60 min at 4°C, and aliquots of the supernatant were combined with 2× SDS-PAGE sample buffer and heated at 99°C to denature the protein. The proteins were then electrophoresed through SDS-PAGE gels (10% [w/v] acrylamide) and transferred to a 0.45-μm polyvinylidene difluoride (PVDF) membrane. After blocking the membrane with 5% (w/v) bovine serum albumin and incubating with primary and secondary antibodies (fluorescence labeled), the fluorescence signal was detected using the Odyssey Infrared Imaging System (LI-COR). Western-blot quantification was performed by measuring band intensity with the ImageJ freeware.
Chlorophyll Assay
Chlorophyll content was measured according to the method described in a previous study (Arnon, 1949). The chlorophyll concentrations were calculated using a dual-beam UV spectrophotometer (UV-2550; Shimadzu).
PPDK Enzyme Assay
The enzyme assay was performed as described (Usuda et al., 1984; Ashton et al., 1990) with some modifications. Soluble protein extracts for enzyme assays were prepared by homogenizing 0.5 g of maize leaves in 500 μL of ice-cold extraction buffer containing 50 mm HEPES-KOH (pH 8.0), 10 mm MgCl2, 0.1 mm EDTA, 5 mm Glc-6-PNa2, 10 mm NaHCO3, 10 mm DTT, 2.5 mm KH2PO4, and 5 mm (NH4)2SO4. The homogenate was clarified by a 20-min, 4°C centrifugation at 20,000g. Aliquots of the extracts were assayed for PPDK activity using a coupled PEP carboxylase/malate dehydrogenase-based spectrophotometric assay at 30°C for 20 min by adding 2 mm pyruvate, 1.25 mm ATP, 0.2 mm NADH, 12 units of malate dehydrogenase, and 0.5 units of PEP carboxylase. NADH oxidation was monitored at 340 nm in a dual-beam UV spectrophotometer (UV-2550; Shimadzu). The activities of recombinant C4PPDK and its mutant proteins in vitro were assayed as described (Chastain et al., 2000).
Gas-Exchange Measurements
Light-response curves for measuring gas exchange were obtained using an open gas-exchange system (GFS-3000; Waltz), equipped with a standard measuring head (3010-S; 8-cm2 leaf area), on the fully expanded fifth leaf of 30-d-old plants. The measurement conditions were 25°C, 360 µL L−1 CO2, and 16,000 µL L−1 water. Measurements were taken from the point at which leaves showed a constant photosynthetic rate at a light intensity of 1,400 µmol m−2 s−1; gas exchange was then similarly recorded when light intensity decreased from 1,400 to 20 µmol m−2 s−1.
Analysis of C4ppdk and Cyppdk Transcript Levels
Levels of mRNAs were measured by quantitative real-time PCR. Total RNA from maize leaves was isolated using a Trizol reagent for eight collection samples (ES + 0, 6, 12, or 24 h and GS + 0, 6, 12, or 24 h). The specific forward and reverse primers for the three genes were as follows: 5′-TAGGGGATCGGAAGGATGGC-3′ and 5′-CCCTGTCCCTGGTGCATTTT-3′ for C4ppdk, 5′-CCATGTTGATCTGAGGTGGC-3′ and 5′-TACACGTCACGTAGTCTGG-3′ for cyppdkZm1, and 5′-TCCATGTGGCCGTTCGTC-3′ and 5′-TCCGTCGACACCGTGAAC-3′ for cyppdkZm2. For each sample, reactions were set up in triplicate to ensure the reproducibility of the results. Negative controls without templates (containing distilled, deionized water) were set up using gene-specific primer pairs, and the internal controls were set up in duplicate using primers for actin and tubulin genes. The quantitative real-time PCR experiments were performed using the Chromo4 Real-Time PCR instrument (ABI 7500) and SYBR Green I (Invitrogen; S-7567). Gene expression was quantified using the comparative cycle threshold method.
In-Gel Digestion and Purification of Phosphopeptides
Spots corresponding to those detected by western blotting were excised from 2DGE gels. In-gel digestion was performed as described (Wang et al., 2006). Briefly, gel pieces were destained and then incubated with trypsin (10 ng μL–1) overnight at 37°C. Peptides were extracted by incubation with 20 μL of 5% (v/v) trifluoroacetic acid (TFA) for 1 h at 37°C, followed by the addition of 20 μL of 2.5% (v/v) TFA/50% (v/v) acetonitrile for 1 h at 37°C. The combined supernatants were dried in a SpeedVac concentrator (Thermo Fisher) and used for MS analysis or phosphopeptide enrichment.
Enrichment of Phosphorylated Peptides by the TiO2 Centrifugation Method
The TiO2 centrifugation enrichment method was developed previously (Baiet al., 2009). First, approximately 2 mg of TiO2 beads in a tube was suspended in 100 μL of acetonitrile and incubated for 5 min at 4°C, centrifuged, and the supernatant was discarded. The TiO2 beads were then equilibrated with 40 μL of loading buffer (80% acetonitrile, 5% [v/v] TFA, and saturated phthalic acid), centrifuged, and the buffer was discarded. The equilibrated TiO2 beads were then mixed with the in-gel-digested peptide mixture, which was dissolved in 30 μL of loading buffer and then agitated gently for 2 h at room temperature. The TiO2 beads were then centrifuged to remove the loading buffer, which contained nonadsorbed peptides. The beads were washed using 100 μL of loading buffer and 100 μL of washing buffer (2% [v/v] TFA and 80% [v/v] acetonitrile) three times to remove the remaining nonspecifically adsorbed peptides. Finally, the phosphopeptides were eluted twice with 40 µL of elution buffer (0.5% [v/v] ammonia solution, pH > 10.5) and dried in a SpeedVac concentrator for MS analysis.
UHPLC-MS/MS Analysis of Phosphopeptides
Automated nanoflow UHPLC-MS/MS analysis was performed using the nanoACQUITY UHPLC system connected to an electrospray ionization-MS/MS system (Waters Synapt HD-MS). Tryptic phosphopeptides were separated by UHPLC with a Symmetry C18, 20-mm × 180-µm × 5-μm precolumn and a 250-mm × 75-μm × 1.7-μm BEH C18 column (Waters) using the nanoACQUITY UHPLC system. The samples were loaded onto the precolumn with 0.1% (v/v) aqueous formic acid (solvent A) at a flow rate of 5 μL min–1 for 4 min. The peptides were eluted with a binary solvent system of solvent A and solvent B (0.1% [v/v] formic acid in acetonitrile) using a linear gradient of 3% to 40% solvent B over 60 min at 200 nL min–1, followed by a rinse for 10 min with 90% solvent B and reequilibration at initial conditions for 20 min. Column temperature was maintained at 35°C.
The internal calibration solution was delivered to the Synapt HD-MS instrument from the auxiliary pump of the nanoACQUITY UHPLC system using a constant flow rate of 300 nL min–1 and a concentration of 100 fmol μL–1 [Glu-1]-Fibrinopeptide B (Sigma-Aldrich). For all measurements, the Synapt HD-MS system was operated in the v-mode with a typical resolving power of at least 10,000 full-width at half-maximum. The time-of-flight analyzer of the mass spectrometer was calibrated with the MS/MS fragment ions of [Glu-1]-Fibrinopeptide B from m/z 50 to 1,600. The reference sprayer was sampled at a frequency of 30 s. Accurate UHPLC-MS/MS data were collected in data-dependent analysis mode by performing MS/MS scans (2-s duration) for the two most intense peaks from each MS scan (1-s duration). Peak lists were generated using ProteinLynx Global Server software version 2.3 (Waters) and automatically combined into a single pkl file for every UHPLC-MS/MS run for use in the database search.
Database searches were performed using the search engine Mascot version 2.2 (http://www.matrixscience.com) with its MS/MS ion search program. The following parameters were used in the search: (1) database, NCBInr (version November 2008; 2,551,671,261 residues, 7,387,702 sequences); (2) taxonomy, Viridiplantae (green plants; version November 2008; 526,858 sequences); (3) enzyme, trypsin; (4) missed cleavages, one; (5) peptide mass tolerance, 20 µL L−1; (6) MS/MS tolerance, 0.2 D; and (7) variable modifications, carboxyamidomethylation of Cys, oxidation of Met, phosphorylation of Ser, Thr, and Tyr, and protein N-terminal acetylation. For phosphorylated peptides, the MS/MS spectra identified by Mascot were manually checked using the following criteria: (1) a series of either four or five of the six sequential b or y ions must have been present; (2) at least one ion with b-98 or y-98 was observed; and (3) the Mascot score was greater than 37 (significance threshold, P < 0.05), and the peptides were required to be the top-ranking peptide matches. For acetylated peptides, the same criteria were used for the manual check except for the second criterion.
N-Terminal Sequencing by Edman Degradation
Target PPDKs in spots were electroblotted onto a 0.45-μm PVDF membrane (GE Healthcare) using a Mini Trans-Blot cell (Bio-Rad) and were detected by Coomassie Brilliant Blue R-250 staining. The gel spots were excised from the membrane and subjected to automatic Edman degradation using the Procise Sequencing System (model 491; Applied Biosystems).
Site-Directed Mutagenesis and in Vitro Phosphorylation
Site-directed mutagenesis was performed using a double-stranded plasmid mutagenesis procedure according to the manufacturer’s protocol (QuickChange Lightning Site-Directed Mutagenesis Kit; Agilent Technologies). Mutations were verified by two independent sequence analyses of the same DNA strand.
The full-length C4PPDK and PDRP coding sequences were amplified from complementary DNA obtained from leaves of wild-type maize. The primers used for gene cloning were as follows: 5′-TACTTCCAATCCAATGCGATGGCGGCATCGGTTTCC-3′ and 5′-TTATCCACTTCCAATGCGCTATCAGACAAGCACCTGAGC-3′ for C4ppdk; 5′-TACTTCCAATCCAATGCGATGATTGGGTGCGCCAAGC-3′ and 5′-TTATCCACTTCCAATGCGCTACTAGTATCGTTTTGATATGC-3′ for PDRP. Recombinant His-C4PPDK, His-PDRP, and 12 mutant PPDKs (T309A, S506A, T527A, T527D, S528A, H529A, S528T, S528C, S528Y, S528D, G525A, and G525P) were expressed in Escherichia coli strain BL21 (RIL) and purified using an immobilized metal affinity resin (Profinity IMAC Ni-Charged Resin; Bio-Rad). The primers for the mutants are shown in Supplemental Table S1. Purified recombinant proteins were incubated with the indicated substrates in the presence of the phosphoryl donor, 1 mm ADP or [γ-32P]ADP, in reaction buffer (50 mm HEPES-KOH [pH 8.3], 10 mm MgCl2, 5 mm DTT, 1 mm bovine serum albumin, and 0.2 mm ATP) for 5 min at 30°C. Proteins were separated by SDS-PAGE (8% [w/v] acrylamide), transferred to a 0.45-μm PVDF membrane, and detected using specific phosphopeptide antibodies or autoradiography.
Molecular Graphics
Molecular graphic representations of PPDK and PEPS were generated from their x-ray structures (Herzberget al., 1996; PDB identifiers 1VBH or 1VBG and 2OLS) using PyMOL (http://www.pymol.org) and UCSF Chimera (Pettersen et al., 2004) and were further modified using Adobe Photoshop CS2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative high-resolution MS/MS ion spectra of the unphosphorylated and phosphorylated peptide GGMTSHAAVVAR (residues 524–535) identified in spots 2 and 4.
Supplemental Figure S2. Crude extracts and chlorophyll contents of maize leaves grown under different illumination regimens.
Supplemental Figure S3. Assessment of the specificity of the anti-phospho-T527 antibody.
Supplemental Figure S4. Numbers of MS/MS spectral counts for phosphopeptides and unphosphorylated peptides containing Thr-527.
Supplemental Figure S5. Correlations between net photosynthesis rates and PPDK activity in maize leaves grown under different levels of light intensity.
Supplemental Figure S6. PPDK activity and the phosphorylation of Thr-527 in PPDK are independent of the circadian clock.
Supplemental Figure S7. Expression profiling of C4ppdk and the two types of Cyppdk in maize leaves under different illumination conditions.
Supplemental Figure S8. MS/MS spectra reveal the structures of the N termini of mature CyPPDKZm1 and CyPPDKZm2.
Supplemental Figure S9. MS/MS spectra of the identified phosphorylated peptides in maize C4PPDK.
Supplemental Figure S10. Phosphorylation levels of PPDK at Ser-528 in maize seedlings illuminated with different light intensities, time courses of PDRP catalyzing the phosphorylation of PPDK at Thr-527 and Ser-528 in vitro, numbers of MS/MS spectral counts of phosphopeptides containing Ser-528, and structural analysis of the side chains of Thr-527 and Ser-528 in maize PPDK (PDB identifier 1VBG).
Supplemental Figure S11. Time course of the phosphorylation of PPDK by PDRP.
Supplemental Figure S12. Analysis of the enzyme activities of recombinant C4PPDK and its site-directed mutant proteins.
Supplemental Table S1. Primers for PPDK mutagenesis.
Supplementary Material
Acknowledgments
We thank Qun He (China Agricultural University) and Pinghua Li (Chinese Academy of Tropical Agriculture Sciences) for helpful discussions, Zhongzhou Chen (China Agricultural University) for the valuable advice on protein structures, Leslie M. Hicks (Donald Danforth Plant Science Center) for critical reading of the manuscript, and Shanghai Invitrogen Biotechnology for generating the antibodies.
Glossary
- PEP
phosphoenolpyruvate
- 2DGE
two-dimensional gel electrophoresis
- UHPLC
ultra-high-performance liquid chromatography
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- MS
mass spectrometry
- GS
green seedlings
- ES
etiolated seedlings
- UTR
untranslated region
- TiO2
titanium dioxide
- PDB
Protein Data Bank
- H-bond
hydrogen bond
- DTT
dithiothreitol
- PVDF
polyvinylidene difluoride
- TFA
trifluoroacetic acid
Footnotes
This work was supported by the State Key Program of National Natural Science of China (grant no. 31030017) and the Program of National Natural Science of China (grant no. 31270347).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agarie S, Kai M, Takatsuji H, Ueno O. (1997) Expression of C3 and C4 photosynthetic characteristics in the amphibious plant Eleocharis vivipara: structure and analysis of the expression of isogenes for pyruvate, orthophosphate dikinase. Plant Mol Biol 34: 363–369 [DOI] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton A, Burnell J, Furbank R, Jenkins C, Hatch M (1990) Enzymes of C4 photosynthesis. In PJ Lea, ed, Methods in Plant Biochemistry, Vol 3. Academic Press, San Diego, pp 39–72 [Google Scholar]
- Ashton AR, Hatch MD. (1983) Regulation of C4 photosynthesis: regulation of pyruvate, Pi dikinase by ADP-dependent phosphorylation and dephosphorylation. Biochem Biophys Res Commun 115: 53–60 [DOI] [PubMed] [Google Scholar]
- Astley HM, Parsley K, Aubry S, Chastain CJ, Burnell JN, Webb ME, Hibberd JM. (2011) The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis are both bifunctional and interact with the catalytic and nucleotide-binding domains of pyruvate, orthophosphate dikinase. Plant J 68: 1070–1080 [DOI] [PubMed] [Google Scholar]
- Baer GR, Schrader LE. (1985) Stabilization of pyruvate, Pi dikinase regulatory protein in maize leaf extracts. Plant Physiol 77: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z, Liu B, Li W, Li P, Wang H, Wang H. (2009) The development of an improved simple titanium dioxide enrichment method for phosphoproteomic research. Rapid Commun Mass Spectrom 23: 3013–3017 [DOI] [PubMed] [Google Scholar]
- Bi YD, Wang HX, Lu TC, Li XH, Shen Z, Chen YB, Wang BC. (2011) Large-scale analysis of phosphorylated proteins in maize leaf. Planta 233: 383–392 [DOI] [PubMed] [Google Scholar]
- Burnell J. (1990) A comparative study of the cold-sensitivity of pyruvate, Pi dikinase in Flaveria species. Plant Cell Physiol 31: 295–297 [Google Scholar]
- Burnell JN. (2010) Cloning and characterization of Escherichia coli DUF299: a bifunctional ADP-dependent kinase-Pi-dependent pyrophosphorylase from bacteria. BMC Biochem 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN, Chastain CJ. (2006) Cloning and expression of maize-leaf pyruvate, Pi dikinase regulatory protein gene. Biochem Biophys Res Commun 345: 675–680 [DOI] [PubMed] [Google Scholar]
- Burnell JN, Hatch MD. (1984) Regulation of C4 photosynthesis: identification of a catalytically important histidine residue and its role in the regulation of pyruvate,Pi dikinase. Arch Biochem Biophys 231: 175–182 [DOI] [PubMed] [Google Scholar]
- Burnell JN, Hatch MD. (1985) Regulation of C4 photosynthesis: purification and properties of the protein catalyzing ADP-mediated inactivation and Pi-mediated activation of pyruvate,Pi dikinase. Arch Biochem Biophys 237: 490–503 [DOI] [PubMed] [Google Scholar]
- Burnell JN, Hatch MD. (1986) Activation and inactivation of an enzyme catalyzed by a single, bifunctional protein: a new example and why. Arch Biochem Biophys 245: 297–304 [DOI] [PubMed] [Google Scholar]
- Casati P, Zhang X, Burlingame AL, Walbot V. (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol Cell Proteomics 4: 1673–1685 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Botschner M, Harrington GE, Thompson BJ, Mills SE, Sarath G, Chollet R. (2000) Further analysis of maize C(4) pyruvate,orthophosphate dikinase phosphorylation by its bifunctional regulatory protein using selective substitutions of the regulatory Thr-456 and catalytic His-458 residues. Arch Biochem Biophys 375: 165–170 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Chollet R. (2003) Regulation of pyruvate, orthophosphate dikinase by ADP-/Pi-dependent reversible phosphorylation in C-3 and C-4 plants. Plant Physiol Biochem 41: 523–532 [Google Scholar]
- Chastain CJ, Failing CJ, Manandhar L, Zimmerman MA, Lakner MM, Nguyen TH. (2011) Functional evolution of C(4) pyruvate, orthophosphate dikinase. J Exp Bot 62: 3083–3091 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Lee ME, Moorman MA, Shameekumar P, Chollet R. (1997) Site-directed mutagenesis of maize recombinant C4-pyruvate,orthophosphate dikinase at the phosphorylatable target threonine residue. FEBS Lett 413: 169–173 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Xu W, Parsley K, Sarath G, Hibberd JM, Chollet R. (2008) The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis possess a novel, unprecedented Ser/Thr protein kinase primary structure. Plant J 53: 854–863 [DOI] [PubMed] [Google Scholar]
- Edwards G, Nakamoto H, Burnell J, Hatch M. (1985) Pyruvate, Pi dikinase and NADP-malate dehydrogenase in C4 photosynthesis: properties and mechanism of light/dark regulation. Annu Rev Plant Physiol 36: 255–286 [Google Scholar]
- Edwards GE, Furbank RT, Hatch MD, Osmond CB. (2001) What does it take to be C4? Lessons from the evolution of C4 photosynthesis. Plant Physiol 125: 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisslthaler B, Meyer G, Bohnert HJ, Schmitt JM. (1995) Age-dependent induction of pyruvate, orthophosphate dikinase in Mesembryanthemum crystallinum L. Planta 196: 492–500 [DOI] [PubMed] [Google Scholar]
- Hatch M. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895: 81–106 [Google Scholar]
- Hatch MD, Slack CR. (1968) A new enzyme for the interconversion of pyruvate and phosphopyruvate and its role in the C4 dicarboxylic acid pathway of photosynthesis. Biochem J 106: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg O, Chen CC, Kapadia G, McGuire M, Carroll LJ, Noh SJ, Dunaway-Mariano D. (1996) Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites. Proc Natl Acad Sci USA 93: 2652–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Quick WP. (2002) Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415: 451–454 [DOI] [PubMed] [Google Scholar]
- Imaizumi N, Ku MS, Ishihara K, Samejima M, Kaneko S, Matsuoka M. (1997) Characterization of the gene for pyruvate,orthophosphate dikinase from rice, a C3 plant, and a comparison of structure and expression between C3 and C4 genes for this protein. Plant Mol Biol 34: 701–716 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S. (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol 143: 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lusin JD, Ye DM, Dunaway-Mariano D, Ames JB. (2006) Examination of the structure, stability, and catalytic potential in the engineered phosphoryl carrier domain of pyruvate phosphate dikinase. Biochemistry 45: 1702–1711 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Ozeki Y, Yamamoto N, Hirano H, Kano-Murakami Y, Tanaka Y. (1988) Primary structure of maize pyruvate, orthophosphate dikinase as deduced from cDNA sequence. J Biol Chem 263: 11080–11083 [PubMed] [Google Scholar]
- McGuire M, Huang K, Kapadia G, Herzberg O, Dunaway-Mariano D. (1998) Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase. Biochemistry 37: 13463–13474 [DOI] [PubMed] [Google Scholar]
- McNaughton GA, Fewson CA, Wilkins MB, Nimmo HG. (1989) Purification, oligomerization state and malate sensitivity of maize leaf phosphoenolpyruvate carboxylase. Biochem J 261: 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Nakatsu T, Matsuoka M, Sakata K, Kato H. (2005) Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion. Biochemistry 44: 1136–1144 [DOI] [PubMed] [Google Scholar]
- Ohta S, Ishida Y, Usami S. (2006) High-level expression of cold-tolerant pyruvate, orthophosphate dikinase from a genomic clone with site-directed mutations in transgenic maize. Mol Breed 18: 29–38 [Google Scholar]
- Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, Wickner R, Salamini F, Leister D. (2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeske CA, Chollet R. (1987) Chemical modification of the bifunctional regulatory protein of maize leaf pyruvate,orthophosphate dikinase: evidence for two distinct active sites. J Biol Chem 262: 12575–12582 [PubMed] [Google Scholar]
- Roeske CA, Chollet R. (1989) Role of metabolites in the reversible light activation of pyruvate, orthophosphate dikinase in Zea mays mesophyll cells in vivo. Plant Physiol 90: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche E, Streubel M, Westhoff P. (1994) Primary structure of the photosynthetic pyruvate orthophosphate dikinase of the C3 plant Flaveria pringlei and expression analysis of pyruvate orthophosphate dikinase sequences in C3, C3-C4 and C4 Flaveria species. Plant Mol Biol 26: 763–769 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1991) Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3: 225–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER, Chock PB. (1977) Superiority of interconvertible enzyme cascades in metabolic regulation: analysis of monocyclic systems. Proc Natl Acad Sci USA 74: 2761–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Ohkawa H, Masumoto C, Fukuda T, Tamai T, Lee K, Sudoh S, Tsuchida H, Sasaki H, Fukayama H, et al. (2008) Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4-like photosynthetic pathway into rice. J Exp Bot 59: 1799–1809 [DOI] [PubMed] [Google Scholar]
- Usuda H, Ku MS, Edwards GE. (1984) Activation of NADP-malate dehydrogenase, pyruvate,Pi dikinase, and fructose 1,6-bisphosphatase in relation to photosynthetic rate in maize. Plant Physiol 76: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda H, Ku MSB, Edwards GE. (1985) Influence of light-intensity during growth on photosynthesis and activity of several key photosynthetic enzymes in a C-4 plant (Zea mays). Physiol Plant 63: 65–70 [Google Scholar]
- Walker R, Acheson R, Técsi L, Leegood R. (1997) Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust J Plant Physiol 24: 459–468 [Google Scholar]
- Walker RP, Leegood RC. (1995) Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett 362: 70–74 [DOI] [PubMed] [Google Scholar]
- Wang BC, Wang HX, Feng JX, Meng DZ, Qu LJ, Zhu YX. (2006) Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6: 2555–2563 [DOI] [PubMed] [Google Scholar]
- Wang D, Portis AR, Jr, Moose SP, Long SP. (2008) Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus × giganteus. Plant Physiol 148: 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. (2009) Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol 10: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Li Z, Ye D, Herzberg O, Dunaway-Mariano D. (2000) Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement. J Biol Chem 275: 41156–41165 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, Van Wijk K. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.