Two glutathione transferase enzymes are involved in making the explosive, environmental pollutant, 2,4,6-trinitrotoluene less toxic in Arabidopsis plants.
Abstract
The explosive 2,4,6-trinitrotoluene (TNT) is a major worldwide military pollutant. The presence of this toxic and highly persistent pollutant, particularly at military sites and former manufacturing facilities, presents various health and environmental concerns. Due to the chemically resistant structure of TNT, it has proven to be highly recalcitrant to biodegradation in the environment. Here, we demonstrate the importance of two glutathione transferases (GSTs), GST-U24 and GST-U25, from Arabidopsis (Arabidopsis thaliana) that are specifically up-regulated in response to TNT exposure. To assess the role of GST-U24 and GST-U25, we purified and characterized recombinant forms of both enzymes and demonstrated the formation of three TNT glutathionyl products. Importantly, GST-U25 catalyzed the denitration of TNT to form 2-glutathionyl-4,6-dinitrotoluene, a product that is likely to be more amenable to subsequent biodegradation in the environment. Despite the presence of this biochemical detoxification pathway in plants, physiological concentrations of GST-U24 and GST-U25 result in only a limited innate ability to cope with the levels of TNT found at contaminated sites. We demonstrate that Arabidopsis plants overexpressing GST-U24 and GST-U25 exhibit significantly enhanced ability to withstand and detoxify TNT, properties that could be applied for in planta detoxification of TNT in the field. The overexpressing lines removed significantly more TNT from soil and exhibited a corresponding reduction in glutathione levels when compared with wild-type plants. However, in the absence of TNT, overexpression of these GSTs reduces root and shoot biomass, and although glutathione levels are not affected, this effect has implications for xenobiotic detoxification.
The containment and cleanup of environmental pollutants is increasingly both a legal requirement and a responsible action in many developed countries. The most commonly used explosives in military weapons are 2,4,6-trinitrotoluene (TNT) and hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), and their continual use, along with production and decommissioning, are progressively contaminating millions of hectares of military land (Rylott and Bruce, 2009). Bioremediation of TNT is particularly challenging, as the electron-withdrawing properties of the three nitro groups render the aromatic ring particularly resistant to oxidative attack and ring cleavage by microbial oxygenases, which in the environment are normally central to the biodegradation of aromatic compounds (Qasim et al., 2007). In the United States, the Environmental Protection Agency and the military are addressing methods by which toxic TNT and RDX can be contained and detoxified on active military training ranges. One way this problem might be tackled is through the use of plants that are adapted to detoxify these compounds. This could be achieved either by traditional breeding programs or by genetic modification, as has been demonstrated previously for both RDX and TNT (Hannink et al., 2001; Rylott et al., 2006; Jackson et al., 2007).
In the majority of species tested so far (tobacco [Nicotiana tabacum], bean [Phaseolus vulgaris], wheat [Triticum aestivum], poplar [Populus spp.], and switchgrass [Panicum virgatum]), with the exception of some conifer trees (Schoenmuth and Pestemer, 2004), TNT is located almost entirely in the roots (Sens et al., 1998, 1999; Hannink et al., 2007; Van Dillewijn et al., 2008; Brentner et al., 2010). Endogenous metabolism of TNT by plants has been characterized (Rylott and Bruce, 2009; Rylott et al., 2011b), with recent research focusing on the model plant species Arabidopsis (Arabidopsis thaliana; Hannink et al., 2001; Van Dillewijn et al., 2008; Rylott et al., 2011a). First, TNT is transformed by nitroreductases to hydroxylamino dinitrotoluenes (HADNTs), with a varying portion further reduced to amino dinitrotoluenes (ADNTs). In Arabidopsis, oxophytodienoate reductases are known to catalyze these steps (Beynon et al., 2009). Plants engineered to express bacterial nitroreductases, which also perform this transformation step, have increased TNT transformation activity and show dramatically enhanced resistance to TNT (Hannink et al., 2001; Rylott et al., 2011a). The additional functionality of HADNTs and ADNTs permits their subsequent conjugation to amino acids, organic acids, and sugars (Bhadra et al., 1999, 2001), and conjugation of HADNT and ADNT isomers to Glc by Arabidopsis glucosyltransferases has been characterized (Gandia-Herrero et al., 2008), with research suggesting that these conjugates are subsequently sequestered within the cell walls (Rylott and Bruce, 2009).
Glutathione transferases (GSTs) are a multigene family of proteins known to conjugate glutathione to electrophilic molecules and, in plants, are involved in the detoxification of herbicide xenobiotics (Cummins et al., 2011). Since GSTs have evolved the ability to catalyze glutathione-linked reactions with thousands of different chemical structures, it has been hypothesized that GSTs should play a central role, alongside glucosyltransferases, in the detoxification of TNT (Mezzari et al., 2005; Brentner et al., 2008). Gene expression studies in poplar (Tanaka et al., 2007; Brentner et al., 2008) and Arabidopsis (Ekman et al., 2003; Mezzari et al., 2005; Gandia-Herrero et al., 2008) have identified GSTs up-regulated in response to TNT; however, to date, the biochemical response of GSTs toward TNT has not been investigated. The overexpression of plant GSTs has been shown to increase resistance to a range of stresses, with some τ class GSTs shown to detoxify herbicides via a conjugation activity (Dixon and Edwards, 2010; Cummins et al., 2011). Many Arabidopsis GSTs (in common with some mammalian and other plant GSTs) exhibit a glutathione-dependent peroxide (GPOX) activity (Dixon and Edwards, 2010), catalyzing the reduction of lipid hydroperoxides to the respective monohydroxyalcohols, an activity that confers tolerance to a number of oxidative stresses (Dixon et al., 1998; Dixon and Edwards, 2010). Here, we expressed, purified, and characterized TNT-responsive Arabidopsis GSTs and investigated their contribution toward the detoxification of TNT in Arabidopsis.
RESULTS
Up-Regulation of GSTs in Response to TNT
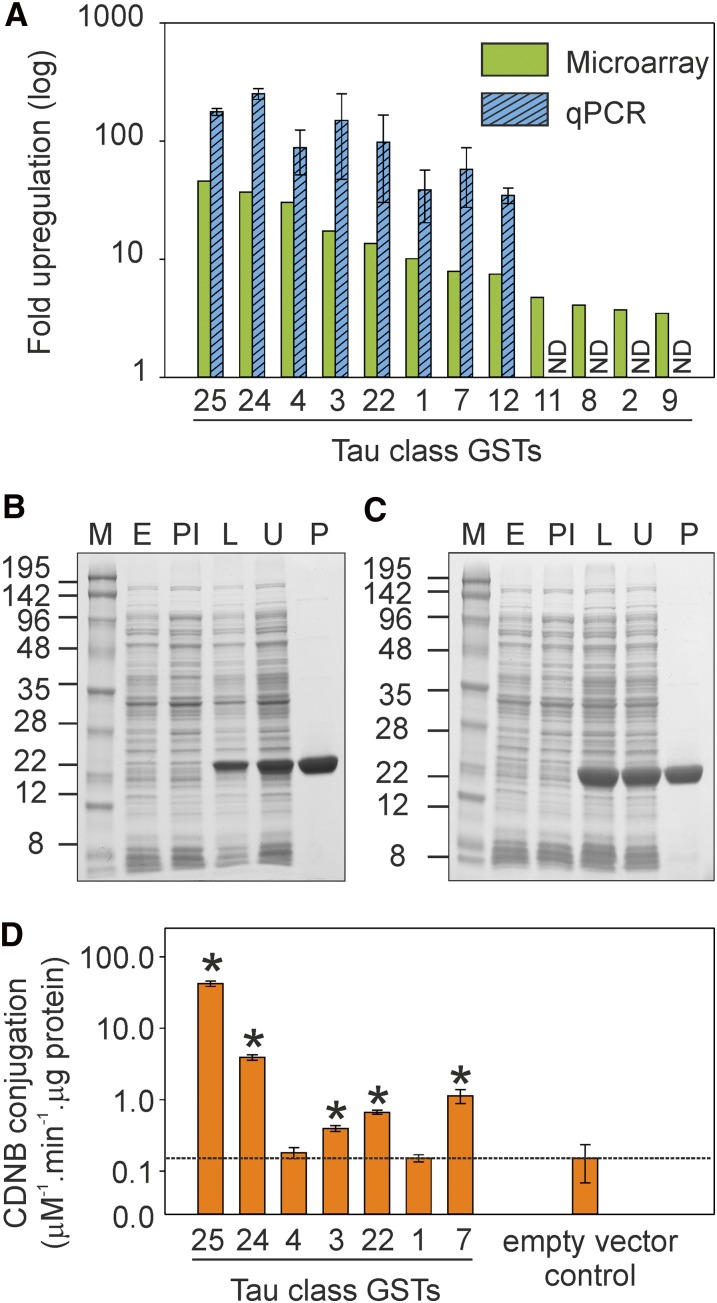
There are currently 55 GSTs identified in the Arabidopsis genome (Dixon and Edwards, 2010). Following microarray analysis of TNT-treated Arabidopsis plants (Gandia-Herrero et al., 2008), eight up-regulated GSTs, all in the τ class (GST-U1, GST-U3, GST-U4, GST-U7, GST-U12, GST-U22, GST-U24, and GST-U25), were selected for further study. Quantitative PCR analysis verified the TNT-responsive up-regulation, with GST-U24 (At1g17180) and GST-U25 (At1g17170) the most up-regulated (252- and 177-fold, respectively; Fig. 1A).
Figure 1.
Gene expression, protein purification, and CDNB activity of GSTs. A, Microarray data showing Arabidopsis GSTs up-regulated more than 2-fold following TNT treatment (P < 0.05 for all values; Gandia-Herrero et al., 2008) and corroborative qPCR data on Arabidopsis plants treated with TNT. The qPCR results were normalized to the ACTIN2 gene, and the results are means of five biological replicates ± se. qPCR data were not determined (ND) for U11, U8, U2, or U9. B and C, SDS-PAGE gels for GST-U24 (B) and GST-U25 (C) showing the expression and purification of recombinant GSTs. M, Molecular mass marker (kD); E, protein extract from E. coli transformed with empty vector; PI, protein extract from E. coli culture prior to induction; L, lysed induced protein extract from E. coli after 60 h of expression time; U, unbound fraction of the purification process; P, purified protein. D, Conjugation activity of purified GSTs toward a CDNB equivalent volume of the purified empty vector lysate. Results are means of three technical replicates ± sd. Asterisks denote samples that were significantly different (P < 0.05) from the empty vector control. [See online article for color version of this figure.]
Activity of Recombinant GSTs
To determine the functional activity of the GSTs toward TNT, the eight GSTs were expressed in Escherichia coli, and we were able to purify seven by affinity chromatography to homogeneity (Fig. 1, B and C; Supplemental Fig. S1). Catalytic activity was assessed spectrophotometrically using 1-chloro-2,4-dinitrobenzene (CDNB; Fig. 1D), a widely used marker for GST activity that had previously been shown to be a substrate for these GSTs (Dixon et al., 2009). The glutathione-CDNB-conjugating activities of GST-U24 and GST-U25 were broadly similar to those reported by Dixon et al. (2009), and the enzyme kinetics are presented in Table I. Activities for GST-U1 and GST-U4 were not significantly higher than those for the empty vector controls, and Dixon et al. (2009) also reported low values for these GSTs using CDNB (1.4% and 0.8%, respectively, of that for GST-U25).
Table I. Kinetic analysis of TNT- and CDNB-conjugating activities by GST-U24 and GST-U25.
Preliminary assays to detect TNT-conjugating activity by the purified GSTs, using HPLC techniques, revealed activity toward TNT by GST-U24 and GST-U25. Measurement of nitrite release, by Griess assay, showed that under the conditions tested (pH 6.5, 5 mm reduced glutathione [GSH], 200 µm TNT, and 100 µg of protein, measured after 24 h), GST-U24 released 7.8 µm nitrite and GST-U25 released 40 µm (Supplemental Fig. S2). These values equate to 3.4% and 20% of the 200 µm starting TNT concentration for GST-U24 and GST-U25, respectively. Nitrite release was not detected for the remaining GSTs under these conditions. Following the preliminary screening of the seven GSTs, our studies focused on the two GSTs with significant activity toward TNT: GST-U24 and GST-U25.
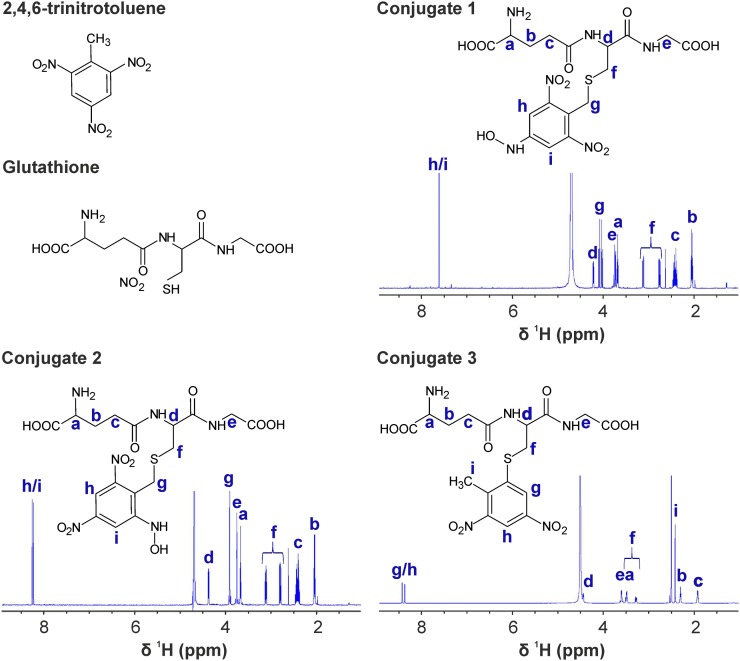
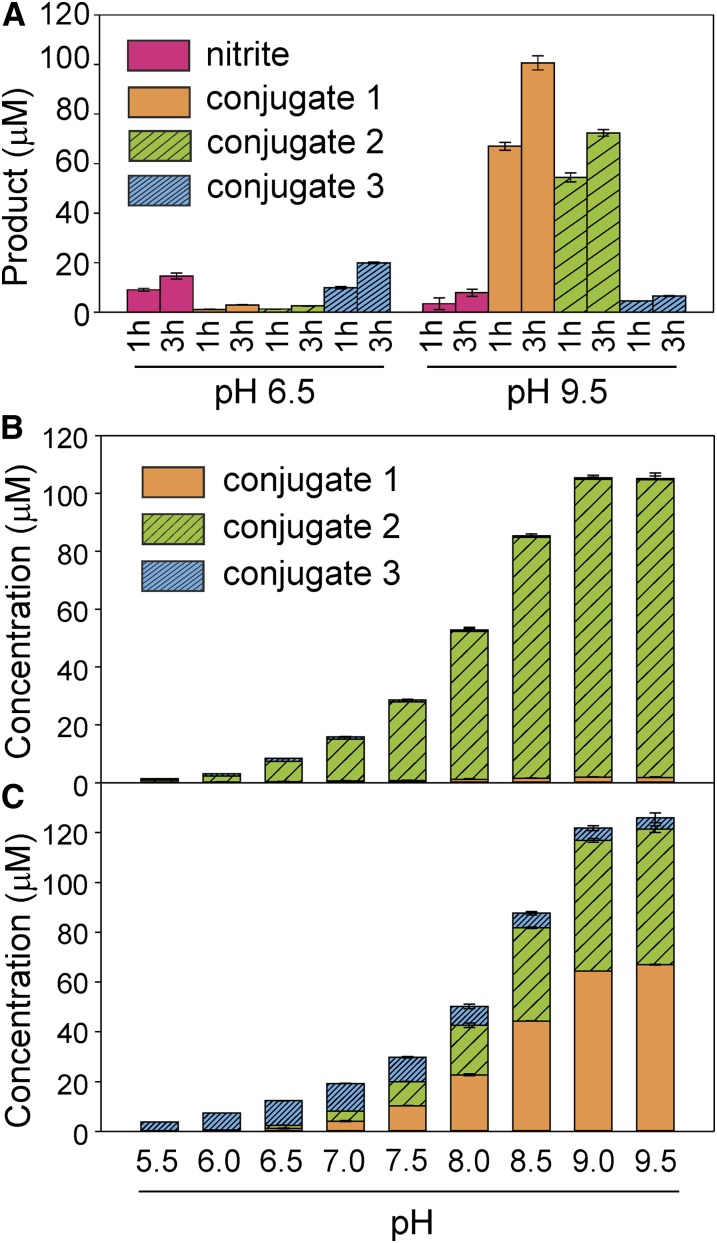
More detailed HPLC-based assays using TNT as a substrate, and subsequent mass spectrometry analyses, revealed that GST-U24 and GST-U25 catalyzed the production of three distinct products (Fig. 2). Conjugates 1 and 2 had similar absorption spectra with slightly different absorption maxima: 239 and 231 nm, respectively. Conjugate 3 had an absorption maximum of 251 nm and a spectrum distinct from conjugates 1 and 2 (Supplemental Fig. S3). Subsequent tandem mass spectrometry analysis showed that both conjugates 1 and 2 produced [M-H]− ions of 517 with identical molecular masses of 518, whereas conjugate 3 had a fragmentation pattern consistent with a desulfo-glutathione [M-H] ion of 274, a dinitrotoluene-sulfur ion [M-H]− of 213, and a molecular mass of 487, indicative of 2-glutathionyl-4,6-dinitrotoluene. Analysis using NMR spectroscopy highlighted the absence of the characteristic methyl resonance of TNT in the 1H spectra of conjugates 1 and 2, suggesting that GSH conjugation occurs via the methyl group. Correspondingly, a resonance attributed to the formation of a methylene bridge between position 1 of the aromatic ring and the sulfur of GSH is apparent in both spectra. Reduction of a nitro group occurs at either position 2/6 or 4 in conjugate 1 or 2, respectively. The asymmetry introduced via nitro group reduction at position 2/6 resolves the degeneracy of the two aromatic proton resonances. Reduction at position 4 maintains the equivalency of the aromatic protons, and a single resonance is observed. For conjugate 3, GSH conjugation occurs via substitution of a nitro group at the 2/6 position. Thus, the methyl group of TNT is preserved and a resonance is observed in the NMR spectrum. Asymmetry of the aromatic protons again gives rise to two distinct peaks. To further support the chemical structures shown in Figure 2, Figure 3A shows that release of nitrite, measured using Griess assays, correlated with the approximately equimolar production of conjugate 3 (nitrite:conjugate 3 of 1:1.2 at pH 6.5, and nitrite:conjugate 3 of 1:1 at pH 9.5). In further HPLC-based assays using HADNT and ADNT as substrates for GST-U24 and GST-U25, there was no detectable conjugation (data not shown).
Figure 2.
1H-NMR spectra and deduced chemical structures of TNT and the three GSH-TNT conjugates. Protons in the chemical structure, and their corresponding resonances, are labeled a to i. Assignments were completed using 1H13C-HMBC, 1H13C-HSQC, and 1H1H-COSY spectra. As mentioned in the text, the key features are the resolved degeneracy of ring proton resonances (h/i and g/h) in conjugate 2 and conjugate 3 compared with conjugate 1 and the presence of a methyl singlet peak (i) in conjugate 3, which is absent in conjugates 1 and 2. [See online article for color version of this figure.]
Figure 3.
A, Levels of nitrite and conjugate produced by GST-U25 at pH 6.5 and 9.5 after 1 and 3 h. B and C, Activities of GST-U24 (B) and GST-U25 (C) with increasing pH after 1 h of incubation. All TNT conjugation reactions were performed at 20°C in 100 mm phosphate buffer with 150 μg of GST, 200 μm TNT, and 5 mm GSH in a total volume of 250 μL. Conjugation products and nitrite were measured by HPLC and Griess assays, respectively. Results are means of three biological replicates ± sd. [See online article for color version of this figure.]
For both GST-U24 and GST-U25, the highest TNT-conjugating activity was observed at the highest pH tested (pH 9.5; Fig. 3, B and C). While GST-U24 produced predominantly conjugate 2, with only trace amounts of conjugates 1 and 3, GST-U25 produced different conjugate ratios dependent on pH; at pH 6.5, GST-U25 produced almost exclusively conjugate 3, with only trace amounts of conjugates 1 and 2, whereas at pH 9.5, mostly conjugates 1 and 2 were made. For consistency, phosphate buffer was used across the pH range tested (pH 5–9.5), and although this exceeded the optimal span for a phosphate buffer, preliminary assays using MOPS and Tris-HCl buffers at the higher (greater than 7) pH values gave rates indistinguishable from those using phosphate buffer (data not shown). It has been reported that phosphate and Tris buffers can increase nonenzymatic conjugation of metolachlor (Irzyk and Fuerst, 1993), chloroacetamide (Leavitt and Penner, 1979), and triazine (Frear and Swanson, 1970) at elevated pH; however, we did not see any significant GSH-TNT conjugates in boiled enzyme controls at any of the pH levels tested. Unlike pH, increasing the assay temperature did not alter the ratio of conjugates produced by either GST-U24 or GST-U25 (Supplemental Fig. S4). For GST-U24, the highest rate of TNT conjugation was observed between 30°C and 37°C, whereas GST-U25 TNT-conjugating activity increased with temperature up to 50°C, the highest temperature tested. Above 60°C, the GSTs were rapidly inactivated.
Using TNT as a substrate, kinetic data were obtained from purified GST-U24 and GST-U25 (Table I). The Km and Vmax values show that both enzymes have similar activities toward TNT, with GST-U25 being the more active of the two enzymes studied. For both GST-U24 and GST-U25, the Km values (1.6 and 1.2 mm, respectively) indicate that, in wild-type plants, the TNT would be present at levels that are likely to be cytotoxic before GST-U24 and GST-U25 would efficiently metabolize and detoxify it. The τ and ϕ class GSTs, have been most studied in cereal crops and weeds for their role in detoxifying agrichemicals. These GSTs have relatively high activities toward chloroacetanilide and triazine herbicides and other chlorinated xenobiotics (Cummins et al., 2011). While the ability to conjugate these synthetic compounds is not the endogenous role of τ class GSTs, it is possible that these xenobiotics might share chemical similarities with endogenous substrates.
To assay the GPOX activity of purified GST-U24 and GST-U25, cumene hydroperoxide was used as a substrate. The results, shown in Table II, revealed activities similar to those reported by Dixon et al. (2009), with GST-U25 exhibiting a 2-fold lower Km and a 4-fold higher Vmax than GST-U24. To measure the effect of TNT on the GPOX activity of the two enzymes, GPOX activity was measured with increasing concentrations of TNT. The resulting Lineweaver-Burk double reciprocal plots demonstrate that for both GST-U24 and GST-U25, the Vmax decreases with increasing TNT concentration. For GST-U25, the Km also decreases, indicative of uncompetitive inhibition. However, for GST-U24, the calculated Km values increase with increasing TNT concentration. While this result is difficult to explain, it indicates that TNT is inhibiting GST-U24 GPOX activity in a different, possibly noncompetitive, way than it is for GST-U25 (Table II; Supplemental Fig. S5).
Table II. Kinetic analysis for GST-U24 and GST-U25 using cumene hydroperoxides to measure GPOX activity with increasing concentrations of TNT.
| GST | TNT Concentration | Vmax | Km |
|---|---|---|---|
| μm | nkat mg−1 | μm | |
| GST-U24 | 0 | 35.5 ± 0.7 | 495.8 ± 23.8 |
| 2.5 | 34.5 ± 0.6 | 1,036.1 ± 34.5 | |
| 5.0 | 33.9 ± 1.0 | 1,322.9 ± 63.5 | |
| 10 | 22.4 ± 1.8 | 1,733.0 ± 221.5 | |
| GST-U25 | 0 | 151.0 ± 2.0 | 259.6 ± 10.4 |
| 2.5 | 116.0 ± 1.5 | 244.0 ± 10.2 | |
| 5.0 | 101.2 ± 1.3 | 222.3 ± 9.0 | |
| 10 | 75.9 ± 1.3 | 167.0 ± 10.0 |
Overexpressing GST-U24 and GST-U25 Arabidopsis Lines
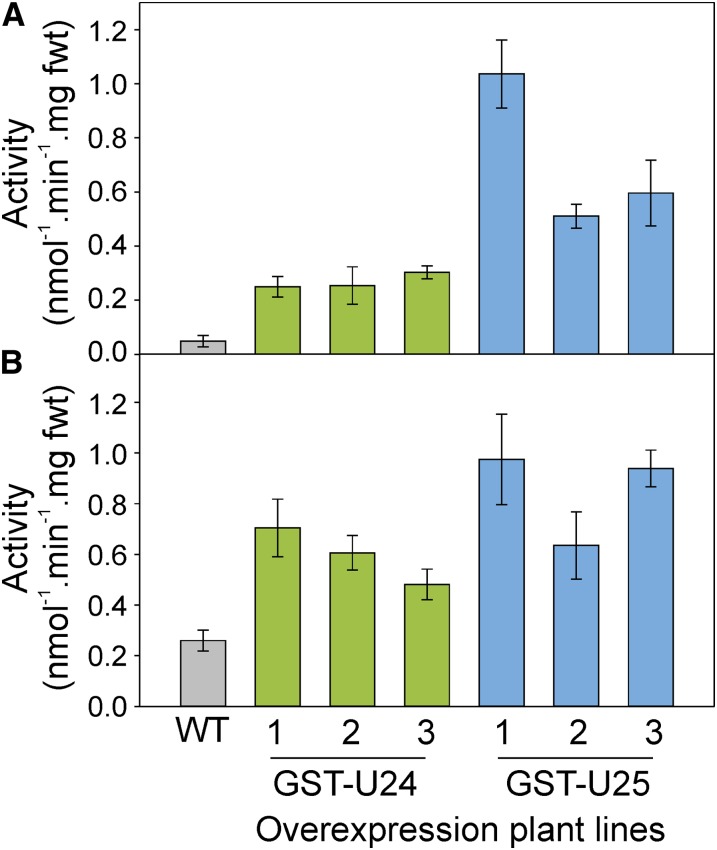
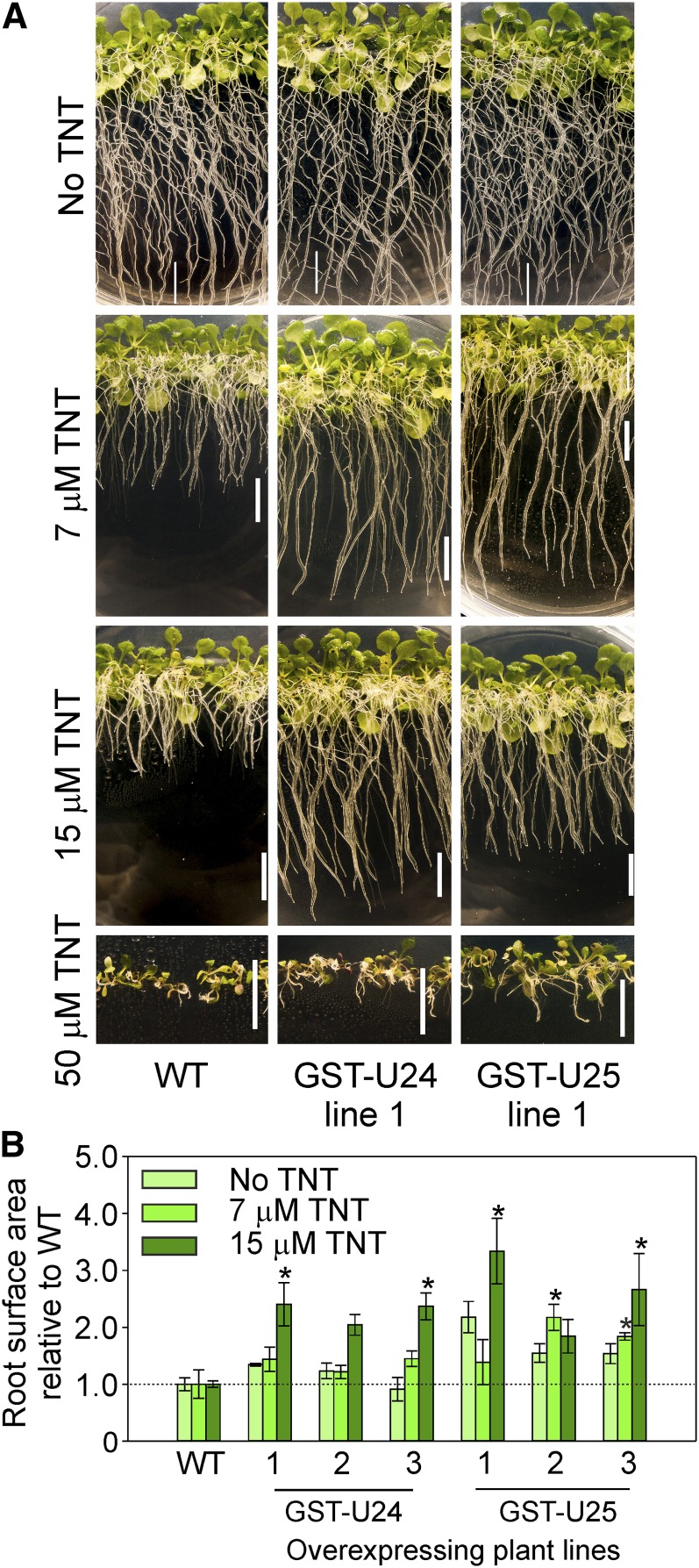
To investigate whether increased levels of GST-U24 and GST-U25 activities confer increased abilities to detoxify TNT and thus withstand the phytotoxic effects of this compound, we conducted a range of hydroponic and soil-based experiments using Arabidopsis lines overexpressing GST-U24 or GST-U25. The near-constitutively expressing cauliflower mosaic virus 35S promoter was used to drive the overexpression of GST-U24 and GST-U25. Plants were assayed for GST activity using CDNB as a substrate, and lines with GST activity significantly higher than the wild type were selected for further analysis (Fig. 4; Supplemental Fig. S6). The response of the GST overexpression lines to TNT was first tested by growing plants on agar plates containing one-half-strength Murashige and Skoog (1/2 MS) medium plus a range of TNT concentrations. After 9 d, the GST-U24- and GST-U25-overexpressing lines had root surface areas similar to wild-type plants, but after 20 d, the overexpression lines had produced significantly more root surface area than wild-type plants (Fig. 5; Supplemental Tables S1 and S2). To see if the in vitro TNT conjugates were produced in vivo, liquid culture-grown, 3-week-old wild-type, GST-U24-, and GST-U25-overexpressing lines were dosed with 200 µm TNT and uptake was monitored over 7 d (Supplemental Fig. S7). Rates of TNT uptake were similar to those of wild-type plants; however, measurement of conjugates within the plant tissues after 24 h using liquid chromatography-mass spectrometry (LC-MS) revealed that conjugate 2 was detected solely in the GST-U24 lines and conjugate 3 only in the GST-U25 lines. Conjugate 1 was not detected (Supplemental Fig. S7C).
Figure 4.
Conjugation activities in leaf (A) and root (B) protein extracts from the Arabidopsis wild type (WT) and GST-overexpressing lines assayed using CDNB substrate. Rosette leaves were from 6-week-old plants grown in uncontaminated soil. Results are means of three biological replicates ± sd. Roots were from 2-week-old plants grown on agar plates containing 1/2 MS medium without TNT. Results are means of five biological replicates ± se. fwt, Fresh weight. [See online article for color version of this figure.]
Figure 5.
Root growth of Arabidopsis grown vertically on agar plates containing 1/2 MS medium and a range of TNT concentrations. A, Appearance of 20-d-old Arabidopsis wild-type (WT) and GST-U24- and GST-U25-overexpressing plants. Bars = 1 cm. B, Ratio of root surface area relative to wild-type plants. Results are combined means of the root lengths of 12 to 15 seedlings grown on each of three replicate plates ± sd. Asterisks denote statistical differences compared with the wild type (P < 0.05). [See online article for color version of this figure.]
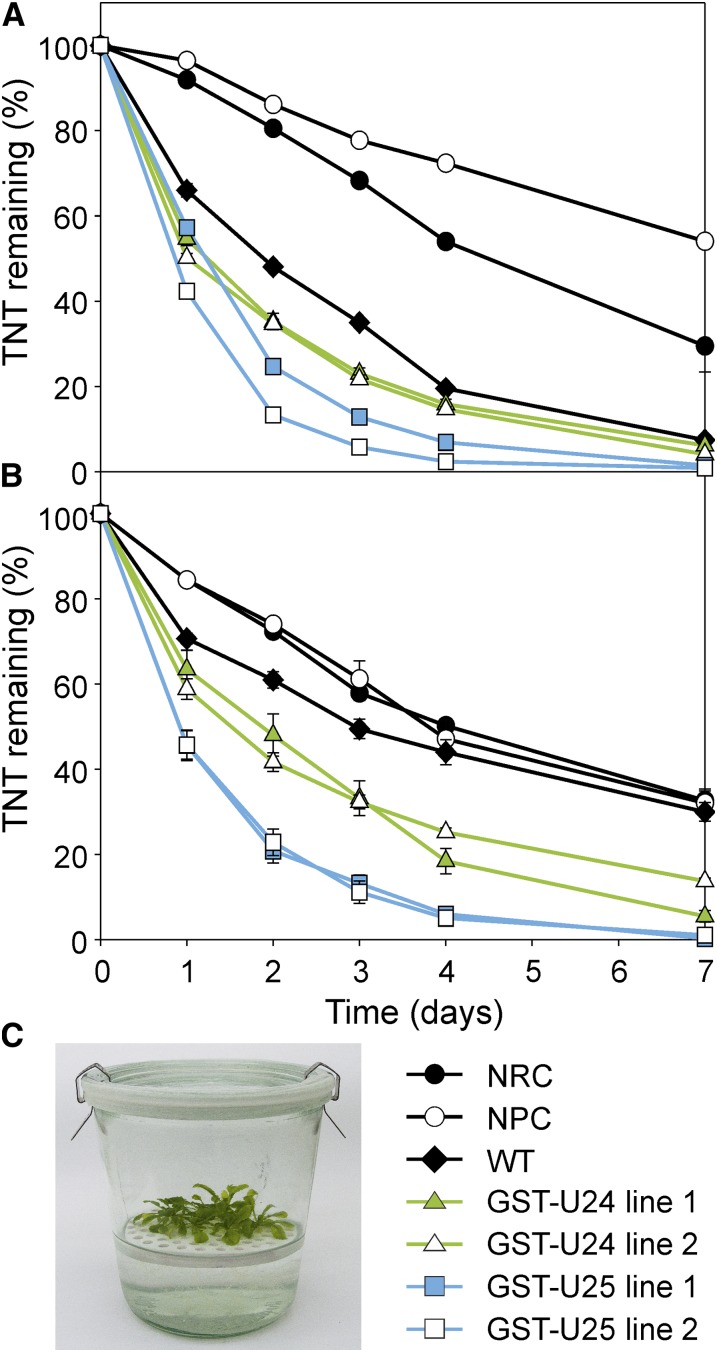
In the natural environment, predominantly the roots of a terrestrial plant would be exposed to TNT, whereas in the liquid culture experiment, the entire plant was submerged in liquid medium. To more closely replicate natural environmental conditions, Arabidopsis lines were grown hydroponically on plastic rafts floating on liquid medium, and after 3 weeks the roots were exposed to TNT by dosing the medium. Uptake of TNT over the following week was recorded (Fig. 6); using the raft-based system, both the GST-U24 and GST-U25 lines removed TNT from liquid medium significantly more quickly than untransformed plants. While conjugates 2 and 3 were detected in the liquid culture experiments, in the raft-based system no such derivatives were identified in medium or plant extracts.
Figure 6.
Rate of TNT uptake from hydroponic culture by wild-type (WT) and GST-U24- and GST-U25-overexpressing Arabidopsis plants. A and B, Three-week-old plants were grown on rafts with roots submerged in 1/2 MS medium, then dosed with 50 µm (A) or 100 µm (B) TNT. C, Experimental setup. NRC, No-raft control; NPC, no-plant control. Results are means of five biological replicates ± se. [See online article for color version of this figure.]
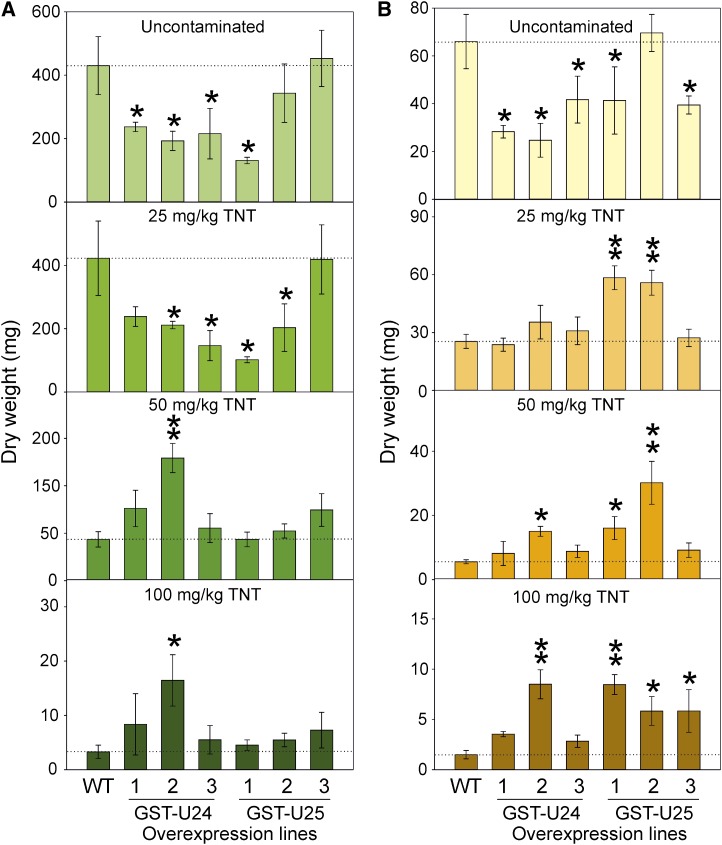
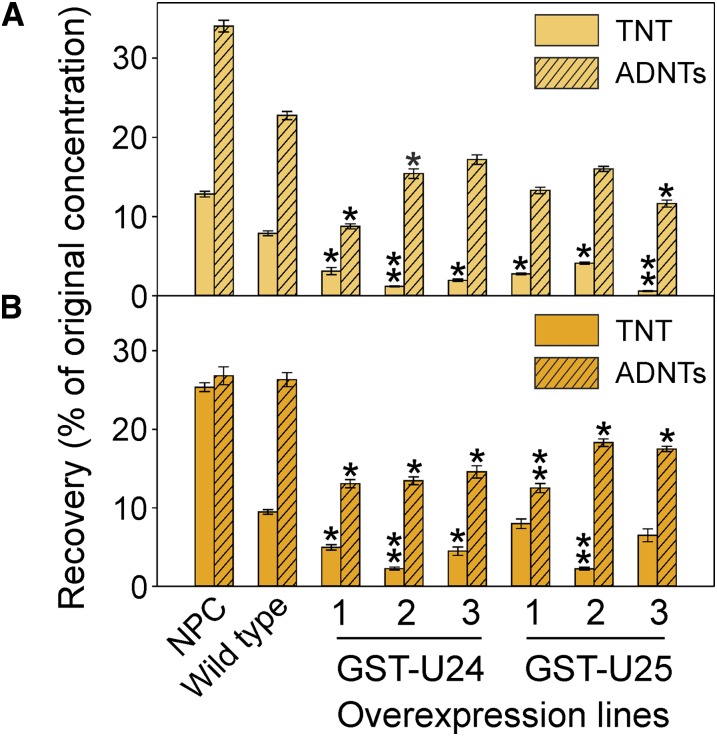
Military training ranges often have areas of contamination containing between 100 and 1,000 mg kg−1 explosive pollutants (Talmage et al., 1999; Jenkins et al., 2006), although the majority are usually below this level. To test the detoxification abilities of the GST-U24 and GST-U25 overexpression lines in a soil-based system, we chose TNT concentrations of 25, 50, and 100 mg kg−1 (levels that unmodified Arabidopsis plants could tolerate while still being representative of the amounts found on training ranges). After 6 weeks of growth in soil, the aerial tissues of the GST-U24- and GST-U25-overexpressing lines appeared slightly smaller than those of unmodified plants when grown in uncontaminated soil. Subsequent dry weight measurements confirmed that both aerial and root tissues were significantly reduced (P < 0.05) in all three of the GST-U24 lines and, to a lesser degree, in the GST-U25-overexpressing lines grown in uncontaminated soil (Fig. 7). In increasing concentrations of TNT-contaminated soil, the tissue dry weights of the GST-U24- and GST-U25-overexpressing lines increased such that they were similar to, or larger than, unmodified plants grown in soil at the same TNT concentration. To account for the reduced biomasses of the GST-U24 and GST-U25 overexpression lines grown in uncontaminated soil, we normalized the data by presenting the biomasses for each line when grown in TNT-contaminated soil relative to the biomass of that line when grown in uncontaminated soil. These normalized data showed that the mean aerial and root biomasses of the combined GST-U24-overexpressing lines grown in 100 mg kg−1 TNT were 6.4- and 7.9-fold, respectively, higher than unmodified plants. Similarly, for the GST-U25-overexpressing lines, the normalized mean shoot and root biomasses were 2.9- and 6.4-fold higher, respectively. The remaining extractable TNT was measured in the soil, along with levels of ADNT resulting from the transformation of TNT by soil-based microbial communities. The soil in which the GST-U24 and GST-U25 lines had been grown for 6 weeks was found to have significantly less TNT and ADNT than soil in which wild-type plants had been grown (Fig. 8). The TNT is readily converted to ADNTs by soil microorganisms. Because the GST-overexpressing lines have removed more TNT than the wild-type lines, there is less TNT available for the soil microorganisms to convert to ADNT. Furthermore, the ratio of TNT to ADNT was lower in the soil from the overexpressing lines than from the wild type, indicating that the overexpressing lines removed TNT in preference to ADNT (TNT:ADNT for the wild type, 0.35; for GST-U24 lines, 0.35, 0.08, and 0.11, respectively; and for GST-U25 lines, 0.21, 0.26, and 0.05 respectively; from soil with an initial contamination of 25 mg kg−1 TNT). This is in agreement with our studies using purified GST-U24 and GST-U25, which show that these enzymes have activity toward TNT but not ADNTs.
Figure 7.
Shoot (A) and root (B) biomasses of Arabidopsis plants grown in TNT-contaminated soil. Wild-type (WT) and GST-U24- and GST-U25-overexpressing Arabidopsis plants were grown in soil contaminated with a range of TNT concentrations for 6 weeks. Results are means of eight biological replicates ± se. Asterisks denote statistically significant differences from the wild type: *P < 0.05, **P < 0.01. [See online article for color version of this figure.]
Figure 8.
Levels of nitrotoluenes recovered from TNT-contaminated soil. Wild-type and GST-U24- and GST-U25-overexpressing Arabidopsis plants were grown alongside no-plant control (NPC) pots of soil contaminated with 25 mg kg−1 (A) or 50 mg kg−1 (B) TNT for 6 weeks, and then TNT and ADNTs were extracted from the soil. Results are means of eight biological replicates ± se. Asterisks denote statistically significant differences from the wild type: *P < 0.05, **P < 0.01. [See online article for color version of this figure.]
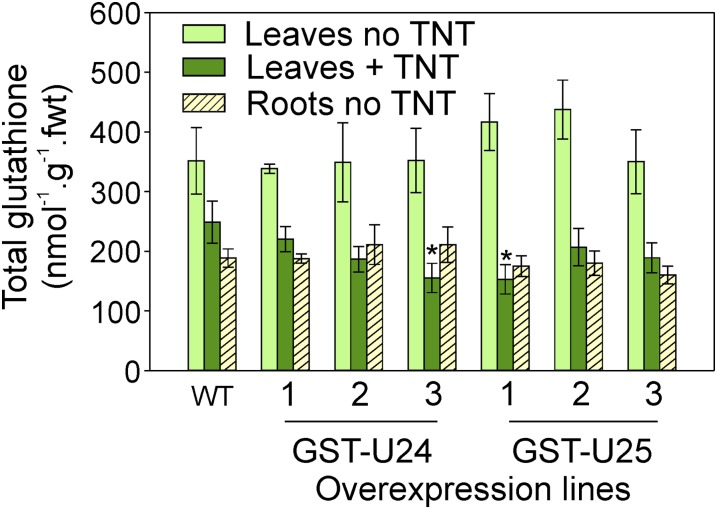
As the GST overexpression lines removed significantly more TNT from soil than unmodified plants, glutathione levels were measured to see if there was a corresponding decrease in glutathione pools resulting from the formation of TNT-GST conjugates. Additionally, glutathione levels were measured to ascertain if the yield-drag effect observed in GST-expressing lines grown in the uncontaminated soil could be explained by glutathione depletion. Levels of glutathione were measured in the rosette leaves of plants grown in soil contaminated with 50 mg kg−1 TNT. Due to the difficulty of extracting soil-grown roots for glutathione measurement, glutathione levels were also determined in the roots and leaves of 2-week-old plants grown on vertical agar plates of uncontaminated, 1/2 MS medium. Figure 9 shows that, when grown on uncontaminated agar plates, the levels of total glutathione were approximately 30% to 50% lower in the roots than in the leaves of all lines, including the wild type. Additionally, the levels of total glutathione in the leaves and roots of the GST overexpression lines were not significantly different from those found in wild-type tissues. While the leaves from wild-type plants grown in TNT-contaminated soil had 29% less total glutathione than those grown in uncontaminated soil, Figure 9 shows that the GST overexpression lines had even less total glutathione when grown in TNT-contaminated soil (GST-U24 lines, 35%, 46%, and 47%, respectively; GST-U25 lines, 53%, 56%, and 65% respectively). There was no significant difference in the ratios of GSH to oxidized glutathione (GSSG) in leaves from the wild type and overexpressing lines (data not shown).
Figure 9.
Glutathione levels in Arabidopsis leaves and roots. Rosette leaves were from plants grown in uncontaminated soil or soil contaminated with 50 mg kg−1 TNT for 6 weeks. Roots were from 2-week-old plants grown on 1/2 MS plus agar plates. Wild-type (WT) and GST-U24- and GST-U25-overexpressing plants were used. Results are means of eight biological replicates ± sd. Asterisks denote statistical differences compared with the wild type (P < 0.05). fwt, Fresh weight. [See online article for color version of this figure.]
DISCUSSION
Glutathione-TNT Products
The GST-U24 and GST-U25 enzymes catalyze the conjugation of TNT with GSH to form three distinct products. For conjugates 1 and 2, conjugation at the methyl is likely to occur through the formation of a quinone methide intermediate. While this has been observed previously in GST conjugation to methyl-substituted aromatic rings (Kassahun et al., 2001; Li et al., 2009), this is, to our knowledge, the first report of such a GST conjugate to TNT. For both GST-U24 and GST-U25, catalytic activity increased with pH, with the optimum appearing to be pH 9.5 or greater. The high catalytic activity at higher pH values could be explained by the fact that the sulfhydryl group of glutathione has a pKa value of 9.4, making the reactive thiolate anion more stable at the higher pH (Dixon and Edwards, 2010). There are few studies that have measured the pH optima for plant GSTs. Irzyk and Fuerst (1993) reported a purified maize GST with a broad pH optimum of 7 to 8 using metolachlor as substrate. Using atrazine, another purified maize GST isozyme had a pH optimum of 8 to 8.5 (Guddewar and Dautermann, 1979). Our data show that the production of conjugate 3 by GST-U25 is favored between pH 6.5 and 7. Within the root, the site of TNT detoxification in dicot and grass species tested (Sens et al., 1998, 1999; Brentner et al., 2010), the pH of the cytosol is estimated to range between 6.5 and 7.9 (Scott and Allen, 1999; Moseyko and Feldman, 2001; Tournaire-Roux et al., 2003). If GST-U25 is expressed in a cytosol with a pH of less than 7, this might explain why only conjugate 3 was detected in the root extracts from GST-U25-overexpressing plants. Alternatively, conjugate 3 may be more stable or metabolized more slowly than the other conjugates.
Following production, GST conjugates are imported to the vacuole, and two glutathione conjugate ATP-binding cassette transporters, Multidrug resistance-associated protein1 (MRP1) and MRP2, are up-regulated in Arabidopsis in response to TNT (2- and 5-fold respectively; Gandia-Herrero et al., 2008). After import, studies on herbicide detoxification by GSTs showed that further processing occurs, releasing cysteinyl-Gly, γ-glutamyl-Cys, and Cys derivatives (Grzam et al., 2006; Ohkama-Ohtsu et al., 2007). The longer term fate of these xenobiotic Cys derivatives, once sequestered into the vacuole, is not known. These derivatives were seen at low levels in the liquid culture experiments and not found in the hydroponic or soil-based studies, and it is possible that, in the case of TNT detoxification, these derivatives are quickly further processed in the vacuole.
Both GST-U24 and GST-U25 have similar rates of activity toward TNT, and, in combination with other endogenous GSTs, there is likely to be considerable overlapping activity toward TNT in vivo. Although not up-regulated in our microarray study (Gandia Herrero et al., 2008), GST-U19, which is closely related to GST-U24 and GST-U25, can also be up-regulated in response to TNT (Ekman et al., 2003), and it is likely that GST-U19 also has activity toward TNT. Root length studies of knockout GST-U24 mutant plants were reported to have a similar response to TNT to wild-type plants (Yoon et al., 2007). The results here suggest that this is because GST-U25 and additional τ class GST activities compensated for the loss of GST-U24. Thus, knockout GST-U25 plants would be predicted to also lack an altered TNT-related phenotype. Due to the close proximity of GST-U24 and GST-U25 (they are 1.3 kb apart on chromosome I), double knockouts would be difficult to achieve.
The reduced biomasses of the GST-U24 and GST-U25 overexpression lines when grown in uncontaminated soil were not expected and have not been reported in studies overexpressing related, τ class GSTs. The yield drag could be the result of deleterious conjugation and/or GPOX activity. While we show that the glutathione pools are unaffected in the overexpressing lines in the absence of TNT, GPOX activity utilizes glutathione to reduce organic hydroperoxides to alcohols. Thus, it is possible that the negative effects on biomass when the GST-U24- and GST-U25-overexpressing plants are grown in uncontaminated soil might be the end result of a depletion of short-chain hydroxylated fatty acid pools. In support of this, Dixon and Edwards (2009) reported the accumulation of short-chain hydroxyl-acylated glutathione conjugates by GST-U25 when expressed in E. coli and, transiently, in tobacco. While we saw a decrease in glutathione pools in plants grown in TNT-contaminated soil, studies using monochlorobimane, which binds glutathione to form a fluorescent compound, indicated that glutathione pools increase in the presence of TNT (Mezzari et al., 2005). Additionally, gene expression studies in Arabidopsis show that glutathione reductases, which are involved in sustaining glutathione pools, can be up-regulated in response to TNT treatment (Ekman et al., 2003).
The GST-overexpressing lines removed more TNT from soil than wild-type plants, with a corresponding decrease in the relative amounts of total glutathione. In addition, we have shown that, in vitro, TNT inhibits GPOX activity in both GST-U24 and GST-U25. Together, these findings indicate that the enhanced TNT tolerance observed in the overexpressing lines is due to direct glutathionylation of TNT rather than enhanced GPOX activity. Chemical treatments associated with the production of active oxygen species have been shown to cause a strong up-regulation of GPOX activity (Dixon et al., 1998; Dixon and Edwards, 2010). In bacteria, TNT can undergo one-electron reduction to produce a nitroanion radical, which reacts with oxygen to form a superoxide radical, with the regeneration of TNT. Plant cells contain flavoproteins capable of one-electron reduction activity, and the formation of reactive oxygen species from the one-electron reduction of TNT is likely to be a major cause of toxicity (French et al., 2001). Overexpression of plant GSTs has been shown to confer resistance to a number of biotic and abiotic stresses (Dixon and Edwards, 2010; Cummins et al., 2011), and it is possible that the GST-U24 and GST-U25 overexpression lines also have resistance to additional xenobiotics; members of the τ class of GSTs are particularly associated with herbicide detoxification (Dixon and Edwards, 2010).
Arabidopsis is not a suitable species for remediating TNT toxicity from the environment, and to develop plants as a means of cleaning up contaminated soil and water, biochemical studies need to be extended to field-applicable species. Such species could include poplar and grasses such as switchgrass; however, while there is a range in tolerance levels, TNT has been found to be toxic at environment-contaminating amounts in all species tested so far (Hannink et al., 2002). Traditional breeding or genetic modification techniques might be a way to enhance the ability of plants to withstand and detoxify TNT. Brentner et al. (2008) have extrapolated the results of a TNT-treated Arabidopsis microarray study (Ekman et al., 2003) to identify GST homologs in poplar. The corresponding poplar homolog (GST173) to GST-U24 was up-regulated in response to TNT. Other TNT-responsive poplar genes have also been identified (Tanaka et al., 2007; Brentner et al., 2008).
CONCLUSION
The fate and toxicity of TNT-transformed conjugates in the environment are not known. The current hypothesis in plants is that TNT is detoxified predominantly via a rate-limiting nitroreductase step followed by glucosyltransferase activities. Here, we determine the contribution of GSTs to the TNT detoxification pathway in Arabidopsis. We show that overexpression of plant GSTs confers enhanced resistance to TNT, along with an increased ability to remove and detoxify this environmental pollutant. GST-U25 can directly conjugate glutathione to TNT, via the removal of a nitro group, producing 2-glutathionyl-4,6-dinitrotoluene. The removal of a nitro group from TNT is highly desirable; it is the presence of the electron-withdrawing nitro groups on the TNT molecule that provides stability to the aromatic ring through resonance (Qasim et al., 2007). The denitration activity of GST-U25 presents the opportunity for subsequent degradation and mineralization of TNT, rather than the indefinite storage of TNT-transformation products in the environment. Our findings demonstrate that selecting or manipulating plants with increased GST activity, particularly for the production of conjugate 3, could significantly contribute toward the ability of the plant to both establish in contaminated soil and clean up TNT from polluted military sites.
MATERIALS AND METHODS
For all plant experiments, Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used.
Cloning
Using the primer sequences presented in Supplemental Table S1, the GSTs were cloned, from Arabidopsis seedlings treated with TNT as described (Gandia-Herrero et al., 2008), into pET-YSBLIC3C (Bonsor et al., 2006) for expression in Escherichia coli. For overexpression in Arabidopsis, complementary DNA (cDNA) sequences were amplified using the primers listed in Supplemental Table S3 and cloned into pART27 (Gleave, 1992) under the control of the cauliflower mosaic virus 35S promoter and octopine synthase terminator. Following transformation, homozygous, independent T3 (for GST-U24) and T4 (for GST-U25) generation plants were used for all experiments.
Gene Expression
Plant RNA was extracted using the Plant RNeasy kit (Qiagen), and cDNA was synthesized using oligo(dT)12-18 using SuperScript II reverse transcriptase (Invitrogen) containing RNAsin (Promega) at 42°C for 2 h before inactivation at 70°C for 15 min. Synthesized cDNA was purified using the Wizard DNA Clean Up System (Promega) and quantified. Quantitative reverse transcription PCR (qPCR), using the primers listed in Supplemental Table S4, was performed using the ABI 7000 Sequence Detection System (Applied Biosystems) with SYBR Green reporter dye. Data were normalized to expression levels of the internal control gene (ACTIN2; At3g18780), and the comparative ΔΔCt method (Livak and Schmittgen, 2001) was used to calculate the mean fold change in gene expression of the candidate genes. The Ct is the cycle number at which the fluorescence generated within a reaction crosses the threshold line. The ΔΔCt method is used to analyze relative changes in gene expression from real-time quantitative PCR experiments. Microarray data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE46958.
Recombinant Expression in E. coli
The pET-YSBLIC3C-GST plasmid was expressed in E. coli BL21 (DE3) cells grown in autoinduction medium (Studier, 2005) at 37°C to an optical density at 600 nm of 0.8 and then at 20°C for 60 h. Cells were centrifuged at 5,000g for 10 min, resuspended in phosphate-buffered saline, sonicated, and then centrifuged at 17,500g for 30 min to remove cell debris. Proteins from supernatants were purified using Glutathione Sepharose 4B resin (GE Healthcare).
CDNB Activity
The GSH-conjugating activity of the purified proteins, and crude extracts from rosette leaves, was assessed using the model substrate CDNB. A 1-mL reaction containing 100 mm phosphate buffer, pH 6.5, 100 µg of GST, and 5 mm GSH was initiated by the addition of 1 mm CDNB, and the increase in A340 was measured spectrophotometrically, as described previously (Colville and Smirnoff, 2008).
TNT-Conjugating Activity
The TNT-conjugating activity was measured in reactions containing 150 μg of enzyme, 200 μm TNT, and 5 mm GSH in 100 mm potassium phosphate buffer and, unless stated otherwise, at pH 9 and 20°C. Reactions were stopped, when still in the linear phase, with 10% (v/v) TCA, to a final concentration of 1% (v/v). Reactions were analyzed by HPLC using a Waters HPLC system (Waters 2695 separator and Waters photodiode array detector) with a Waters X-Bridge C18 column (300 × 4.5 mm, 5 μm). The mobile phases for the gradient conditions were as follows: mobile phase A, acetonitrile; mobile phase B, water plus 0.1% (v/v) formic acid. The gradient ran as follows: 0 min, 5% A and 95% B; 5 min, 5% A and 95% B; 25 min, 40% A and 60% B; 30 min, 100% A and 0% B; and 35 min, 5% A and 95% (v/v) B.
Nitrite Production
Nitrite production was assayed according to the method of French et al. (1998) with modifications. Using a microtiter plate, 200 μL of sample, 200 μL of 10 mg of sulfanilamide (10 mg mL−1), and 40 μL of 10 mg of N-(1-naphthyl)ethylenediamine (10 mg mL−1) were added per well. Samples were incubated at 20°C for 10 min, and then A540 was measured.
GPOX Assays
The assays were based on the method of Edwards and Dixon (2005) with modifications as follows. The reaction mix contained 50 mm Tris-HCl, pH 6.5, 0.5 mm EDTA, 5 mm GSH, 0.25 mm NADPH, 0.6 units mL−1 glutathione reductase, 5 to 1,500 µm cumene hydroperoxide, 2.5 to 10 µm TNT, and 5 and 30 µg of enzyme for GST-U25 and GST-U24, respectively, in a final volume of 190 µL. The reaction was initiated by the addition of cumene hydroperoxide, and the decrease in A340 was measured spectrophotometrically.
Kinetic Assays
GST assays were performed using 100 mm phosphate buffer, pH 9.5, at 42°C with 150 μg of enzyme. The concentration of TNT ranged from 25 to 5,000 μm, and GSH concentration ranged between 5 and 45 mm. The aqueous solubility limit of TNT is approximately 500 µm at 25°C. To ensure that TNT was solubilized to a concentration at which Vmax could be measured, stock TNT concentrations were prepared in dimethyl sulfoxide (DMSO), with the final DMSO concentration constituting 5% (v/v) of the total volume of the reaction. This amount of DMSO was found to have no effect on GST protein activity. To determine the Vmax value, TNT was solubilized up to 5,000 µm. Experiments were performed in triplicate, and Michaelis-Menten parameters were calculated using SigmaPlot version 12.0.
In all enzyme assays, boiled enzyme controls were included to detect any nonenzymic reactions. Activity in the boiled controls was subtracted from the enzymatic rates, although significant rates were not detected in the boiled controls under any of the assay conditions tested. There was no significant effect of 10% (v/v) TCA, used to stop the enzyme reactions, on the stability of TNT- or GSH-conjugated products.
Purification of Conjugation Products in Vitro
The conjugation products were produced using purified GST-U25 in 100 mm potassium phosphate buffer, pH 9.5, at 20°C for 6 h, using 1.5 mg of purified GST-U25 with 2 mm TNT, 25 mm GSH, and 50 μL of DMSO in a total volume of 500 μL. The TNT conjugates were purified from the reaction by HPLC (using the conditions described above). The freeze-dried purified compounds were weighed, resuspended in water, and stored at −80°C for use as HPLC standards and NMR.
Measurement of Conjugation Products in Planta
Fresh plant tissues were ground in liquid nitrogen, and metabolites were extracted using 1 mL 10 mg−1 fresh weight methanol:water (60:40). After centrifugation, the supernatant was collected, evaporated, and resuspended in methanol:water (60:40) at a concentration of 1 µL mg−1 fresh weight. Samples were analyzed by HPLC and LC-MS and quantified using the standards generated as described above.
LC-MS Analysis of Conjugation Products
Mass spectrometry of the conjugated TNT products was performed using a Finnigan Surveyor Autosampler 1.4 (Thermo Electron), Finnigan Surveyor LC pump 1.4 SP1, Finnigan Surveyor PDA detector 1.0, and LCQ detector Finnigan MAT 2.0 with a Waters X-Bridge C18 column (250 × 4.6 mm, 5 μm), and HPLC conditions were as described for TNT conjugates above. The LCQ detector was tuned using TNT standard. Data were collected for peaks from 5.11 to 32.77 min. Electrospray ionization was used to produce ions in negative mode with a mass range of 100 to 1,000. Data were analyzed using Excalibur 2.0 SUR 1 software.
NMR Analysis of Conjugation Products
The NMR samples were prepared by dissolving 4 mg of the freeze-dried, HPLC-purified conjugates 1 and 2 separately in 700 µL of deuterium oxide or in 50% (v/v) fully deuterated (d6-) DMSO in deuterium oxide for conjugate 3. Two-dimensional 1H-1H-COSY (correlation spectroscopy), H-C HSQC (heteronuclear single quantum coherence), and H-C HMBC (heteronuclear multiple bond correlation) and one-dimensional COSY, HC-HSQC, HC-HMBC, and 1H spectra were acquired at 25°C on a Bruker Avance II 700 MHz spectrometer. Spectra were processed using TopSpin 3.0 (Bruker BioSpin) and assigned via standard procedures and using the previously published assignments of TNT and GSH 1H spectra.
TNT-Contaminated Media Studies
Root Surface Area Studies
Arabidopsis seeds were germinated on agar plates containing 1/2 MS (Murashige and Skoog, 1962) medium plus 20 mm Suc with TNT dissolved in DMSO (final DMSO concentration, 0.05% [v/v]). Root surface areas were determined using Image Pro 6.2 (Media Cybernetics).
Liquid Culture Experiments
Eight 7-d-old seedlings were transferred to 100-mL conical flasks containing 20 mL of 1/2 MS medium plus 20 mm Suc. Plants were grown for 14 d under 20 μmol m−2 s−1 light on a rotary shaker at 130 rpm. After this time, the medium was replaced with 20 mL of 20 mm Suc containing 200 μm TNT.
Hydroponic Uptake Studies
The hydroponic studies were based on the method of Kumari et al. (2008) with modifications as follows. Rafts were made from circular, lightweight plastic, 70 mm in diameter and 6 mm thick, with approximately 100 holes (3- to 4-mm diameter) drilled into each disk. Sterile, stratified Arabidopsis seeds (eight seeds per raft) were pipetted into the holes filled with 1/2 MS agar and placed in sterile glass jars containing 1/2 MS medium. The plants were grown for 20 d in 100 μmol m−2 s−1 light with a 16-h photoperiod with 21°C and 18°C, respectively, day and night temperatures. After this time, the medium was replaced with 30 mL of 1/2 MS medium containing 50 or 100 μm TNT. Samples were taken every 24 h over 7 d and analyzed by HPLC.
TNT-Contaminated Soil Studies
The soil studies were conducted as described (Rylott et al., 2011a). Briefly, five 5-d-old seedlings were planted into each pot containing 18 g of TNT-contaminated soil and grown for 6 weeks at 180 μmol m−2 s−1 light with a 12-h photoperiod and 21°C light and 18°C dark temperatures. Nitrotoluenes were extracted from 2 g of dried, ground soil by mixing in a glass tube with 10 mL of acetonitrile in a cooled ultrasonic bath for 18 h. Following centrifugation, 5 mL of supernatant was transferred to a fresh tube, evaporated, and then resuspended in 500 µL of methanol prior to HPLC analysis.
Glutathione Measurements
Plant tissue was frozen in liquid nitrogen and then disrupted in 1 mL 100 mg−1 tissue fresh weight of 0.2 n HCl using a bead mill. Samples were centrifuged at 13,000 rpm for 10 min at 6°C, and then supernatant pH was adjusted to between 5 and 6. Glutathione levels were assayed in samples and standards using a plate-reader protocol, as described in detail by Queval and Noctor (2007). Briefly, the rate of 5,5′-dithiobis(2-nitrobenzoic acid) reduction to thionitrobenzoic acid by GSH was measured by following the increase in A412 in a system containing glutathione reductase to recycle GSSG. To measure GSSG only, GSH was first removed from the assay by complexing with 2-vinylpyridine.
Statistical Analysis
Data were analyzed for statistical significance using ANOVA, with posthoc Tukey’s honestly significant difference, using SPSS software. Significance is denoted by asterisks (*P < 0.05 and **P < 0.01). For Figure 1D, ANOVA was calculated from three technical replicates. For Figure 5, data were the combined means of the root lengths of 12 to 15 seedlings grown on each of three replicate plates. For Figures 8 and 9, ANOVA was conducted on plants grown separately in eight replicate pots.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_101578 and NM_101579.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDS-PAGE analysis showing expression and purification of recombinant glutathione transferases.
Supplemental Figure S2. Griess assays using purified GSTs to detect nitrite production.
Supplemental Figure S3. HPLC and spectrophotometric analysis of GSH-TNT conjugates.
Supplemental Figure S4. Change in TNT-conjugating activity of GSTs with increasing temperature.
Supplemental Figure S5. Lineweaver-Burk double reciprocal plots for GST-U24 and GST-U25.
Supplemental Figure S6. Conjugation activity in protein extracts from Arabidopsis rosette leaves using 1-chloro-2,4,-dinitrobenzene substrate.
Supplemental Figure S7. Rate of TNT uptake and TNT-conjugate formation by Arabidopsis in liquid culture.
Supplemental Table S1. Root surface area of GST-U24 overexpressing lines relative to wild type.
Supplemental Table S2. Root surface area of GST-U25-overexpressing lines relative to wild type.
Supplemental Table S3. Primer sequences used during cloning of GSTs for expression in E. coli and Arabidopsis.
Supplemental Table S4. Primer sequences used for qPCR of GSTs in Arabidopsis.
Supplementary Material
Glossary
- TNT
2,4,6-trinitrotoluene
- RDX
hexahydro-1,3,5-trinitro-1,3,5-triazine
- HADNT
hydroxylamino dinitrotoluene
- ADNT
amino dinitrotoluene
- GPOX
glutathione-dependent peroxide
- CDNB
1-chloro-2,4-dinitrobenzene
- GSH
reduced glutathione
- 1/2 MS
one-half-strength Murashige and Skoog
- LC-MS
liquid chromatography-mass spectrometry
- cDNA
complementary DNA
- qPCR
quantitative reverse transcription PCR
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the Strategic Environmental Research and Development Program of the U.S. Department of Defense, the Biotechnology and Biological Sciences Research Council (to H.S. and E.J.J.), and a Burgess studentship (to K.T.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Beynon ER, Symons ZC, Jackson RG, Lorenz A, Rylott EL, Bruce NC. (2009) The role of oxophytodienoate reductases in the detoxification of the explosive 2,4,6-trinitrotoluene by Arabidopsis. Plant Physiol 151: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra R, Wayment DG, Hughes JB, Shanks JV. (1999) Confirmation of conjugation processes during TNT metabolism by axenic plant roots. Environ Sci Technol 33: 446–452 [Google Scholar]
- Bhadra R, Wayment DG, Williams RK, Barman SN, Stone MB, Hughes JB, Shanks JV. (2001) Studies on plant-mediated fate of the explosives RDX and HMX. Chemosphere 44: 1259–1264 [DOI] [PubMed] [Google Scholar]
- Bonsor D, Butz SF, Solomons J, Grant S, Fairlamb IJ, Fogg MJ, Grogan G. (2006) Ligation independent cloning (LIC) as a rapid route to families of recombinant biocatalysts from sequenced prokaryotic genomes. Org Biomol Chem 4: 1252–1260 [DOI] [PubMed] [Google Scholar]
- Brentner LB, Mukherji ST, Merchie KM, Yoon JM, Schnoor JL, Van Aken B. (2008) Expression of glutathione S-transferases in poplar trees (Populus trichocarpa) exposed to 2,4,6-trinitrotoluene (TNT). Chemosphere 73: 657–662 [DOI] [PubMed] [Google Scholar]
- Brentner LB, Mukherji ST, Walsh SA, Schnoor JL. (2010) Localization of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) and 2,4,6-trinitrotoluene (TNT) in poplar and switchgrass plants using phosphor imager autoradiography. Environ Pollut 158: 470–475 [DOI] [PubMed] [Google Scholar]
- Colville L, Smirnoff N. (2008) Antioxidant status, peroxidase activity, and PR protein transcript levels in ascorbate-deficient Arabidopsis thaliana vtc mutants. J Exp Bot 59: 3857–3868 [DOI] [PubMed] [Google Scholar]
- Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R. (2011) Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev 43: 266–280 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Cummins L, Cole DJ, Edwards R. (1998) Glutathione-mediated detoxification systems in plants. Curr Opin Plant Biol 1: 258–266 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. (2009) Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J Biol Chem 284: 21249–21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. (2010) Glutathione transferases. The Arabidopsis Book 8: e0131, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60: 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP. (2005) Plant glutathione transferases. Methods Enzymol 401: 169–186 [DOI] [PubMed] [Google Scholar]
- Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JF. (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol 133: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frear DS, Swanson HR. (1970) Biosynthesis of S-(4-ethyl-amino-6-isopropyl-amino-2-S-triazino) glutathione: partial purification and properties of a glutathione-S-transferase from corn. Phytochemistry 9: 2123–2132 [Google Scholar]
- French CE, Nicklin S, Bruce NC. (1998) Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl Environ Microbiol 64: 2864–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CE, Rosser SJ, Bruce NC. (2001) Biotransformations of explosives. Biotechnol Genet Eng Rev 18: 171–217 [DOI] [PubMed] [Google Scholar]
- Gandia-Herrero F, Lorenz A, Larson T, Graham IA, Bowles DJ, Rylott EL, Bruce NC. (2008) Detoxification of the explosive 2,4,6-trinitrotoluene in Arabidopsis: discovery of bifunctional O- and C-glucosyltransferases. Plant J 56: 963–974 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grzam A, Tennstedt P, Clemens S, Hell R, Meyer AJ. (2006) Vacuolar sequestration of glutathione S-conjugates outcompetes a possible degradation of the glutathione moiety by phytochelatin synthase. FEBS Lett 580: 6384–6390 [DOI] [PubMed] [Google Scholar]
- Guddewar MB, Dautermann WC. (1979) Purification and properties of a glutathione-S-transferase from corn which conjugates S-triazine herbicides. Phytochemistry 18: 735–740 [Google Scholar]
- Hannink NK, Rosser SJ, Bruce NC. (2002) Phytoremediation of explosives. Crit Rev Plant Sci 21: 511–538 [Google Scholar]
- Hannink NK, Rosser SJ, French CE, Basran A, Murray JA, Nicklin S, Bruce NC. (2001) Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat Biotechnol 19: 1168–1172 [DOI] [PubMed] [Google Scholar]
- Hannink NK, Subramanian M, Rosser SJ, Basran A, Murray JA, Shanks JV, Bruce NC. (2007) Enhanced transformation of TNT by tobacco plants expressing a bacterial nitroreductase. Int J Phytoremediation 9: 385–401 [DOI] [PubMed] [Google Scholar]
- Irzyk GP, Fuerst EP. (1993) Purification and characterization of a glutathione S-transferase from benoxacor-treated maize (Zea mays). Plant Physiol 102: 803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Rylott EL, Fournier D, Hawari J, Bruce NC. (2007) Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XplA/B. Proc Natl Acad Sci USA 104: 16822–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TF, Hewitt AD, Grant CL, Thiboutot S, Ampleman G, Walsh ME, Ranney TA, Ramsey CA, Palazzo AJ, Pennington JC. (2006) Identity and distribution of residues of energetic compounds at army live-fire training ranges. Chemosphere 63: 1280–1290 [DOI] [PubMed] [Google Scholar]
- Kassahun K, Pearson PG, Tang W, McIntosh I, Leung K, Elmore C, Dean D, Wang R, Doss G, Baillie TA. (2001) Studies on the metabolism of troglitazone to reactive intermediates in vitro and in vivo: evidence for novel biotransformation pathways involving quinone methide formation and thiazolidinedione ring scission. Chem Res Toxicol 14: 62–70 [DOI] [PubMed] [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK. (2008) Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol Genet Genomics 279: 339–357 [DOI] [PubMed] [Google Scholar]
- Leavitt JC, Penner D. (1979) In vitro conjugation of glutathione and other thiols with acetanilide herbicides and EPTC sulfoxide and the action of the herbicide antidote R-225788. J Agric Food Chem 27: 533–536 [Google Scholar]
- Li X, He Y, Ruiz CH, Koenig M, Cameron MD, Vojkovsky T. (2009) Characterization of dasatinib and its structural analogs as CYP3A4 mechanism-based inactivators and the proposed bioactivation pathways. Drug Metab Dispos 37: 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mezzari MP, Walters K, Jelínkova M, Shih MC, Just CL, Schnoor JL. (2005) Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds: a phytoremediation approach. Plant Physiol 138: 858–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ. (2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–496 [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ. (2007) Glutathione conjugates in the vacuole are degraded by gamma-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J 49: 878–888 [DOI] [PubMed] [Google Scholar]
- Qasim M, Moore B, Taylor L, Gorb L, Leszczynski J, Honea P. (2007) Structural characteristics and reactivity relationships of nitroaromatic and nitramine explosives: a review of our computational chemistry and spectroscopic research. Int J Mol Sci 8: 1234–1264 [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Bruce NC. (2009) Plants disarm soil: engineering plants for the phytoremediation of explosives. Trends Biotechnol 27: 73–81 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Budarina MV, Barker A, Lorenz A, Strand SE, Bruce NC. (2011b) Engineering plants for the phytoremediation of RDX in the presence of the co-contaminating explosive TNT. New Phytol 192: 405–413 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith HM, Rathbone DA, Strand SE, Bruce NC. (2006) An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat Biotechnol 24: 216–219 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Lorenz A, Bruce NC. (2011a) Biodegradation and biotransformation of explosives. Curr Opin Biotechnol 22: 434–440 [DOI] [PubMed] [Google Scholar]
- Scott AC, Allen NS. (1999) Changes in cytosolic pH within Arabidopsis root columella cells play a key role in the early signaling pathway for root gravitropism. Plant Physiol 121: 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens C, Scheidemann P, Klunk A, Werner D. (1998) Distribution of 14C-TNT and derivatives in different biochemical compartments of Phaseolus vulgaris. Environ Sci Pollut Res Int 5: 202–208 [DOI] [PubMed] [Google Scholar]
- Sens C, Scheidemann P, Werner D. (1999) The distribution of 14C-TNT in different biochemical compartments of the monocotyledonous Triticum aestivum. Environ Pollut 104: 113–119 [DOI] [PubMed] [Google Scholar]
- Schoenmuth BW, Pestemer W. (2004) Dendroremediation of trinitrotoluene (TNT). Part 2. Fate of radio-labelled TNT in trees. Environ Sci Pollut Res Int 11: 331–339 [DOI] [PubMed] [Google Scholar]
- Studier FW. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Talmage SS, Opresko DM, Maxwell CJ, Welsh CJ, Cretella FM, Reno PH, Daniel FB. (1999) Nitroaromatic munition compounds: environmental effects and screening values. Rev Environ Contam Toxicol 161: 1–156 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Brentner LB, Merchie KM, Schnoor JL, Yoon JM, Van Aken B. (2007) Analysis of gene expression in poplar trees (Populus deltoides × nigra, DN34) exposed to the toxic explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX). Int J Phytoremediation 9: 15–30 [DOI] [PubMed] [Google Scholar]
- Tournaire-Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu DT, Bligny R, Maurel C. (2003) Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425: 393–397 [DOI] [PubMed] [Google Scholar]
- Van Dillewijn P, Couselo JL, Corredoira E, Delgado A, Wittich RM, Ballester A, Ramos JL. (2008) Bioremediation of 2,4,6-trinitrotoluene by bacterial nitroreductase expressing transgenic aspen. Environ Sci Technol 42: 7405–7410 [DOI] [PubMed] [Google Scholar]
- Yoon JM, Oliver DJ, Shanks JV. (2007) Phytotoxicity and phytoremediation of 2,6-dinitrotoluene using a model plant, Arabidopsis thaliana. Chemosphere 68: 1050–1057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.