The transcriptional Mediator complex plays an important role in regulating root system architecture through auxin-related mechanisms in Arabidopsis.
Abstract
Root system architecture is a major determinant of water and nutrient acquisition as well as stress tolerance in plants. The Mediator complex is a conserved multiprotein complex that acts as a universal adaptor between transcription factors and the RNA polymerase II. In this article, we characterize possible roles of the MEDIATOR8 (MED8) and MED25 subunits of the plant Mediator complex in the regulation of root system architecture in Arabidopsis (Arabidopsis thaliana). We found that loss-of-function mutations in PHYTOCHROME AND FLOWERING TIME1 (PFT1)/MED25 increase primary and lateral root growth as well as lateral and adventitious root formation. In contrast, PFT1/MED25 overexpression reduces these responses, suggesting that PFT1/MED25 is an important element of meristematic cell proliferation and cell size control in both lateral and primary roots. PFT1/MED25 negatively regulates auxin transport and response gene expression in most parts of the plant, as evidenced by increased and decreased expression of the auxin-related reporters PIN-FORMED1 (PIN1)::PIN1::GFP (for green fluorescent protein), DR5:GFP, DR5:uidA, and BA3:uidA in pft1-2 mutants and in 35S:PFT1 seedlings, respectively. No alterations in endogenous auxin levels could be found in pft1-2 mutants or in 35S:PFT1-overexpressing seedlings. However, detailed analyses of DR5:GFP and DR5:uidA activity in wild-type, pft1-2, and 35S:PFT1 seedlings in response to indole-3-acetic acid, naphthaleneacetic acid, and the polar auxin transport inhibitor 1-N-naphthylphthalamic acid indicated that PFT1/MED25 principally regulates auxin transport and response. These results provide compelling evidence for a new role for PFT1/MED25 as an important transcriptional regulator of root system architecture through auxin-related mechanisms in Arabidopsis.
The indeterminate growth of the plant root system through continuous cell division and elongation processes can be profoundly affected by nutrient and water availability as well as various stress conditions, such as extreme temperatures, drought, and/or salt stress (López-Bucio et al., 2003; Malamy, 2005). Therefore, the plasticity of the root system is of critical importance for the plant to compete for resources and adapt to constantly changing growth conditions. The Arabidopsis (Arabidopsis thaliana) root system architecture, which consists of a primary root, lateral roots, and root hairs, determines the exploratory ability of roots in the soil. Lateral roots initiate from a few pericycle cells that acquire the attributes of founder cells and then divide asymmetrically to give rise to lateral root primordia (LRP), which continue to grow and eventually emerge from the primary root (Dubrovsky et al., 2000, 2008). Finally, the new apical meristem is established and takes over the control of the growth of mature lateral roots (Malamy and Benfey, 1997).
The phytohormone auxin (indole-3-acetic acid [IAA]) plays an important role during all stages of lateral root formation (Casimiro et al., 2003; De Smet et al., 2006; Fukaki et al., 2007). Application of IAA or synthetic auxins such as naphthaleneacetic acid (NAA) stimulates lateral root formation (Celenza et al., 1995), whereas treatment with polar auxin transport inhibitors prevents lateral root initiation (Casimiro et al., 2001; Himanen et al., 2002). Consistently, Arabidopsis mutants with increased auxin levels, such as rooty and its alleles aberrant lateral root formation1 and superroot1, have increased numbers of lateral roots (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995), while mutants such as AUXIN RESISTANT1 (aux1), AUXIN RESPONSE1 (axr1), DARK OVEREXPRESSION OF CAB1 (doc1), SOLITARY ROOT1, and AUXIN-RESPONSE FACTORS7/19, with defective auxin transport, perception, and/or signaling, show reduced lateral root formation (Lincoln et al., 1990; Gil et al., 2001; Swarup et al., 2001; Fukaki et al., 2002). Auxin is unique among plant hormones in being actively and directionally transported from the place of synthesis in young apical parts to distant tissues. The auxin-efflux regulators PIN-FORMED (PIN) are crucial for auxin distribution throughout the plant. PIN proteins have been shown to mediate various developmental processes. For instance, vascular tissue and flower development is regulated by PIN1 (Gälweiler et al., 1998), tropisms are regulated by PIN2 and PIN3 (Friml et al., 2002b), and the patterning of the root is regulated by PIN4 (Friml et al., 2002a).
Auxin can interact with other plant hormones to orchestrate root development. Recently, it has been found that CORONATINE-INSENSITIVE1 (COI1), the jasmonic acid (JA) receptor, is required to mediate lateral root formation in response to JA, a canonical defense signal, thus indicating the participation of downstream signaling components integrating the responses of two or more plant hormones into plant organogenesis (Raya-González et al., 2012).
The Mediator complex is a large multiprotein complex conserved in all eukaryotes, from yeast to human. Mediator acts as a bridge between the RNA polymerase II complex and the myriad transcription factors present within the cell (Kim et al., 1994; Koleske and Young, 1994). Mediator fine-tunes diverse regulatory inputs and presents a balanced output to the RNA polymerase II to initiate transcription by binding to distal activators/repressors as well as general transcription factors at the promoter site (Malik and Roeder, 2005). In Saccharomyces cerevisiae, Mediator is composed of 25 subunits, of which 22 are at least partially conserved among eukaryotes (Boube et al., 2002; Bourbon et al., 2004). Specific Mediator subunits are required for growth and development in organisms such as Drosophila melanogaster, zebrafish, mouse, and Caenorhabditis elegans (Lehner et al., 2006; Wang et al., 2006; Loncle et al., 2007; Jiang et al., 2010).
Plant growth is regulated by both cell number and cell size, which in turn are controlled by coordinated cell proliferation and expansion events during organogenesis (Mizukami, 2001; Sugimoto-Shirasu and Roberts, 2003). Recently, the participation of the Mediator complex in plant organ size control and cell differentiation was evidenced through the identification of 21 conserved and six putative plant-specific Mediator subunits in Arabidopsis and the analysis of Arabidopsis mutants defective in MEDIATOR14 (MED14) and MED25 (Autran et al., 2002; Xu and Li, 2011; Sundaravelpandian et al., 2013). Prior to its identification as Mediator subunit 14, STRUWWELPETER was found to regulate cell number and shoot meristem development (Autran et al., 2002). Arabidopsis MED25 was originally identified as PHYTOCHROME AND FLOWERING TIME1 (PFT1), a nuclear protein acting in a photoreceptor pathway that induces flowering in response to suboptimal light conditions (Cerdán and Chory, 2003). In addition, Arabidopsis mutants compromised in the kinase module of the Mediator complex, MED12, MED13, and CYCLIN-DEPENDENT KINASE8, all show developmental phenotypes due to altered cell differentiation (Wang and Chen, 2004; Gillmor et al., 2010; Ito et al., 2011). The med12 and med13 mutants are affected in the transition of embryos from globular to heart stage, due to a delay in the expression of KANADI1 and KANADI2 transcription factors during early development (Gillmor et al., 2010). The effect of the med13 mutation on cell differentiation has also been explained by a defective response to the hormone auxin (Ito et al., 2011). Even though MED8 and MED25 play important roles in JA signaling, stress responses, and plant development such as root hair formation and flowering time, at present, possible roles of these and other Mediator complex subunits on root system architecture and auxin signaling are unknown.
In this study, we tested the possible participation of MED8 and PFT1/MED25 subunits of Mediator in root development and auxin signaling in Arabidopsis. While med8 mutants did not show any evident alteration of root architecture, the pft1 mutants had increased primary root growth and root branching. Analysis of the cell cycle marker CycB1:uidA, cell size measurements, auxin transport, and auxin-responsive reporter gene expression in pft1-2 and 35S:PFT1 seedlings before and after treatments with IAA, NAA, and the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) further indicated that the Mediator subunit PFT1/MED25 acts as a negative regulator of cell proliferation, lateral root formation, auxin transport, and auxin-responsive gene expression in Arabidopsis.
RESULTS
The PFT1/MED25 Subunit of Mediator Regulates Root Architecture in Arabidopsis
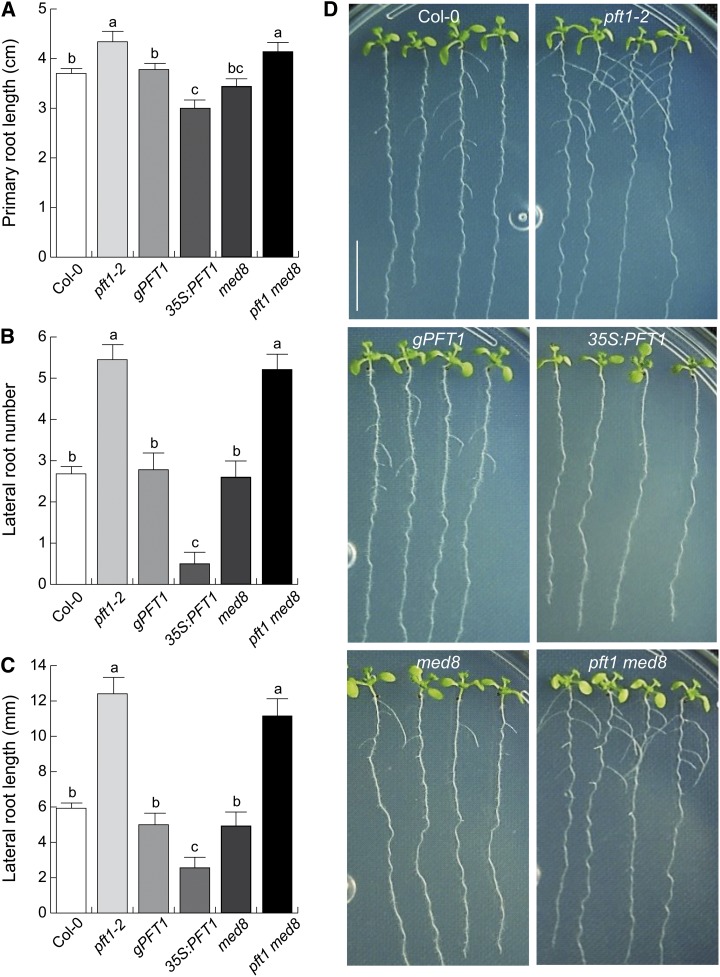
To determine the participation of the two subunits of Mediator, MED8 and PFT1/MED25, as regulators of Arabidopsis root architecture, we compared root growth phenotypes of the wild type (Columbia-0 [Col-0]), pft1-2 and med8 single mutants, the pft1 med8 double mutant, as well as pft1-1 mutants that were transformed with a genomic copy of PFT1 (G1 complementation line [gPFT1]; Cerdán and Chory, 2003) and PFT1/MED25 overexpression seedlings (35S:PFT1). The seedlings were grown on agar-solidified 0.2× Murashige and Skoog (MS) medium, and primary root lengths and lateral root numbers and lengths were quantified 8 d after germination. We found that pft1-2 and pft1 med8 seedlings had longer primary roots than wild-type seedlings, whereas med8 and gPFT1 plants were not affected in primary root growth (Fig. 1A). In contrast, 35S:PFT1 plants had shorter primary roots than wild-type plants (Fig. 1A). Interestingly, pft1-2 and pft1 med8 seedlings showed nearly 2-fold increases in lateral root numbers and lateral root lengths when compared with wild-type, gPFT1, and med8 seedlings (Fig. 1, B–D). In contrast, 35S:PFT1 seedlings showed fewer and shorter lateral roots than wild-type and gPFT1 seedlings (Fig. 1, B–D). An increase in primary root growth and lateral root formation in pft1-1, pft1-2, and pft1-3 mutants relative to the wild type was confirmed in additional experiments (Supplemental Fig. S1). Together, these results indicate that PFT1/MED25 regulates root system architecture in Arabidopsis.
Figure 1.
PFT1/MED25 regulates root system architecture in Arabidopsis. Wild-type (Col-0), pft1-2, gPFT1, 35S:PFT1, med8, and pft1 med8 seeds were germinated and seedlings were grown for 8 d on agar-solidified 0.2× MS medium. A, Primary root length. B, Lateral root number. C, Lateral root length. Root traits were scored as indicated in “Materials and Methods.” Error bars represent se from 30 seedlings. Different letters indicate statistical differences at P < 0.05. D, Photographs of representative wild-type (Col-0), pft1-2, gPFT1, 35S:PFT1, med8, and pft1 med8 seedlings. Note that an opposite response is seen in lateral root formation and lengths between the pft1-2 mutant and PFT1-overexpressing seedlings. The experiment was repeated twice with similar results. Bar = 1 cm. [See online article for color version of this figure.]
Adventitious roots originate from stem tissue and provide an increased ability for the plant to explore soil for water and nutrients. To evaluate the participation of PFT1/MED25 and MED8 in adventitious root formation, we obtained stem explants from wild-type, pft1-2, 35S:PFT1, med8, and pft1 med8 seedlings grown in dark conditions. These explants were cultured for 5 d over the surface of petri plates containing agar-solidified 0.2× MS medium, and both first- and second-order adventitious roots were quantified. In these experiments, pft1-2 and pft1 med8 seedlings had 2- and 5-fold increases in first- and second-order adventitious root numbers, respectively, when compared with wild-type seedlings, while the formation of second-order adventitious roots was drastically reduced in 35S:PFT1 seedlings (Supplemental Fig. S2). These results suggest that PFT1/MED25 also regulates processes associated with adventitious root development in Arabidopsis.
PFT1/MED25 Controls Cell Division and Elongation
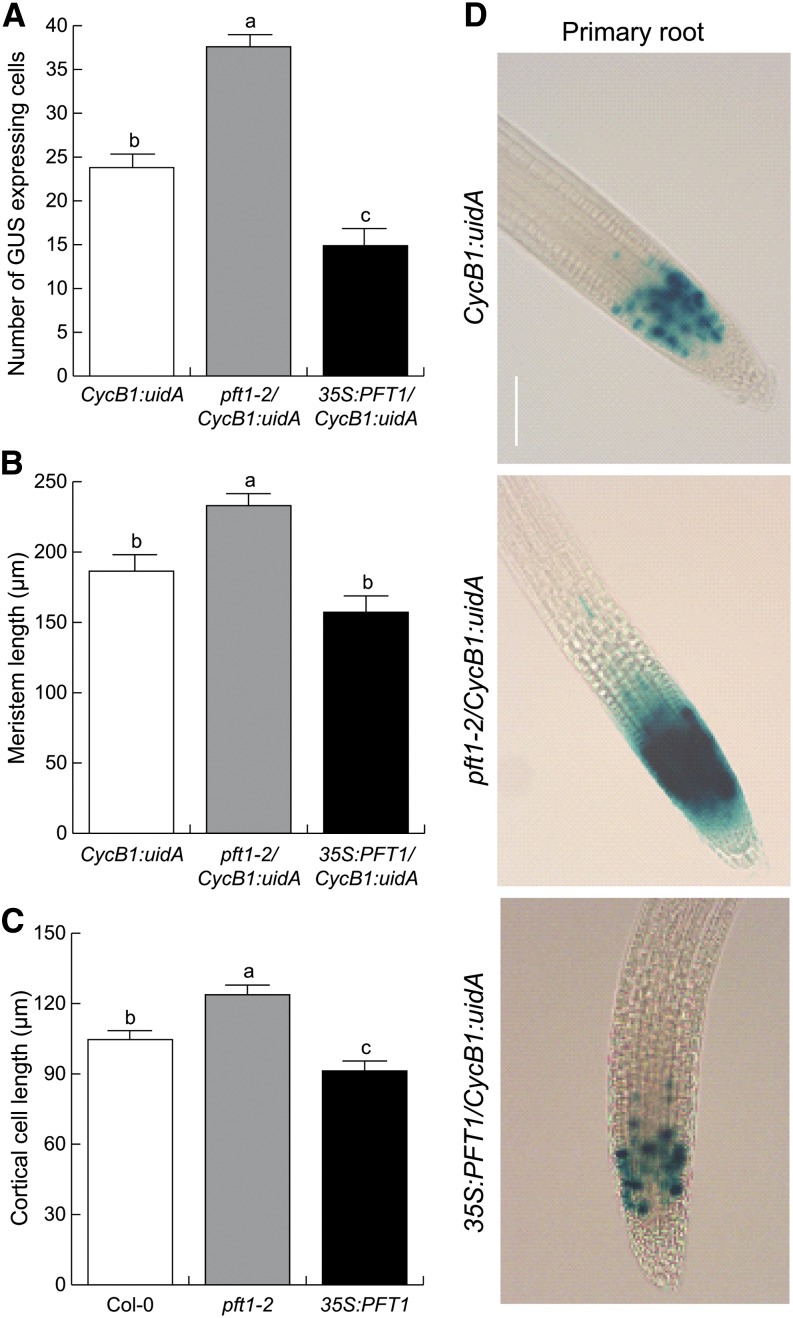
Primary root growth depends on two basic processes: cell division in the root apical meristem and elongation of divided cells that subsequently leave the root meristem (Blilou et al., 2002). Because root architecture was altered in pft1 mutants, we next explored the role of PFT1/MED25 on cell division and elongation of primary and lateral roots. For this aim, we crossed pft1-2 and 35S:PFT1 plants with Arabidopsis plants expressing the cell cycle marker CycB1:uidA (Colón-Carmona et al., 1999), which monitors cell cycle progression in the root meristem. We found that pft1-2/CycB1:uidA root meristems contain 40% more dividing cells than wild-type meristems, as revealed by the increased numbers of blue spots in the GUS expression domain of pft1-2/CycB1:uidA primary root meristems (Fig. 2, A and D), which were also larger than CycB1:uidA and 35S:PFT1/CycB1:uidA root meristems (Fig. 2B). In contrast, 35S:PFT1/CycB1:uidA seedlings had fewer dividing cells and smaller root meristems than CycB1:uidA seedlings (Fig. 2, A, B, and D). Contrasting expression of CycB1:uidA was also observed in emerged lateral roots of pft1-2/CycB1:uidA and 35S:PFT1/CycB1:uidA seedlings (Supplemental Fig. S3). To determine the involvement of PFT1/MED25 in cell elongation, we measured fully developed cortical cells of the primary and lateral roots of CycB1:uidA, pft1-2/CycB1:uidA, and 35S:PFT1/CycB1:uidA seedlings and found that cortical cells of pft1-2/CycB1:uidA seedlings were, on average, 15% longer than those of CycB1:uidA seedlings (Fig. 2C; Supplemental Fig. S3). In contrast, cortical cells of 35S:PFT1/CycB1:uidA seedlings were significantly shorter than those of pft1-2/CycB1:uidA seedlings (Fig. 2C). Taken together, these data indicate that PFT1/MED25 acts as a repressor of cell division and elongation during primary and lateral root growth in Arabidopsis.
Figure 2.
PFT1/MED25 represses cell division and elongation in primary root apical meristems. Wild-type (Col-0), pft1-2, and 35S:PFT1 Arabidopsis seeds harboring the CycB1:uidA gene construct were germinated and seedlings were grown for 7 d on agar-solidified 0.2× MS medium. A, Number of GUS-positive spots per root meristem. B and C, Meristem length (B) and cortical cell length (C) were scored as indicated in “Materials and Methods.” D, Primary roots of young seedlings were stained for GUS activity and cleared to show the expression of CycB1:uidA. Photographs show representative individuals from 15 GUS-stained seedlings. Error bars represent se from 15 GUS-stained seedlings analyzed. Different letters indicate means statistically different at P < 0.05. The experiment was repeated two times with similar results. Bar = 100 µm. [See online article for color version of this figure.]
PFT1/MED25 Regulates LRP Development through Auxin Signaling
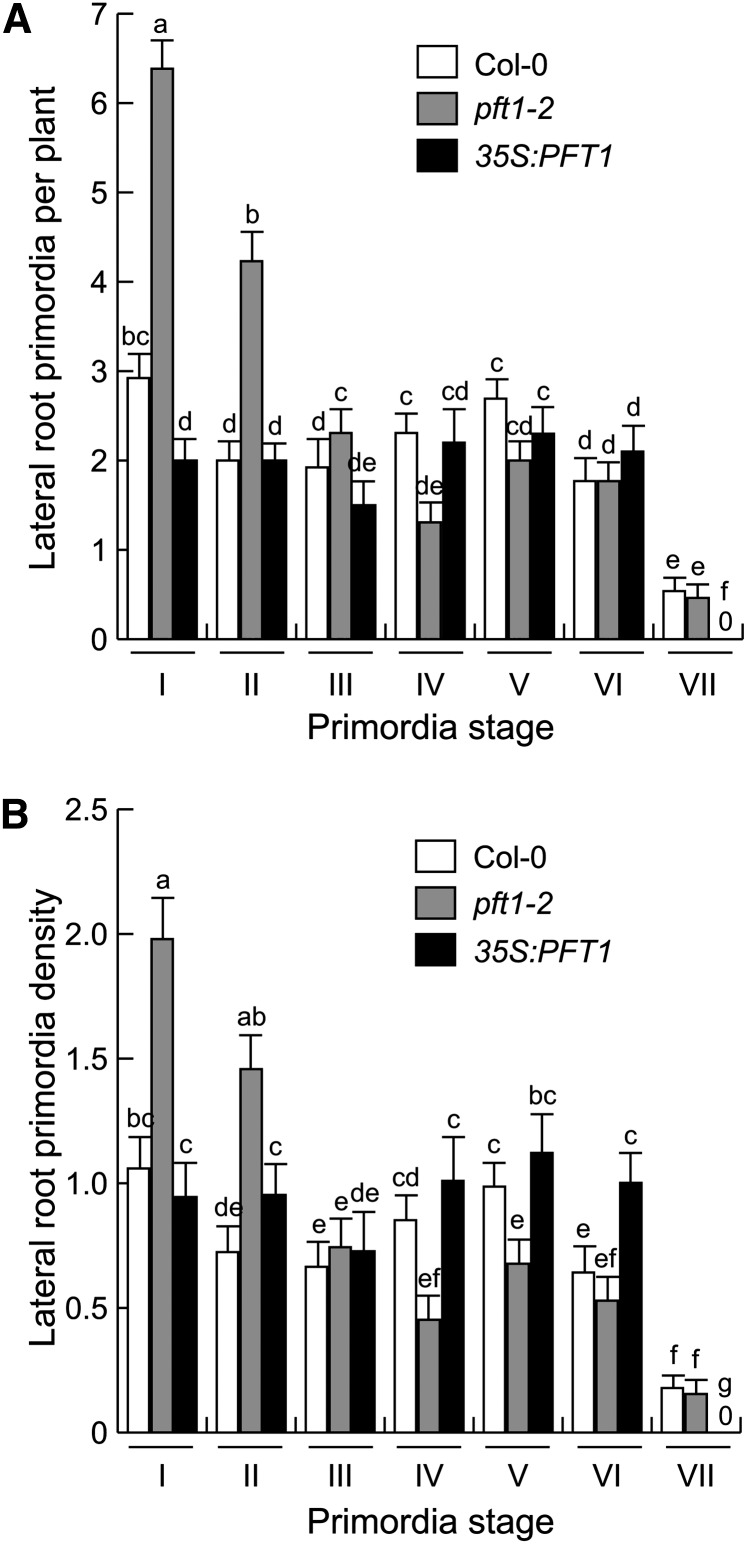
To understand the role played by PFT1/MED25 during lateral root formation and its possible relationship with auxin signaling, which regulates this organogenesis process, we analyzed LRP originating from the primary roots of wild-type, pft1-2, and 35S:PFT1 seedlings, according to Malamy and Benfey (1997). In 7-d-old pft1-2 roots, the number of stage I and II LRP were 2-fold higher than in wild-type roots (Fig. 3), suggesting that pft1-2 primary roots are more branched because they produce more de novo LRP from pericycle cells. Given that lateral root formation is a process regulated by auxin signaling, we then evaluated the expression of the auxin-responsive marker DR5:uidA during LRP development in DR5:uidA, pft1-2/DR5:uidA, and 35S:PFT1/DR5:uidA seedlings. DR5:uidA expression in pft1-2/DR5:uidA seedlings was stronger at all LRP developmental stages and in mature lateral roots than in DR5:uidA and 35S:PFT1/ DR5:uidA seedlings (Supplemental Figs. S4 and S5). These data suggest that PFT1/MED25 modulates lateral root formation by regulating auxin signaling during LRP development.
Figure 3.
PFT1/MED25 modulates LRP formation. Wild-type (Col-0), pft1-2, and 35S:PFT1 Arabidopsis seeds were germinated and seedlings were grown for 7 d on agar-solidified 0.2× MS medium and the development of root primordia was evaluated. A, LRP per plant. B, LRP density (LRP per cm). LRP stages were recorded according to Malamy and Benfey (1997). Error bars represent se from 15 GUS-stained seedlings analyzed. Different letters indicate statistical differences at P < 0.05. The experiment was repeated two times with similar results.
PFT1/MED25 Regulates Auxin-Responsive Reporter Gene Expression in Roots and Shoots
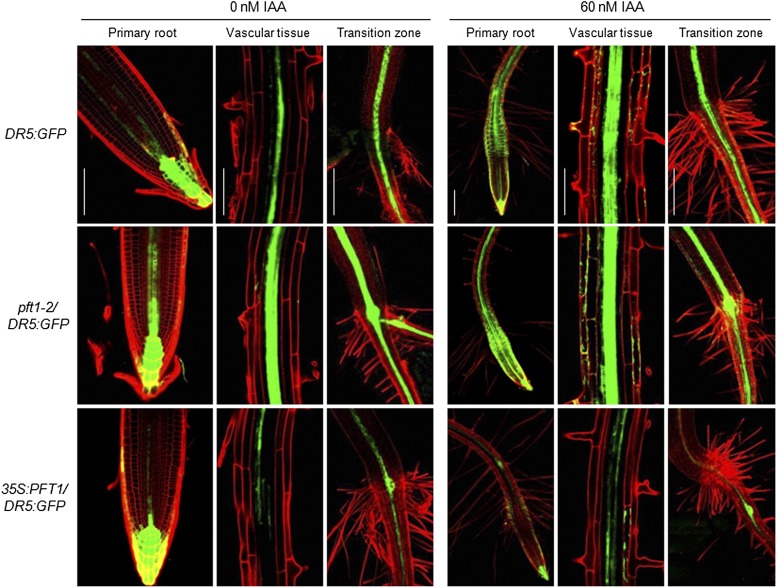
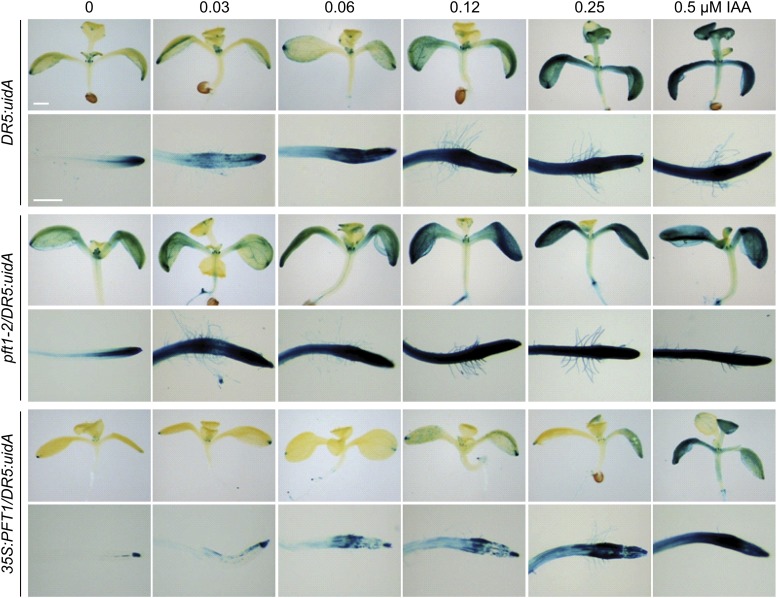
Auxin signaling has been implicated in many development processes in both root and shoot systems. To determine whether PFT1/MED25-mediated alterations in auxin responses could also occur in other plant tissues, we evaluated reporter gene activity in wild-type as well as pft1-2 and 35S:PFT1 seedlings harboring the DR5:GFP, DR5:uidA, or BA3:uidA gene construct. First, seedlings harboring the DR5:GFP construct were grown for 7 d on agar-solidified 0.2× MS medium with or without 60 nm IAA, and different parts of the plants, including root tips, vascular tissues, and root/shoot transition zones, were analyzed by confocal microscopy. GFP expression was higher in pft1-2/DR5:GFP but lower in 35S:PFT1/DR5:GFP in all three regions analyzed (Fig. 4). Exogenous auxin further induced GFP expression in pft1-2/DR5:GFP and DR5:GFP, while no such induction was evident in 35S:PFT1/DR5:GFP seedling roots (Fig. 4). We next analyzed the DR5:uidA-driven GUS activity in cotyledons, young leaves, shoot meristems, the stem/root transition zone, and the primary root tip. GUS activity was present in the primary root tip region and in leaves of untreated DR5:uidA seedlings (Supplemental Fig. S6). However, an increase in GUS activity was evident in most tissues of pft1-2/DR5:GUS seedlings, including cotyledons, petioles, the stem/root transition zone, lateral roots, and primary root tips. In contrast, GUS activity was much lower in 35S:PFT1 seedlings than in DR5:GUS and particularly in pft1-2/DR5:GUS in all tissues analyzed (Supplemental Fig. S6).
Figure 4.
PFT1 modulates auxin-responsive gene expression. Wild-type, pft1-2, and 35S:PFT1 seeds harboring the DR5:GFP gene construct were germinated and seedlings were grown for 7 d in agar-solidified 0.2× MS medium and then transferred for 8 h to liquid medium supplemented with or without IAA. Photographs show representative individuals of at least 15 seedlings analyzed by confocal microscopy. Note that pft1-2 and 35S:PFT1 seedlings show stronger and weaker DR5:GFP reporter expression, respectively, than wild-type seedlings in all regions analyzed. Bars = 100 µm. [See online article for color version of this figure.]
PFT1/MED25 Regulates the Auxin Response
The observation of contrasting root growth phenotypes resulting from the loss and gain of PFT1/MED25 function, together with the contrasting expression of auxin reporter genes in pft1-2 and 35S:PFT1 seedlings, suggest a role for PFT1/MED25 in the auxin signaling pathway. As IAA is a major regulator of root architecture, PFT1/MED25-mediated effects in root development could be due to potential alterations in auxin biosynthesis, auxin transport, and/or auxin response. To determine whether pft1 mutants could overaccumulate auxin, we first measured free IAA levels in wild-type, pft1-2, and 35S:PFT1 whole seedlings by gas chromatography-mass spectrometry. No significant differences in auxin accumulation were observed between wild-type, pft1-2, and 35S:PFT1 seedlings (Supplemental Fig. S7), suggesting that PFT1/MED25 is not a regulator of auxin biosynthesis.
To assess whether the observed root phenotypes could be due to changes in auxin-responsive gene expression, we performed experiments with wild-type, pft1-2, and 35S:PFT1 seedlings harboring the DR5:uidA or BA3:uidA gene construct after treatments with IAA. Arabidopsis seedlings were grown for 7 d on 0.2× MS medium solidified with agar and then transferred to liquid 0.2× MS medium supplemented with either the solvent only or different concentrations of IAA and incubated for 8 h at 22°C. In solvent-treated DR5:uidA seedlings, GUS expression was present in leaves and primary roots (Fig. 5). As expected, DR5:uidA seedlings treated with IAA showed a dose-dependent increase in GUS activity (Fig. 5). In contrast, the GUS activity in response to IAA was higher and lower in pft1-2/DR5:uidA and 35S:PFT1/DR5:uidA seedlings, respectively, than in DR5:uidA (Fig. 5).
Figure 5.
Effects of IAA on auxin-responsive reporter gene expression in wild-type, pft1-2, and 35S:PFT1 seedlings. Wild-type, pft1-2, and 35S:PFT1 seeds harboring the DR5:uidA gene construct were germinated and seedlings were grown for 7 d in agar-solidified 0.2× MS medium and then transferred for 8 h to liquid medium supplemented with or without IAA. Photographs show representative individuals of at least 15 GUS-stained seedlings analyzed. Note that pft1-2 and 35S:PFT1 seedlings show stronger and weaker GUS activity, respectively, than wild-type seedlings. Bars = 200 µm. [See online article for color version of this figure.]
We further evaluated the IAA response in wild-type, pft1-2, and 35S:PFT1 seedlings harboring the BA3:uidA marker. We found that this marker is strongly expressed in petioles and vascular tissues of pft1-2/BA3:uidA seedlings under standard growth conditions (Supplemental Fig. S8). In response to IAA, pft1-2/BA3:uidA even showed a stronger GUS activity than the wild type in petioles, vascular tissues, and primary root elongation zones (Supplemental Fig. S8). In contrast, 35S:PFT1/BA3:uidA seedlings showed a weaker GUS activity than BA3:uidA in petioles and primary roots both in the absence of any treatment and in response to IAA (Supplemental Fig. S8). These results indicate that PFT1/MED25 modulates the auxin response in Arabidopsis.
PFT1/MED25 Affects Lateral Root Formation in Response to Auxin
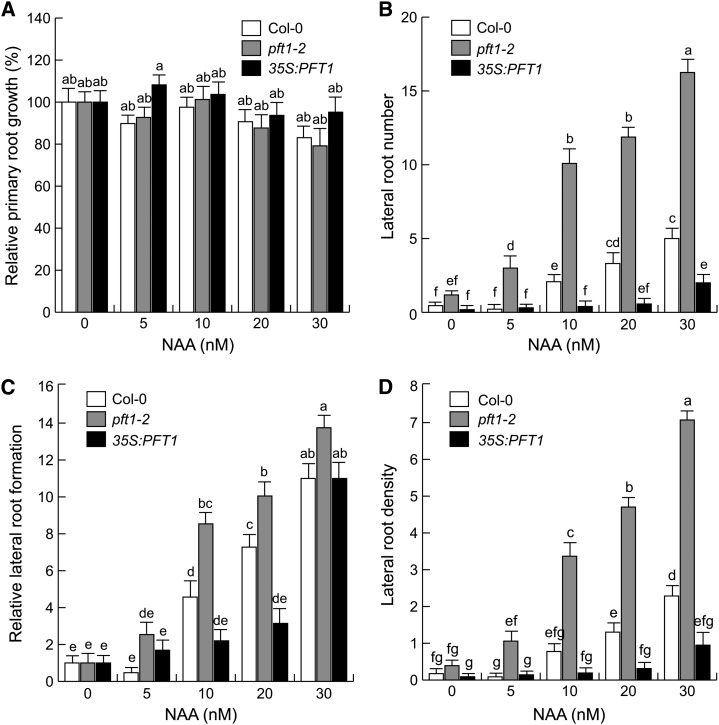
Auxin has been shown to inhibit the elongation of the primary root and to stimulate lateral root formation, whereas auxin transport inhibitors (e.g. NPA) antagonize lateral root formation (Blakely et al., 1988; Muday and Haworth, 1994; Casimiro et al., 2001). To determine whether auxin transport is an important determinant of the root developmental changes mediated by PFT1/MED25, we evaluated root architectural responses of wild-type, pft1-2, and 35S:PFT1 seedlings grown on petri plates containing 0.2× MS medium supplied with low concentrations of NAA, a synthetic auxin that enters the root cells via diffusion. Primary root growth was similarly inhibited in wild-type and pft1-2 seedlings in response to 5 to 30 nm NAA (Fig. 6A). Interestingly, pft1-2 seedlings showed an increased response to NAA, as evidenced by 2- to 3-fold increases in the number and density of lateral roots in all evaluated NAA concentrations (Fig. 6, B–D). In contrast, in response to NAA, 35S:PFT1 seedlings produced lower numbers and density of lateral roots than wild-type and pft1-2 seedlings (Fig. 6, B–D). When the lateral root data are normalized to the value obtained in the untreated control for each genotype, the fold increases in lateral root numbers appear clearly different in all three genotypes, with a nearly 2-fold higher relative lateral root formation in pft1-2 seedlings than in the wild type (Fig. 6C). Together, these observations suggest that PFT1/MED25 regulates pericycle cells to divide in response to auxin.
Figure 6.
Effects of NAA on root system architecture of wild-type (Col-0), pft1-2, and 35S:PFT1 seedlings. Arabidopsis seeds were germinated and seedlings were grown for 7 d with or without NAA. A, Relative primary root growth. B, Lateral root number. C, Relative lateral root formation (fold induction). D, Lateral root density. Primary root lengths at 0 nm NAA were 26 mm for Col-0, 30 mm for pft1-2, and 19 mm for 35S:PFT1. Error bars represent se from 15 seedlings. Different letters indicate statistical differences at P < 0.05. The experiment was repeated two times with similar results.
PFT1/MED25 Regulates the Expression and Distribution of the Auxin Transporter PIN1
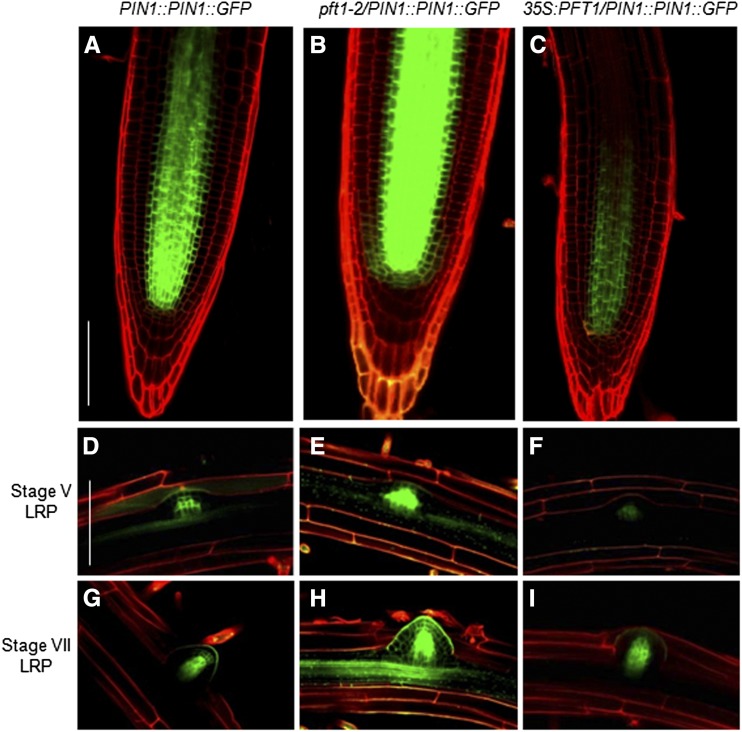
Auxin positively influences the PIN family of auxin transporters in a tissue-specific manner through an AUX/IAA-dependent signaling pathway (Vieten et al., 2005). PIN1 and PIN2 play important roles in lateral root formation and auxin-mediated gravitropism, respectively (Benková et al., 2003). To test whether PFT1/MED25 could regulate primary root growth and/or lateral root formation through differential expression of PIN1 or PIN2, we analyzed the spatial pattern of PIN1 and PIN2 localization in wild-type, pft1-2, and 35S:PFT1 seedlings. In primary roots of seedlings expressing PIN1::PIN1:GFP (Vieten et al., 2005), the GFP fluorescence was detected in the stele and endodermis cells (Fig. 7A). In pft1-2 primary roots, the GFP fluorescence was stronger than in PIN1::PIN1:GFP and extended toward the root differentiation zone (Fig. 7B), while in 35S:PFT1 seedlings, GFP fluorescence was weaker than in PIN1::PIN1:GFP and pft1-2/PIN1::PIN1:GFP and remained somewhat restricted (Fig. 7C). An analysis of PIN1 localization during lateral root initiation showed that in wild-type plants, stage V and VII primordia displayed the typical localization of PIN1 in most external cell layers, while increased and decreased PIN1 expression was evident in stage V and VII primordia of pft1-2/PIN1::PIN1:GFP and 35S:PFT1/PIN1::PIN1:GFP seedlings, respectively (Fig. 7, D–i). In contrast to the differential expression of PIN1 in pft1, PIN2 expression was similarly detected in cortex and epidermal cells of wild-type, pft1-2, and 35S:PFT1 primary roots (Supplemental Fig. S9). These findings suggest that PFT1/MED25 specifically regulates the expression and distribution of the PIN1 auxin transporter.
Figure 7.
PIN1 expression in wild-type, pft1-2, and 35S:PFT1 seedlings. PIN1::PIN1::GFP, pft1-2/PIN1::PIN1::GFP, and 35S:PFT1/PIN1::PIN1::GFP seeds were germinated and seedlings were grown on agar-solidified 0.2× MS medium. Seven days after germination, the seedlings were stained with propidium iodide and analyzed by confocal microscopy. A to C, Primary root apical meristems. D to F, Stage V LRP. G to I, Stage VII LRP. Representative photographs of primary roots and LRP are shown (n = 10). Note the increase and decrease of PIN1::PIN1::GFP in pft1-2 and 35S:PFT1 backgrounds, respectively. Bars = 100 μm. [See online article for color version of this figure.]
PFT1/MED25 Modulates the Response of Root Architecture to NPA
To further analyze the participation of PFT1/MED25 in auxin transport, we evaluated the effects of the polar auxin transport inhibitor NPA on root architecture in wild-type, pft1-2, and 35S:PFT1 seedlings. Arabidopsis seedlings were grown side by side for 8 d on agar plates containing 0.2× MS medium supplied either with 0.25 to 4 µm NPA or without NPA.
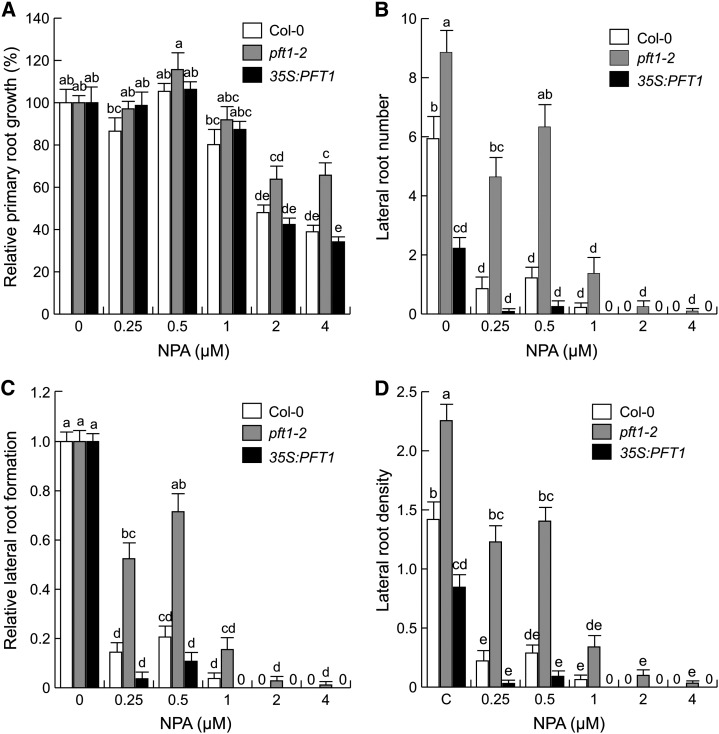
NPA at 4 µm inhibited primary root growth approximately 60% in wild-type seedlings when the data for root lengths were normalized to the values obtained in their untreated counterparts. Relative to the response we observed in wild-type seedlings, pft1-2 seedlings showed reduced responses to 2 and 4 µm NPA (Fig. 8A), while 35S:PFT1 seedlings had a wild-type-like response (Fig. 8A). As expected, NPA dramatically inhibited lateral root formation in wild-type seedlings (Fig. 8, B–D). However, lateral root formation in pft1-2 seedlings was less sensitive while in 35S:PFT1 seedlings it was more sensitive to NPA than in the wild type. Together, these contrasting responses of pft1-2 and 35S:PFT1 seedlings to an IAA efflux inhibitor in terms of both primary root growth and lateral root formation suggest a role for PFT1/MED25 in modulating auxin transport and response.
Figure 8.
Effects of NPA on root architecture in wild-type (Col-0), pft1-2, and 35S:PFT1 seedlings. Arabidopsis seeds were germinated and seedlings were grown for 8 d with or without NPA. A, Relative primary root growth. B, Lateral root number. C, Relative lateral root formation. D, Lateral root density. Primary root lengths at 0 µm NPA were 38 mm for Col-0, 40 mm for pft1-2, and 27 mm for 35S:PFT1. Error bars represent se from 15 seedlings. Different letters indicate statistical differences at P < 0.05. The experiment was repeated three times with similar results.
Root Architectural Responses of pft1 and coi1 Mutants to JA
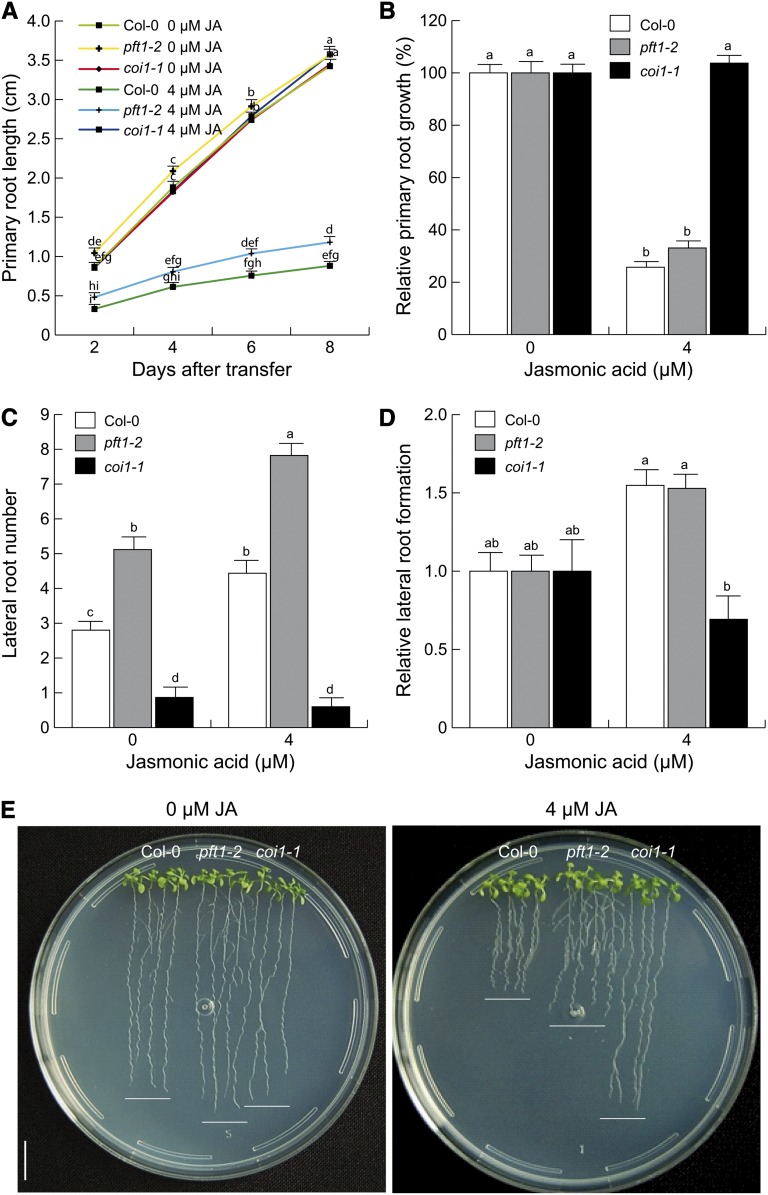
Recent reports have shown that PFT1/MED25 is required for JA-mediated defense gene expression (Kidd et al., 2009). Because JA affects both auxin signaling and root architecture (Raya-González et al., 2012), it was important to test the function of PFT1/MED25 in JA-mediated root architecture regulation. Primary root growth and lateral root formation were analyzed in wild-type, pft1-2, and coi1-1 seedlings grown side by side. In these experiments, pft1-2 showed a wild-type-like phenotype in its relative response to JA inhibition of primary root growth (Fig. 9, A and B). Also, a similar increase in lateral root numbers to wild-type seedlings in response to 4 µm JA was evident in pft1-2 seedlings (Fig. 9, C–E). In contrast, as expected, coi1-1 seedlings were highly resistant to primary root inhibition and lateral root promotion by JA (Fig. 9, B and E). Thus, these data show that PFT1/MED25 likely acts independently of the jasmonate receptor COI1 and JA to regulate lateral root formation in Arabidopsis.
Figure 9.
Effects of JA on root development of wild-type (Col-0), pft1-2, and coi1-1 seedlings. Wild-type and pft1-2 seeds were germinated and seedlings were grown for 4 d on 0.2× MS medium, and homozygous coi1-1 seedlings were selected from a coi1-1/COI1 segregating population in medium supplemented with 4 µm JA. A, Four-day-old seedlings were transferred and grown side by side over the surface of 0.2× MS agar plates supplied or not with 4 µm JA, and primary root growth was measured every 2 d. B, Relative primary root growth in response to 4 µm JA. C, Lateral root number. D, Relative lateral root formation. Error bars represent se from 15 seedlings. Different letters indicate statistical differences at P < 0.05. E, Photographs of representative wild-type, pft1-2, and coi1-1 seedlings illustrating the phenotype in response to JA. Relative primary root growth and lateral root formation were analyzed 8 d after transfer to JA. The experiment was repeated three times with similar results. Bar = 1 cm. [See online article for color version of this figure.]
DISCUSSION
PFT1/MED25 Plays a Role in Root Development
The root system, which plays an important role in anchoring the plant to the soil and in water and nutrient acquisition, exhibits an amazing architectural diversity manifested through changes in root hair, lateral root, and adventitious root formation and primary root growth (López-Bucio et al., 2003; Nibau et al., 2008). Engineering of the root architecture of crop plants can be of value for increasing plant stress tolerance, but this requires a thorough understanding of complex and interacting endogenous and exogenous factors that control individual aspects of root system configuration.
In this study, we investigated possible roles of MED8 and PFT1/MED25 Mediator subunits as regulators of the root system architecture of Arabidopsis seedlings. To the best of our knowledge, the Mediator complex has not been implicated so far in lateral root development and auxin signaling, despite its requirement as an essential component of gene transcription in all eukaryotes. Our results show that while med8 single mutants had root architecture similar to wild-type seedlings, the loss and gain of function of PFT1/MED25 showed opposite effects on lateral and adventitious root development and on primary root growth, indicating that PFT1/MED25 functions as a key modulator of cellular processes that control root architecture configuration.
The root phenotypes observed in pft1-2 and 35S:PFT1 seedlings correlated with changes in CycB1:uidA expression and cell expansion in primary and lateral roots (Fig. 2; Supplemental Fig. S3). Therefore, it is plausible that PFT1/MED25 regulates root development by inhibiting cell division and expansion in the roots. Through microarray analyses conducted on wild-type and pft1 roots, Sundaravelpandian et al. (2013) found that the genes implicated in growth- and cell cycle-associated processes were differentially expressed between pft1-2 and wild-type seedlings. For instance, the genes associated with the indole butyric acid-related processes were found to be differentially expressed in pft1 roots. In the light of our results, this is of particular interest, as genetic evidence in Arabidopsis suggests that indole-3-butyric acid converted into IAA by peroxisomal β-oxidation plays an important role in root hair formation and other developmental events by regulating cell expansion (Strader et al., 2010). Recently, Xu and Li (2011), through the use of transgenic Arabidopsis plants harboring the MED25 promoter::GUS fusion (pMED25::GUS) construct, showed that MED25 is expressed throughout the plant. The expression of PFT1/MED25 in roots is particularly relevant to our findings, which suggest that both cell proliferation and elongation are affected by PFT1/MED25 in the Arabidopsis root system. Also, given that PFT1/MED25 was first described as a positive regulator of shade avoidance and later as a regulator of basal defense and abiotic stress responses (Cerdán and Chory, 2003; Bäckström et al., 2007; Wollenberg et al., 2008; Kidd et al., 2009; Elfving et al., 2011; Chen et al., 2012), it seems likely that PFT1/MED25 represents a molecular node for the integration of distinct environmental and developmental cues.
The PFT1/MED25 protein is highly conserved across diverse eukaryotes. Remarkably, MED25 in other eukaryotes is also associated with several developmental processes. For example, the RNA interference-mediated suppression of MED25 expression in D. melanogaster results in the failure of extension of some axons that affect the embryonic nervous system (Koizumi et al., 2007). In zebrafish, morpholino-mediated knockdown of MED25 induces palatal malformation, suggesting an important role for MED25 in cartilage development (Nakamura et al., 2011). Remarkably, through the characterization of mutants and complemented and overexpressing lines of PFT1/MED25, we obtained evidence in this study that PFT1/MED25 functions in the development of organs and tissues ubiquitous to plants, such as lateral and adventitious roots.
PFT1/MED25 Is a Negative Regulator of LRP Development
Lateral root formation is initiated when the pericycle cells respond to auxin and acquire the status of founder cells and, through subsequent asymmetric cell divisions, give rise to new LRP (Boerjan et al., 1995; Malamy and Benfey, 1997; Dubrovsky et al., 2008). The loss of PFT1/MED25 function leads to the formation of increased numbers of LRP, particularly in stages I and II. In contrast, the overexpression of PFT1/MED25 resulted in decreased primordium formation (Fig. 3). These results indicate that PFT1/MED25 modulates root branching in Arabidopsis by inducing the de novo formation of LRP from pericycle cells. Kidd et al. (2009) showed that pft1 seedlings form more rosette leaves than wild-type plants. In plants, lateral root formation and leaf development are both regulated by auxin. Increased and reduced lateral root formation in pft1-2 and 35S:PFT1 seedlings, respectively, were correlated with alterations in DR5:uidA-driven reporter gene activity detected in all stages of LRP and emerged lateral roots (Supplemental Fig. S4). Benková et al. (2003) showed that auxin accumulates at developing primordia. Subsequently, an auxin gradient is established with its maximum at the tip of the forming lateral root, in which the auxin transporter PIN1 plays an important role (Benková et al., 2003). Therefore, both increased distribution of auxin from producing cells and efficient transport by PIN1 may explain why the pft1-2 mutant shows accelerated lateral root formation and greater proliferative capacity in pericycle cells than wild-type seedlings. Our results indicate that PFT1/MED25 negatively regulates lateral root initiation and development, probably by modulating an initial step required for the establishment of an auxin response maximum in lateral root founder cells.
PFT1 Modulates Auxin-Inducible Gene Expression
The plant growth hormone auxin has been implicated in regulating many developmental and cellular processes by altering basic patterns of gene expression (Kende and Zeevaart, 1997). The availability of well-established auxin markers such as DR5:GFP, DR5:uidA, and BA3:uidA provides important genetic tools to study the involvement of auxin signaling in plant development, as different markers show different sensitivities to endogenous and applied auxins. Importantly, we found differential expression of all three markers, DR5:GFP, DR5:uidA, and BA3:uidA, in pft1-2 and 35S:PFT1 seedlings (Fig. 4; Supplemental Figs. S5 and S6). These results indicate that PFT1/MED25 might regulate either auxin distribution and/or response. PFT1/MED25 was initially identified as a nuclear protein that acts in the phytochrome B pathway that induces flowering in response to suboptimal light conditions. pft1 mutants display defects in hypocotyl elongation under both red and far-red light conditions (Cerdán and Chory, 2003). The involvement of auxin in photomorphogenesis, including the shade avoidance response, has long been known (Shinkle et al., 1998; Steindler et al., 1999; Gil et al., 2001). It is noteworthy that axr1, doc1/tir3, and other auxin-related mutants with altered responses to light and shade avoidance manifest important alterations in root system architecture, including root hair and lateral root formation (Lincoln et al., 1990; Pitts et al., 1998; López-Bucio et al., 2005). The expression of AXR1 is localized to zones of active cell division and elongation and in epidermal cells that differentiate the root hairs. This expression pattern is correlated with a defect in root hair elongation observed in axr1 mutant seedlings (Pitts et al., 1998; del Pozo et al., 2002). Therefore, it is possible that the altered auxin responses observed in the pft1 mutant could contribute to developmental phenotypes such as altered flowering time, response to light quality, and root development.
Through an analysis of the microarray data reported by Kidd et al. (2009), we identified several auxin-associated genes, such as ANTHRANILATE SYNTHASE1 (ASA1), TRYPTOPHAN SYNTHASE BETA-SUBUNIT2, CHORISMATE MUTASE3, several auxin-responsive GH3 family genes, IAA17, AUX/IAA, an amino acid permease, and a gene similar to AUX1 that were differentially expressed between wild-type and pft1 seedlings. Although our analyses did not show any alteration in free auxin levels in the pft1-2 mutants (Supplemental Fig. S7), the gene expression data are overall in agreement with the detailed experiments reported here, with the plants expressing well-established auxin reporters in pft1 and 35S:PFT1 backgrounds.
The phytohormone JA is a crucial component of the plant defense signaling system. JA and its metabolites, collectively called jasmonates, are lipid-derived signals produced during defense responses against insects and pathogens but also under exposure to ozone, UV light, wounding, and other abiotic stresses (Wasternack, 2007). Reduction in root growth and carbon allocation patterns in several plant species upon mechanical wounding or by herbivory was ascribed to JA. In Arabidopsis, treatment with jasmonates strongly inhibits primary root growth and promotes lateral root formation (Sun et al., 2009; Raya-González et al., 2012). JA promoted lateral root formation through auxin biosynthesis and transport by directly inducing the auxin biosynthesis gene ASA1 (Sun et al., 2009). Emerging evidence suggests that jasmonate and auxin signaling contain many common components (for review, see Cuéllar Pérez and Goossens, 2013). However, the exact cellular/tissue responses to jasmonates during Arabidopsis root system remodeling are currently not understood. Given the involvement of PFT1/MED25 in the regulation of both JA (Kidd et al., 2009) and auxin responses (this study), it was important to test the function of this protein in JA-mediated root architecture. A comparison of the root architecture of wild-type, pft1-2, and coi1-1 seedlings indicates that the increased lateral root formation in pft1-2 is likely independent of COI1 and JA signaling, in which a loss-of-function mutation in COI1 renders the plants highly resistant to JA in both primary root growth inhibition and lateral root formation (Fig. 9).
It is worth noting that auxin itself positively feeds back on PIN gene expression in a tissue-specific manner through an AUX/IAA-dependent signaling pathway. Vieten et al. (2005) suggested a positive effect of IAA on PIN1 expression. Our data suggest that both auxin response and transport rather than auxin accumulation might be important factors involved in root system remodeling in pft1 mutants. Indeed, an analysis of the spatial pattern and abundance of PIN1 localization in the wild type, pft1-2, and 35S:PFT1 revealed an increased GFP fluorescence in the stele and endodermal cells of pft1-2 primary roots (Fig. 7, A–C) and in LRP (Fig. 7, D–I). In contrast, PIN2 was detected only in the cortex and epidermal cells in a similar manner in wild-type, pft1-2, and 35S:PFT1 primary roots (Supplemental Fig. S9). These findings suggest that PFT1/MED25 regulates the expression and distribution of the PIN1 auxin transporter and may explain why pft1 seedlings show an amplified response to exogenous auxin based on enhanced auxin-responsive gene expression and associated root developmental phenotypes. These results are also informative in explaining why pft1 seedlings are more resistant than the wild type and 35S:PFT1 to NPA-mediated inhibition of lateral root formation.
PFT1 Regulates Lateral Root and Root Hair Formation through Different Mechanisms
Auxin is important for a multitude of physiological processes and regulates plant development through its biosynthesis and transport. The ability of plant cells to respond to this phytohormone in an appropriate manner is also critical for auxin-mediated plant development (Okushima et al., 2007). Our analysis by gas chromatography-mass spectrometry revealed that PFT1/MED25 is unlikely to be involved in general auxin biosynthesis (Supplemental Fig. S7). The activity of the DR5:GFP and DR5:uidA markers may not necessarily reflect global auxin levels but the sensitivity of tissues to IAA or other auxins (Benková et al., 2003). Our results suggest that auxin distribution and/or response could be involved in the activation of auxin-inducible genes involved in lateral root formation modulated by PFT1/MED25. Detailed analysis of DR5:uidA and BA3:uidA in the wild type, pft1-2, and 35S:PFT1 in response to IAA revealed that pft1-2 and 35S:PFT1 were more sensitive and resistant, respectively, to auxin (Fig. 5; Supplemental Fig. S8). This indicates that PFT1/MED25 modulates the auxin response rather than auxin biosynthesis and auxin transport through the regulation of the PIN1 auxin transporter. This hypothesis is supported by the finding that pft1-2 and 35S:PFT1 seedlings had opposite responses to NAA during lateral root formation (Fig. 6). The increased and reduced NAA responses shown by pft1-2 and 35S:PFT1 seedlings, respectively, suggest that PFT1/MED25 is a key element that controls pericycle cell activation during lateral root formation, which is modulated by the auxin signaling pathway. Similarly, 35S:PFT1 seedlings showed developmental alterations and auxin-responsive gene expression consistent with a decreased auxin transport that correlates with their increased sensitivity to NPA (Figs. 4–6 and 8). This suggests that PFT1/MED25 might be involved in auxin transport, possibly by mediating the transcriptional regulation and/or distribution of PIN1 (Fig. 7).
In a recent work, Sundaravelpandian et al. (2013) reported that PFT1/MED25 controls root hair differentiation through reactive oxygen species distribution. Both pft1-2 and pft1-3 Arabidopsis mutants showed a short-root-hair phenotype that was correlated with perturbations in hydrogen peroxide and superoxide distribution. Supply of hydrogen peroxide or potassium cyanide rescued the pft1 mutant phenotype, indicating that PFT1/MED25 regulates root hair differentiation through reactive oxygen species. The short-root-hair phenotype of pft1 mutants could be reproduced in our research. Furthermore, we also found that 35S:PFT1 shows the opposite phenotype, with root hairs longer than wild-type and pft1 seedlings (Supplemental Fig. S10). Considering the positive effect of auxin on root hair growth (Pitts et al., 1998), the short-root-hair phenotype of pft1 mutants indicates that the role of PFT1/MED25 in epidermal cell differentiation most likely occurs through an auxin-independent mechanism.
In conclusion, our results have shown that (1) gain and loss of PFT1/MED25 function lead to opposite responses in primary root growth and lateral and adventitious root development; (2) PFT1/MED25 negatively regulates cell division and elongation processes that are important in modulating the configuration of the root system; (3) PFT1/MED25 regulates auxin-responsive gene expression during LRP development; and (4) PFT1/MED25 modulates auxin responses to endogenous and supplied auxin and in lateral root formation, which seems to be independent from JA signaling. Emerging evidence indicates that PFT1/MED25 plays multiple roles in a number of essential plant processes, including light signaling and flowering time (Cerdán and Chory, 2003; Wollenberg et al., 2008; Iñigo et al., 2012; Klose et al., 2012), JA-mediated pathogen defense (Kidd et al., 2009), organ growth (Xu and Li, 2011), abiotic stress (Elfving et al., 2011), and JA and abscisic acid signaling (Çevik et al., 2012; Chen et al., 2012). This is consistent with the expectation that individual Mediator subunits recognize and respond to a subset of the approximately 1,500 transcription factors present in the Arabidopsis genome. Therefore, determining which transcription factor(s) interacts with PFT1/MED25 to coordinate auxin responses in pericycle cells should provide important information about the roles of PFT1/MED25 and the Mediator complex in root morphogenesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0, the transgenic Arabidopsis lines 35S:PFT1 (Cerdán and Chory, 2003), CycB1:uidA (Colón-Carmona et al., 1999), DR5:uidA (Ulmasov et al., 1997), DR5:GFP (Ottenschläger et al., 2003), BA3:uidA (Oono et al., 1998), PIN1::PIN1::GFP (Benková et al., 2003), and PIN2::PIN2::GFP (Blilou et al., 2005), and the mutant lines pft1-1 (Cerdán and Chory, 2003), pft1-2 (SALK_129555), pft1-3 (SALK_059316), med8 (SALK_092406), pft1 med8 (Kidd et al., 2009), and coi1-1 (Feys et al., 1994) were used for the experiments reported here. To generate the wild type, pft1-2, and 35S:PFT1 lines expressing auxin reporter gene constructs, crosses were made between the respective lines, and the lines homozygous for both loci were used in subsequent experiments. Seeds were surface sterilized with 95% (v/v) ethanol for 5 min and 20% (v/v) bleach for 7 min. After five washes in distilled water, seeds were germinated and grown on agar plates containing 0.2× MS medium. The MS medium (Murashige and Skoog Basal Salts Mixture; catalog no. M5524) was purchased from Sigma. Phytagar (commercial grade) was purchased from Gibco-BRL. Plates were placed vertically at an angle of 65° to allow root growth along the agar surface and to allow unimpeded aerial growth of the hypocotyls. Plants were placed in a plant growth chamber (Percival AR-95L) with a photoperiod of 16 h of light/8 h of darkness, light intensity of 300 μmol m−2 s−1, and temperature of 22°C.
Chemicals
IAA, NAA, and NPA were purchased from Sigma and dissolved in dimethyl sulfoxide. In control treatments, the solvents were used in equal amounts as present in the greatest concentration of each compound tested.
Analysis of Growth
Arabidopsis root system and primary root meristem integrity were analyzed with a stereoscopic microscope (Leica MZ6). All lateral roots emerged from the primary root and observed with the 3× objective were taken into account for lateral root number data. Images were captured with a Samsung SCC 131-A digital color camera adapted to the microscope. Primary root length was determined for each root using a ruler. Lateral root number was determined by counting the lateral roots per seedling, and lateral root density was determined by dividing the lateral root number value by the primary root length values for each analyzed seedling. For all experiments with wild-type and mutant lines, the overall data were statistically analyzed using the SPSS 10 program. Univariate and multivariate analyses with Tukey’s posthoc test were used for testing the differences in growth and root development responses. Different letters were used to indicate means that differ significantly (P < 0.05).
Determination of the Developmental Stages of LRP
LRP were quantified 7 d after germination. Seedling roots were first cleared to enable primordia at early stages of development to be visualized and counted. Each primordium was classified according to its stage of development as reported by Malamy and Benfey (1997). The developmental stages are as follows. Stage I, LRP initiation. In the longitudinal plane, approximately eight to 10 short pericycle cells are formed. Stage II, the primordium is divided into two layers by a periclinal division. Stage III, the outer layer of the primordium divides periclinally, generating a three-layer primordium. Stage IV, a primordium with four cell layers. Stage V, the primordium is midway through the parent cortex. Stage VI, the primordium has passed through the parent cortex layer and penetrated the epidermis. It begins to resemble the mature root tip. Stage VII, the primordium appears to be just about to emerge from the parent root.
Histochemical Analysis
For histochemical analysis of GUS activity, Arabidopsis seedlings were incubated overnight at 37°C in a GUS reaction buffer (0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 100 mm sodium phosphate, pH 7). The stained plants were cleared and fixed with 0.24 n HCl in 20% (v/v) methanol and incubated for 60 min at 62°C. The solution was substituted by 7% (w/v) NaOH in 60% (v/v) ethanol for 20 min at room temperature. Plants were dehydrated with ethanol treatments at 40%, 20%, and 10% (v/v) for a 24-h period each and fixed in 50% (v/v) glycerol. The processed roots were placed on glass slides and sealed with commercial nail varnish. For each marker line and each treatment, at least 15 transgenic plants were analyzed.
Propidium Iodide Staining and GFP Detection
For fluorescent staining with propidium iodide, plants were transferred from the growth medium to 10 mg mL−1 propidium iodide solution for 1 min. Seedlings were rinsed in water and mounted in 50% (v/v) glycerol on microscope slides. The same sample was recorded separately at wavelengths specific to both propidium iodide fluorescence, with a 568-nm excitation line and an emission window of 585 to 610 nm, and GFP emission, with a 500- to 523-nm emission filter (488-nm excitation line), using a confocal microscope (Olympus FV1000), after which the two images were merged to produce the final image.
Free IAA Determination
The determination of IAA was from whole plants grown on agar-solidified 0.2× MS medium for 10 d, and free IAA was quantified as described by Edlund et al. (1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Fig. S1. PFT1/MED25 mutations affect root architecture.
Supplemental Fig. S2. PFT1/MED25 modulates adventitious root development.
Supplemental Fig. S3. PFT1/MED25 represses cell division and elongation in lateral roots.
Supplemental Fig. S4. PFT1/MED25 regulates auxin-responsive gene expression in young roots.
Supplemental Fig. S5. PFT1/MED25 modulates auxin responses in whole plants.
Supplemental Fig. S6. PFT1/MED25 is involved in auxin responses in roots and shoots.
Supplemental Fig. S7. IAA contents in Col-0, pft1-2, and 35S:PFT1 seedlings.
Supplemental Fig. S8. PFT1/MED25 modulates auxin sensitivity.
Supplemental Fig. S9. PFT1/MED25 regulates PIN2 auxin transporter.
Supplemental Fig. S10. PFT1/MED25 is involved in root hair formation.
Supplementary Material
Acknowledgments
We thank Peter Doerner, Pablo D. Cerdán, Athanasios Theologis, and Thomas J. Guilfoyle for providing the seeds of transgenic and mutant lines, Jorge Molina Torres and Enrique Ramírez Chávez for help in auxin determinations, and Juan José Valdez Alarcón for permission to use the confocal microscope.
Glossary
- LRP
lateral root primordia
- IAA
indole-3-acetic acid
- NAA
naphthaleneacetic acid
- JA
jasmonic acid
- NPA
1-N-naphthylphthalamic acid
- Col-0
Columbia-0
- MS
Murashige and Skoog
Footnotes
This work was supported by the Consejo Nacional de Ciencia y Tecnología (grant nos. 60999 and 177775), the Consejo de la Investigación Científica (grant no. CIC 2.26), and the Marcos Moshinsky Foundation.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakely LM, Blakely RM, Colowit PM, Elliott DS. (1988) Experimental studies on lateral root formation in radish seedling roots. II. Analysis of the dose-response to exogenous auxin. Plant Physiol 87: 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Frugier F, Folmer S, Serralbo O, Willemsen V, Wolkenfelt H, Eloy NB, Ferreira PC, Weisbeek P, Scheres B. (2002) The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev 16: 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube M, Joulia L, Cribbs DL, Bourbon HM. (2002) Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151 [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. (2004) A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Cuéllar Pérez A, Goossens A. (2013) Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ 36: 2071–2084 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T. (2006) Lateral root initiation or the birth of a new meristem. Plant Mol Biol 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL. (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Eklöf S, Sundberg B, Moritz T, Sandberg G. (1995) A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, et al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Okushima Y, Tasaka M. (2007) Auxin-mediated lateral root formation in higher plants. Int Rev Cytol 256: 111–137 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J. (2001) BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev 15: 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS. (2010) The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD. (2012) PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Ito J, Sono T, Tasaka M, Furutani M. (2011) MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol 52: 539–552 [DOI] [PubMed] [Google Scholar]
- Jiang P, Hu Q, Ito M, Meyer S, Waltz S, Khan S, Roeder RG, Zhang X. (2010) Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc Natl Acad Sci USA 107: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Zeevaart JAD. (1997) The five “classical” plant hormones. Plant Cell 9: 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. (1995) A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7: 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Büche C, Fernandez AP, Schäfer E, Zwick E, Kretsch T. (2012) The mediator complex subunit PFT1 interferes with COP1 and HY5 in the regulation of Arabidopsis light signaling. Plant Physiol 160: 289–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Higashida H, Yoo S, Islam MS, Ivanov AI, Guo V, Pozzi P, Yu SH, Rovescalli AC, Tang D, et al. (2007) RNA interference screen to identify genes required for Drosophila embryonic nervous system development. Proc Natl Acad Sci USA 104: 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. (1994) An RNA polymerase II holoenzyme responsive to activators. Nature 368: 466–469 [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. (2006) Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38: 896–903 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. (2007) Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J 26: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L. (2005) An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis: identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137: 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. (2005) Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Mizukami Y. (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4: 533–539 [DOI] [PubMed] [Google Scholar]
- Muday GK, Haworth P. (1994) Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32: 193–203 [PubMed] [Google Scholar]
- Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, et al. (2011) Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun 2: 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Chen QG, Overvoorde PJ, Köhler C, Theologis A. (1998) age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Raya-González J, Pelagio-Flores R, López-Bucio J. (2012) The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J Plant Physiol 169: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Shinkle JR, Kadakia R, Jones AM. (1998) Dim-red-light-induced increase in polar auxin transport in cucumber seedlings. I. Development of altered capacity, velocity, and response to inhibitors. Plant Physiol 116: 1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B. (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaravelpandian K, Chandrika NNP, Schmidt W. (2013) PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol 197: 151–161 [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X. (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang N, Uno E, Roeder RG, Guo S. (2006) A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci USA 103: 17284–17289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Li Y. (2011) Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.