Image analysis facilitates high-resolution phenotyping and complements dense genetic marker data to improve quantitative trait locus mapping of inflorescence traits in rice.
Abstract
Variation in inflorescence development is an important target of selection for numerous crop species, including many members of the Poaceae (grasses). In Asian rice (Oryza sativa), inflorescence (panicle) architecture is correlated with yield and grain-quality traits. However, many rice breeders continue to use composite phenotypes in selection pipelines, because measuring complex, branched panicles requires a significant investment of resources. We developed an open-source phenotyping platform, PANorama, which measures multiple architectural and branching phenotypes from images simultaneously. PANorama automatically extracts skeletons from images, allows users to subdivide axes into individual internodes, and thresholds away structures, such as awns, that normally interfere with accurate panicle phenotyping. PANorama represents an improvement in both efficiency and accuracy over existing panicle imaging platforms, and flexible implementation makes PANorama capable of measuring a range of organs from other plant species. Using high-resolution phenotypes, a mapping population of recombinant inbred lines, and a dense single-nucleotide polymorphism data set, we identify, to our knowledge, the largest number of quantitative trait loci (QTLs) for panicle traits ever reported in a single study. Several areas of the genome show pleiotropic clusters of panicle QTLs, including a region near the rice Green Revolution gene SEMIDWARF1. We also confirm that multiple panicle phenotypes are distinctly different among a small collection of diverse rice varieties. Taken together, these results suggest that clusters of small-effect QTLs may be responsible for varietal or subpopulation-specific panicle traits, representing a significant opportunity for rice breeders selecting for yield performance across different genetic backgrounds.
Flowering is arguably the pinnacle of plant development. For plants, floral morphology and complex inflorescence architectures are evolutionary embellishments on an age-old process: reproduction and the transfer of genes. For humans, flowers fulfill diverse roles in both food crops and ornamental plant production. This is especially true for cereals (Poaceae), which have remarkably diverse inflorescence architectures that have been the target of selection for 10,000 years (Doust, 2007; Kellogg, 2007).
As the first crop species to be sequenced and a major source of grain for human consumption, domesticated Asian rice (Oryza sativa) represents an excellent system in which to study inflorescence development. The rice inflorescence is classified as a panicle, a conically shaped compound raceme in which spikelets are borne on long lateral branches instead of directly on the rachis axis (Ikeda et al., 2004). The beginning of rice inflorescence development, known as panicle initiation (PI), occurs when the shoot apical meristem transitions into an inflorescence meristem (IM). Over approximately 7 d, the IM undergoes several transitions into primary branch meristems (PBMs) before ultimately aborting (McSteen, 2006; Yoshida and Nagato, 2011). Rice PBMs are capable of dividing into secondary or tertiary branch meristems before transitioning into spikelet and floret meristems, resulting in iterative branching morphologies. After IM abortion, floret development and elongation of axes occur over roughly 30 d until heading, the point at which the panicle emerges to begin reproduction (International Plant Genetic Resources Institute and Association, 2007).
Every aspect of panicle development in rice is pleiotropically interconnected, coordinated by a host of transcription factors and hormones that are responsive to environmental fluctuation. Variation in plant spacing, daylength, temperature, nutrient availability, and water supply can significantly affect panicle development, stunt branch formation, reduce spikelet number, and/or result in sterility long before a farmer or breeder even sees the first panicle at heading (Kobayashi et al., 2003; Li et al., 2003; Latif et al., 2005; Liu et al., 2008; Thakur et al., 2010). Because anthesis and grain filling occur in a wave of development, beginning with spikelets located farthest away from the rachis axis at the tips of panicle branches and working inward, the position of a spikelet within a panicle determines the time of both fertilization and starch deposition during grain filling. Thus, panicle size, shape, and branching patterns are thought to directly impact grain uniformity and quality (Yoshida and Nagato, 2011). Additionally, tradeoffs between sink and source tissues in rice often translate into direct compensation in the number of seeds or grains per plant (Ohsumi et al., 2011). For example, although large panicles often carry more spikelets, overall seed and grain sizes usually diminish. Panicle length (PL) and panicle number are also usually negatively correlated. All of these developmental characteristics are directly dependent on the genetic background of the variety and the environment in which it is grown. Currently, the best way to ensure optimal panicle development in rice, and thus stable yield performance, is to strictly control irrigation and fertilizer regimens from several weeks before PI begins until grain filling initiates (Chen et al., 2007). This is true even in modern varieties.
The inherent genetic and morphological diversity for many traits within rice is partitioned into distinct subpopulations. Asian rice is composed of two subspecies, Indica and Japonica, which can be further divided into five subpopulations (indica, aus, tropical japonica, temperate japonica, and aromatic; Garris et al., 2005; Zhao et al., 2011). In many cases, novel polymorphisms (and sometimes entirely novel genes) are subpopulation specific (Xu et al., 2006; Fukao et al., 2009; Hattori et al., 2009; Gamuyao et al., 2012). Abundant quantitative variation for panicle traits is known to exist both within and between subpopulations (Ikeda et al., 2004; Zhao et al., 2011). The International Rice Research Institute Gene Bank reports panicles with numerous branching phenotypes and lengths ranging from 10 to 43 cm (R.S. Hamilton, personal communication). Without a quick and accurate method of measuring panicle morphology, how can we hope to identify and leverage the genetic mechanisms underlying this spectrum of diversity? Is there an optimal panicle architecture to enhance yield in rice, and is it specific to a particular subpopulation or environment? If so, can we maximize productivity and resource use efficiency by selectively tweaking reproductive development?
The Green Revolution gene SEMIDWARF1 (SD1), which increased harvest index by reducing the ratio of vegetative to reproductive tissue, is an excellent example of how breeding for changes in rice plant architecture can significantly improve yield potential (Hedden, 2003; Asano et al., 2011). However, despite a sizable body of molecular and physiological research on rice panicle development (Wang and Li, 2011; Yoshida and Nagato, 2011), breeding for panicle architecture per se has had little success with regard to improving yield. This may be due in part to phenotyping gaps within rice breeding pipelines, which often use composite traits like PL that are not always developmentally or botanically derived.
Numerous studies have used two-dimensional image-based phenotyping to measure rice root architecture (Famoso et al., 2010, 2011; Iyer-Pascuzzi et al., 2010; Clark et al., 2011) and grain traits (Tanabata et al., 2012). Two software packages have previously been developed to measure panicle-related traits, PASTAR/PASTA Viewer and P-TRAP (Ikeda et al., 2010; AL-Tam et al., 2013). PASTAR/PASTA Viewer extracts length and count traits from panicle images by identifying and connecting the top and bottom termini of branches (Ikeda et al., 2010). This platform was tested in a quantitative trait locus (QTL) mapping study (Ikeda et al., 2010), yet it is under license and not freely available. The second platform, P-TRAP, is open source, yet it has not been validated in a genetic mapping study and was released after being tested on only 26 images (AL-Tam et al., 2013). P-TRAP converts a panicle image into a mathematical graph and treats branch junctions as vertices (AL-Tam et al., 2013). Although P-TRAP can also quantify several grain traits directly from a panicle image, its skeleton calculation is fundamentally similar to that of PASTAR/PASTA Viewer: vertices are connected using a series of straight-line segments (AL-Tam et al., 2013). Straight-line skeletons can only approximate the true nature of curved axes like panicle branches, limiting the resolution of length phenotypes. Manually arranging a panicle before imaging can partially overcome this limitation, yet it significantly increases the time cost for trait evaluation. Additionally, neither PASTAR/PASTA Viewer nor P-TRAP can automatically eliminate awns, hair-like extensions on rice spikelets that can be misconstrued as branches.
The objectives of this study were (1) to establish a flexible phenotyping methodology and platform capable of quantitatively measuring primary panicle architecture in rice, (2) to determine whether increased phenotyping resolution improved our ability to identify panicle-related QTLs in a population of recombinant inbred lines (RILs), and (3) to test our approach on a diverse set of rice varieties grown under different planting regimens in a controlled environment.
RESULTS
Development and Implementation of the Phenotyping Platform PANorama
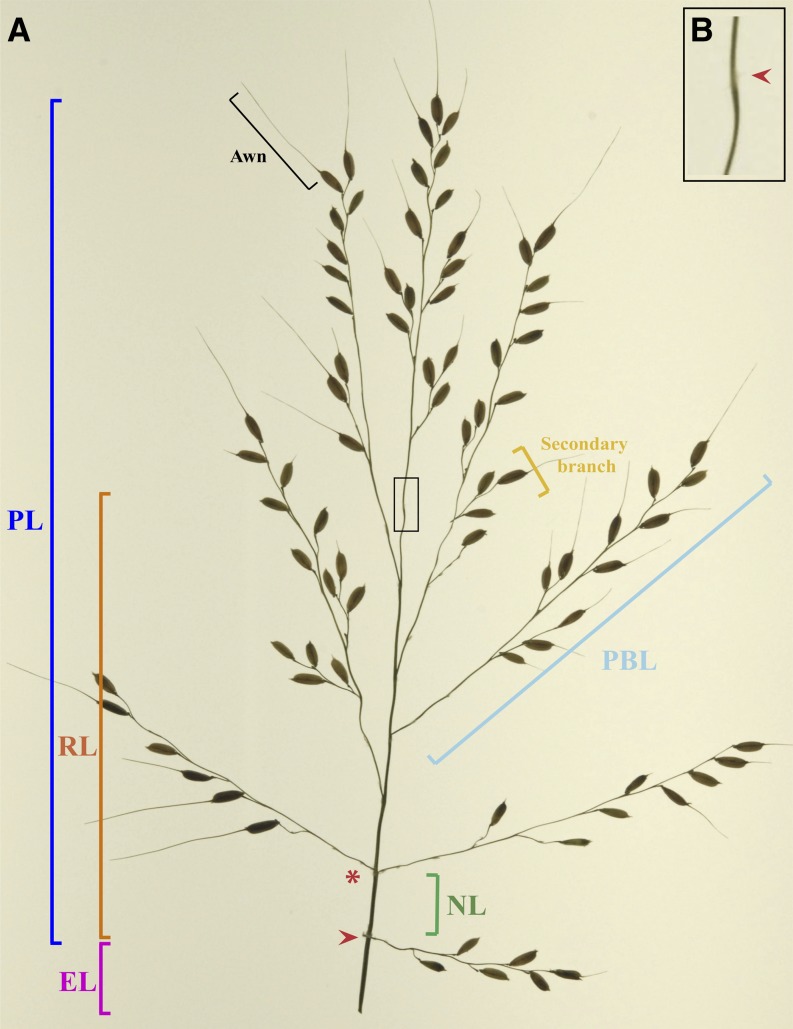
Within plants, meristem transition points are developmentally important and should be captured and separated functionally. In a rice panicle, meristem transitions along the main axis are relatively obvious because of the presence of trace bracts. The traditional breeding phenotype PL not only ignores meristem transitions but also lumps in the length of the rachis axis with that of the terminal primary branch (Fig. 1; Ikeda et al., 2004, 2010). Following this logic, measuring rachis length (RL) using the IM abortion point is also not ideal, as the rachis can be considered a compound phenotype. Every point at which a primary branch forms represents a functional meristem transition during PI, from the IM to a PBM. Thus, count measurements are representative of the number of times a meristem transition occurs in development, and when multiple PBM transitions occur closely together they should be clustered into internodes along the rachis axis (Fig. 1). Accordingly, length measurements may represent the amount of time between each transition and/or hormonal expansion after PI is completed, while width measurements may relate to physiological traits such as vascular architecture.
Figure 1.
Inflorescence architecture in rice. A, Rice panicles are characterized by hierarchical transitions between meristems. Plant breeders often use the composite phenotype PL, even though it combines the length of the rachis axis (RL) with that of the terminal primary branch. Thus, several structural components should be accounted for when using image analysis to measure rice panicles. The panicle notch (red arrowhead) and IM abortion point (red arrowhead in B) denote the beginning and end of IM division, respectively. Multiple IM-to-branch meristem transitions can occur at a single internode point (red asterisk), and the distance between two such points can thus be defined as the NL. Measuring PBL is also important but can be complicated by the presence of awns and secondary branches. The vegetative phenotype EL is calculated as the distance from the flag leaf ligule to the panicle notch and is reflective of how far the panicle emerges from the leaf sheath during heading.
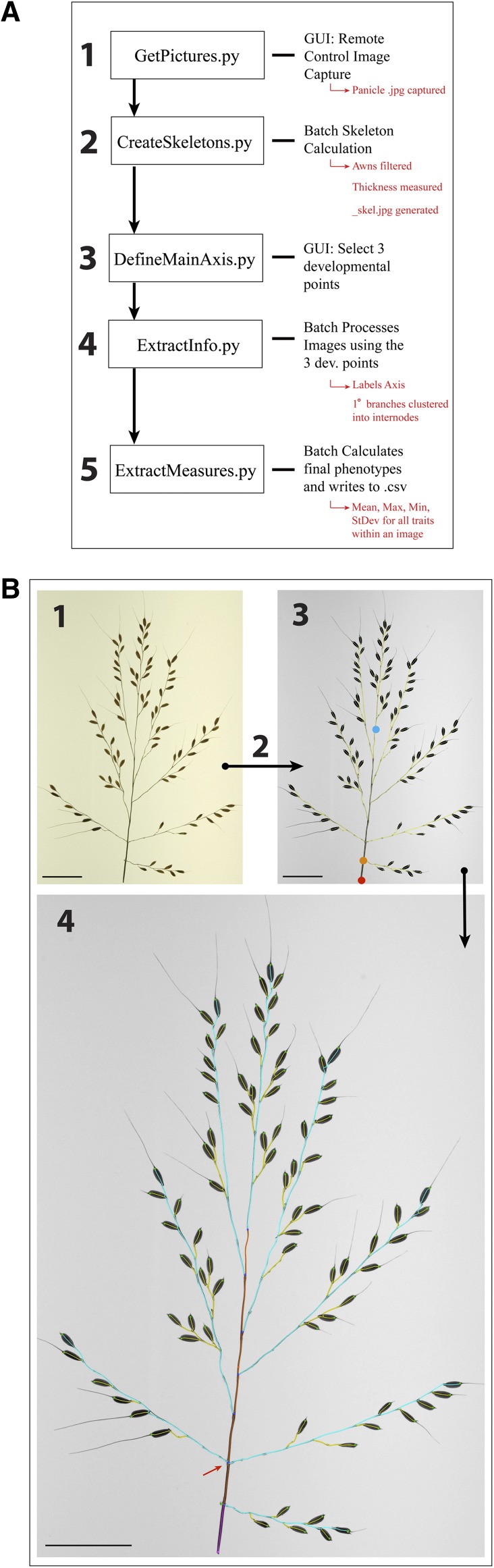
When using two-dimensional image-analysis and skeletonization techniques to accurately measure length, width, and count traits, an ideal skeleton should be a thinner representation of actual panicle axes. Previous studies have computed skeletons based on the exact morphological erosions of shapes using the image foresting transform algorithm (Falcão et al., 2002, 2004). Within this study, we incorporate this skeletonization technique into a new plant phenotyping pipeline, PANorama. PANorama is a collection of Python scripts that photograph, skeletonize, and extract multiple phenotypes from images. Its source code is written in C, and the scripts essentially call the stand-alone C programs. There are five major steps in the PANorama pipeline, which are performed using a combination of automated batch processing and user-directed input via graphical user interfaces (GUIs; Fig. 2). Scripts are executed at different levels within a structured directory (Supplemental Fig. S1; Supplemental Videos S1–S3), and multiple parameters that control the implementation of the pipeline are stored in one file, iftPANorama.h (Supplemental Table S1). Thus, a user can edit iftPANorama.h based on the qualities of images in a directory and recompile the program before running the pipeline. The following sections discuss how the parameters in iftPANorama.h have been calibrated across the pipeline for use on rice panicle images.
Figure 2.
PANorama pipeline. PANorama generates a skeletonized panicle image based on the exact morphology of the panicle. A, Image capture, skeletonization, processing, and phenotype extraction are controlled by five scripts. B, Panicle images from different stages of the pipeline. Numbers correspond to the steps listed in A. 1, A panicle image captured and saved using GetPictures.py. 2, Execution of CreateSkeletons.py batch generates skeletons for every panicle image. 3, A _skel.jpg version of the original panicle image generated by CreateSkeletons.py. The skeleton is depicted in yellow, superimposed over the panicle axes. Using the DefineMainAxis.py GUI, three developmental checkpoints are selected using a mouse: EP (red circle), IMAP (orange circle), and FMAP (blue circle). 4, The _skel.jpg image after ExtractInfo.py has been executed. Using the three developmental checkpoints selected, ExtractInfo.py labels the skeleton and defines axes: exsertion axis (pink), RL (orange), primary branches (blue), and secondary and tertiary axes (yellow). Branches are collapsed into internodes during this step (red arrow). Bars = 500 pixels.
Image Acquisition (GetPictures.py)
Image capture represents a large source of human error and directly limits the speed and accuracy of high-throughput phenotyping platforms. Regardless of the software platform used (Ikeda et al., 2010; AL-Tam et al., 2013), two-dimensional image analysis requires panicles to be positioned within a camera’s field of view so that branches are nonoverlapping. To reduce the need for postimage capture processing, the GetPictures.py GUI was incorporated into the PANorama pipeline. GetPictures.py interacts with the open-access software gPhoto2, allowing a user to remotely control image capture via his/her computer. All imaging in this study was conducted using a fixed camera mount and a light box, so that images are of the same exposure and scale (see “Materials and Methods”). After image capture, GetPictures.py generates a preview of a panicle’s skeleton so that the user can correct misarranged branches and retake the photograph. Images are saved as a .jpg file within a directory titled “originals,” and image names are stored in the .txt file imagefiles.txt (Supplemental Fig. S1). Every other script in the pipeline uses imagefiles.txt to access images stored within the originals directory (Supplemental Video S1).
Skeletonization (CreateSkeletons.py)
The second step of the pipeline, CreateSkeletons.py, uses a combination of image-processing operators to segment the panicle from the image background, remove awns, and calculate a skeleton for each image within the originals directory. For every original image, two new files are generated with the same root name. The first is a _skel.jpg file, containing a grayscale version of the original image with the final skeleton superimposed on the panicle (Fig. 2B); the second is an .inf extension file containing information associated with skeleton calculation. Both of these files are stored in a parallel directory named “skeletons” (Supplemental Fig. S1). The entire process is fast and takes approximately 10 s for a 2,592- × 3,872-pixel image (Supplemental Video S2).
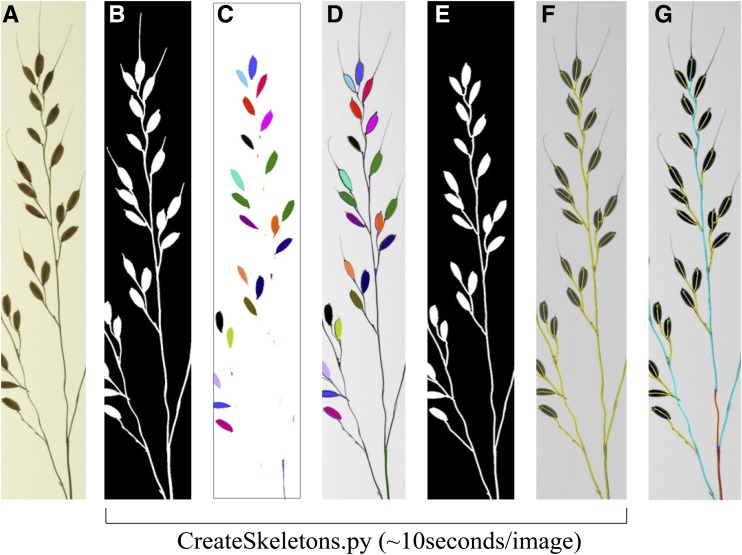
Given that some panicles have awns, we incorporated a preprocessing step into CreateSkeletons.py that is automatically performed before final skeleton calculation (Fig. 3, A–E). First, the Euclidean distance transform (Falcão et al., 2002) is used to compute an exact erosion of 0.032 cm. The erosion eliminates awns but also breaks the panicle mask into several connected components. Because the pixels from panicle axes and adjacent connected components (i.e. seeds) appear dark in the brightness image, these components are labeled (Fig. 3C) and reconnected by paths that contain a minimum sum of pixel values between each pair of distinctly labeled components (Fig. 3D). A final binary mask for skeletonization is computed by exact dilation at 0.032 cm, followed by a morphological closing of holes. Although this generates a mask with a single contour (Fig. 3E), the holes within panicle branches are still used to define the final skeleton (Supplemental Fig. S2). This is important when calculating skeletons for panicles in which primary branches have not been arranged to have open secondary or tertiary branches (see below). Euclidean distance transform (exact erosions), path reconnection, and closing of holes are all computed by the image foresting transform algorithm (Falcão et al., 2004) during the execution of CreateSkeletons.py.
Figure 3.
Schematic depicting awn removal using exact morphological erosions. A, Raw image of a panicle branch. B, Initial binary image after segmentation. C, Erosion of the image at 0.032 cm using the Euclidean distance transform. The binary components of the image object that remain after erosion are labeled with distinct colors. D, Binary components are superimposed over the original panicle image and reconnected by the darkest path (i.e. branches and rachis axis). E, After path reconnection, the awnless binary image is used to calculate the final skeleton. F, Panicle skeleton (yellow) superimposed over axes. All segmentation and skeletonization steps (B–F) are controlled by the CreateSkeletons.py script. G, Final panicle skeleton after the use of DefineMainAxis.py and ExtractInfo.py.
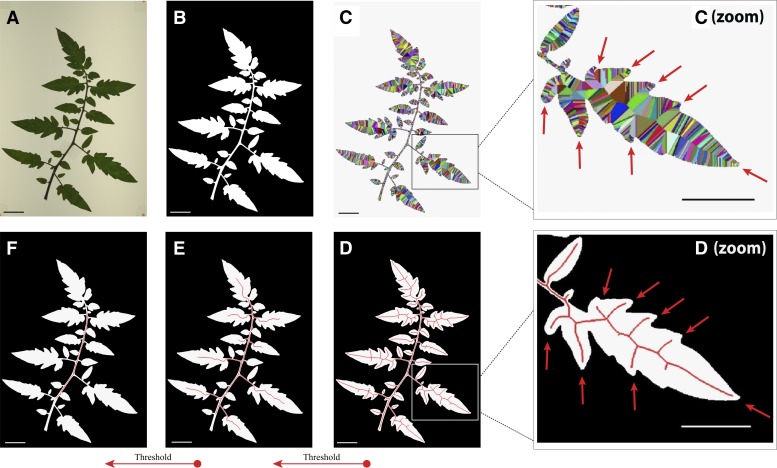
Figure 4 depicts skeleton calculation using a tomato (Solanum lycopersicum) leaf, which is wider and easier for visualization than a rice panicle. First, a Red, Green, Blue (RGB) color image (Fig. 4A) is segmented and thresholded to generate a binary image (Fig. 4B). This is done using morphological erosion with a circular structuring element of radius 0.083 mm on its brightness values to enhance the dark pixels that represent a panicle and a fixed factor of 1.2 on the Otsu’s optimum threshold. To eliminate spurious structures that are not physically connected to panicles (dust particles or detached seeds), the resulting binary mask contains only the largest connected component (Fig. 4B). The skeletonization process proposed by Falcão et al. (2002) uses the Euclidean distance transform to propagate the closest contour pixel (called the root) to each internal pixel of the panicle shape. This process can be visualized as wave fronts of distinct colors, which propagate from each contour pixel and meet one another in the center of the image at pixels equidistant to the contour (Fig. 4C, arrows). For each internal pixel, the method computes the maximum among the geodesic distances along the contour between the root of the pixel and the roots of its four neighbors. These measurements are assigned to each internal pixel and represent multiscale skeletons, which can be simplified by increasing thresholds on the scale (Supplemental Materials and Methods S1). The generated skeletons are all 1 pixel wide and fully connected (Fig. 4, D–F), which makes it very simple to identify nodes and extract accurate length measurements from a panicle. Additionally, skeletons are in the center of the shape at meeting points between distinct wave fronts (Fig. 4D, arrows). Because of this, distances to the contour can be doubled to accurately estimate thickness measurements along panicle segments (Supplemental Materials and Methods S1). For rice images, we set the scale threshold at 0.1% of the contour length using the SKELTHRES parameter in iftPANorama.h.
Figure 4.
Skeleton calculation using the image foresting transform algorithm. Using an RGB color image of a tomato leaf (A), a binary mask is generated from the largest eight-connected component (B). A Voronoi discrete map (C) is calculated using the Euclidean distance transform. This process is visualized as wave fronts of distinct colors, which propagate inward from each contour pixel (C). When pixels meet in the center of the shape, they generate central contours (C zoom, arrows). These contours are used to define the skeleton of an image object (D zoom, arrows). By weighting the importance of pixels with respect to their positions along the length of the skeleton (i.e. along the scales), a skeleton can be thresholded using the exact morphology of the shape (E and F). For rice images, this scale parameter is set at 0.1%, as observed in D.
Image Orientation (DefineMainAxis.py)
After obtaining accurate skeletons from panicle images, it is necessary to define the orientation of an image prior to phenotype extraction. Executing the DefineMainAxis.py script opens a GUI that allows a user to load individual _skel.jpg files. The user can select, move, and save three points on top of a panicle skeleton: exsertion point (EP), initial main axis point (IMAP), and final main axis point (FMAP; Fig. 2). For rice panicles, we identify EP as the point at which the flag leaf ligule attaches, whereas IMAP and FMAP are selected based on the presence of the panicle notch and the IM abortion point, respectively (Fig. 1). Manual selection of these points is essential, as their positions are variable for every image. For each _skel.jpg analyzed, the selection points are stored within a .txt file of the same root name within the skeletons directory (Supplemental Fig. S1; Supplemental Video S2). This file is not edited if a user needs to recalculate a skeleton using CreateSkeletons.py, so that the three points only have to be selected once for each image.
Phenotype Extraction (ExtractInfo.py and ExtractMeasures.py)
Phenotypes are automatically calculated using two scripts: ExtractInfo.py and ExtractMeasures.py. ExtractInfo.py assigns and labels the orientation of panicle axes for each _skel.jpg image using the three points identified with DefineMainAxis.py. After running ExtractInfo.py, the _skel.jpg image depicts the final, labeled skeleton (Fig. 2B). Several preset thresholds stored in the iftPANorama.h file are important for this step, as ExtractInfo.py filters some phenotypic traits before final measurements are calculated (Fig. 2). For example, branches are excluded from final measurement calculation if they are below a certain length, and multiple primary branches are collapsed into single internodes if they attach to the rachis axis within a certain distance of one another (Supplemental Materials and Methods S1; Supplemental Table S1; Supplemental Video S3).
The final thresholded and labeled _skel.jpg files are used by ExtractMeasures.py to calculate phenotypes for primary panicle architecture. ExtractMeasures.py operates on every image across multiple directories and takes approximately 5 to 7 s per image (Supplemental Fig. S1; Supplemental Video S3). Table I lists phenotypes and how they are generally defined within the pipeline. Details regarding the algorithms used are stored within Supplemental Materials and Methods S1. ExtractMeasures.py outputs two .csv files; the first contains mean, maximum, minimum, and sd values for phenotypes within each image (PanicleMeasures.csv), whereas the second contains the raw phenotypic measures (AllPanicleMeasures.csv). Length and width measurements are derived using a pixel-to-length conversion, which is dependent on the scale of the image (see “Materials and Methods”).
Table I. Phenotype calculations in PANorama.
The following phenotypes are written to a .csv file after running ExtractMeasures.py using the PANorama phenotype designation. For simplicity, we refer to rice phenotypes in subsequent tables using the QTL abbreviations given here. All parameters described within the table are controlled by the iftPANorama.h file.
| PANorama Phenotype Designation | Calculation | QTL Abbreviation |
|---|---|---|
| LengthOfExsertion (cm) | The distance between the EP and the IMAP. In rice, we select these points at the flag leaf ligule attachment point and the panicle notch, respectively. | EL |
| LengthOfMainAxis (cm) | The distance between the IMAP and the FMAP. In rice, we select these points at the panicle notch and inflorescence abortion point, respectively. | RL |
| InflorescenceLength (cm) | The distance between the IMAP and the tip of the longest terminal branch. In rice, the IMAP is selected at the panicle notch. | PL |
| NumberOfPrimaryBranches | A primary branch is any distinct skeleton segment that attaches (i.e. shares a neighboring pixel with the main axis) between the IMAP and the FMAP. The MINBRANCSZ parameter defines the minimum length requirement of a primary branch. | PBN |
| MeanLengthOfPrimaryBranches (cm) | The distance from the first pixel in a primary branch that borders the rachis skeleton, to the eight-neighbor pixel at the tip of a terminal seed. PBL is the average of all primary branches within a panicle. | PBL |
| MaxLengthOfPrimaryBranches (cm) | The maximum length of a primary branch within a panicle. | xPBL |
| MinLengthOfPrimaryBranches (cm) | The minimum length of a primary branch within a panicle. | nPBL |
| StdevLengthOfPrimaryBranches (cm) | The sd of the mean length of primary branches. | |
| NumberOfNodesOnMainAxis | Nodes are defined as anywhere a meristem transition has occurred. The pixels at which primary branches attach are considered nodes. If a branch forms at the IMAP and FMAP, they are also treated as nodes. Branches that attach within a specified distance of one another are automatically collapsed into a single internode point using the MINJCTDIST parameter, and the pixel closest to the geometric center of the cluster is considered the true internode point. NN is defined is as the total number of nodes in an image. | NN |
| MeanNumberOfBranchesPerNode | The average number of branches per node for all nodes within a panicle. Branches are collapsed into single nodes using the MINJCTDIST parameter, and the pixel closest to the geometric center of the cluster is considered the true node. | BpN |
| MaxNumberOfBranchesPerNode | The maximum number of branches to occur at a single node within a panicle. | xBpN |
| StdevNumberOfBranchesPerNode | The sd when calculating the number of branches per node. | |
| MeanLengthBetweenNodes (cm) | The distance between two nodes, after collapsing of primary branches. NL is the average length of all internodes in an image. | NL |
| MaxLengthBetweenNodes (cm) | The maximum distance between two nodes. | xNL |
| MinLengthBetweenNodes (cm) | The minimum distance between two nodes. | nNL |
| StdevLengthBetweenNodes (cm) | The sd of the mean length between nodes. | |
| MeanThicknessOfMainAxis (cm) | Thickness is calculated as 2× the distance between a skeleton point and its closest pixel on the image contour. Distances are obtained directly from the Euclidean distance map of the panicle mask during the skeletonization step. MeanThicknessOfMainAxis is the average of all thickness measures between the IMAP and the FMAP. | TR |
| MaxThicknessOfMainAxis (cm) | The maximum thickness between the IMAP and the FMAP. | xTR |
| StdevThicknessOfMainAxis (cm) | The sd of the mean thickness of the main axis. | |
| MeanThicknessOfExsertion (cm) | Thickness is calculated as 2× the distance between a skeleton point and its closest pixel on the image contour. Distances are obtained directly from the Euclidean distance map of the panicle mask during the skeletonization step. MeanThicknessOfExsertion is the average of all thickness measures between the EP and the IMAP. | TE |
| MaxThicknessOfExsertion (cm) | The maximum of all thickness measures between the EP and the IMAP. | xTE |
| MinThicknessOfExsertion (cm) | The minimum thickness measurement between the EP and the IMAP. | nTE |
| StdevThicknessOfExsertion (cm) | The sd of the mean thickness of exsertion. |
Error Correction (CorrectBranches.py and RebuildImageFiles.py)
Two supplementary scripts are included in PANorama for error correction. The RebuildImageFiles.py script regenerates the imagefiles.txt file that lists the name of every image stored within originals. Rebuilding imagefiles.txt is necessary if a user edits the originals directory by adding or removing images and wishes to reanalyze the edited directory using the other scripts in the pipeline (Supplemental Video S1). Executing CorrectBranches.py opens a GUI that allows users to add, remove, or shorten primary branches that were incorrectly arranged during image capture. The edits are saved directly within the .inf file of the image being edited (Supplemental Video S3).
Quality Testing of the PANorama Pipeline
Comparison of Manual Measurements, PANorama, and P-TRAP
To compare manual measures with PANorama and P-TRAP measures, we used GetPictures.py to photograph 30 panicles from the parents of the RIL mapping population in this study (IR64 and Azucena). Each panicle was photographed two times using different arrangements within an image (n = 60 images): once with secondary and tertiary branches closed and once with all branching arranged openly (Supplemental Fig. S2). Manual measurements were taken by hand (see “Materials and Methods”). To test awn filtering on primary branches, PANorama skeletons were calculated with the AWNSTHICK parameter in iftPANorama.h turned on and off (on = 0.032 cm, off = 0.0 cm). Panicle images were also processed using the open-source software P-TRAP (AL-Tam et al., 2013). For PANorama and P-TRAP, the panicle notch and IM abortion points were selected in order to extract phenotypes. However, skeletons were not corrected in any other way.
Table II reports squares of Pearson’s correlation coefficients (r2 values), mean absolute percentage deviations (MAPDs), and mean percentage errors (MPEs) between manually measured phenotypes and the phenotypes calculated by each of the software packages. Measuring opened panicle branches in PANorama decreased MAPD from manual measurements for primary branch length (PBL) by approximately 1% when compared with measuring closed panicle branches. Additionally, MAPDs were always lower for PBL when the awn filter was turned on (AWNSTHICK = 0.032 cm), regardless of whether branches were open or closed (Table II). This suggests that awn filtering always increases the accuracy of primary branch measures, even in varieties like IR64 and Azucena that have very small awns (Supplemental Fig. S2). The squares of correlation coefficients between PANorama phenotypes were high for every reported phenotype (Supplemental Table S2), confirming that the AWNSTHICK parameter accurately filters awns without negatively impacting other phenotypes.
Table II. Comparison of manual, PANorama, and P-TRAP measurements.
Squares of Pearson’s correlation coefficient (r2 values), MAPDs, and MPEs were calculated using 30 images between manual measurements and phenotypes calculated by each software platform. RL and PBN are not controlled by parameters (NA). Awn filtering and number of branches per node (BpN) are not supported (NS) in P-TRAP version 20130213220. Total processing time for 60 images includes skeletonization and measurement calculation but excludes the selection of meristem transition points.
| Trait | PANorama | P-TRAP | |||||
|---|---|---|---|---|---|---|---|
| Parameter | r2 | MAPD | MPE | r2 | MAPD | MPE | |
| RL | NA | 0.998 | 0.78 | −0.004 | 0.06 | 59.51 | −26.20 |
| PBN | NA | 0.99 | 0.42 | 0.004 | 0.77 | 3.16 | −2.25 |
| NN | MINJCTDIST (0.25 cm) | 0.86 | 5.76 | −0.050 | 0.42 | 28.26 | −20.10 |
| BpN | MINJCTDIST (0.25 cm) | 0.83 | 6.35 | 0.065 | NS | NS | NS |
| PBL (open) | AWNSTHICK (0.032 cm) | 0.996 | 1.42 | 0.002 | NS | NS | NS |
| PBL (closed) | AWNSTHICK (0.032 cm) | 0.998 | 2.54 | −0.019 | NS | NS | NS |
| PBL (open) | AWNSTHICK (0.0 cm) | 0.996 | 2.59 | −0.021 | 0.93 | 9.55 | −3.26 |
| PBL (closed) | AWNSTHICK (0.0 cm) | 0.996 | 3.69 | −0.032 | 0.90 | 6.48 | 2.85 |
| Total processing time (60 images) | 10 min, 30 s | 1 h, 10 min | |||||
When compared with manual measurements, PANorama phenotypes always showed higher r2 values, lower MAPDs, and MPEs closer to zero than P-TRAP. Additionally, PANorama required much less computational time to generate skeletons and calculate phenotypes (Table II). When comparing PANorama and P-TRAP skeletons visually, we found that P-TRAP was less accurate at identifying curved axes (Supplemental Fig. S3A). This could partially explain why P-TRAP misrepresented RL and internode number (NN), resulting in high MAPDs and strongly negative MPEs for both traits (Table II). P-TRAP also could not distinguish awns from branches (Supplemental Fig. S3B).
PANorama in Other Species
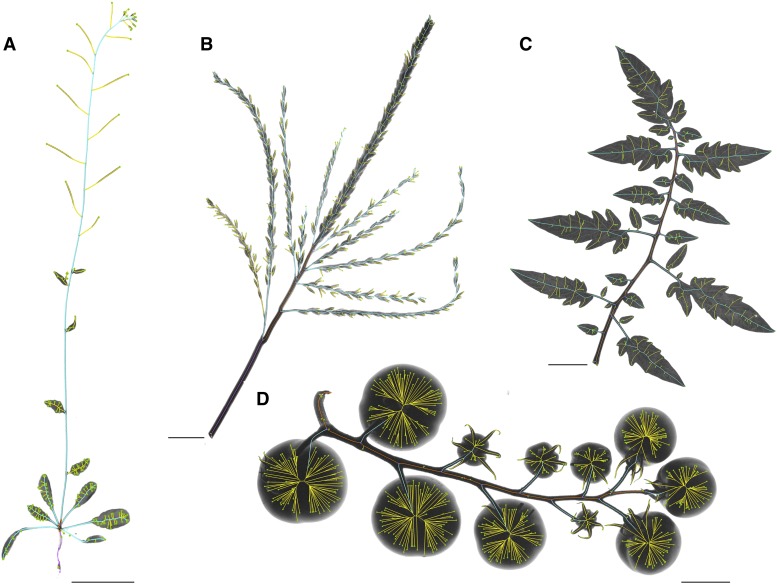
In order to test the applicability of PANorama in other organisms, we photographed and analyzed the following plant structures: a male inflorescence from maize (Zea mays), a compound leaf and an inflorescence from tomato, and an entire Arabidopsis (Arabidopsis thaliana) plant (Fig. 5). With minimal tweaking to the parameters within iftPANorama.h, we were able to calculate phenotypes across these differently shaped image objects. The three points selected in DefineMainAxis.py were chosen in order to measure different biological structures within each organism (see “Materials and Methods”; Supplemental Table S3).
Figure 5.
PANorama in other species. With minimal adjustments to parameters stored in the iftPANorama.h file, PANorama can be easily adapted to measure structures from other organisms, including total plant architecture in Arabidopsis (A), the male inflorescence of maize (B), and the compound leaf (C) and inflorescence (D) of tomato. Phenotypic values are listed in Supplemental Table S3. Bars = 500 pixels.
Validation of PANorama Phenotypes in Rice
QTL Analysis
To test the biological validity of PANorama phenotypes, we performed a QTL analysis using a population of RILs derived from a wide cross between IR64 (Indica) and Azucena (Japonica). In any phenotype-mapping study, there are tradeoffs between the number of replicates, the number of traits, and the resolution of measurements that are feasible to obtain. Because the MAPDs of open versus closed primary branches differed by approximately 1% (Table II), we phenotyped primary architecture in high replication (n = 1,522 images) and forewent the manual arrangement of higher branching (Supplemental Fig. S2A). QTL mapping was performed following the methods described previously (Spindel et al., 2013) using 30,984 single-nucleotide polymorphisms generated via genotyping by sequencing (GBS). Numerous QTLs were detected throughout the genome for length, width, and count measurements (Table III; Supplemental Table S4). We also detected several QTLs for two reproductive traits that were measured without using PANorama: days to heading (DTH) and panicle number (PN; Supplemental Table S4; see “Materials and Methods”). Several regions of the genome showed clusters of QTLs for PANorama panicle traits and/or these independently measured reproductive phenotypes (Supplemental Table S4).
Table III. Panicle trait QTLs identified using PANorama phenotypes.
| Trait | Trait Type | Chromosome | Total PVE | No. of QTLs |
|---|---|---|---|---|
| EL | Length | 1, 4, 6, 8 | 49.77 | 5 |
| RL | Length | 1, 8 | 32.50 | 2 |
| PL | Length | 1 | 17.26 | 1 |
| NL | Length | 1, 5, 6, 9, 10 | 56.15 | 6 |
| xNL | Length | 1, 5 | 23.25 | 2 |
| nNL | Length | 1, 3, 9, 11 | 42.39 | 4 |
| PBL | Length | 1, 3, 4, 8, 11 | 40.26 | 5 |
| xPBL | Length | 4 | 10.80 | 1 |
| nPBL | Length | 6 | 9.99 | 1 |
| PBN | Count | 3, 4 | 35.86 | 3 |
| NN | Count | 3, 8 | 25.19 | 2 |
| BpN | Count | 1, 4 | 28.76 | 2 |
| xBpN | Count | 1, 3, 4, 7 | 44.30 | 4 |
| TR | Width | 1, 4, 7 | 30.86 | 3 |
| xTR | Width | 4 | 17.02 | 1 |
| TE | Width | 4 | 10.86 | 1 |
| xTE | Width | 1, 4 | 20.01 | 2 |
| nTE | Width | 2, 8 | 22.65 | 2 |
| Total No. of PANorama QTLs identified | 47 | |||
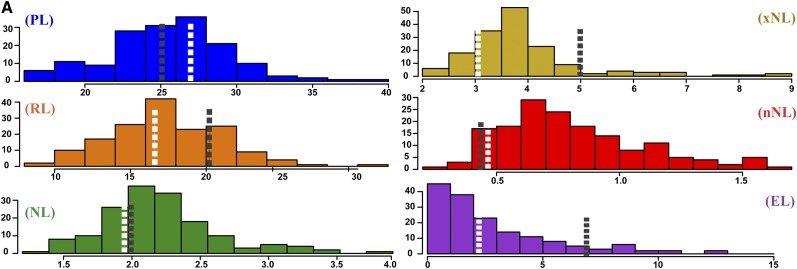
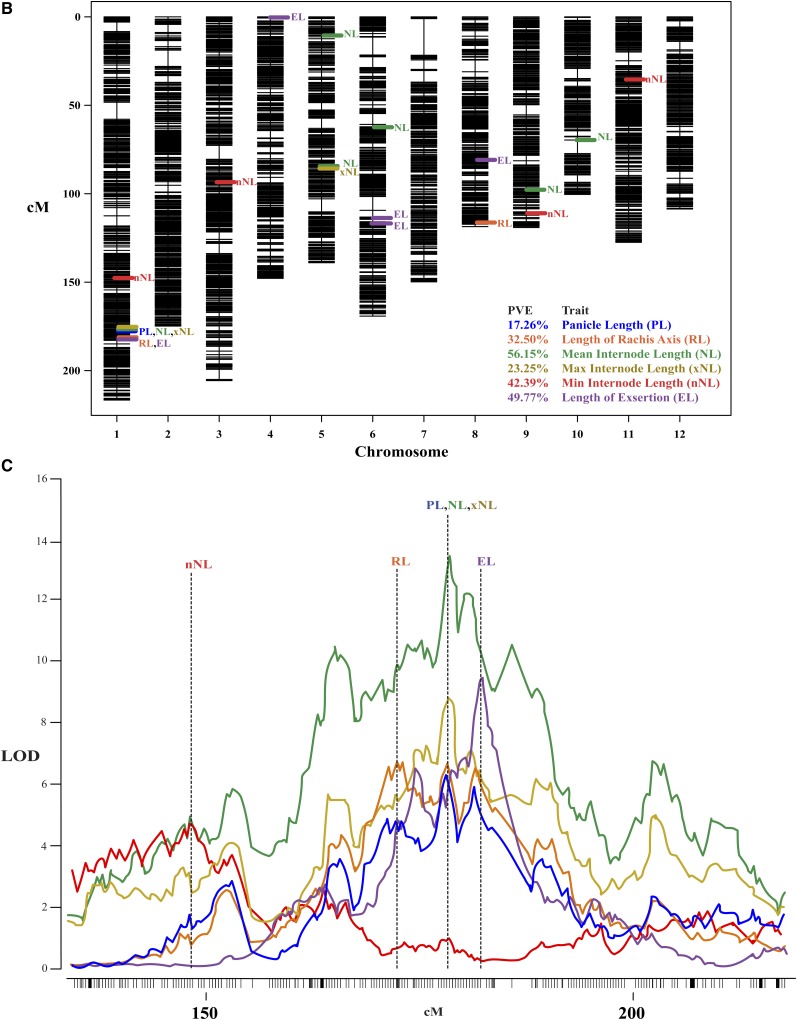
Figure 6 and Table IV summarize results for six traits, including one vegetative phenotype, exsertion length (EL), and multiple phenotypes that represent components of the composite trait PL. All of these phenotypes display transgressive variation in the RIL population compared with the parents (Fig. 6A). Notably, despite using PANorama, PL had a single QTL on chromosome 1 explaining 17.26% of the phenotypic variation (Table IV). By dividing PL into RL, mean internode length (NL), maximum internode length (xNL), and minimum internode length (nNL) traits, we identified 16 additional QTLs at 11 distinct locations throughout the genome and increased the total percentage variance explained (Fig. 6B; Table III). Of particular interest, nearly every length phenotype within this study, vegetative and reproductive, mapped to a large region on the distal end of chromosome 1 (Supplemental Table S4). Figure 6C depicts the log10 of the odds (LOD) profiles for a few of these traits from approximately 140 centimorgans (cM) to the end of chromosome. Notably, peak markers for PL, xNL, and NL show increasing LOD scores of 6.38, 8.56, and 13.53, respectively. Additionally, within the region, the peak markers for RL and EL are shifted several centimorgans up and downstream, respectively (Fig. 6C; Table IV). These results demonstrate that biologically precise phenotyping using PANorama complements dense genetic marker data, improving the resolution of QTL peaks at a single genomic locus.
Figure 6.
PANorama improved our ability to measure and map for quantitative panicle traits. A, Frequency distributions of phenotypic line means for six traits, calculated using 158 RILs and the two parents of the mapping population. The total number of panicle images was 1, 522. The means for IR64 and Azucena are labeled using black or white dashed lines, respectively. The x axes depict length measurements (cm) for each phenotype. B, Genetic map of the 12 chromosomes in rice, constructed with 30,984 single-nucleotide polymorphisms. PVE indicates total PVE for all QTLs within a trait. C, QTL peaks at the distal end of chromosome 1, shown from approximately 140 cM to the end of the chromosome. The y axis depicts the LOD score calculated for each marker across the region. QTL mapping was performed using Haley-Knott regression and 1,000 permutations to determine a significance threshold for each phenotype; the average threshold for all 6 phenotypes is 3.42 (not shown). LOD profiles show the refined QTL locations after forward selection and backward elimination were used to probe the model space. The peak marker for each trait is depicted using a vertical dashed line.
Table IV. Dissection of PL into different internode measurements using PANorama increases the number of QTL identified in an IR64 x Azucena RIL Population.
Several of these peaks have been identified in previous studies (Pr. id.), but many are novel. Percent variance explained (PVE) was calculated for each peak. IR64 additive effects (Add. Eff.) are reported in units for the trait, except for non-normal phenotypes transformed using the natural logarithm (*). Nonparametric phenotypes are denoted with (np).
| Pheno | Chr | Peak(cM) | Peak Marker | Left, Right Markers | LOD | PVE | Add. Eff. | Pr. id. |
|---|---|---|---|---|---|---|---|---|
| nNL | 1 | 148.44 | S1_30057829 | S1_26453459, S1_31817994 | 4.53 | 8.31 | −0.05* | f |
| PL | 1 | 178.17 | S1_37484178 | S1_35922333, S1_38642939 | 6.38 | 17.26 | −1.66 cm | a, b |
| NL | 1 | 178.5 | S1_37521762 | S1_37370036, S1_38150186 | 13.53 | 21.7 | −0.06* | a, b |
| xNL | 1 | 178.5 | S1_37521762 | S1_37314452, S1_37838629 | 8.56 | 20.48 | −0.55 cmnp | a, b |
| RL | 1 | 172.11 | S1_36198766 | S1_35691398, S1_38679377 | 6.65 | 14.75 | −1.58 cm | a, b |
| EL | 1 | 182.15 | S1_38527519 | S1_38116619, S1_38642939 | 9.38 | 16.15 | −1.2 cmnp | a, c, e |
| nNL | 3 | 94.16 | S3_16076715 | S3_15765689, S3_16438463 | 6.62 | 12.51 | 0.06* | |
| EL | 4 | 2.32 | S4_405005 | S4_309716, S4_13379128 | 4.09 | 6.49 | 0.89 cm | |
| NL | 5 | 11.35 | S5_1442539 | S5_1247072, S5_1568034 | 5.93 | 8.45 | −0.04* | |
| NL | 5 | 85.13 | S5_21207727 | S5_20587858, S5_21705936 | 3.79 | 5.23 | −0.05* | |
| xNL | 5 | 85.13 | S5_21207727 | S5_1279922, S5_22515124 | 3.39 | 7.48 | −0.34 cm | |
| NL | 6 | 62.63 | S6_10319721 | S6_8821624, S6_12046394 | 4.31 | 5.99 | −0.03* | |
| EL | 6 | 114.93 | S6_22200268 | S6_21980490, S6_22254152 | 3.97 | 6.29 | −0.29 cmnp | |
| EL | 6 | 116.58 | S6_22260180 | S6_22200268, S6_22410496 | 6.46 | 10.64 | −0.57 cmnp | |
| EL | 8 | 79.91 | S8_22603837 | S8_22484223, S8_23108392 | 6.5 | 10.64 | −0.89 cmnp | a, c, e |
| RL | 8 | 112.69 | S8_27305892 | S8_27213374, S8_28389376 | 7.29 | 16.32 | −1.67 cm | d |
| NL | 9 | 96.74 | S9_18987864 | S9_16451344, S9_20174149 | 3.94 | 5.44 | −0.03* | |
| nNL | 9 | 110.9 | S9_21397331 | S9_20403572, S9_22503375 | 4.34 | 7.94 | −0.04* | |
| NL | 10 | 67.94 | S10_19095358 | S10_18448365, S10_19941941 | 5.47 | 7.74 | 0.03* | |
| RL | 10 | 67.94 | S10_19095358 | S10_18437567, S10_21920802 | 3.69 | 5.06 | 1.08 cm | |
| nNL | 11 | 34.31 | S11_5453415 | S11_4362683, S11_5644994 | 7.33 | 14.02 | 0.04* |
aPrevious identification citations are as follows: a, Hittalmani et al. (2003); b, Kobayashi et al. (2003); c, Li et al. (2003); d, Ikeda et al. (2010); e, Asano et al. (2011); and f, Zhao et al. (2011).
Comparison of Panicle Traits across Genetic Lines
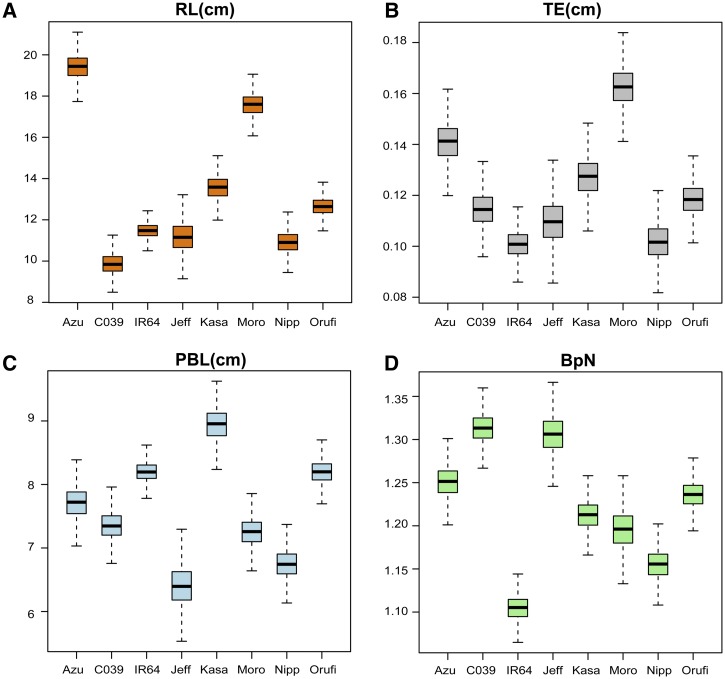
To establish the extent of morphological variation in rice panicle architecture, both within a variety and across different genetic backgrounds, we grew a small panel of eight genetic lines in varying pot sizes under controlled environmental conditions (n = 1,139 images). The eight lines represent the parents of four mapping populations currently available in the international rice community, including the IR64 × Azucena RIL population used in the QTL mapping portion of this study. They come from diverse genetic backgrounds, representing seven domesticated varieties, four of the rice subpopulations, and one wild relative: Azucena, Jefferson, and Moroberekan (tropical japonica); Nipponbare (temperate japonica); IR64 and CO39 (indica); Kasalath (aus); and Oryza rufipogon IRGC 105491 (wild). For many of the measurements, each line shows different means and distributions. The variation observed within varieties was much smaller than that observed between varieties, even when plants were grown under different pot sizes (Fig. 7; Supplemental Table S5). Although we detected subtle variation within a genetic line across pot sizes, the differences were not statistically significant (Supplemental Table S6).
Figure 7.
Quantitative variation in rice panicle traits. Phenotypic distributions are shown for seven distinct genetic lines of rice and one wild relative: Azucena (Azu), CO39, IR64, Jefferson (Jeff), Kasalath (Kasa), Moroberekan (Moro), Nipponbare (Nipp), and O. rufipogon IRGC 105491 (Orufi). Phenotypes include RL (A), exsertion thickness (TE; B), PBL (C), and BpN (D). Distributions were calculated using the methods described by Greenberg et al. (2011). Briefly, Markov chain Monte Carlo line means were calculated using 1,000 permutations and the panicle phenotypes from plants grown in four different pot sizes (2, 3, 4, and 6 inches). The total number of panicle images was 1,139.
DISCUSSION
The rice inflorescence is a complex organ, developmentally and physiologically, and yet plant breeders have continued to measure panicles using very blunt tools. This is out of step with the major advances in sequencing and genotyping technology that have greatly increased the dissection of complex traits by QTL mapping and/or genome-wide association studies (GWAS). The major goal of this study was to develop a platform and methodology that facilitates high-resolution phenotyping of rice panicles.
PANorama Is an Accurate and Flexible Phenotyping Platform
For branched structures like the rice panicle, image analysis represents an accurate and high-throughput method to obtain multiple phenotypes simultaneously. However, phenotyping via image analysis has its own limitations, often requiring special preparation of plant tissue or time-intensive, manual arrangement of samples before a photograph can be taken. Within this study, we demonstrate that PANorama represents an improvement in efficiency, flexibility, and accuracy when phenotyping primary panicle axis traits. Table V summarizes the major differences in software implementation, testing, and phenotyping capabilities between PANorama and P-TRAP, an open-source software platform released in August 2013 (AL-Tam et al., 2013).
Table V. Comparison of the PANorama and P-TRAP software platforms.
Features that are not supported in PANorama or P-TRAP version 20130213220 are labeled with (NS).
| Parameter | Feature | PANorama | P-TRAP |
|---|---|---|---|
| Software implementation | Image acquisition | Built directly into the pipeline with skeleton preview | NS |
| Skeletonization | Based on exact morphological erosions of a shape | Axes connected with straight-line segments | |
| Skeletonization | Potential for future software releases to target differently shaped image objects | Generates skeletons for branching structures only | |
| Axis definition | Internode/awn filtering built directly into skeleton calculation with adjustable parameters | Internodes and awns must be manually edited using a GUI | |
| Axis definition | User divides the primary axis three times | User divides the primary axis two times | |
| Axis definition | Skeleton points are selected on top of a _skel.jpg image, generating an accessory file | Points selected within a skeleton graph and require manual editing of the skeleton | |
| Software execution | Linear pipeline controlled with scripts | Self-contained GUI | |
| Compatibility | Linux | Windows, Mac OSX, and Linux | |
| Beta testing | Image number | More than 2,600 images in this study | 26 images |
| Genetic mapping | QTL mapping using 154 RILs in rice | NS | |
| Phenotyping differences | Primary axis internodes | Length of individual rachis internodes | Mean internode length only |
| Primary axis thickness | Mean and maximum thickness measures | Mean thickness only | |
| Branches per internode | Calculated at every internode (mean and maximum) | NS | |
| Exsertion phenotypes | Length and thickness | NS | |
| Secondary/tertiary axes | NS | Branch length, count, and number of internodes | |
| Spikelet traits | NS | Count, length, and shape |
The PANorama pipeline is broken into distinct steps that are executed linearly using a small collection of scripts (Fig. 2; Supplemental Fig. S1; Supplemental Videos S1–S3). The most demanding tasks are computed in batch, including awn filtering, skeletonization, and data extraction. Meanwhile, basic orientation of axes and error correction for images are flexibly controlled for each image via stand-alone GUIs. All parameters controlling implementation of the pipeline are stored in a single editable file (Supplemental Table S1). PANorama skeletons are calculated using the exact morphology of panicle axes, which accurately excludes awns and requires no editing by the user (Figs. 2–4). When comparing PANorama phenotypes and manual measurements, we found that using the awn filter always increased the accuracy of PBL measurements, even in varieties with developmentally small awns (Table II; Supplemental Fig. S2). For landraces and wild relatives of rice, which have long awns of varying length (Fig. 2), this feature was crucial when measuring PBL in high replication (Fig. 7). When identifying developmental transition points in an image using the PANorama GUI DefineMainAxis.py, points are selected on top of a skeleton and their positions are stored in an accessory .txt file separately from the skeleton (Supplemental Fig. S1; Supplemental Video S3). This allows a user to flexibly edit any parameters controlling the pipeline (Supplemental Table S1) and rerun skeleton calculation without having to reselect developmental transition points for every image in a directory. Selecting three points during this step also allows division of the main axis into two types of tissue: in this study, vegetative (EL) and reproductive (RL).
In contrast, the P-TRAP platform calculates skeletons using straight-line segments and operates entirely within a single GUI, requiring users to manually edit skeletons using a mouse (Table V). The P-TRAP GUI implementation is flexible and facilitates large-scale error correction of an image skeleton, but it comes at a great cost in terms of the accuracy of skeletons and overall processing time, defeating the purpose of high-throughput image analysis. When comparing PANorama and P-TRAP on the same computer using 60 images, we found that skeleton calculation and phenotype extraction were 8 times faster in PANorama (Table II). Additionally, rerunning skeleton calculation within P-TRAP required that the IM initiation and abortion points be reselected for every image, increasing manual processing time when replicating results (data not included in time calculations). P-TRAP could not distinguish curved panicle axes, cluster branches into internodes automatically, or filter awns (Supplemental Fig. S3). This resulted in decreased accuracy (low r2 values, large MPEs) and increased error (large MAPDs) for all P-TRAP phenotypes (Table II).
The flexible implementation of skeletonization and awn filtering within PANorama directly facilitated our ability to perform, to our knowledge, the first large-scale, image-based quantification of primary panicle architecture in rice using thousands of images (Table V). In contrast, P-TRAP was only tested on 26 images before being released (AL-Tam et al., 2013). We found our methods to be robust, even when photographing panicles from different varieties and subpopulations. With minimal tweaking to parameters within iftPANorama.h (see “Materials and Methods”), we were also able to generate skeletons for plant organs from other species (Fig. 5; Supplemental Table S3). For structures like the maize male inflorescence, which is similar to rice, processing only required that the AWNSTHICK parameter be turned off. However, while we could accurately generate skeletons for the Arabidopsis and tomato images (Fig. 5), we could only extract phenotypes that were similar in structure to a rice panicle (Supplemental Table S3). By releasing the PANorama source code freely to the public, other research groups can expand on the skeleton implementation used within the pipeline and fully customize PANorama to extract measurements from differently shaped image objects.
Quantitative Panicle Phenotypes Are Necessary to Leverage High-Resolution Genotype Data
To our knowledge, this study represents the first QTL analysis to use both high-density genetic marker data and high-resolution phenotypes to characterize primary panicle architecture in rice. In doing so, we were able to detect the largest number of panicle and exsertion QTLs ever reported in a single study (Table III). Several of these QTLs have been identified in previous studies using different mapping populations, but many are novel (Table IV; Supplemental Table S4). Using PANorama, we were able to measure many traits simultaneously in high replication, identify new QTLs for known traits that have never been identified, and measure several novel phenotypes that are otherwise difficult to capture, such as internode composition of the main rachis axis. Additionally, we identified several regions of the genome that show pleiotropic clustering of panicle QTLs and confirmed the overlap of PANorama QTLs with two independently measured reproductive traits (DTH and PN). Both of these findings are consistent with previous studies in rice (Kobayashi et al., 2003; Ando et al., 2008; Liu et al., 2008; Ikeda et al., 2010), adding further biological validity to our QTL analysis. Taken together, these results attest to both the efficiency and accuracy of the PANorama pipeline.
When using high-resolution GBS marker data to map PL (Fig. 1), we were only able to detect one QTL on chromosome 1. However, by dissecting this composite phenotype into its internode components using PANorama, we detected 12 additional QTLs at 8 distinct locations throughout the genome (Fig. 6B; Table IV). Although some of these QTLs overlap with one another, several are unique to different rachis measures and together are responsible for larger percentage variances explained (PVEs; Table III). These findings not only validate our decision to treat the rachis axis as a composite phenotype but suggest that high-resolution phenotyping is necessary to dissect pleiotropic panicle traits (Fig. 6B; Table IV). Interestingly, the PL, RL, NL, and xNL phenotypes all have QTLs that tightly cluster at the distal end of chromosome 1, in the same region as a QTL for the vegetative trait EL (Fig. 6B; Table IV). A partial measurement of the culm (Fig. 1), EL has become an important phenotype with the advent of hybrid rice breeding. Nearly every male-sterile line available to breeders has a short EL, which severely reduces outcrossing because panicles remain trapped within the leaf sheath. Even though a gene controlling panicle exsertion has been cloned on chromosome 5, ELONGATED UPPERMOST INTERNODE, many breeders still rely on exogenously applied gibberellic acid to ensure full panicle exsertion during crossing schemes (Luo et al., 2006).
The EL LOD profile across chromosome 1 is relatively narrow and highly significant, with one major peak near the gibberellin oxidase gene SD1 (Fig. 6C; Table III). During the Green Revolution, breeders incorporated an allele of SD1 into diverse indica varieties that significantly increased harvest index by reducing the ratio of vegetative to reproductive tissue (Hedden, 2003). This was accomplished by selecting for the phenotype “plant height,” measured as the total length of a panicle bearing culm, from the soil level at the base of the vegetative stem to the tip of the longest panicle (International Plant Genetic Resources Institute and Association, 2007). This measurement integrates multiple internodes, vegetative and reproductive, into a single composite trait that encompasses all exsertion and panicle phenotypes. As such, it is not surprising that previous QTL studies have mapped PL and plant height to the distal end of chromosome 1, using a population of similar genetic background; IR64 is one of the indica varieties that received the dwarfing SD1 allele, while Azucena is a tall tropical japonica landrace. Breeders have always assumed that the SD1 locus was pleiotropically responsible for length phenotypes that map to this region, even though these studies were conducted using low-density marker coverage and low-resolution phenotyping (Hittalmani et al., 2003; Li et al., 2003).
By comparing QTLs for PL, RL, and the various internode measures surrounding SD1 within our study, we note significant changes with respect to both the overall LOD scores and the distribution of the LOD profiles. Specifically, with increasing resolution of PL phenotypes (from PL to RL to NL), the LOD profiles resolve from one amorphous peak into several small-effect QTL peaks (Fig. 6C). This suggests that that there are likely multiple, linked loci across the region involved in panicle development surrounding the large-effect SD1 gene, which are easier to detect using a combination of dense marker data and high-resolution panicle phenotype data. Although our ability to identify specific candidate genes within complex QTL regions is limited by the number of recombination events within this relatively small RIL population, mapping numerous internode-derived traits to this region is consistent with the recent breeding history at this locus. These results present compelling evidence that high-resolution panicle phenotyping will facilitate the dissection of complex loci and complement other high-resolution genotype data sets, such as those used in GWAS.
Rice Panicle Architecture Varies across Different Genetic Backgrounds
The high-resolution phenotypes we generated using PANorama allowed us to detect subtle variation in panicle architecture, which may otherwise have been lost using standard phenotyping methodologies. Many panicle traits have distinctly different distributions within a genetic line, despite being grown in varying pot sizes (Fig. 7; Supplemental Tables S5 and S6). These results support the findings observed in a previous study, which identified subpopulation-specific polymorphisms associated with PL in rice using GWAS (Zhao et al., 2011). However, it is interesting that although three of the lines in our study (Azucena, Moroberekan, and Jefferson) are from the same subpopulation, tropical japonica, their panicle traits are distinctly different from one another. This confirms that quantitative variation exists within multiple stages of panicle development, at the variety, subpopulation, and rice species level.
Within the QTL mapping portion of this paper, we identify several genomic regions that have overlapping QTLs for panicle traits, PN, and/or DTH phenotypes (Supplemental Table S4). The concept of pleiotropy in panicle development has been confirmed numerous times within other QTL studies (Kobayashi et al., 2003; Ando et al., 2008; Liu et al., 2008; Ikeda et al., 2010). However, limited genotype and phenotype resolution has always prevented the dissection of these complex loci, which could be composed of single large-effect genes or arrays of linked, small-effect genes, as we suggest exist on chromosome 1 (Fig. 6C). In either case, without performing high-resolution panicle phenotyping on a larger set of rice germplasm, it is difficult to interpret whether multitrait loci observed within experimental genetic mapping populations are actually responsible for variety- and subpopulation-specific panicle traits, such as those observed within our small collection of genetic lines (Fig. 7; Supplemental Table S5). For breeders working with diverse sets of germplasm, the existence of such loci could have significant implications when selecting for panicle-related traits that affect yield and grain quality.
CONCLUSION
PANorama is a unique and flexible phenotyping pipeline that improved our ability to quantitatively measure rice panicles in both a QTL mapping study and a collection of diverse genetic lines. Although breeding for rice panicle morphology to boost yield potential is promising in theory, quantitative phenotyping must be done on a larger set of germplasm in order to tease apart the pleiotropic nature of panicle phenotypes observed in genetic mapping populations. By releasing PANorama freely to the public, it is our hope that others will be able to adapt it to their own phenotyping needs. The use of a single phenotyping platform across multiple research groups could represent a significant step forward in streamlining comparative genetic studies both within and across species.
MATERIALS AND METHODS
Plant Material and Harvesting
The set of rice (Oryza sativa) genetic lines used in the pot size experiment are the parents of four mapping populations currently available within the international rice community. Genotypes include Azucena, CO39, IR64, Jefferson, Kasalath, Moroberekan, Nipponbare, and one wild relative (Oryza rufipogon IRGC 105491). The IR64 × Azucena RIL population, composed of 176 F10 to F12 lines, was originally developed at Institut de Recherche pour le Développement in Montpellier, France, using single-seed descent under greenhouse conditions. For the comparison of manual measures, PANorama, and P-TRAP, IR64 and Azucena varieties from Rice Diversity Panel 1 were used.
Seeds were surface sterilized using 20% bleach for 15 to 20 min and germinated directly in soil at approximately 1 cm depth. A 1:1 soil mixture of Promix:clay was used in both studies. Panicles were harvested by cutting the culm at the flag leaf ligule so as to include the EL measurement. For panicles that still remained wrapped within the leaf sheath of the culm, panicles were cut just below the panicle notch (Fig. 1) so as to include all reproductive phenotypes. Samples were stored in 70% ethanol to maintain the flexibility of tissue until the time of imaging. PN was measured as the total number of panicles on a plant after grain filling had finished. DTH was recorded when the first panicle on a plant emerged 50% from the leaf sheath.
Planting Conditions
Growth Chamber
Plants were grown in the Weill Growth Chamber Facility at Cornell University. Chamber settings were as follows: 300 μmol m−2 s−1 on a 12-h day/night cycle; day/night temperature of 30°C/25°C; and humidity between 75% and 85%. The eight genetic lines were planted in four pot sizes (2, 3, 4, and 6 inches) and replicated (n = 40 plants per variety; 10 replicates per pot size). To reduce shading effects, plants were grown in a pseudorandomized block design. The lines were grouped by plant height (short and tall), and pots were also clustered into two major groups based on size (4- and 6-inch pots together and 2- and 3-inch pots together). The positions of plants within clusters were randomized, and clusters of different sized pots were randomized throughout the chamber. When available, five panicles per plant replicate were photographed (total panicles in study, n = 1,139).
Greenhouse
For comparison of manual measures, PANorama, and P-TRAP, IR64 and Azucena were grown in summer 2013 in the Gutterman Greenhouse Facility at Cornell University (n = 3 plants per variety). Five panicles were harvested from each replicate (n = 15 panicles per variety). For the QTL mapping study, the RILs were grown in summer 2012 in the Gutterman Greenhouse Facility. In order to reduce shading or greenhouse positional effects, three replicates (n = 3 plants per line) were grown in a pseudorandomized block design. Briefly, plants were grown in 6-inch pots and clustered by height; plant positions within a cluster and the positions of overall clusters within the greenhouse were both randomized at the start of the study. Several weeks after tillering phase began, plants with excessively short phenotypes or narrow leaves were moved into groups outside of the randomized design to prevent shading. When available, five panicles per plant replicate were photographed using PANorama (total panicles in study, n = 1,522).
Sample Imaging Protocol
Rice
Before photographing, ethanol-soaked panicles were air dried for 15 to 24 h to improve the rigidity of the tissue without resulting in brittleness. Samples were photographed using a Nikon D200 digital SLR camera, a 60-mm Macro lens, and a 0.3 neutral density filter. Camera control in PANorama is mediated by gPhoto2 (version 2.4.14). During imaging, panicles were arranged so that primary branches were spread open so as not to overlap within the image view. The camera was mounted on a fixed copy stand, and color images of samples were acquired on a light box to improve contrast. Images were saved at 2,592 × 3,872 pixels. The scale parameter within iftPANorama.h was adjusted for each experiment to ensure that pixel-to-length conversions were accurate (comparison of manual measures, PANorama, and P-TRAP = 116 pixels cm−1; QTL mapping = 120 pixels cm−1; and measurement of eight genetic lines = 120 pixels cm−1). To determine the scale, an image of a ruler was acquired using GetPictures.py, and the open-source software ImageJ (version 1.46a-1) was used to measure the length of 1 cm in pixels at several places within the image. All image processing was performed within the PANorama pipeline using the parameters listed in Supplemental Table S1.
Maize, Tomato, and Arabidopsis
Images for maize (Zea mays), tomato (Solanum lycopersicum), and Arabidopsis (Arabidopsis thaliana) were acquired using PANorama and saved at 2,592 × 3,872 pixels (scale = 120 pixels cm−1). Several adjustments were made to the iftPANorama.h file before skeleton calculation. The segmentation and multiscale parameters used by CreateSkeletons.py were not altered, but the awn filter was turned off (AWNSTHICK = 0.0 cm). All other parameters were kept the same for the maize and tomato inflorescences. For the Arabidopsis plant and tomato leaf, the minimum junction distance and minimum primary branch length parameters were altered (Arabidopsis, MINJCTDIST = 0.5, MINBRANCSZ = 0.1; tomato leaf, MINJCTDIST = 0.1, MINBRANCSZ = 0.1).
The EP, IMAP, and FMAP points selected in the DefineMainAxis.py GUI were carefully chosen to highlight different structures within each image (Fig. 5, color differences). Measurements are listed in Supplemental Table S3. To measure total Arabidopsis plant architecture, EP was selected at the tip of the longest root, IMAP at the base of the inflorescence, and FMAP at the tip of the inflorescence (Fig. 5A). To measure Arabidopsis inflorescence traits, the EP was selected at the base of the inflorescence, the IMAP just below the terminal floret cluster, and the FMAP at the tip of the inflorescence (data not shown). For the maize inflorescence, EP was selected at the flag leaf ligule attachment point, IMAP at the first tassel branch, and FMAP at the base of the terminal spike (Fig. 5B). For the tomato inflorescence, FMAP was selected at the base of the last fruit on the cyme. For the tomato leaf, FMAP was selected at the base of the terminal leaflet. In both tomato images, EP and IMAP were selected directly on top of one another at the base of the axis in order to exclude pink exsertion measurements (Fig. 5, C and D).
Comparison of Manual Measures, PANorama, and P-TRAP
Manual measurements were taken for the following phenotypes by hand: RL, primary branch number (PBN), NN, and average number of branches per node (BpN). Average PBL was calculated by manually measuring every branch within an image using the Segmented Line Tool in ImageJ (version 1.46a-1; pixel conversion = 116 pixels cm−1). Measurements were taken using images that contained openly arranged panicle branches to ensure that curved primary branches could be measured as accurately as possible. Phenotypes were calculated from raw skeletons in both PANorama and P-TRAP (version 20130213220) and used to calculate all reported statistics. No images were corrected after skeleton calculation, except to select the meristem transition points. Because P-TRAP had large percentage deviations from manual measures for both RL and NN, we reran the analysis two times with P-TRAP freshly installed on two separate computers (Linux and Macintosh). However, doing so did not significantly improve the r2, MAPD, or MPE. Total processing time for PANorama includes the execution of the following scripts for 60 images: CreateSkeletons.py, ExtractInfo.py, and ExtractMeasures.py. Total processing time for P-TRAP includes skeletonization (Find Structure) and phenotype calculation (Collect Results) for 60 images. The amount of time required to select meristem points was not included in either calculation. Additionally, P-TRAP grain quantification (Find Grains) was not included in the time calculation. Processing times were measured on the same computer for both platforms.
QTL Analysis
QTL mapping within this study was performed using 154 of the IR64 × Azucena RIL lines and the software package R/QTL (R version 2.15.1; R/qtl package 1.24.9) following the methods described (Spindel et al., 2013). Briefly, we performed an initial scan for QTLs using Haley-Knott regression and 1,000 permutations to determine the LOD significance threshold. We then rescanned for additional QTLs, conditioning on peaks detected in the initial scan. Finally, forward selection and backward elimination were used to probe the model space and refine QTL locations. For phenotypes that failed the Shapiro-Wilks test for normality, a natural log transformation of the raw data was performed before QTL mapping was conducted. The distributions for two of the phenotypes (EL and xNL) remained highly non-normal even after the natural log transformation; thus, a nonparametric QTL model was used (Table IV; Supplemental Table S4).
Statistical Analysis of Diverse Lines
To construct distributions of line means and pot size effects on each panicle phenotype, we implemented a multivariate Bayesian hierarchical model (Greenberg et al., 2011). The data contained between one and five individual data points per plant and between 24 and 32 plants (grown in various pot sizes) per genotype. We analyzed all phenotypes (Fig. 5) simultaneously. The posterior distributions of line means were constructed using Markov chain Monte Carlo as described (Greenberg et al., 2011). We ran 500 iterations of burn in and saved 25,000 samples each from four independent chains. Convergence was verified by inspecting trace plots of the chains.
Supplemental Data
The following materials are available in the online version of this article. The open source software PANorama is free to download at ricediversity.org, and videos can be viewed using from the links below.
Supplemental Figure S1. Directory structure used in the PANorama pipeline.
Supplemental Figure S2. Final skeletons in closed versus open rice panicles.
Supplemental Figure S3. Screenshots comparing PANorama with the open-source platform P-TRAP.
Supplemental Table S1. Parameters stored within the iftPANorama.h file.
Supplemental Table S2. Correlation coefficients between PANorama phenotypes with and without awn filtering.
Supplemental Table S3. Phenotypes from other species.
Supplemental Table S4. Panicle trait QTLs identified in an IR64 × Azucena RIL population.
Supplemental Table S5. Phenotypic grand means for eight genetic lines, calculated using all pot sizes.
Supplemental Table S6. Phenotypic raw means for each genetic line, calculated per pot size.
Supplemental Video S1. Using GetPictures.py and RebuildImageFiles.py (https://vimeo.com/87497670).
Supplemental Video S2. Using CreateSkeletons.py and DefineMainAxis.py (https://vimeo.com/87497348).
Supplemental Video S3. Using ExtractInfo.py, CorrectBranches.py, and ExtractMeasures.py (https://vimeo.com/87497347).
Supplemental Materials and Methods S1. Image-analysis methods for high-resolution inflorescence phenotyping.
Supplementary Material
Acknowledgments
We thank the laboratories of Michael Scanlon, Jian Hua, and Joceyln Rose for supplying plant tissue specimens from maize, Arabidopsis, and tomato, respectively; Randy Clark for support in setting up a plant imaging station; Sandra Harrington and Gen Onishi for support with greenhouse management; and Joseph LeCates for assistance in harvesting for the growth chamber experiment.
Glossary
- PI
panicle initiation
- IM
inflorescence meristem
- PBM
primary branch meristem
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- PL
panicle length
- GUI
graphical user interface
- EP
extrusion point
- IMAP
initial main axis point
- FMAP
final main axis point
- MAPD
mean absolute percentage deviation
- MPE
mean percentage error
- PBL
primary branch length
- NN
internode number
- DTH
days to heading
- PN
panicle number
- EL
exsertion length
- LOD
log10 of the odds
- cM
centimorgan
- NL
internode length
- GWAS
genome-wide association studies
- PVE
percentage variance explained
- RL
rachis length
- BpN
average number of branches per node
- PBN
primary branch number
Footnotes
This work was supported by the National Science Foundation (Plant Genome Research Program grant no. 1026555 to S.R.M. and Graduate Research Fellowship to S.C.) and by the Fundação de Amparo à Pesquisa do Estado de São Paulo (grant no. 2011/03110–6 to A.X.F.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- AL-Tam F, Adam H, dos Anjos A, Lorieux M, Larmande P, Ghesquière A, Jouannic S, Shahbazkia HR. (2013) P-TRAP: a panicle trait phenotyping tool. BMC Plant Biol 13: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Yamamoto T, Shimizu T, Ma XF, Shomura A, Takeuchi Y, Lin SY, Yano M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor Appl Genet 116: 881–890 [DOI] [PubMed] [Google Scholar]
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, et al. (2011) Artificial selection for a Green Revolution gene during japonica rice domestication. Proc Natl Acad Sci USA 108: 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Xiao YH, Tang WB, Lei DY. (2007) Practices and prospects of super hybrid rice breeding. Rice Sci 14: 71–77 [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV. (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A. (2007) Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcão AX, da Fontoura Costa L, Da Cunha BS. (2002) Multiscale skeletons by image foresting transform and its application to neuromorphometry. Pattern Recognition 35: 1571–1582 [Google Scholar]
- Falcão AX, Stolfi J, de Alencar Lotufo R. (2004) The image foresting transform: theory, algorithms, and applications. IEEE Trans Pattern Anal Mach Intell 26: 19–29 [DOI] [PubMed] [Google Scholar]
- Famoso AN, Clark RT, Shaff JE, Craft E, McCouch SR, Kochian LV. (2010) Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol 153: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Harris T, Bailey-Serres J. (2009) Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann Bot (Lond) 103: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AJ, Hackett SR, Harshman LG, Clark AG. (2011) Environmental and genetic perturbations reveal different networks of metabolic regulation. Mol Syst Biol 7: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hedden P. (2003) The genes of the Green Revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang JY, Zheng KL, Liu GF, Wang GC, Sidhu JS, et al. (2003) Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor Appl Genet 107: 679–690 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y. (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54: 147–156 [Google Scholar]
- Ikeda M, Hirose Y, Takashi T, Shibata Y, Yamamura T, Komura T, Doi K, Ashikari M, Matsuoka M, Kitano H. (2010) Analysis of rice panicle traits and detection of QTLs using an image analyzing method. Breed Sci 60: 55–64 [Google Scholar]
- International Plant Genetic Resources Institute and Association (2007) Descriptors for Wild and Cultivated Rice (Oryza spp.). Biodiversity International, Rome [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS, Benfey PN. (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. (2007) Floral displays: genetic control of grass inflorescences. Curr Opin Plant Biol 10: 26–31 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Fukuta Y, Sato T, Osaki M, Khush GS. (2003) Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor Appl Genet 107: 1350–1356 [DOI] [PubMed] [Google Scholar]
- Latif MA, Islam MR, Ali MY, Saleque MA. (2005) Validation of the system of rice intensification (SRI) in Bangladesh. Field Crops Res 93: 281–292 [Google Scholar]
- Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CHM, Liu GF, Wang GC, Shashidhar HE, et al. (2003) QTL × environment interactions in rice. I. heading date and plant height. Theor Appl Genet 108: 141–153 [DOI] [PubMed] [Google Scholar]
- Liu GL, Mei HW, Yu XQ, Zou GH, Liu HY, Hu SP, Li MS, Wu JH, Chen L, Luo LJ. (2008) QTL analysis of panicle neck diameter, a trait highly correlated with panicle size, under well-watered and drought conditions in rice (Oryza sativa L.). Plant Sci 174: 71–77 [Google Scholar]
- Luo A, Qian Q, Yin H, Liu X, Yin C, Lan Y, Tang J, Tang Z, Cao S, Wang X, et al. (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol 47: 181–191 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2006) Branching out: the ramosa pathway and the evolution of grass inflorescence morphology. Plant Cell 18: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi A, Takai T, Ida M, Yamamoto T, Arai-Sanoh Y, Yano M, Ando T, Kondo M. (2011) Evaluation of yield performance in rice near-isogenic lines with increased spikelet number. Field Crops Res 120: 68–75 [Google Scholar]
- Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, Lorieux M, Ahmadi N, McCouch S. (2013) Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet 126: 2699–2716 [DOI] [PubMed] [Google Scholar]
- Tanabata T, Shibaya T, Hori K, Ebana K, Yano M. (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol 160: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur AK, Rath S, Roychowdhury S, Uphoff N. (2010) Comparative performance of rice with system of rice intensification (SRI) and conventional management using different plant spacings. J Agron Crop Sci 196: 146–159 [Google Scholar]
- Wang Y, Li J. (2011) Branching in rice. Curr Opin Plant Biol 14: 94–99 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y. (2011) Flower development in rice. J Exp Bot 62: 4719–4730 [DOI] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.