Oxygen and phosphate deficiency trigger overlapping changes in gene expression, including genes for galactolipid metabolism, which are mediated by a common transcription factor.
Abstract
Plant responses to biotic and abiotic stresses are often very specific, but signal transduction pathways can partially or completely overlap. Here, we demonstrate that in Arabidopsis (Arabidopsis thaliana), the transcriptional responses to phosphate starvation and oxygen deficiency stress comprise a set of commonly induced genes. While the phosphate deficiency response is systemic, under oxygen deficiency, most of the commonly induced genes are found only in illuminated shoots. This jointly induced response to the two stresses is under control of the transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1), but not of the oxygen-sensing N-end rule pathway, and includes genes encoding proteins for the synthesis of galactolipids, which replace phospholipids in plant membranes under phosphate starvation. Despite the induction of galactolipid synthesis genes, total galactolipid content and plant survival are not severely affected by the up-regulation of galactolipid gene expression in illuminated leaves during hypoxia. However, changes in galactolipid molecular species composition point to an adaptation of lipid fluxes through the endoplasmic reticulum and chloroplast pathways during hypoxia. PHR1-mediated signaling of phosphate deprivation was also light dependent. Because a photoreceptor-mediated PHR1 activation was not detectable under hypoxia, our data suggest that a chloroplast-derived retrograde signal, potentially arising from metabolic changes, regulates PHR1 activity under both oxygen and phosphate deficiency.
Sessile organisms such as plants suffer from the simultaneous exposure to multiple stresses due to the inability to escape. In plants, different stresses can cause similar symptoms as a result of partially overlapping signal transduction pathways. For example, cold, drought, and excess salinity lead to water deficiency, osmotic stress, and oxidative stress in plant cells, which turn on related signal transduction pathways (Vinocur and Altman, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). Here, we report an overlapping signal transduction pathway that is activated under both phosphate and oxygen deficiency. Furthermore, recent studies on plant stress responses frequently suggest posttranslational control of signaling components, including protein phosphorylation (Ghelis 2011; Tena et al., 2011), thiol-disulfide exchange reactions (Spoel and Loake, 2011; König et al., 2012), and proteasome-mediated protein degradation (Sadanandom et al., 2012; Shan et al., 2012). In this study, we present evidence that the phosphate and oxygen deficiency responses could be mutually controlled by posttranslational mechanisms.
Oxygen deficiency can naturally occur within plants but is also dependent on environmental conditions. Bulky plant organs that usually lack a distribution system for oxygen can develop cores with low oxygen concentrations (hypoxia) when mitochondrial activity is high and diffusion of oxygen into the plant tissue is limited. Also, oxygen supply can decrease during partial (waterlogging) or complete (submergence) flooding of plants. Hypoxia causes an inhibition of mitochondrial respiration due to the lack of the final electron acceptor, leading to an energy crisis that influences growth and biosynthetic processes (Drew, 1997; Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012). Under these circumstances, Arabidopsis (Arabidopsis thaliana) plants activate the expression of a set of about 49 core genes to cope with the stress, at least for a limited period of time (Mustroph et al., 2009). This set includes genes encoding transcription factors as well as metabolism-related proteins, such as enzymes involved in glycolysis and fermentation.
Recently, the signal transduction pathway that regulates the expression of most of the hypoxia core genes has been described (Gibbs et al., 2011; Licausi et al., 2011). Members of the group VII Ethylene Response Factor (ERF) family are processed by Met aminopeptidases, resulting in the exposure of an N-terminal Cys that can be oxidized enzymatically by plant cysteine oxidases (PCOs) depending on oxygen as a cosubstrate (Weits et al., 2014) and/or in a nitric oxide-dependent manner (Gibbs et al., 2014). This modified Cys makes the transcription factors accessible to the N-end rule pathway of protein degradation (Graciet et al., 2009; Holman et al., 2009; Weits et al., 2014). When oxygen becomes limiting, the group VII ERF transcription factors, such as Related to APETALA2.2 (RAP2.2) and RAP2.12, accumulate and activate hypoxia-responsive genes (Licausi et al., 2011). When oxygen is resupplied, the transcriptional response is turned off by oxygen-dependent degradation of the transcription factors. Mutants defective in the N-end rule pathway, namely ate1/ate2 (arginyl-tRNA:protein arginyltransferases), PROTEOLYSIS6 (prt6), and pco1/pco2, show a constitutive hypoxic response (Gibbs et al., 2011; Licausi et al., 2011; Weits et al., 2014).
Phosphate deficiency in plants originates from poor fertilization of the soil in agriculture but also occurs in natural environments (Yang and Finnegan, 2010; Plaxton and Tran, 2011; Lambers et al., 2012). Phosphate is one of the macronutrients required for plant growth and is specifically important for energy transfer (i.e. ATP) and biosynthesis of DNA, RNA, and other molecules. Furthermore, phospholipids are a major sink for phosphate and are abundant constituents of plant membranes, except the chloroplast membranes, which mainly consist of galactolipids.
Upon phosphate starvation, plants induce a number of genes that function in recycling and mobilization of phosphate such as phosphate transporters, acid phosphatases, and nucleases (Nilsson et al., 2010; Plaxton and Tran, 2011). Furthermore, expression of genes coding for enzymes involved in galactolipid and sulfolipid biosynthesis is induced. The two main galactolipids are monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG). Galactolipids are essential for plastid biogenesis, consequently photosynthesis, and, under normal growth conditions, are produced by Monogalactosyldiacylglycerol synthase1 (MGD1) and Digalactosyldiacylglycerol synthase1 (DGD1) in Arabidopsis (Dörmann et al., 1999; Awai et al., 2001; Kobayashi et al., 2007). During phosphate deficiency, the expression of MGD2 and MGD3 as well as DGD1 and DGD2, encoding MGDG synthases and DGDG synthases, respectively, is induced, and DGDG is produced to replace phospholipids in plant membranes to save phosphate (Härtel et al., 2000; Kobayashi et al., 2009; Shimojima and Ohta, 2011).
The regulation of gene expression under phosphate deficiency is not completely understood, but many components of the signal transduction chain have been identified. One important regulator of the phosphate deficiency response is the MYB transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1; At4g28610; Rubio et al., 2001; Bustos et al., 2010). Arabidopsis phr1 knockout mutant plants show a reduced induction of phosphate-responsive genes (Rubio et al., 2001; Gaude et al., 2008; Bustos et al., 2010; Nilsson et al., 2012). However, the expression of PHR1 per se is not influenced by phosphate deficiency (Rubio et al., 2001; Gaude et al., 2008). It has been suggested that sumoylation by SIZ1 (At5g60410) is one important posttranslational modification of Arabidopsis PHR1 (Miura et al., 2005). However, apart from the phosphate deficiency response, SIZ1 activity is generally involved in stress responses (Castro et al., 2012). In addition, other transcription factors such as AtMYB62, ZINC FINGER OF ARABIDOPSIS THALIANA6, BASIC HELIX-LOOP-HELIX32, and AtWRKY75, posttranslational regulatory factors (e.g. belonging to the SYG1/Pho81/XPR1 [SPX] domain family), and microRNA399 are associated with signal transduction under phosphate starvation (Nilsson et al., 2010; Yang and Finnegan, 2010; Jain et al., 2012).
At first glance, it is not expected that plants respond similarly toward oxygen and phosphate deficiency. However, it has been reported that the two stresses share common response pathways (Plaxton and Podestá, 2006). First, ATP is a key metabolite, and its content can be compromised upon exposure to the two stresses. During oxygen deficiency, ATP becomes limiting due to a deficit in mitochondrial respiration (Mustroph et al., 2005, 2006a, 2006b; Branco-Price et al., 2008). A lack of phosphate also limits ATP synthesis (Dancer et al., 1990; Rao and Terry, 1995; Morcuende et al., 2007). Second, the metabolic responses to oxygen and phosphate deprivation show some overlap in a number of plant species. In particular, alternative energy pathway reactions, including those catalyzed by pyrophosphate-Fru-6-phosphate phosphotransferase, pyruvate-orthophosphate dikinase, or Suc synthase, were activated under the two stress conditions (Mustroph et al., 2005; Plaxton and Podestá, 2006; Huang et al., 2008; Plaxton and Tran, 2011). Lastly, root flooding as well as phosphate deficiency can also lead to the accumulation of anthocyanins in leaves, indicating an additional resemblance to high-light acclimation responses (Steyn et al., 2002).
Arabidopsis seedlings display a shoot-specific response to whole-plant hypoxia, as depicted by the differential gene expression of about 50 genes (Mustroph et al., 2009). Interestingly, the whole-seedling response to phosphate deficiency (Misson et al., 2005; Bustos et al., 2010; Woo et al., 2012) overlaps with this response to hypoxia. The group of similarly regulated genes includes several genes associated with galactolipid synthesis. To unravel common response pathways between oxygen and phosphate deficiency in detail, we first studied the signal transduction pathway leading to the gene induction under hypoxia and, secondly, more specifically, the significance of the induction of galactolipid-related genes under oxygen deficiency. Our findings reveal a new regulatory function of PHR1 under oxygen deficiency stress and therefore provide new insights into the regulatory mechanism of PHR1 under phosphate deficiency.
RESULTS
A Part of the Hypoxic Response Is Light Dependent
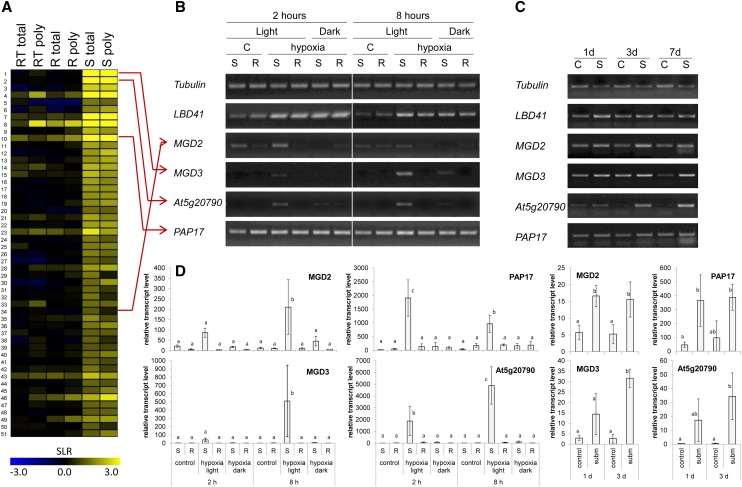
In a previous large-scale microarray analysis, a group of genes was found to be specifically up-regulated in shoot-specific cell types under hypoxia but not in roots or root tips (Mustroph et al., 2009). For further characterization, we selected genes that were hypoxia induced in shoots by use of the following criteria: first, an at least 2-fold induction in shoots (hypoxia [H] versus aeration [C], signal-log ratio > 1, adjusted P value [Padj.] < 0.01), second, an at least 2-fold higher expression in hypoxic shoots compared with hypoxic roots (signal-log ratio > 1, Padj. < 0.01), and third, the ratio of gene induction during hypoxia in shoots was significantly higher than in roots ([shoot H/shoot C]/[root H/root C] > 2, Padj. < 0.1). In total, 51 genes met these criteria (Fig. 1; Supplemental Table S1). Interestingly, several genes in this group showed either an involvement in galactolipid and sulfolipid metabolism (At2g11810, MGD3; At5g20410, MGD2; and At5g01220, sulfoquinovosyldiacylglycerol synthase2 [SQD2]) or another association with phosphate deficiency (At3g47420, phosphate starvation-induced gene3 [G3Pp1] and At3g17790, purple acid phosphatase17 [PAP17]).
Figure 1.
Identification and characterization of a set of genes with shoot-specific response to hypoxia. A, Heat map of 51 genes showing shoot-specific induction during 2 h of hypoxia. Microarray data are signal-log ratios of a hypoxia versus control comparison and were taken from Mustroph et al. (2009), representing root tip (RT), root (R), and shoot (S) tissue, including a change in total RNA (total) and polysomal RNA (poly). Yellow color represents a signal-log ratio of greater than 0, and blue represents a signal-log ratio of less than 0. The list of genes is presented in Supplemental Table S1. B, Expression of selected hypoxia-responsive genes after treatment of seedlings with hypoxia or control (C) for 2 and 8 h in either light or darkness. Roots (R) and shoots (S) were harvested separately. One representative experiment out of eight independent replicates is shown. The photo shows the RT-PCR bands separated by agarose gel electrophoresis and after staining with Serva DNA stain G. C, Expression of selected hypoxia-responsive genes after treatment of plants with submergence (S, subm) for 1, 3, and 7 d in light or without treatment (control [C]). Only leaves were harvested and analyzed. One representative experiment out of five independent replicates is shown. The photo shows the RT-PCR bands separated by agarose gel electrophoresis and after staining with Serva DNA stain G. D, qRT-PCR analysis of selected hypoxia-responsive genes of three independent replicates from B and C. Means and sds are shown for each sample. Transcript level was normalized to Elongation factor 1α (EF1α). Different letters represent significant differences at P < 0.05 (Tukey’s honestly significant difference mean-separation test).
The shoot specificity of gene induction under hypoxia was confirmed for several genes of this set (Fig. 1), and additionally, the effect of light treatment was analyzed. MGD2, MGD3, PAP17, and At5g20790 showed a higher expression in light-exposed shoots after 2 and 8 h of hypoxia than under aerated control conditions but not in darkness or in roots (Fig. 1, B and D). This expression pattern was also observed for G3Pp1, SQD2, and SENESCENCE-RELATED GENE3 (Supplemental Fig. S1). By contrast, the expression of LOB domain-containing protein41 (LBD41), a gene that is generally hypoxia inducible (Mustroph et al., 2009), increased in roots and shoots independently from illumination during hypoxia (Fig. 1B).
Plants were subjected to hypoxic stress by gassing the seedlings with nitrogen gas. When the nitrogen flow during the hypoxic treatment was supplemented with CO2, it was shown that the gene induction was solely dependent on oxygen deficiency rather than on a lack of CO2 in the atmosphere (Supplemental Fig. S2). We also analyzed gene expression in response to submergence-induced hypoxia during a short-day light regime. The expression of the four candidate genes MGD2, MGD3, PAP17, and At5g20790 was induced in leaves after 1, 3, and 7 d of submergence, with transcript levels of MGD3 and At5g20790 increasing more strongly with longer duration of the stress (Fig. 1, C and D).
The Lipid Composition of Membranes Is Modified under Hypoxia
As light-exposed Arabidopsis seedlings showed an increased expression of genes involved in galactolipid and sulfolipid metabolism during hypoxia, we determined the galactolipid and sulfolipid contents of leaves and examined mutants of galactolipid synthesis (Kobayashi et al., 2009) for their survival capacity under oxygen deficiency stress.
Previous experiments on phosphate deficiency demonstrated induction of galactolipid and sulfolipid synthesis genes within 24 to 48 h (Essigmann et al., 1998; Awai et al., 2001; Yu et al., 2002; Kelly et al., 2003; Misson et al., 2005; Kobayashi et al., 2009; Bustos et al., 2010; Woo et al., 2012), while changes in galactolipid and sulfolipid contents were generally monitored only after several days of phosphate deficiency. Because hypoxic incubation in our system caused death of the seedlings after more than 24 h, prolonged stress treatments were only performed under submergence conditions.
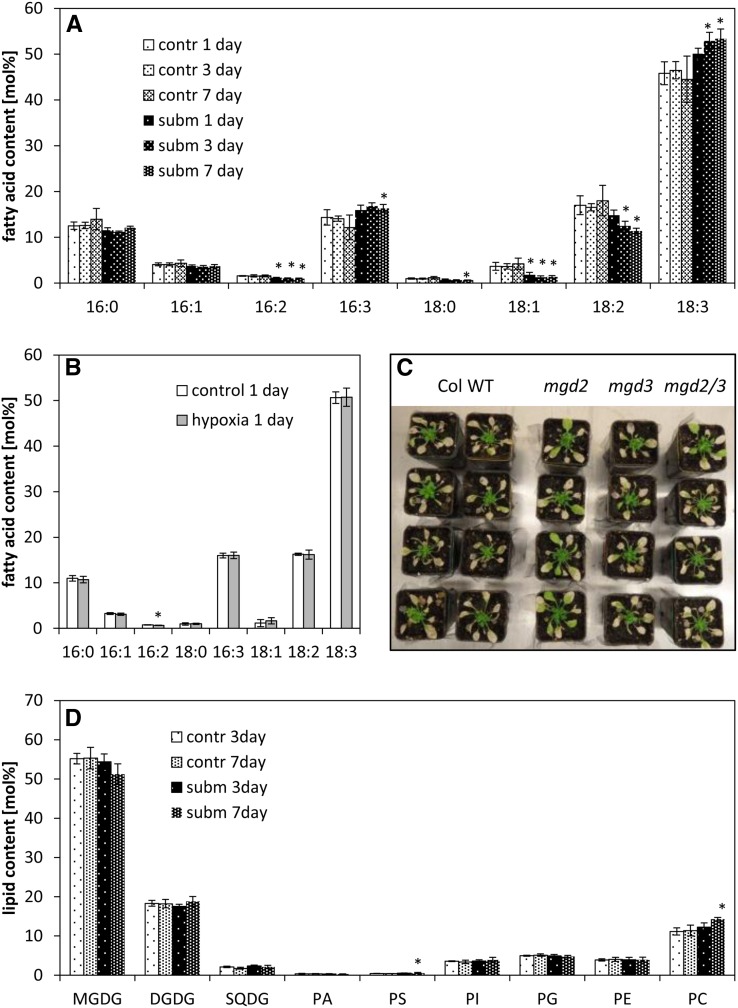
The measurement of fatty acid composition can reveal subtle changes in membrane lipid composition. A 24-h hypoxia treatment did not affect seedling fatty acid composition (Fig. 2B), despite increased transcript levels of MGD2, MGD3, and SQD2. By contrast, 3 and 7 d of submergence caused a shift of the fatty acid composition of rosette leaves toward an increase of desaturated C16 and C18 fatty acids compared with aerated control leaves (Fig. 2A).
Figure 2.
Membrane lipid contents and composition under hypoxia or submergence. A, Fatty acid composition (mol %) of leaves from plants that were exposed to submerged (subm) or control (contr) conditions for 1 to 7 d. Data are means ± sd for four independent biological replicates. Asterisks mark significant differences between control and submerged conditions (P < 0.05, Tukey’s honestly significant difference mean-separation test). B, Fatty acid composition (mol %) of seedlings treated with hypoxic conditions for 24 h. Data are means ± sd for three independent biological replicates. Asterisks mark significant differences between control and hypoxic conditions (P < 0.05, Tukey’s honestly significant difference mean-separation test). C, Survival of the wild type (WT) and mgd2/mgd3 mutants under submergence. Four-week-old plants were submerged for 4 weeks in a short-day rhythm. Photos were taken after 2 weeks of recovery. The experiment was repeated three times with similar results. D, Lipid composition (mol %) of leaves from plants exposed to submerged (subm) or control (contr) conditions for 3 and 7 d. Data are means ± sd for five replicates. Asterisks mark significant differences between control and submerged conditions (P < 0.05, Tukey’s honestly significant difference mean-separation test). Fatty acid composition of each lipid is presented in Supplemental Figure S3. PA, Phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SQDG, sulfoquinovosyldiacylglycerol.
A more detailed analysis of the lipid composition under submergence via quadrupole time-of-flight (Q-TOF) tandem mass spectroscopy measurements revealed no increase in the MGDG content but an increase in phosphatidylcholine (PC; Fig. 2D). Furthermore, submergence changed the molecular species composition of the different lipids. In the phospholipids, particularly in PC and phosphatidylethanolamine, the degree of desaturation was strongly elevated (Supplemental Fig. S3). Interestingly, the content of 34:6 (18:3, 16:3) MGDG slightly increased during submergence, accompanied with the decrease in 36:6 (18:3, 18:3) MGDG, while at the same time, 36:6 (18:3, 18:3) DGDG showed a small increase and 34:3 (18:3, 16:0) DGDG was decreased. These changes in galactolipid composition point toward a redistribution of molecular lipid species originating from the endoplasmic reticulum (ER) and from the chloroplast. The content and composition of sulfolipid (sulfoquinovosyldiacylglycerol) did not change during submergence.
In accordance with the detection of only subtle changes in the galactolipid content of plants exposed to submergence, the survival of the Arabidopsis galactolipid mutants mgd2, mgd3, and the mgd2/mgd3 double mutant under submergence was comparable to the wild type or even better for the mgd2 mutant in some experiments (Fig. 2C). Therefore, despite the induction of MGD2 and MGD3 expression, these genes were not essential for survival under submergence.
The Hypoxic Response Overlaps with the Phosphate Deficiency Response
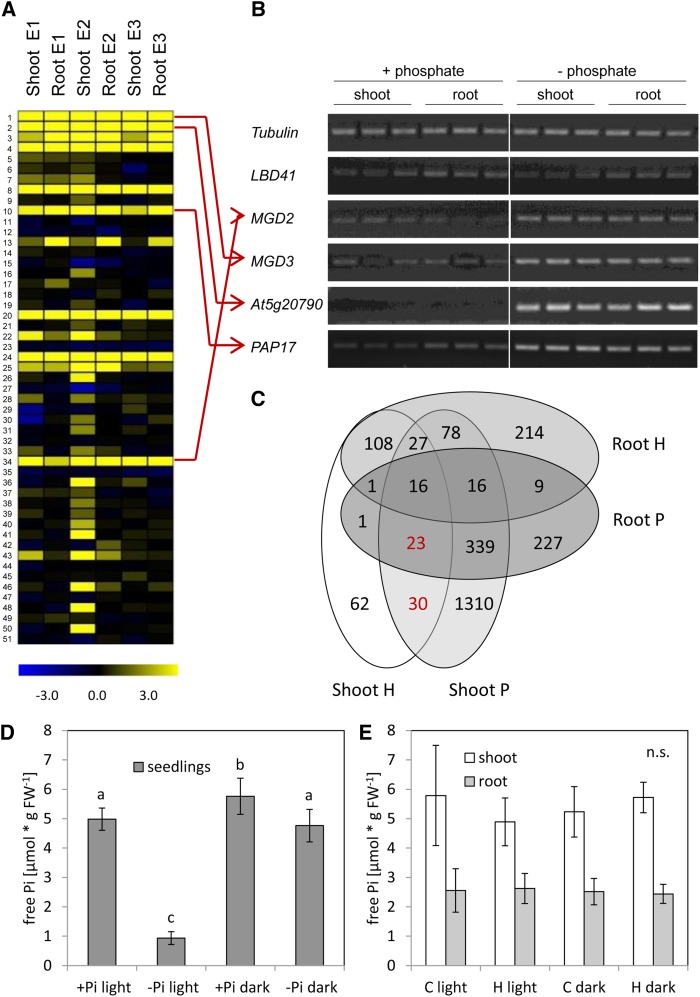
A comparison of the 51 shoot-specific hypoxia-induced genes (Fig. 1A) with published transcriptome analyses performed under phosphate deficiency (Misson et al., 2005; Bustos et al., 2010; Woo et al., 2012) revealed that about 50% of the genes are also induced by phosphate deficiency (Fig. 3A; Supplemental Table S2). Furthermore, a survey of expression data for the 51 genes in Genevestigator (https://www.genevestigator.com/gv/plant.jsp; Hruz et al., 2008) revealed increased expression in various phosphate deficiency experiments under hypoxia and cold treatments (Supplemental Fig. S4). However, many of the genes are induced by phosphate deficiency not only in shoots, but also in roots (Misson et al., 2005; Bustos et al., 2010; Woo et al., 2012), as confirmed here for the candidate genes MGD2, MGD3, At5g20790, and PAP17 (Fig. 3B). To find additional coregulated genes, a direct comparison of transcriptome analyses under oxygen and phosphate deficiency was performed (Mustroph et al., 2009; Bustos et al., 2010). Thus, a group of 53 genes was found to be induced upon hypoxia solely in the shoots and upon phosphate starvation in roots and/or shoots, including PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1, SPX2, and SPX3 (Fig. 3C; Supplemental Table S2).
Figure 3.
Overlap of gene expression observed under hypoxia/submergence and the phosphate deficiency response. A, Heat map of the same 51 genes from Figure 1, containing data from microarrays after phosphate deficiency stress. Data are signal-log ratios of three independent experiments of phosphate deficiency versus control. Experiment 1 (E1) was taken from Misson et al. (2005), experiment 2 (E2) was taken from Bustos et al. (2010), and experiment 3 (E3) was taken from Woo et al. (2012). Yellow represents a signal-log ratio of greater than 0, and blue represents a signal-log ratio of less than 0. The list of genes is presented in Supplemental Table S1. B, Expression of selected genes after treatment of seedlings with phosphate deficiency for 7 d. Roots and shoots were harvested separately. Three independent biological replicates are shown. The photo shows the RT-PCR bands separated by agarose gel electrophoresis and after staining with Serva DNA stain G. C, Overlap of hypoxia-responsive (H; Mustroph et al., 2009) and phosphate deficiency-responsive (P; Bustos et al., 2010) genes. Selection criteria for significantly induced genes were a signal-log ratio greater than 1 and an adjusted P < 0.01 (Supplemental Table S2). D and E, Free phosphate content in seedlings after treatment with phosphate deficiency (–Pi) for 48 h (D) or with hypoxia (H) for 2 h in comparison with aeration (C; E) in light or darkness. For hypoxia, roots and shoots were harvested separately, and for phosphate deficiency, whole seedlings were harvested. Data are mean values ± sd of five to seven replicates. Values with different letters represent significant differences between four treatment conditions for each tissue type (P < 0.05, Tukey’s honestly significant difference mean-separation test). n.s., No significant changes; FW, fresh weight.
Several low-phosphate-responsive genes (Misson et al., 2005; Bustos et al., 2010; Woo et al., 2012), which were not responsive in the hypoxia microarray experiment (Mustroph et al., 2009), were tested for hypoxic induction to extend our gene expression analysis to a larger number of genes. An intermediate time point of 4 h of hypoxic stress was chosen, and transcript levels were compared to an early time point (48 h) of phosphate deficiency stress (Supplemental Fig. S5). All genes examined, β-amylase5, Glc-6-phosphate transporter2, RNase1, FAR-RED-ELONGATED HYPOCOTYL1-LIKE, SQD1, DGD1, and DGD2, were responsive to intermediate hypoxia and short-term phosphate deficiency, while the hypoxia-responsive genes LBD41 and Pyruvate Decarboxylase1 were only induced by hypoxic treatment. These results point to a general activation of the low-phosphate response under illuminated hypoxia.
To rule out that the overlap of gene expression data is due to a change in the phosphate status of plants under hypoxia, the free phosphate (Pi) content in roots and shoots was measured after 2 h of low-oxygen stress, when changes in expression of many low-phosphate-responsive genes were already detectable (Fig. 1, A and D). For comparison, we included a time point of 48 h after transfer to phosphate-free medium. No changes in the amount of Pi were detectable in roots or shoots after 2 h of hypoxia, while the Pi content dramatically decreased by 80% in plants after 48 h of phosphate depletion (Fig. 3, D and E). Interestingly, the level of Pi only decreased by 15% in dark-incubated seedlings on phosphate-free medium compared with phosphate-containing medium.
Gene Expression under Illuminated Hypoxia Depends on PHR1
Although phosphate limitation and oxygen deficiency share a set of commonly regulated genes, the responsiveness of roots and shoots to the two stresses was different. We therefore asked how changes in gene expression in different organs under the two stress conditions were regulated. Previous studies highlighted an involvement of the group VII ERF transcription factors in low-oxygen sensing (Gibbs et al., 2011; Licausi et al., 2011) and of the MYB transcription factors PHR1 (At4g28619) and PHR1-like1 (PHL1; At5g29000) in the low-phosphate response (Rubio et al., 2001; Bustos et al., 2010). Therefore, we analyzed the involvement of these transcription factors in the induction of expression of the four candidate genes under hypoxic stress by use of Arabidopsis mutants.
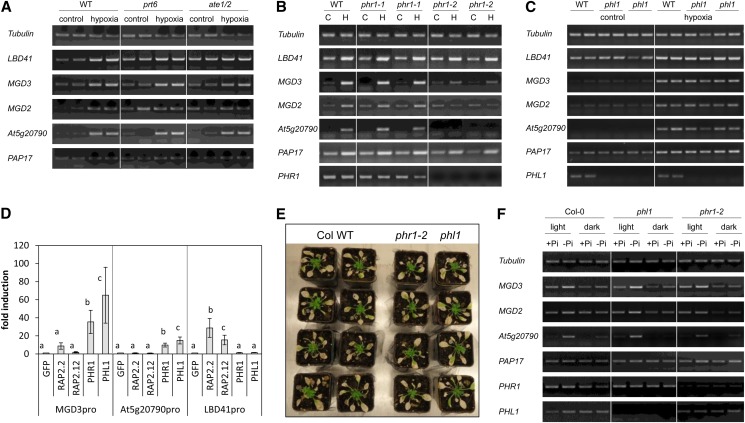
The N-end rule pathway mutants prt6 and ate1/ate2 show a constitutive hypoxic response through constitutive accumulation of group VII ERF transcription factors (Gibbs et al., 2011; Licausi et al., 2011). While these mutants exhibited the expected high expression of the hypoxia-responsive marker gene LBD41 in aerated and hypoxic conditions, none of the four galactolipid-related genes were constitutively expressed but were induced under hypoxia as in the wild type (Fig. 4A). It was concluded that the N-end rule pathway and the group VII ERF transcription factors do not play a critical role in the induction of low-phosphate-responsive genes under illuminated hypoxia. These gene expression data were further confirmed by the evaluation of available microarray data on N-end rule pathway mutants and an overexpression line for the group VII ERF transcription factor RAP2.12 showing only a low number of induced genes that were also low-phosphate responsive (Supplemental Fig. S6).
Figure 4.
The shoot-specific hypoxia response is dependent on PHR1. A, Expression of selected hypoxia-responsive genes after treatment of seedlings with hypoxia for 4 h in light. One prt6 and one ate1/ate2 mutant line (Gibbs et al., 2011) were compared to the wild type (WT). One representative experiment of three independent experiments is shown with two individual samples. B, Expression of selected hypoxia-responsive genes after treatment of seedlings with hypoxia (H) for 4 h in light or without hypoxia treatment (control [C]). Two different phr1 mutant lines were compared to the wild type, and seeds from two individual homozygous plants were used for each mutant line. One representative experiment out of three independent experiments is shown. C, Expression of selected hypoxia-responsive genes after treatment of seedlings with hypoxia for 4 h in light. One phl1 mutant line with seeds from two different homozygous plants was compared to the wild type. One representative experiment out of two independent experiments is shown with two individual samples. D, Promoter activity assay for two shoot-specific hypoxia-responsive promoters (MGD3pro and At5g20790pro) and one core hypoxic promoter (LBD41pro). Protoplasts were transformed with a promoter::Luciferase construct and either GFP or a transcription factor. Luciferase activity was normalized to the reference vector p70SRUC, and the fold change was calculated in comparison to GFP. Different letters represent significant differences at P < 0.05 from eight biological replicates (Tukey’s honestly significant difference mean-separation test). E, Survival of the wild type and phr1-2 or phl1 mutants under submergence. Four-week-old plants were submerged for 4 weeks in a short-day rhythm. Photos were taken after 2 weeks of recovery. The experiment was performed together with the one shown in Figure 2. The experiment was repeated three times with similar results. F, Expression of selected hypoxia-responsive genes after growth of wild-type (WT), phl1, and phr1-2 seedlings on phosphate-deficient medium in light or darkness. The experiment was performed seven (wild type) or two (mutants) times, and one representative experiment is shown. In A, B, D, and F, the photos show the RT-PCR bands separated by agarose gel electrophoresis and after staining with Serva DNA stain G.
Analysis of two independent transfer DNA (T-DNA) insertion alleles for PHR1 revealed a wild-type-like hypoxic response in phr1-1 and a drastically reduced induction of MGD2, MGD3, and At5g20790 in phr1-2, while the hypoxic induction of LBD41 was similar in the wild type and the two phr1 allelic mutants (Fig. 4B). Interestingly, only phr1-2 also showed dramatically reduced mRNA levels of PHR1, while in phr1-1, the expression level of PHR1 was only slightly reduced. Thus, phr1-1 apparently is not a null mutant allele, and we focused on phr1-2 for subsequent experiments. Under aerated conditions, no differences in expression of the four galactolipid biosynthesis genes were observed in phr1-2 compared with the wild type. PHL1 represents another transcription factor involved in the phosphate deficiency response (Bustos et al., 2010). The phl1 knockout mutant plants did not show differences in the response to normoxic or hypoxic conditions compared with the wild type (Fig. 4C).
Promoter transactivation assays were performed to confirm that light-dependent hypoxic gene induction is dependent on PHR1 but not on group VII ERF transcription factors. As shown in Figure 4D, PHR1 and PHL1 were able to induce expression of a luciferase reporter construct under control of the promoters of MGD3 and At5g20790 compared with the control protein GFP, which caused no transcriptional activation. However, the promoter of LBD41, which is solely induced by hypoxia, was not induced by either PHR1 or PHL1 expression. On the other hand, pLBD41::LUCIFERASE (Luc) was responsive to RAP2.2 and RAP2.12 (group VII ERFs) expression, indicating their involvement in the hypoxic response. By contrast, these two transcription factors could not induce the promoters of MGD3 or At5g20790 (Fig. 4D).
PHR1 was essential for the shoot-specific hypoxic response of Arabidopsis (Fig. 4B), but neither the phr1-2 nor the phl1 mutants showed a modified sensitivity to submergence compared with the wild type (Fig. 4E).
The Phosphate Deficiency Response Is Partially Light Dependent
Due to the involvement of light and PHR1 in the hypoxic induction of galactolipid synthesis genes, it was hypothesized that also the phosphate deficiency response could, at least partially, be dependent on illumination. To address this question, 7-d-old light-grown seedlings were transferred to medium with or without phosphate and immediately exposed to light or kept in darkness. After 48 h of darkness, the expression and phosphate starvation-triggered induction of MGD3, MGD2, and At5g20790 were lower than that under illumination (Fig. 4F). However, the gene induction was not completely inhibited under phosphate deficiency in darkness. For PAP17, the trend was similar but not as clear as for the other three genes.
The dark exposure of phr1-2 and phl1 mutants during phosphate deficiency did not completely inhibit the gene induction either. Especially, expression of At5g20790 and MGD3 was still slightly induced compared with the phosphate-supplemented control in darkness (Fig. 4F). Furthermore, the phosphate deficiency response under illumination was still pronounced in the two mutants (Fig. 4F), in contrast to the hypoxia response in phr1-2 (Fig. 4B). These results confirm that PHR1 and PHL1 can partially replace each other under phosphate deficiency (Supplemental Fig. S6; Bustos et al., 2010).
PHR1 Activity Is Potentially Regulated by a Chloroplast-Derived Retrograde Signal
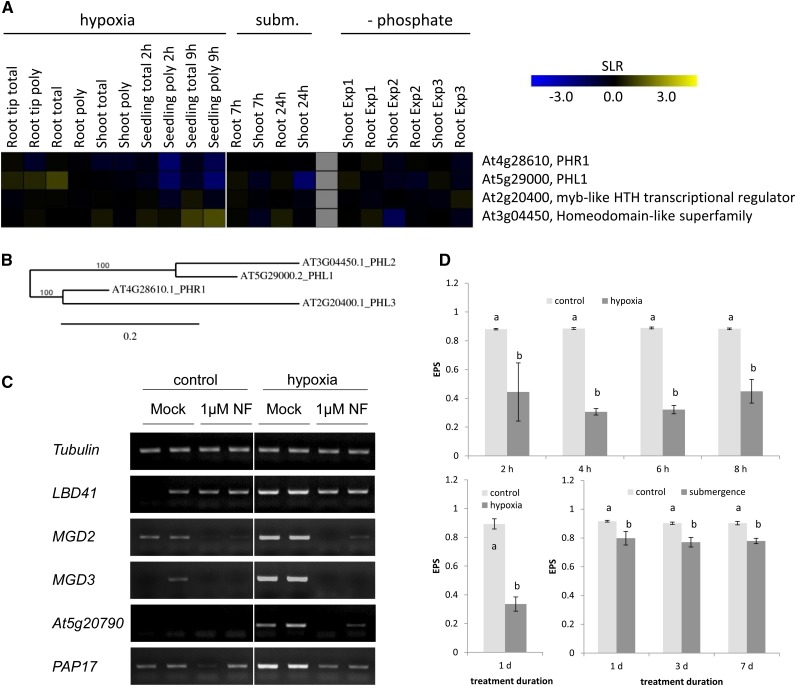
The results presented above demonstrated that part of the hypoxic and phosphate deficiency responses are dependent on PHR1 and illumination. However, the regulatory mechanisms of PHR1 activity are still elusive. A survey of microarray expression data revealed that neither PHR1 nor PHR1-like transcription factors are induced under these two stress conditions (Fig. 5, A and B). Also, translation of these transcription factors under hypoxic conditions is not modified compared with aerated controls (Branco-Price et al., 2008). It is therefore hypothesized that the proteins are regulated at the posttranslational level under stress. The observation of the light dependency of gene induction suggests the involvement of photoreceptors or of a chloroplast-derived retrograde signal that responds to a change in the metabolic status of the chloroplast.
Figure 5.
Evidence for posttranslational regulation of PHR1 activity. A, Heat map for the expression of PHR1 and related genes under oxygen and phosphate deficiency stress. Data are signal-log ratios (SLRs) of a comparison of stress treatment versus control and were taken from the following sources: Mustroph et al. (2009), including a change in total RNA (total) and polysomal RNA (poly; nos. 1–6); Branco-Price et al. (2008; nos. 7–10); Lee et al. (2011; nos. 11–14); Misson et al. (2005; experiment 1, nos. 15 and 16); Bustos et al. (2010; experiment 2, nos. 17 and 18); and Woo et al. (2012; experiment 3, nos. 19 and 20). Yellow represents a signal-log ratio of greater than 0, and blue represents a signal-log ratio of less than 0. B, Phylogenetic tree of MYB transcription factors related to PHR1, constructed by the platform http://www.phylogeny.fr (Dereeper et al., 2008). Protein sequences were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org). C, Expression of selected hypoxia-responsive genes in shoots after treatment of seedlings with hypoxia for 4 h in light. Before stress treatment, plants were placed on norflurazon (NF)-containing medium or control medium for 4 d to inhibit carotenoid biosynthesis. One representative experiment of three independent experiments is shown, with two separate samples each. The photo shows the RT-PCR bands separated by agarose gel electrophoresis and after staining with Serva DNA stain G. D, Xanthophyll epoxidation status (EPS) in leaves from plants exposed to submerged or control conditions for 1, 3, and 7 d and in shoots of seedlings treated with hypoxia for 2 to 8 h or 24 h. The 8-h value is derived from a sample harvested 1 h after the end of the day. Data are means ± sd for four biological replicates. Different letters show significant differences between control and submerged conditions (P < 0.05, Tukey’s honestly significant difference mean-separation test). The pigment contents are presented in Supplemental Table S3.
Therefore, the contribution of the three main groups of photoreceptors to the light-dependent induction of the four candidate genes under hypoxia was tested in double mutants for phototropins (phot1-5 phot2-1; de Carbonnel et al., 2010), cryptochromes (cry2-1 cry1-304; Duek and Fankhauser, 2003), and phytochromes (phyA-211 phyB-9; Cerdán and Chory, 2003). The genes MGD3, MGD2, PAP17, and At5g20790 showed no difference in their expression during normoxic or hypoxic conditions in the photoreceptor mutants compared with the wild type (Supplemental Fig. S7), indicating that photoreceptors are not involved in this signaling pathway.
Second, the involvement of retrograde signaling was analyzed. To this end, the herbicide norflurazon, an inhibitor of phytoene desaturase (Susek et al., 1993; Gray et al., 2003; Saini et al., 2011), was applied to seedlings, and 4 d later, the response to hypoxia was tested. Norflurazon inhibits the synthesis of carotenoids and causes a disintegration of the photosynthetic apparatus. The pretreatment with norflurazon almost completely blocked the induction of all four genes under illuminated hypoxia (Fig. 5C), but it only slightly compromised the phosphate deficiency response of plants, because an induction of the four candidate genes was still visible (Supplemental Fig. S8). This result might be in line with the observation that the phosphate deficiency response was not completely blocked in darkness either (Fig. 4F) or that the pretreatment with norflurazon for only 2 d and the subsequent 2-d low-phosphate treatment under norflurazon addition did not completely block photosynthesis. However, a 4-d pretreatment with norflurazon (as for hypoxia) followed by a 2-d low-phosphate treatment including norflurazon could not be applied, as this caused plant death.
The expression analysis of norflurazon-treated plants suggests a role of a signal coming from active photosynthesis in the regulation of gene expression. Presumably, stress-related modifications in the chloroplast could lead to posttranslational modifications of PHR1. Therefore, the pigment composition of plants exposed to hypoxic conditions was analyzed to monitor photosynthesis-related stress. The chlorophyll and β-carotene content of aerated and hypoxic leaves was unchanged by hypoxic stress, but a dramatic decrease in the epoxidation status of xanthophylls ([violaxanthin + 0.5 × antheraxanthin]/[violaxanthin + antheraxanthin + zeaxanthin]) was observed (Fig. 5D). Zeaxanthin accumulated to about 30 nmol g–1 fresh weight under hypoxic conditions, while its levels were almost undetectable under control conditions (Supplemental Table S3). At the same time, violaxanthin levels dropped by two-thirds. This effect was already visible within 2 h of hypoxic stress and remained at this level for at least 24 h. Submergence caused a similar but less pronounced decrease of the epoxidation state (Fig. 5D). One explanation for this effect is the lack of oxygen required for the epoxidation of zeaxanthin back to violaxanthin. Second, the formation of zeaxanthin points to an increased light sensitivity under hypoxia, although the plants were grown under similar light conditions as the aerated controls. It is suggested that light stress in hypoxia-treated plants initiates a signal to the nucleus via PHR1.
We also analyzed selected metabolites in seedlings treated with hypoxia or phosphate deficiency to detect metabolic changes that could be a simple cause for the common alterations in gene expression. For this, 2-h-hypoxia-treated plants and 48-h-low-phosphate-treated plants were compared to their respective controls (Supplemental Fig. S9). Significant changes in response to stress were observed for the level of ATP, but only for roots under hypoxia, and after a 48-h-dark treatment irrespective of the phosphate status. Only slight modifications in the content of soluble sugars were observed in leaves under hypoxia or in seedlings under phosphate deficiency, which were mainly due to the illumination status. Among the phosphorylated metabolites analyzed, 3-phosphoglycerate (3-PGA) showed a significant increase in hypoxic leaves compared with unstressed leaves and a slight decrease in seedlings under phosphate deficiency. Hexose phosphates were not significantly modified by the stress treatments. The starch content significantly decreased after 48-h darkness by 95% but remained unchanged in response to the phosphate status of the plants or in response to hypoxia (Supplemental Fig. S9). In conclusion, short-term hypoxia or phosphate deficiency only modify the metabolic state of the plant slightly.
DISCUSSION
Oxygen Deficiency and Phosphate Starvation Share a Set of Stress-Induced Genes
We initially observed an organ-specific stress response to oxygen deficiency, consistent with the scenario that roots and leaves have specific and very distinct functions under normoxic conditions. While roots are heterotrophic and are responsible for water and nutrient uptake, the shoots perform photosynthesis and supply photoassimilates for heterotrophic tissues. Therefore, organ-specific responses were expected in hypoxia-treated plants. This is in agreement with previous studies, which already revealed drastic differences in the responses and survival rates of different plant organs under oxygen deficiency stress (Ellis et al., 1999; Mustroph et al., 2006b, 2009; Lee et al., 2011), with photosynthetic organs being generally more tolerant of oxygen deficiency than roots. The light-dependent oxygen generation of photosynthesis was shown to be very important for plant survival, because illumination strongly increased the survival rate of hypoxia-treated or submerged plants (Nabben et al., 1999; Mustroph et al., 2006b; Vashisht et al., 2011).
Several shoot-specific up-regulated genes were identified from a previous comprehensive survey on responses of different cell types and organs of Arabidopsis to hypoxia (Mustroph et al., 2009; Fig. 1). Among these, galactolipid synthesis genes were found, indicating a specific regulation of the chloroplast membranes under hypoxia. Interestingly, a part of this set of genes is known to be induced by phosphate deficiency (Misson et al., 2005; Bustos et al., 2010; Woo et al., 2012; Fig. 3C; Supplemental Table S2). Our experiments demonstrate a tight link between the two stresses, as they result in the PHR1- and illumination-dependent induction of the same signal transduction pathway.
Galactolipids Seem to Play No Major Role in the Survival under Hypoxia
One of the initial questions addresses the role of galactolipids under oxygen deficiency stress. Expression analyses confirmed that the genes MGD2, MGD3, and At5g20790 were induced in shoots under hypoxia in a light-dependent manner but not in roots (Fig. 1). Under submergence, these genes were also induced, albeit more slowly and less pronounced than under artificial nitrogen gassing. The induction of this set of genes during hypoxia or submergence was not observed previously, because most experiments were performed in darkness or with roots to exclude photosynthetically produced oxygen [i.e. hypoxia (Loreti et al., 2005; Bond et al., 2009; Christianson et al., 2009; van Dongen et al., 2009), submergence (Lee et al., 2011), and flooding (Hsu et al., 2011)]. However, when plants were illuminated during hypoxic conditions, expression of these genes was induced (Branco-Price et al., 2008; Mustroph et al., 2009). Consistently, submergence also led to the induction of an ortholog of MGD2 and MGD3 in illuminated rice (Oryza sativa) plants (Qi et al., 2004). This gene induction was not only dependent on illumination, but also observed under additional stresses such as salt and drought stress. MGD genes are induced in rice also under long-term phosphate deficiency (Secco et al., 2013), making this flooding-tolerant plant an ideal model for future studies.
Despite the induction of expression of the galactolipid synthase genes MGD2 and MGD3 and the association of their transcripts with ribosomes indicating active translation under hypoxia (Branco-Price et al., 2008; Mustroph et al., 2009), almost no differences in the galactolipid content were observed under hypoxia or during submergence, although the molecular species compositions of MGDG and DGDG were slightly altered (Fig. 2; Supplemental Fig. S3). Hypoxia causes decreased amounts of 36:6 MGDG but an increase in 34:6 MGDG. On the other hand, 36:6 DGDG accumulated, while 34:3 DGDG decreased (Supplemental Fig. S3). The diacylglycerol moieties employed for galactolipid synthesis in Arabidopsis are derived from two different pathways; 36:6 molecular species are ER derived (eukaryotic lipids), and 34:6/34:3 molecular species are, in general, derived from the chloroplast (prokaryotic lipids). Eukaryotic lipid moieties transported from the ER to the chloroplast outer envelope are readily converted into MGDG by the two MGDG synthases MGD2 and MGD3, both localized in the outer envelope (Awai et al., 2001). Because the major DGDG synthase DGD1 also is located in the outer envelope (Froehlich et al., 2001), 36:6 DGDG can be produced from 36:6 MGDG. Therefore, activation of MGD2 and MGD3 is expected to result in the increased production of 36:6 MGDG, which could be readily converted into 36:6 DGDG during hypoxia by DGD1. The DGD1 gene is slightly induced under intermediate hypoxia (Supplemental Fig. S5). As the total amounts of MGDG and DGDG are under strict control (Dörmann et al., 1995), prokaryotic 34:6 MGDG accumulates, because its conversion into DGDG is reduced. Thus, the induction of MGD2 and MGD3 expression contributes to a balanced galactolipid biosynthetic pathway to meet the altered fluxes of lipid precursors through the prokaryotic or eukaryotic lipid-synthesizing pathways. In contrast to this scenario, upon phosphate deprivation, MGD2 and MGD3 together with DGD1 and DGD2 are strongly expressed to produce large amounts of DGDG for plastidial and extraplastidial membranes (Härtel et al., 2000; Kelly et al., 2003; Li et al., 2006). During phosphate deprivation, expression of two sulfolipid synthesis genes, SQD1 and SQD2, is induced, resulting in the strong increase in sulfolipid content (Essigmann et al., 1998; Yu et al., 2002), but these alterations were not observed under hypoxia (Supplemental Fig. S3).
The finding that the induction of MGD2, MGD3, and other galactolipid synthesis genes does not result in galactolipid accumulation upon hypoxia is in line with the observation that the survival rates of the mgd2 and mgd3 mutants under submergence were not altered (Fig. 2C). So far, only a few studies on the effect of low oxygen concentrations on the fatty acid or lipid compositions of leaves were reported. DGDG was suggested to accumulate in some plant species upon hypoxia, as shown for Amaranthus paniculatus (Knacker and Schaub, 1984). However, these plants were treated only with mild hypoxia (4% oxygen) for an extended period of 21 d.
In addition to changes in galactolipids, the degree of desaturation of the acyl groups in the phospholipids was increased during hypoxia, in particular, PC and phosphatidylethanolamine. Interestingly, a similar modification in fatty acid composition of phospholipids and galactolipids was reported for crown galls produced in Arabidopsis by infection with Agrobacterium tumefaciens. In this hypoxic tissue, the relative amounts of desaturated fatty acids accumulated, which was related to an increased expression of the STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 (SAD6; Klinkenberg et al., 2014). SAD6 belongs to the hypoxic core response (Mustroph et al., 2009), and the induction of its expression is most likely also the cause for the increased degree of lipid desaturation in our analysis (Supplemental Fig. S3).
Taken together, although galactolipids do not seem to play an essential role in plant survival under submergence, they might still be involved in the long-term adaptation to oxygen deficiency under less severe stress conditions. Furthermore, we cannot exclude that galactolipids could play a role in reoxygenation, a severe stress that occurs after the waters recede (Biemelt et al., 1998; Blokhina et al., 1999; Branco-Price et al., 2008). However, in our survival assays, this stress was also present and did not cause a different fitness of the mutants (Figs. 2C and 4E).
Hypoxic Induction of Galactolipid-Related Genes Provides Insight into the Signal Transduction Mechanism under Phosphate Deficiency
Although our results did not provide evidence for an essential role of the induced expression of MGD2, MGD3, and coexpressed genes under hypoxia, the analysis of the control mechanisms of induced gene expression revealed interesting new facts in signaling. Analysis of mutants of the hypoxia signaling pathway (prt6 and ate1/ate2; Gibbs et al., 2011; Licausi et al., 2011) and of the low-phosphate signaling pathway (phr1 and phl1; Rubio et al., 2001; Gaude et al., 2008; Bustos et al., 2010; Nilsson et al., 2012) revealed that only the phosphate-dependent signal transduction pathway, namely the one depending on PHR1, induces MGD2, MGD3, and At5g20790 under illuminated hypoxia, as inferred from mutant transcript analyses and protoplast transactivation assays (Fig. 4).
The involvement of PHR1 in the light-dependent induction of genes under hypoxia suggests also a light dependency of the low-phosphate response. Expression of the tested genes showed a much lower induction under low phosphate in darkness than in light (Fig. 4F), although this response was not completely absent as under hypoxic conditions in darkness (Fig. 1B). A light dependency of the low-phosphate response has been previously observed for lupine (Lupinus albus; Liu et al., 2005) and Arabidopsis (Karthikeyan et al., 2007). Interestingly, in these and other studies (Müller et al., 2005), the effect of light was related to the carbohydrates provided via photosynthesis, because sugar addition mimicked or partially restored the low-phosphate response, even in darkness. In our dark-treated plants, sugars were added to the medium, and a partial response to the stress was observed (Fig. 4F), but this effect was not seen for hypoxic stress (Fig. 1B). Furthermore, phosphate deficiency under continuous darkness did not lead to a massive reduction in the level of Pi in the plants as under illuminated conditions (Fig. 3D). This effect was presumably related to the strong decrease in primary metabolites and starch during a 48-h darkness treatment (Supplemental Fig. S9), thus releasing Pi and therefore ameliorating the phosphate availability. These results, together with the observation that metabolites were not significantly altered due to changes in the phosphate status in our short-term experiments (Supplemental Fig. S9), suggest that the phosphate or sugar level of the cell itself does not alone trigger the transcriptional responses addressed in this study.
We concluded that the signal transduction pathway under illuminated hypoxia and phosphate starvation are in part similar. Most likely, the shoot-specific hypoxia response depends almost solely on PHR1 and a light-dependent signal, while the phosphate deficiency response is only partially dependent on illumination through PHR1. The phosphate deficiency response is not solely limited to shoots and illumination as the hypoxia response (Fig. 1B versus Figs. 3B and 4F), and phr1 mutants respond to oxygen and phosphate deficiency in different ways (Fig. 4B versus Fig. 4F). This is in agreement with earlier observations that phr1 mutants can still partially respond to phosphate starvation (Gaude et al., 2008; Bustos et al., 2010). The remaining part of the response to phosphate deficiency could be either due to PHL1 or to other closely related transcription factors, namely At2g20400 (PHL3) and At3g04450 (PHL2; Fig. 5B), which could also mediate the phosphate deficiency response. Therefore, further experiments with multiple knockouts are required to completely understand the function of this transcription factor family under phosphate deficiency. Additionally, other transcription factors were suggested to be involved in the low-phosphate response (Nilsson et al., 2010; Yang and Finnegan, 2010; Jain et al., 2012).
Although PHR1 and related transcription factors are involved in the signal transduction chain, their expression or translation is not affected by phosphate deficiency or hypoxia (Fig. 5A; Misson et al., 2005; Mustroph et al., 2009; Bustos et al., 2010; Woo et al., 2012). But, until now, no mechanism has been suggested as to how PHR1 responds to low phosphate. Based on the results presented here, we suggest that PHR1 is regulated by light or a light-dependent modification at the posttranslational level, possibly by phosphorylation, ubiquitination, sumoylation, or redox modification in response to a modification of the metabolic status of the chloroplast.
The light dependency of the signal transduction chain might be mediated by (1) a photoreceptor or (2) the status of the chloroplast, generally termed as retrograde signaling. The analysis of photoreceptor mutants revealed that the hypoxia response is independent of known photoreceptors (Supplemental Fig. S7), although we cannot rule out the involvement of additional photoreceptors. As photoreceptors usually detect changes in light quality and quantity, it is unlikely that they could play a major role during oxygen or phosphate deficiency responses because of the similar light conditions used. However, under submergence, the quality and quantity of light could be modified (Sand-Jensen and Krause-Jensen, 1997; Voesenek et al., 2006; Pedersen et al., 2013).
The involvement of retrograde signaling in this signal transduction pathway is more likely. The rate of photosynthesis under hypoxia and submergence strongly differs from the situation in aerated plants (Krause et al., 1985; Mommer et al., 2005; Mustroph et al., 2006b; Colmer et al., 2011). Already 2 h after the onset of hypoxic stress, the epoxidation status of xanthophylls dramatically declined, indicating strong perturbation of photosynthesis (Fig. 5D). The increase in zeaxanthin is an indication for enhanced nonphotochemical quenching due to excessive excitation energy (Wright et al., 2011). Because of the closure of stomata upon oxygen deficiency (Else et al., 1996), CO2 rapidly becomes limiting, and therefore, the Calvin-Benson cycle is strongly inhibited. As a consequence, NADPH from the light reaction accumulates. Because NADH also accumulates in the mitochondria due to the lack of oxygen, compensatory mechanisms such as the malate valve are not functional, and the entire photosynthetic process is disturbed.
Under these conditions, it is likely that retrograde signals are transmitted from the chloroplast to the nucleus to adjust nuclear gene expression. This process is largely independent of CO2 deficiency in the plant external environment because CO2 supplementation to the nitrogen flow did not suppress the induction of the pathway (Supplemental Fig. S2). Furthermore, submergence, where CO2 in the surrounding water is still available in limited amounts (Pedersen et al., 2013), caused a similar but lower response (Fig. 1C). One indication for retrograde signaling is provided with the norflurazon-mediated repression of hypoxia-induced expression of MGD2, MGD3, and At5g20790 (Fig. 5C). We further cannot rule out the involvement of mitochondrial retrograde signaling, which would indicate mitochondrial disturbances that are likely under the two stress conditions, but those would be also restricted to illumination conditions.
Under phosphate deficiency, photosynthesis was also inhibited (Fredeen et al., 1990; Jacob and Lawlor, 1992, 1993; Nilsson et al., 2012), although this response generally occurred at later time points than under hypoxia. Therefore, the induction of responsive genes was also slower (<2 h after the beginning of hypoxia versus >24 h after the treatment start of phosphate deficiency). For example, in Glycine max, inhibition of the Calvin-Benson cycle as well as an increase in the ratio of NADPH/NADP was observed after 19 d of phosphate deficiency (Fredeen et al., 1990), probably because regulation of the enzymes partially depends on phosphate and other phosphate-related metabolites. Similarly, changes in the NADPH/NADP ratio and/or in chlorophyll fluorescence indicated the down-regulation of photosynthesis during long-term phosphate deprivation in sunflower (Helianthus annuus) and maize (Zea mays; Jacob and Lawlor, 1993), bean (Phaseolus vulgaris; Juszczuk and Rychter, 1997), and sugar beet (Beta vulgaris; Rao and Terry, 1995). Furthermore, Chlamydomonas reinhardtii cells showed a similar decrease in the epoxidation state of xanthophylls under phosphate deficiency (Wykoff et al., 1998), as shown here for Arabidopsis under hypoxia (Fig. 5D). Therefore, a chloroplast-derived retrograde signal is likely transmitted to the nucleus to adapt the photosynthetic process to phosphate deficiency conditions as well.
The inhibition of photosynthesis under stress conditions also may lead to changes in metabolites. We could only detect minor changes in sugars and hexose phosphates and observed a stable ATP content in leaves after 2-h hypoxia or after 48 h of phosphate deficiency (Supplemental Fig. S9), when modifications in gene expression were already detectable. Therefore, the induction of the signal transduction pathway cannot be easily associated with a modification in primary metabolites or a change in Pi under hypoxia (Fig. 3E). Possibly, the ratio of 3-PGA/Pi could serve as a signal, as it increased under the two stress conditions. Levels of 3-PGA were significantly increased under hypoxia, while they tended to decrease under phosphate deficiency, a trend observed previously (low Pi, Fredeen et al., 1990; Jacob and Lawlor, 1992; Rao and Terry, 1995; and hypoxia, Biemelt et al., 1999; Shingaki-Wells et al., 2011; Ampofo-Asiama et al., 2014).
Recent studies with Arabidopsis indicate that PHR1 is mainly responsible for the adaptation of photosynthesis to low-phosphate conditions, including the accumulation of anthocyanins (Nilsson et al., 2012). Therefore, it is hypothesized that short-term hypoxia and long-term phosphate deficiency result in a comparable disturbance of photosynthesis that leads to the modification of expression of genes, including galactolipid-related genes, via the activation of PHR1.
CONCLUSION
The data presented here on the shoot-specific hypoxia response of Arabidopsis, including the induction of galactolipid synthesis genes, are only associated with a minor modification of galactolipid levels under hypoxia or submergence. The data suggest that the transcription factor PHR1 is a new component of the plant response to illuminated hypoxia. It is hypothesized that PHR1 is regulated at the posttranslational level via a retrograde signal derived from the chloroplast, originating from the impaired photosynthesis during oxygen or phosphate deficiency. The complete elucidation of the complex nature of signaling mechanisms as well as the signal transduction to the transcription factor PHR1 remains to be solved.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. Mutant alleles of mgd2-1, mgd3-1 (SALK_084186), and mgd2 mgd3 were described previously (Kobayashi et al., 2009). T-DNA insertions of the genes PHR1 (At4g28610; phr1-1, SALK_112862, containing a T-DNA insertion in intron 2 of PHR1; phr1-2, SALK_067629C, T-DNA insertion in exon 2 of PHR1) and PHL1 (At5g29000; SAIL_731_B09) in the Col-0 background were obtained from the Nottingham Arabidopsis Stock Centre. The N-end rule pathway mutants prt6 and ate1/ate2 have been described previously (Graciet et al., 2009; Holman et al., 2009), as well as photoreceptor mutants (phyA-211 phyB-9, Cerdán and Chory, 2003; phot1-5 phot2-1, de Carbonnel et al., 2010; cry2-1 cry1-304, Duek and Fankhauser, 2003).
phr1-1, phr1-2, and phl1 mutant lines were screened for homozygous plants using specific primers (Supplemental Table S4). The positions of the T-DNA insertions were confirmed by sequencing the PCR fragment downstream of the T-DNA border.
For all growth conditions, surface-sterilized seeds were incubated for 3 d at 4°C in darkness after seeding them on Murashige and Skoog (MS) petri dishes (1× MS salts, 0.8% [w/v] agar, and 1% [w/v] Suc, pH 5.7) and sealing the dishes with Leucopore tape (Duchefa). Germination and seedling growth was carried out in growth chambers at 23°C with a 16-h photoperiod and about 100 µmol photons m–2 s–1. For phosphate starvation experiments, seedlings grown under control conditions on solidified MS medium were transferred to solidified control (2.5 mm Pi) or Pi-deficient (0 mm Pi) medium (Härtel et al., 2000) at the age of 7 d and grown for another 2 to 10 d. For light/dark treatments on Pi-differing medium, seedlings were transferred to the stress conditions 2 h after the onset of the photoperiod, and for dark incubation, plates were wrapped in aluminum foil immediately after transfer of the seedlings. Growth was continued under long-day conditions for indicated time periods.
Hypoxia Treatments
Seven-day-old seedlings were used for oxygen deprivation experiments starting 2 h after the onset of the photoperiod. The sealing of the petri dishes was removed, and plants were either treated with a pure nitrogen atmosphere or kept under ambient air control conditions. For the nitrogen treatments, plants were placed into a desiccator and continuously flushed with nitrogen gas that was passed through a water phase before entering the environment of the plants and being drained off the desiccator at the top of the vessel. Because the nitrogen flow cannot remove the oxygen inside the plants within the first minutes, and oxygen production via photosynthesis is well possible, this treatment is termed hypoxia rather than anoxia. For experiments with differing light conditions, hypoxia treatments were performed either with the desiccator covered with lightproof black tissue or under illumination with a fluorescent lamp of 11 W. Control experiments were conducted under the same light conditions as illuminated hypoxia treatments. After treatments for the times as indicated, plant material was collected and immediately frozen in liquid nitrogen. If needed, roots of the seedlings were separated from the shoots using a razor blade. In case of carbon dioxide supply during the hypoxic treatment, the water phase on bottom of the desiccator was replaced by a freshly prepared solution of 0.5 m K2CO3 and 1.5 m KHCO3.
Submergence of Plants
For submergence experiments, seedlings were transplanted to separate pots containing soil, vermiculite, and sand in a 2:1:1 (soil:vermiculite:sand) volume mixture at 7 d after germination. The pots were covered with a thin black mesh, and plants were transplanted through a small hole in the center of the mesh. Before the onset of the experiments, growth under control conditions was continued as described above for 4 weeks, with the exception that the photoperiod was changed to 8 h to prevent flowering of the plants. Plants were submerged in plastic tubs filled with temperature-adjusted water 3 h after the start of the photoperiod. Controls were kept under air conditions and normal irrigation. The light source and photoperiod were not changed during the treatments. For subsequent gene expression and lipid analyses, rosette leaves from submerged and air-treated plants were collected after submergence for the times indicated. For survival assays, pots with plants were taken out of the tubs after 3 and 4 weeks of submergence, and the plants were allowed to recover in air for 2 weeks in the same growth chamber. Plants were considered dead or alive based on the ability to develop and maintain green tissue within this time period.
RNA Isolation and Reverse Transcription (RT)-PCR
RNA was extracted from rosette leaves of Arabidopsis plants or from dissected root and shoot material of seedlings using the phenol-based reagent TRIsure (Bioline) according to the manufacturer’s instructions. RNA quality and quantity were assessed by screening the A260 and A280. In semiquantitative PCR analysis, 1 to 3 µg of total RNA was reverse transcribed with oligo(dT) using RevertAid Reverse Transcriptase (Fermentas). The optimal cycle number was determined for each primer pair to avoid saturation of the PCR reaction, and results of selected experiments were also verified by quantitative reverse transcription (qRT)-PCR (Fig. 1). For qRT-PCR analysis, 1 µg of total RNA was used for the synthesis of complementary DNA and diluted by a factor of 50 before use as a template in the quantitative PCR. The quantitative PCR was performed with the SensiFAST SYBR and Fluorescein Mix (Bioline) on a MyiQ Single-Color Real-time PCR Detection System (Bio-Rad) and analyzed via the iQ5 Optical System Software (Bio-Rad). EF1α was used as a reference gene, and the primers were described previously (Vert et al., 2003). For EF1α, MGD2, and MGD3, the primers were designed in such a way that they do not anneal with genomic DNA. The primer sequences for all candidate genes are listed in Supplemental Table S4. The relative expression was determined using the following formula: relative transcript level = 1,000 × 2–∆CT, with CT as the threshold cycle (CT) value and dCT as the difference between the CT of the target gene and the CT of EF1α (Becher et al., 2004).
Norflurazon Treatments
For the analysis of hypoxia response upon influence of norflurazon, seedlings were grown on MS control plates for 4 d and subsequently transferred to MS plates containing 1 µm norflurazon or 0.05% (v/v) ethanol as a mock control. After an additional 4 d of growth, hypoxia treatments were performed as described above. For phosphate deficiency, 4 d of pretreatment with norflurazon were not applicable because an additional 2 d of low-phosphate treatment on norflurazon-containing plates almost completely killed the plants. Therefore, 5-d-old seedlings were pretreated for only 2 d on MS plates containing 1 µm norflurazon or 0.05% (v/v) ethanol and then transferred for another 2 d on phosphate deficiency medium with or without norflurazon.
Lipid Extraction and Analysis
Plant material was used for the preparation of fatty acid methyl esters with 1 n methanolic HCl according to Browse et al. (1986). Methyl esters were quantified by gas chromatography using pentadecanoic acid (15:0) as internal standard (Browse et al., 1986).
Phospholipids, sulfolipid, and galactolipids were quantified by Q-TOF mass spectrometry. Briefly, fresh leaf material (approximately 100 mg) from submerged or control plants was collected after 3 and 7 d of treatment and immediately boiled for 15 min in glass vials containing 1 mL of water to destroy any lipase activity. Lipids from boiled leaves were extracted in two steps by vigorous shaking and subsequent centrifugation with CHCl3:CH3OH (1:2) and CHCl3:CH3OH (2:1), respectively. CHCl3 fractions were pooled, and CH3OH:NaCl was added to a final CHCl3:CH3OH:NaCl ratio of 2:1:0.75. After centrifugation (13,000g, room temperature, 1.5 min), the lipid fraction was collected and stored at –20°C until use. Lipids were introduced into the Q-TOF mass spectrometer (Agilent Q-TOF) by nanospray infusion, and phospholipids and galactolipids were quantified by tandem mass spectroscopy experiments using internal standards (Welti et al., 2002; Gasulla et al., 2013).
Metabolite Extraction and Analysis
Plant material was ground in liquid nitrogen, and metabolites were extracted with perchloric acid as previously described (Mustroph et al., 2006a). The contents of Glc, Fru, Suc, starch, and adenylates were determined as in Mustroph et al. (2006a) and phosphorylated metabolites as in Burrell et al., (1994). The content of Pi was determined as described by Itaya and Ui (1966).
Vector Construction and Purification
The fusion constructs with N-terminal hemagglutinin (HA)-tagged effectors p35S:PHR1-HA, p35S:PHL1-HA, p35S:HA-RAP2.2, and p35S:HA-RAP2.12 as well as all cloning vectors have been described elsewhere (Ehlert et al., 2006; Wehner et al., 2011). To create an HA-tagged effector control, the GFP coding sequence was recombined into the Gateway vector p35S:HA-GW (Ehlert et al., 2006). For the construction of the Firefly-Luciferase reporters pBT10-MDG3pro:LUC, pBT10-At5g20790pro:LUC, and pBT10-LBD41pro:LUC, the 5′ upstream sequences (promoter) of MGD3, At5g20790, and LBD41 (–1,231, –719, and –1,649 bp from ATG, respectively) were amplified from genomic DNA of Arabidopsis Col-0, using specific PCR primer pairs (Supplemental Table S4). The MGD3pro PCR product was digested with EcoRI and NcoI, whereas At5g20790pro and LBD41pro were digested with NcoI and BamHI for insertion into the vector pBT10GAL4UAS (Wehner et al., 2011). For each construct, the same respective restriction enzymes were used to cut the vector and remove the GAL4UAS before ligation. For normalization of gene expression, the vector p70SRUC (Stahl et al., 2004) was used. Plasmids were purified using the NucleoBond PC 500 Midi Kit (Macherey-Nagel) and stored at –20°C until use.
Protoplast Transformation and Luminescence Measurement
For mesophyll protoplast isolation and transient transfection, all buffers and solutions were prepared as described previously (Wehner et al., 2011) and used at room temperature. Sixty rosette leaves from Arabidopsis Col-0 plants, grown for 3 to 4 weeks on soil under an 8-h-light/16-h-dark cycle (about 100 µmol m–2 s–1), were sliced into 1-mm strips and submerged in 10 mL of enzyme solution. A vacuum (10 mm Hg) was applied and maintained for 4 h without any perturbation. After slow reaeration, the cell suspension was placed on a rocking table for 5 min at 300 rpm and filtered through a 63-µm steel mesh to separate protoplasts from remaining tissues. Cells were pelleted at 120g for 2 min in a 50-mL tube and washed in 10 mL of W5 (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES, pH 5.7) solution. After repetition of the wash step, protoplasts were pelleted at 120g for 1 min and resuspended in MMG (0.4 m mannitol, 15 mm MgCl2, and 4 mm MES, pH 5.7) solution to a final concentration of 3.5 × 105 cells mL–1. For each transformation, the reporter plasmid, the effector plasmid, and the normalization vector p70SRUC (Stahl et al., 2004) were premixed in a 2-mL round-bottom reaction tube. For basal promoter activity measurement, the effector was replaced with a 35S:HA-GFP-containing vector.
Two hundred microliters of protoplast suspension was added and mixed with the DNA by inverting six times. Subsequently, 220 µL of polyethylene glycol solution (40% polyethylene glycol 4000, 0.2 m mannitol, and 0.1 m CaCl2) was added and mixed by inverting 15 times. After 20-min incubation, the reaction was stopped by dilution with 800 µL of W5 solution. Cells were centrifuged at 120g for 1 min and resuspended in 300 µL of WI (0.5 m mannitol, 20 mm KCl, and 4 mm MES, pH 5.7) solution. To allow for protein synthesis, protoplasts were incubated for 18 h overnight in a climate chamber with a 16-h-light/8-h-dark cycle.
Measurements of the Firefly Luciferase reporter and Renilla Luciferase normalization vector activities were performed using the Beetle-Juice BIG KIT and the Renilla-Juice BIG KIT, respectively (PJK). WI supernatant was removed and replaced by 100 µL of 1× Lysis-Juice2 at 4°C. Lysis was supported by repeated pipetting before the extract was centrifuged for 1 min at 13,000g at room temperature. Twenty microliters of the supernatant was mixed with 100 µL of the respective substrate solution in a transparent polystyrol vial, directly before measurement. Light emission was measured with the Lumac Biocounter M2010 within an integration time of 10 s. The Firefly to Renilla Luciferase (fLUC-rLUC) ratio was calculated for each sample (Pape et al., 2010), and fold induction values are fLUC-rLUC ratios in relation to the basal promoter control without addition of a transcription factor but with GFP.
Pigment Extraction and Analysis
Frozen tissues were homogenized (frequency, 30 s–1, 2 × 1 min) under frozen conditions using the Mill MM300 (Retsch) with two stainless steel beads in each sample. The powdered samples were suspended in extraction buffer (90% acetone; 10% NH4OH [0.1 m]) under low light on ice. After centrifugation, supernatants were transferred to new collection tubes and kept on ice in darkness.
Chlorophyll a, chlorophyll b, and carotenoids were separated by a HPLC system (Agilent 1100) on a Prontosil 200-3-C30 (Bischoff chromatography) column (3 μm; 250 × 4.6 mm; 21°C) at a flow rate of 1 mL min–1 and eluted with a gradient of solvent A (90% acetonitrile, 10% water, and 0.1% triethylamine) and solvent B (100% ethyl acetate). The eluted samples were monitored by a diode array detector at an absorption wavelength of 440 nm (peak width, 10 Hz; slit width, 4 nm). Pigments were identified and quantified by their absorption and compared with authentic standards.
Arabidopsis accession numbers of the genes studied in this article are presented in Supplemental Table S4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of additional genes with shoot-specific response to hypoxia.
Supplemental Figure S2. The gene expression changes are not due to carbon dioxide deficiency.
Supplemental Figure S3. Lipid composition of membranes after submergence.
Supplemental Figure S4. Overlap of gene expression with other stresses.
Supplemental Figure S5. Expression analysis of additional low-Pi-responsive genes under hypoxia and phosphate deficiency.
Supplemental Figure S6. Gene expression in different signaling mutants.
Supplemental Figure S7. Photoreceptors are not important for hypoxic gene induction under illumination.
Supplemental Figure S8. The effect of norflurazon-mediated inhibition of photosynthesis on the phosphate deficiency response.
Supplemental Figure S9. Metabolite levels under hypoxia and phosphate deficiency.
Supplemental Table S1. Expression data for the 51 genes of the heat maps in Figures 1 and 3.
Supplemental Table S2. Selection of oxygen or phosphate deficiency response genes presented in Figure 3C.
Supplemental Table S3. Modification of photosynthesis pigments under submergence.
Supplemental Table S4. Primers and Arabidopsis accession numbers used in this study.
Supplementary Material
Acknowledgments
We thank Wolfgang Dröge-Laser for providing the vectors for the luciferase assay, Christian Fankhauser for seeds for the photoreceptor mutants, Julia Bailey-Serres for the N-end rule pathway mutants, Mie Shimojima for the galactolipid mutants, and Rashmi Sasidharan for critical reading of the manuscript.
Glossary
- Pi
inorganic phosphate
- RT
reverse transcription
- qRT
quantitative reverse transcription
- ER
endoplasmic reticulum
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- PC
phosphatidylcholine
- T-DNA
transfer DNA
- 3-PGA
3-phosphoglycerate
- Col-0
ecotype Columbia
- MS
Murashige and Skoog
- Q-TOF
quadrapole time-of-flight
Footnotes
This work was supported by the German Research Foundation (grant no. MU 2755/4–1) and the Stifterverband für die Deutsche Wissenschaft (grant no. H140 5409 9999 15625).
The online version of this article contains Web-only data.
References
- Ampofo-Asiama J, Baiye VMM, Hertog MLATM, Waelkens E, Geeraerd AH, Nicolai BM. (2014) The metabolic response of cultured tomato cells to low oxygen stress. Plant Biol (Stuttg) 16: 594–606 [DOI] [PubMed] [Google Scholar]
- Awai K, Maréchal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT. (2012) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U. (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Biemelt S, Hajirezaei MR, Melzer M, Albrecht G, Sonnewald U. (1999) Sucrose synthase activity does not restrict glycolysis in roots of transgenic potato plants under hypoxic conditions. Planta 210: 41–49 [DOI] [PubMed] [Google Scholar]
- Biemelt S, Keetman U, Albrecht G. (1998) Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiol 116: 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina OB, Fagerstedt KV, Chirkova TV. (1999) Relationships between lipid peroxidation and anoxia tolerance in a range of species during post-anoxic reaeration. Physiol Plant 105: 625–632 [Google Scholar]
- Bond DM, Wilson IW, Dennis ES, Pogson BJ, Jean Finnegan E. (2009) VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J 59: 576–587 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Burrell M, Mooney P, Blundy M, Carter D, Wilson F, Green J, Blundy K, Rees T. (1994) Genetic manipulation of 6-phosphofructokinase in potato tubers. Planta 194: 95–101 [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Tavares RM, Bejarano ER, Azevedo H. (2012) SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol Life Sci 69: 3269–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Winkel A, Pedersen O. (2011) A perspective on underwater photosynthesis in submerged terrestrial wetland plants. AoB Plants 2011: plr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer J, Veith R, Feil R, Komor E, Stitt M. (1990) Independent changes of inorganic pyrophosphate and the ATP/ADP or UTP/UDP ratios in plant cell suspension cultures. Plant Sci 66: 59–63 [Google Scholar]
- de Carbonnel M, Davis P, Roelfsema MRG, Inoue SI, Schepens I, Lariguet P, Geisler M, Shimazaki KI, Hangarter R, Fankhauser C. (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol 152: 1391–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Balbo I, Benning C. (1999) Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284: 2181–2184 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48: 223–250 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W. (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46: 890–900 [DOI] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else MA, Tiekstra AE, Croker SJ, Davies WJ, Jackson MB. (1996) Stomatal closure in flooded tomato plants involves abscisic acid and a chemically unidentified anti-transpirant in xylem sap. Plant Physiol 112: 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredeen AL, Raab TK, Rao IM, Terry N. (1990) Effects of phosphorus nutrition on photosynthesis in Glycine max (L.) Merr. Planta 181: 399–405 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Benning C, Dörmann P. (2001) The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J Biol Chem 276: 31806–31812 [DOI] [PubMed] [Google Scholar]
- Gasulla F, Vom Dorp K, Dombrink I, Zähringer U, Gisch N, Dörmann P, Bartels D. (2013) The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach. Plant J 75: 726–741 [DOI] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P. (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Ghelis T. (2011) Signal processing by protein tyrosine phosphorylation in plants. Plant Signal Behav 6: 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet E, Walter F, Ó’Maoiléidigh DS, Pollmann S, Meyerowitz EM, Varshavsky A, Wellmer F. (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA 106: 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C. (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman TJ, Jones PD, Russell L, Medhurst A, Ubeda Tomás S, Talloji P, Marquez J, Schmuths H, Tung SA, Taylor I, et al. (2009) The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 106: 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Peng HP, Chou SJ, Shih MC. (2011) Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 6: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Itaya K, Ui M. (1966) A new micromethod for the colorimetric determination of inorganic phosphate. Clin Chim Acta 14: 361–366 [DOI] [PubMed] [Google Scholar]
- Jacob J, Lawlor DW. (1992) Dependence of photosynthesis of sunflower and maize leaves on phosphate supply, ribulose-1,5-bisphosphate carboxylase/oxygenase activity, and ribulose-1,5-bisphosphate pool size. Plant Physiol 98: 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Lawlor DW. (1993) In vivo photosynthetic electron transport does not limit photosynthetic capacity in phosphate-deficient sunflower and maize leaves. Plant Cell Environ 16: 785–795 [Google Scholar]
- Jain A, Nagarajan VK, Raghothama KG. (2012) Transcriptional regulation of phosphate acquisition by higher plants. Cell Mol Life Sci 69: 3207–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczuk IM, Rychter AM. (1997) Changes in pyridine nucleotide levels in leaves and roots of bean plants (Phaseolus vulgaris L.) during phosphate deficiency. J Plant Physiol 151: 399–404 [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. (2007) Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225: 907–918 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Froehlich JE, Dörmann P. (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg J, Faist H, Saupe S, Lambertz S, Krischke M, Stingl N, Fekete A, Mueller MJ, Feussner I, Hedrich R, et al. (2014) Two fatty acid desaturases, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 and FATTY ACID DESATURASE3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol 164: 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knacker T, Schaub H. (1984) The effect of low oxygen concentration on the acyl-lipid and fatty-acid composition of the C4 plant Amaranthus paniculatus L. Planta 162: 441–449 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H. (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. (2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Muthuramalingam M, Dietz KJ. (2012) Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr Opin Plant Biol 15: 261–268 [DOI] [PubMed] [Google Scholar]
- Krause GH, Köster S, Wong SC. (1985) Photoinhibition of photosynthesis under anaerobic conditions studied with leaves and chloroplasts of Spinacia oleracea L. Planta 165: 430–438 [DOI] [PubMed] [Google Scholar]
- Lambers H, Cawthray GR, Giavalisco P, Kuo J, Laliberté E, Pearse SJ, Scheible WR, Stitt M, Teste F, Turner BL. (2012) Proteaceae from severely phosphorus-impoverished soils extensively replace phospholipids with galactolipids and sulfolipids during leaf development to achieve a high photosynthetic phosphorus-use-efficiency. New Phytol 196: 1098–1108 [DOI] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Li M, Welti R, Wang X. (2006) Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol 142: 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Liu J, Samac DA, Bucciarelli B, Allan DL, Vance CP. (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41: 257–268 [DOI] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommer L, Pons TL, Wolters-Arts M, Venema JH, Visser EJ. (2005) Submergence-induced morphological, anatomical, and biochemical responses in a terrestrial species affect gas diffusion resistance and photosynthetic performance. Plant Physiol 139: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Müller R, Nilsson L, Nielsen LK, Hamborg Nielsen T. (2005) Interaction between phosphate starvation signalling and hexokinase-independent sugar sensing in Arabidopsis leaves. Physiol Plant 124: 81–90 [Google Scholar]
- Mustroph A, Albrecht G, Hajirezaei M, Grimm B, Biemelt S. (2005) Low levels of pyrophosphate in transgenic potato plants expressing E. coli pyrophosphatase lead to decreased vitality under oxygen deficiency. Ann Bot (Lond) 96: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJJ, Harren FJM, Albrecht G, Grimm B. (2006a) Organ-specific analysis of the anaerobic primary metabolism in rice and wheat seedlings. I. Dark ethanol production is dominated by the shoots. Planta 225: 103–114 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJJ, Harren FJM, Pörs Y, Grimm B. (2006b) Organ specific analysis of the anaerobic primary metabolism in rice and wheat seedlings. II. Light exposure reduces needs for fermentation and extends survival during anaerobiosis. Planta 225: 139–152 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabben RHM, Blom CWPM, Voesenek LACJ. (1999) Resistance to complete submergence in Rumex species with different life histories: the influence of plant size and light. New Phytol 144: 313–321 [Google Scholar]
- Nilsson L, Lundmark M, Jensen PE, Nielsen TH. (2012) The Arabidopsis transcription factor PHR1 is essential for adaptation to high light and retaining functional photosynthesis during phosphate starvation. Physiol Plant 144: 35–47 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Pape S, Thurow C, Gatz C. (2010) The Arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1. Plant Physiol 154: 1805–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen O, Colmer TD, Sand-Jensen K. (2013) Underwater photosynthesis of submerged plants: recent advances and methods. Front Plant Sci 4: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Podestá FE. (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Plaxton WC, Tran HT. (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Yamauchi Y, Ling J, Kawano N, Li D, Tanaka K. (2004) Cloning of a putative monogalactosyldiacylglycerol synthase gene from rice (Oryza sativa L.) plants and its expression in response to submergence and other stresses. Planta 219: 450–458 [DOI] [PubMed] [Google Scholar]
- Rao IM, Terry N. (1995) Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet (IV. Changes with time following increased supply of phosphate to low-phosphate plants). Plant Physiol 107: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LLE, Clarke AK, Straus M, Apel K. (2011) ‘happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65: 690–702 [DOI] [PubMed] [Google Scholar]
- Sand-Jensen K, Krause-Jensen D. (1997) Broad-scale comparison of photosynthesis in terrestrial and aquatic plant communities. Oikos 80: 203 [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J. (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Yan J, Xie D. (2012) Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol 15: 84–91 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Ohta H. (2011) Critical regulation of galactolipid synthesis controls membrane differentiation and remodeling in distinct plant organs and following environmental changes. Prog Lipid Res 50: 258–266 [DOI] [PubMed] [Google Scholar]
- Shingaki-Wells RN, Huang S, Taylor NL, Carroll AJ, Zhou W, Millar AH. (2011) Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol 156: 1706–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Loake GJ. (2011) Redox-based protein modifications: the missing link in plant immune signalling. Curr Opin Plant Biol 14: 358–364 [DOI] [PubMed] [Google Scholar]
- Stahl DJ, Kloos DU, Hehl R. (2004) A sugar beet chlorophyll a/b binding protein promoter void of G-box like elements confers strong and leaf specific reporter gene expression in transgenic sugar beet. BMC Biotechnol 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D, Hesselink A, Pierik R, Ammerlaan JMH, Bailey-Serres J, Visser EJW, Pedersen O, van Zanten M, Vreugdenhil D, Jamar DCL, et al. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C. (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinocur B, Altman A. (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16: 123–132 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJM. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Wehner N, Hartmann L, Ehlert A, Böttner S, Oñate-Sánchez L, Dröge-Laser W. (2011) High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J 68: 560–569 [DOI] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F. (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. (2002) Profiling membrane lipids in plant stress responses. Role of phospholipase D α in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH. (2012) The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AH, DeLong JM, Gunawardena AHLAN, Prange RK. (2011) The interrelationship between the lower oxygen limit, chlorophyll fluorescence and the xanthophyll cycle in plants. Photosynth Res 107: 223–235 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR. (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM. (2010) Regulation of phosphate starvation responses in higher plants. Ann Bot (Lond) 105: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.