A multiple-gene locus in soybean conferring disease resistance has evolved three locus types defined by copy number, protein coding, expression, and DNA methylation differences.
Abstract
Copy number variation of kilobase-scale genomic DNA segments, beyond presence/absence polymorphisms, can be an important driver of adaptive traits. Resistance to Heterodera glycines (Rhg1) is a widely utilized quantitative trait locus that makes the strongest known contribution to resistance against soybean cyst nematode (SCN), Heterodera glycines, the most damaging pathogen of soybean (Glycine max). Rhg1 was recently discovered to be a complex locus at which resistance-conferring haplotypes carry up to 10 tandem repeat copies of a 31-kb DNA segment, and three disparate genes present on each repeat contribute to SCN resistance. Here, we use whole-genome sequencing, fiber-FISH (fluorescence in situ hybridization), and other methods to discover the genetic variation at Rhg1 across 41 diverse soybean accessions. Based on copy number variation, transcript abundance, nucleic acid polymorphisms, and differentially methylated DNA regions, we find that SCN resistance is associated with multicopy Rhg1 haplotypes that form two distinct groups. The tested high-copy-number Rhg1 accessions, including plant introduction (PI) 88788, contain a flexible number of copies (seven to 10) of the 31-kb Rhg1 repeat. The identified low-copy-number Rhg1 group, including PI 548402 (Peking) and PI 437654, contains three copies of the Rhg1 repeat and a newly identified allele of Glyma18g02590 (a predicted α-SNAP [α-soluble N-ethylmaleimide–sensitive factor attachment protein]). There is strong evidence for a shared origin of the two resistance-conferring multicopy Rhg1 groups and subsequent independent evolution. Differentially methylated DNA regions also were identified within Rhg1 that correlate with SCN resistance. These data provide insights into copy number variation of multigene segments, using as the example a disease resistance trait of high economic importance.
Vascular plants experienced a rapid diversification following land colonization, overcoming biotic and abiotic stresses to occupy diverse niches in a process that continues to the present and includes human-guided plant breeding (Kenrick and Crane, 1997; Steemans et al., 2009; Oh et al., 2012). One mechanism of genetic variation is diversification of the physical genome, at scales broader than isolated DNA base pair changes. This genome structural variation (Feuk et al., 2006) is increasingly recognized for having significant impacts on phenotypes and evolution (Aitman et al., 2006; Perry et al., 2008; Maron et al., 2013). Recent advances in plant genomics have highlighted the role of structural variation in plant adaptation to environmental stress (DeBolt, 2010; Dassanayake et al., 2011; Wu et al., 2012; Olsen and Wendel, 2013).
Copy number variation is an important type of structural variation because of its varied evolutionary impacts, facilitating neofunctionalization, subfunctionalization, and gene dosage effects (Ohno, 1970; Moore and Purugganan, 2005; Flagel and Wendel, 2009; Marques-Bonet et al., 2009). While the majority of duplicated genes are not retained, undergo pseudogenation, or exhibit distinct negative effects (Lynch and Conery, 2000; Demuth and Hahn, 2009; Tang and Amon, 2013), gene duplication has facilitated evolution in diverse organisms (Kondrashov et al., 2002; Conant and Wolfe, 2008). For one of the simplest types of copy number variation, gene duplication, a wide range of resulting adaptations to changing local environmental conditions has been characterized (Triglia et al., 1991; Labbé et al., 2007; Schmidt et al., 2010; Dassanayake et al., 2011; Heinberg et al., 2013; for review, see Kondrashov, 2012). Single gene copy number amplification has also been observed as an adaptive response to selective pressures (Bass and Field, 2011).
Epigenetic modifications, prominently including differential cytosine methylation, can also significantly impact organismal phenotypes (Chen, 2007; Gohlke et al., 2013; Hernando-Herraez et al., 2013). While the term epigenetic indicates heritable changes in gene activity not caused by changes in DNA sequence, there is increasing appreciation not only of the extent of methylation and other epigenetic marks throughout genomes, but also of the plasticity of these marks (Schmitz et al., 2013b; Ziller et al., 2013).
Domesticated soybean (Glycine max) is an important world commodity, accounting for a majority of the world’s protein-meal and oilseed production (soystats.com). The most economically damaging pathogen of soybean is the soybean cyst nematode (SCN), Heterodera glycines (Niblack et al., 2006). SCNs are obligate endoparasites that cause disease by reprogramming host root cells to form specialized feeding cells termed syncytia, robbing the plant of carbon and adversely affecting yield (Lauritis et al., 1983; Endo, 1984; Young, 1996; Sharma, 1998). SCN is found in all major soybean-growing states in the United States and cannot feasibly be removed (Niblack, 2005). Because the primary control strategies for SCN are crop rotation and planting resistant varieties, significant attention has been focused on the identification, development, and use of soybean germplasm that exhibits resistance to SCN (Diers et al., 1997; Concibido et al., 2004; Brucker et al., 2005b; Wrather and Koenning, 2009; Kim et al., 2010a, 2011). The Rhg1 (for Resistance to Heterodera glycines) locus, sometimes in combination with Rhg4, makes the greatest contribution to resistance in the vast majority of the commercially utilized soybean cultivars that exhibit SCN resistance (Caldwell et al., 1960; Matson and Williams, 1965; Webb et al., 1995; Li et al., 2004; Brucker et al., 2005b; Tylka et al., 2012).
We recently discovered that the SCN resistance conferred by Rhg1 is mediated by a 31-kb segment of DNA that contains four open reading frames and exhibits substantial copy number variation (Cook et al., 2012). A commercial soybean line containing the most widely utilized version of the Rhg1 locus, derived from plant introduction (PI) 88788, contains 10 tandem repeat copies of the 31-kb segment. Only a single copy of this 31-kb block was detected in the SCN-susceptible line Williams 82 and three other SCN-susceptible lines. It is particularly intriguing that three distinct genes within the 31-kb repeat were shown to contribute to SCN resistance (Cook et al., 2012). These genes are Glyma18g02580 (encoding a predicted amino acid transporter), Glyma18g02590 (encoding a predicted α-SNAP [α-soluble N-ethylmaleimide–sensitive factor attachment protein] vesicle-trafficking protein), and Glyma18g02610 (encoding a protein lacking a predicted function). The predicted protein sequences of Glyma18g02580 and Glyma18g02610 were invariant between the examined SCN-resistant and SCN-susceptible alleles, and experimental evidence suggests that these two genes contribute to resistance via enhanced expression arising through copy number variation. The SCN-resistant line derived from PI 88788 did contain an alternative allele of Glyma18g02590, which was also more highly expressed in SCN-resistant lines relative to susceptible lines. In addition to PI 88788, the other primary source of Rhg1-mediated SCN resistance in commercially cultivated soybean varieties is PI 548402 (commonly and throughout this article referred to as Peking). We found that the Peking Rhg1 contains three copies of the 31-kb region, but nucleotide sequences of the genes in Peking Rhg1 were not determined (Cook et al., 2012).
A well-documented epistasis occurs in Peking-derived SCN resistance, in which Peking Rhg1 has low efficacy relative to the Rhg1 from PI 88788, but only if Peking Rhg4 is not simultaneously present (Brucker et al., 2005a; Liu et al., 2012). The responsible gene at Rhg4 was recently discovered to encode a Ser hydroxymethyltransferase (Liu et al., 2012). Peking and PI 437654 (the source of the less used but commercially relevant Hartwig or CystX resistance) contain an Rhg4 allele whose product exhibits altered enzyme kinetics. Impacts of Rhg4 on SCN resistance are difficult to detect when deployed together with the high-copy-number rhg1-b from PI 88788 (Brucker et al., 2005a). It is intriguing and of high economic relevance that SCN populations arise that partially overcome the resistance mediated by certain sources of Rhg1 while remaining sensitive to the resistance conferred by other Rhg1 sources (Niblack et al., 2002; Colgrove and Niblack, 2008). In addition to understanding the biology of trait variation caused by copy number variation, and of traits in multicellular eukaryotes that are conferred by tightly linked blocks of distinct genes, there is substantial practical interest in understanding the variation in SCN resistance caused by different sources of Rhg1, and in the potential to predict, discover, and/or develop more effective versions of Rhg1.
Here, we use quantitative PCR (qPCR), fiber-fluorescence in situ hybridization (fiber-FISH), whole-genome sequencing, and DNA methylation analyses to investigate the major SCN resistance locus Rhg1 from a diverse population of soybean lines. We sequenced and analyzed the genomes of six Hg Type Test soybean lines that are widely used to characterize SCN field populations for their capacity to overcome different sources of SCN resistance (Niblack et al., 2002) and also analyzed whole-genome sequence data from 35 diverse soybean lines that are in use as parents in a separate soybean nested association mapping (SoyNAM) project. We discovered three classes of the Rhg1 locus that can be differentiated by gene dosage, copy number, and coding sequence. We also observed differential DNA methylation between resistant and susceptible Rhg1 haplotypes at genes impacting SCN resistance. The collective data allow clearer inferences to be drawn regarding the evolutionary history of the locus and provide a detailed analysis of one of the few confirmed examples in plant or animal biology in which copy number variation of a small multigene segment contributes to a defined adaptive trait.
RESULTS
Commonly Used Sources for Rhg1 Resistance Possess Either a Low Copy Number or a High Copy Number of Rhg1 Repeats as Compared with the Wild-Type Single Copy
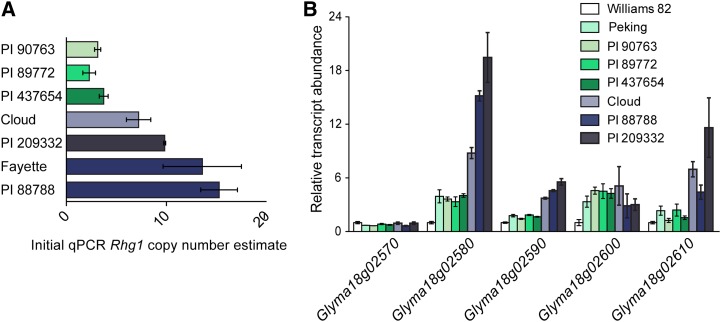
To assess the natural variation present at Rhg1, beyond the previous determination that there are 10 and three copies of the 31-kb Rhg1 repeat in two previously studied lines (Cook et al., 2012), we analyzed five other SCN-resistant lines. Together with PI 88788 and Peking, these seven soybean lines comprise the diagnostic test set in the established Hg Type Test that describes the capacity of SCN populations to overcome different sources of SCN resistance (Niblack et al., 2002). Initial characterization of Rhg1 copy number, using qPCR on genomic DNA, revealed three copy number classes: single copy, low copy (two to four copies), and high copy (more than six copies; Fig. 1A). For lines estimated to contain more than six copies, qPCR produced variable results and unreliable absolute copy number estimates, possibly because it is difficult to reduce qPCR variation below approximately 50% (half of one PCR cycle) between replicate tissue samples. Copy number estimates based on qPCR, however, did consistently identify two different classes for Rhg1 repeats.
Figure 1.
Estimates of copy number and transcript abundance suggest different types of Rhg1 loci. A, Initial Rhg1 copy number estimates, obtained using qPCR to amplify genomic DNA, identify two groups of SCN-resistant lines: low-copy number, between two and four repeats (PI 90763, PI 89772, and PI 437654), and high-copy number, estimated to have greater than seven repeats (Cloud, PI 209332, Fayette, and PI 88788). Copy number is expressed as the ratio of qPCR template abundance estimates for Rhg1 repeat junction and for a nonduplicated neighboring gene. B, Transcript abundance, relative to SCN-susceptible Williams 82, indicates the presence of two expression groups of Rhg1 loci in SCN-resistant lines. Roots from lines identified in subsequent work as having three copies of the 31-kb Rhg1 repeat (Peking, PI 90763, PI 89772, and PI 437654) exhibit lower transcript abundance than lines with seven, nine, or 10 Rhg1 copies (Cloud, PI 88788, and PI 209332). The one complete copy of Glyma18g02570, located immediately outside of the Rhg1 repeat region (Cook et al., 2012; this work), is expressed at a similar level across all the tested lines. The expression level of Glyma18g02600 is near the detection limit for qPCR.
To determine the impact that varying Rhg1 copy number has on constitutive transcription, we quantified root transcript abundance using qPCR in the Hg Type Test lines (Niblack et al., 2002). The four genes encoded within the previously identified Rhg1 repeat, Glyma18g02580, Glyma18g02590, Glyma18g02600, and Glyma18g02610, are more highly expressed in each of the seven tested Hg Type Test SCN resistance lines relative to SCN-susceptible Williams 82 (Fig. 1B). The transcript abundance of an adjacent gene that is outside of the 31-kb repeat, Glyma18g02570, had similar transcript abundance across all tested SCN-resistant and SCN-susceptible genotypes. Four of the SCN-resistant genotypes, Peking, PI 90763, PI 89772, and PI 437654, showed similar levels of elevated expression of the repeated genes, while expression was even more elevated in Cloud (PI 548316), PI 88788, and PI 209332 (Fig. 1B). These groupings were the same as those identified for qPCR estimates of DNA copy number and indicate that transcript abundance for these genes scales with gene copy number. One gene in the repeat, Glyma18g02600, was more highly expressed in SCN-resistant lines, but the expression level was similar between genotypes in different copy number classes. However, transcript abundance for this gene was close to the limit of detection for qPCR, was also detected only at very low levels in published RNA sequencing experiments (Severin et al., 2010), and no contribution of this gene to SCN resistance has yet been demonstrated (Cook et al., 2012). The soybean line Cloud, which was placed in the high-copy-number class but estimated to have fewer Rhg1 copies than PI 88788 and PI 209332, also showed lower transcript abundance of Glyma18g02580 and Glyma18g02590 than the other two lines in the high-copy class (Fig. 1B).
Copy Number at the Rhg1 Locus in the High-Copy Lines Is Dynamic
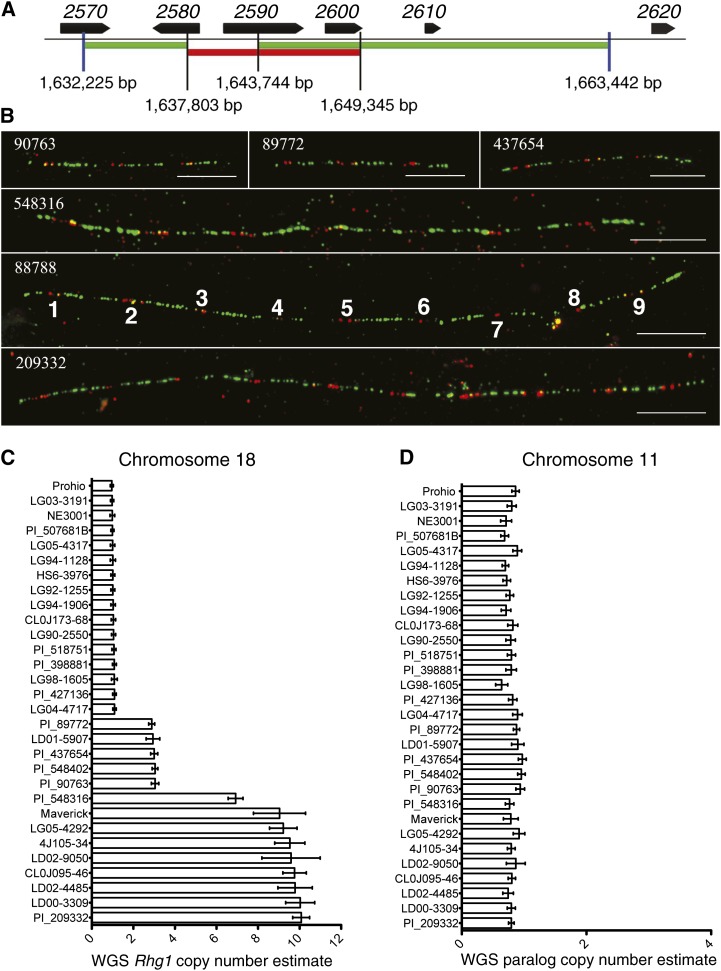
To definitively determine Rhg1 copy number in the Hg Type Test lines, we performed fiber-FISH using a diagnostic pair of DNA probes that span the repeat junction and partially overlap (Cook et al., 2012; Walling and Jiang, 2012). Representative fiber-FISH images for soybean lines PI 90763, PI 89772, and PI 437654 shown in Figure 2B (top row) summarize the finding that all three lines contain three copies of the 31-kb Rhg1 locus per haplotype, arranged as head-to-tail direct repeats. These results confirm the copy number estimates from qPCR. More importantly, for soybean lines in the high-copy Rhg1 class, fiber-FISH precisely determined the presence of seven Rhg1 copies in Cloud, nine copies in PI 88788, and 10 copies in PI 209332 (Fig. 2B, bottom three rows). We had previously used fiber-FISH to determine that Fayette, a soybean variety containing an Rhg1 locus originally from PI 88788, carries 10 copies of the Rhg1 repeat (Cook et al., 2012). Hence, the number of Rhg1 repeats varies not only between haplotype classes but also within the high-copy class and between lines with recent shared ancestry.
Figure 2.
Whole-genome resequencing and fiber-FISH define the copy number of Rhg1 in Hg Type Test lines. A, Diagram of red (11.5 kb) and green (25.3 kb) DNA probes used to detect Rhg1 repeats in fiber-FISH. Gene and base-pair numbers are from chromosome 18 of the soybean Williams 82 reference genome. B, Representative fiber-FISH images collected from six Hg Type Test soybean lines. As documented previously for three soybean lines (Cook et al., 2012), the Rhg1 locus is present as multiple direct repeats on single DNA fibers. These data indicate a copy number of three for PI 90763, PI 89772, and PI 437654 and copy numbers of seven, nine, and 10 for PI 548316 (Cloud), PI 88788, and PI 209332, respectively. The repeats are labeled for clarity for the representative fiber shown in box 88788 (PI 88788). Bars = 10 μm. C, Rhg1 copy number for 30 soybean lines based on whole-genome sequence read depth analysis. Average read depth was determined for 1-kb bins across the Rhg1 repeat and for 30 kb on each side of the Rhg1 repeat region. Data for the flanking single-copy regions from a given line were used to normalize the read depth data of 1-kb bins within the Rhg1 repeat to determine copy number (means ± se). D, Copy numbers determined as in C but for the Rhg1 paralog locus on chromosome 11.
Read Depth Analysis from Whole-Genome Sequencing Identifies Rhg1 Copy Number and Predicts SCN Resistance
To further discover the nature of the diversity within soybean Rhg1, we performed whole-genome sequencing for six of the seven Hg Type Test soybean lines: Peking, PI 90763, PI 89772, PI 437654, PI 209332, and Cloud (Niblack et al., 2002). Derivatives of PI 88788 had been sequenced previously (Cook et al., 2012). In addition, we analyzed whole-genome shotgun sequence data for 35 diverse soybean lines, generated as part of the recently initiated SoyNAM project, and analyzed previously published Illumina sequencing data from an undomesticated Glycine soja accession (Kim et al., 2010b). Supplemental Tables S1 and S2 provide details regarding the sequencing data sets. For this study, we focused on in-depth analysis of Rhg1 on chromosome 18 and its paralogous locus on chromosome 11.
To initially uncover structural variation at Rhg1, we screened the SoyNAM genome sequence data sets by aligning Illumina reads to a portion of the Williams 82 reference genome corresponding to Rhg1 on soybean chromosome 18 and similar loci (see “Materials and Methods”). This screen determined that eight of 35 SoyNAM lines contain an estimated Rhg1 copy number greater than one, based on read depth across the known repeat and flanking regions (Supplemental Table S3). To further investigate the extent of copy number variation in this set of diverse soybean genomes and to eliminate possible mapping bias that might arise from the use of a limited reference sequence region, Illumina sequencing reads were remapped to the entire reference genome for 24 of the SoyNAM lines based on the results of the rapid alignment and sequencing depth. This provided more precise Rhg1 copy number estimates based on read depth. Along with the SoyNAM lines, six Hg Type Test lines sequenced as part of this work and the available G. soja genome sequence were included for in-depth analysis. As shown in Figure 2C, the estimated copy numbers based on read depth for the six Hg Type Test lines are in agreement with the results from qPCR estimates and fiber-FISH. Lines Peking, PI 90763, PI 89772, and PI 437654 contain three copies, while Cloud has seven and PI 209332 has 10 (Fig. 2C). A soybean line derived from PI 88788 was previously estimated to carry 10 copies of the Rhg1 repeat, using read-depth analysis of whole-genome sequence data (Cook et al., 2012). The majority of the soybean lines chosen for the SoyNAM study were found to carry a single copy of the Rhg1 locus (Fig. 2C; Supplemental Table S3) and have not been reported to exhibit SCN resistance where information is publicly available from the Germplasm Resources Information Network (USDA, 2014). Seven other SoyNAM lines contain nine to 10 copies of the Rhg1 locus, while one line contains an estimated three copies (Fig. 2C). These results are in agreement with pedigree information where it is publicly available. The above results indicate that increased copy number at Rhg1 is not a common phenomenon in soybean accessions and likely can be traced to a limited number of parental lines. There is also no indication that structural variation has occurred at the paralogous locus on chromosome 11 (Fig. 2D).
Sequence Analysis Reveals Extensive Rhg1 Locus DNA Sequence Variation, But Amino Acid Polymorphisms Are Only Present in the Predicted α-SNAP
The whole-genome sequence data of the SoyNAM and Hg Type Test lines were analyzed for Rhg1 nucleic acid and derived amino acid variations. Genomic DNA sequence variations, including single-nucleotide polymorphisms (SNPs) and small insertions and deletions relative to the Williams 82 reference genome, were identified using the Genome Analysis Toolkit (GATK) pipeline (McKenna et al., 2010; DePristo et al., 2011). A total of 409 DNA variant sites across the 31.2-kb Rhg1 repeat interval (chromosome 18, bp 1,632,223–1,663,500 of the Williams 82 soybean reference genome, version 1.1) were identified in at least one of the 31 genomes. The average number of Rhg1 variant sites per soybean line was 251 ± 40 (mean ± se) for the low-copy Rhg1 lines, 260 ± 10 for the high-copy lines, and 23 ± 29 for the lines estimated to contain a single copy of Rhg1 (Table I). As described below, within any single accession, the sequences of individual repeats were largely identical to the other repeats. Hence, there were approximately 250 polymorphisms per 31-kb repeat in the SCN-resistant genotypes but zero to 81 polymorphisms in the corresponding 31-kb Rhg1 region of the investigated single-copy lines.
Table I. Summary statistics for DNA variant analysis at Rhg1 from whole-genome sequencing shows higher rates of polymorphism in SCN-resistant lines.
Copy No., Rhg1 copy number estimated from sequencing read depth or from fiber-FISH (asterisks). Variant Class, Whole-genome sequencing for 30 soybean lines and one G. soja line was analyzed for DNA variants and classified as SNPs, insertion, or deletion relative to the Williams 82 reference genome. The total number of DNA variants of each type across the 31-kb Rhg1 sequence are reported. Total, Sum of the SNP, insertion, and deletion variants. Variant Location columns report numbers of variants in each type of genome region. UTR, Untranscribed region.
| Genotype | Variant Class |
Variant Location |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Copy No. | SNPs | Insertion | Deletion | Total | Exon | Intron | Intergenic | UTR | |
| LD00-3309 | 10.03 | 190 | 36 | 32 | 258 | 6 | 24 | 220 | 8 |
| LG05-4292 | 9.23 | 195 | 38 | 34 | 267 | 6 | 25 | 228 | 8 |
| 4J105-34 | 9.53 | 196 | 35 | 32 | 263 | 6 | 24 | 225 | 8 |
| CL0J095-46 | 9.76 | 195 | 36 | 32 | 263 | 6 | 24 | 225 | 8 |
| LD02-4485 | 9.79 | 189 | 36 | 33 | 258 | 6 | 25 | 219 | 8 |
| LD02-9050 | 9.56 | 177 | 32 | 28 | 237 | 6 | 24 | 199 | 8 |
| Maverick | 9.04 | 187 | 33 | 31 | 251 | 6 | 24 | 213 | 8 |
| Cloud | 7* | 194 | 35 | 32 | 261 | 6 | 25 | 222 | 8 |
| PI 209332 | 10* | 197 | 35 | 33 | 265 | 6 | 27 | 224 | 8 |
| PI 437654 | 3* | 193 | 37 | 35 | 265 | 5 | 27 | 224 | 9 |
| Peking | 3* | 200 | 38 | 33 | 271 | 6 | 25 | 232 | 8 |
| PI 89772 | 3* | 194 | 37 | 36 | 267 | 5 | 25 | 229 | 8 |
| PI 90763 | 3* | 192 | 37 | 34 | 263 | 5 | 25 | 224 | 9 |
| LD01-5907 | 2.95 | 146 | 18 | 17 | 181 | 5 | 24 | 146 | 6 |
| NE3001 | 0.99 | 63 | 3 | 6 | 72 | 4 | 23 | 42 | 3 |
| LG05-4317 | 1.01 | 72 | 4 | 5 | 81 | 1 | 19 | 60 | 1 |
| LG94-1128 | 1.02 | 45 | 2 | 3 | 50 | 1 | 15 | 33 | 1 |
| LG92-1255 | 1.02 | 46 | 6 | 3 | 55 | 0 | 3 | 51 | 1 |
| PI 518751 | 1.07 | 44 | 2 | 3 | 49 | 0 | 0 | 49 | 0 |
| LG94-1906 | 1.03 | 26 | 0 | 2 | 28 | 0 | 0 | 28 | 0 |
| LG03-3191 | 0.98 | 28 | 0 | 0 | 28 | 0 | 0 | 28 | 0 |
| CL0J173-68 | 1.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HS6-3976 | 1.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LG04-4717 | 1.10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PI 398881 | 1.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PI 427136 | 1.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PI 507681B | 1.00 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LG90-2550 | 1.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Prohio | 0.97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LG98-1605 | 1.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G. soja | 1.10 | 42 | 3 | 4 | 49 | 1 | 3 | 45 | 0 |
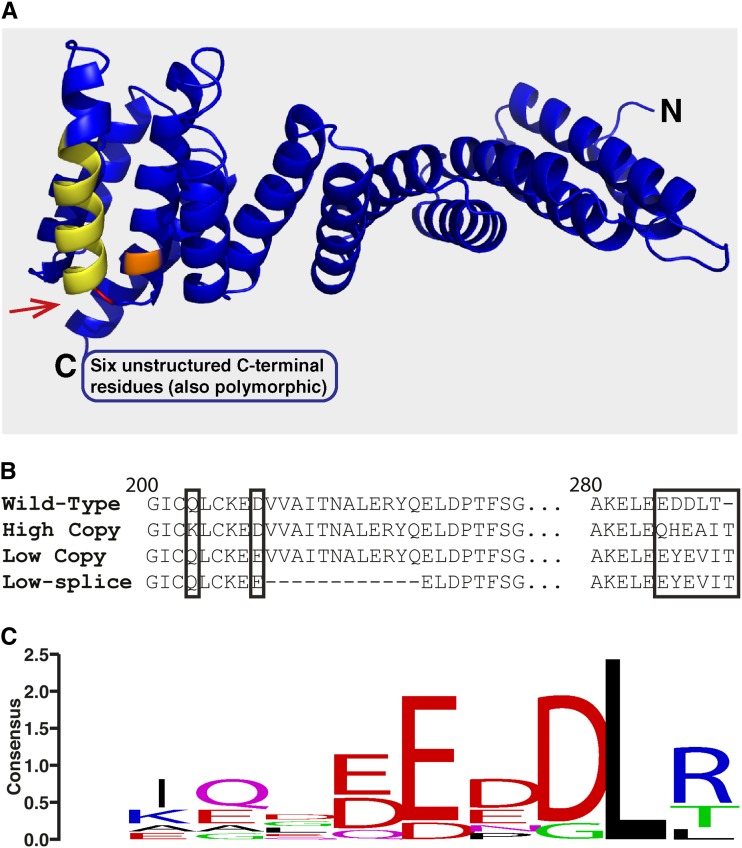
Despite the high number of sequence polymorphisms found within each Rhg1 repeat in SCN-resistant lines, few affected protein-coding sequences. We did not detect any polymorphisms resulting in an altered amino acid sequence for Glyma18g02610 or Glyma18g02580 in any of the SCN-resistant lines. Curiously, in the derived amino acid sequences of Glyma18g02590, two SCN-resistant allele types were observed that carry distinct mutations but that impact similar protein sites (Fig. 3A). The gene Glyma18g02590 encodes a predicted α-SNAP; in other organisms, these proteins have the canonical function of stimulating N-ethylmaleimide-sensitive factor ATPase activity to assist the disassembly of SNARE components following vesicle-mediated transport (Morgan et al., 1994; Barnard et al., 1997; Rice and Brunger, 1999). Amino acid sequence alignment of the available 17 Rhg1 single-copy soybean lines including the Williams 82 reference genome revealed an invariant primary sequence of Glyma18g02590. One type of alternative allele was found in all tested high-copy Rhg1 haplotypes, including the previously reported sequence from Fayette, and a new allele was found in all tested lines carrying the low-copy Rhg1 haplotype associated with SCN resistance (Fig. 3B). The novel alleles of α-SNAP found in SCN-resistant lines have amino acid polymorphisms changing the final five or six amino acids, the residues that otherwise have the strongest consensus sequence across eukaryote α-SNAPs (Fig. 3C), including a substitution for the Leu at the penultimate C-terminal amino acid. The presence of different amino acid substitutions at similar positions between the low- and high-copy-class 2590 alleles suggests a functional importance of these sites for SCN disease resistance. Mutations at these C-terminal residues are unexpected given previous findings that these residues are essential for stimulating N-ethylmaleimide-sensitive factor ATPase activity in other organisms (Barnard et al., 1996). Together, these findings suggest that the SCN resistance-associated Glyma18g02590 proteins may not possess classical α-SNAP functions and may instead promote SCN disease resistance through a novel mechanism.
Figure 3.
Resistant-type Rhg1 classes encode unique α-SNAP alleles with polymorphisms in highly conserved residues localized at the C terminus. A, Glyma18g02590 from Williams 82 modeled to the crystal structure of yeast α-SNAP (sec17p; Protein Data Bank no. 1QQE). The Q203K substitution unique to high-copy Rhg1-encoded α-SNAPs is colored orange. The D208E substitution present only in low-copy Rhg1 α-SNAPs is shown in red. An alternative splice isoform detected in low-copy Rhg1 classes removes 12 residues from the full-length protein (displayed in yellow). Two distinct polymorphisms in the final five to six C-terminal residues are found in the Rhg1 resistant-class α-SNAPs (C-terminal residues are not modeled; they are predicted to be unstructured). B, Amino acid sequence of Glyma18g02590 from Williams 82 (wild-type; SCN-susceptible) aligned to predicted amino acid sequences of both high- and low-copy class Rhg1 α-SNAPs. Note that a second mRNA splice isoform found in low-copy Rhg1 lines is predicted to exclude residues 209 to 220. No predicted amino acid polymorphisms in Glyma18g02590 from the sequenced SCN-susceptible lines have been detected. C, Logo displaying the consensus sequence for the final 10 C-terminal residues of α-SNAP from eight diverse eukaryotes (Homo sapiens, Drosophila melanogaster, Saccharomyces cerevisiae, Caenorhabditis elegans, Danio rerio, Bos taurus, Arabidopsis, and soybean). Strikingly, the unique C-terminal polymorphisms discovered in Rhg1 resistant-type α-SNAPs occur at these five most highly conserved residues.
For Glyma18g02590, we performed 3′ RACE and sequenced at least seven independent complementary DNA (cDNA) clones for each of the Hg Type Test lines and Williams 82. The novel (non-Williams 82) Glyma18g02590 alleles predicted from genomic DNA sequences were present in cDNA from the respective lines carrying the low- or high-copy Rhg1 haplotypes (Supplemental Table S4). Interestingly, a small proportion of the cDNA clones sequenced from PI 88788 and Cloud (high-copy Rhg1 lines) contained Williams 82-type Glyma18g02590 sequences, consistent with the identification of a single Williams 82-type genomic DNA sequence in one of the copies of the 31-kb Rhg1 repeats (described below). We did not detect any Williams 82-type Glyma18g02590 sequences in cDNAs from lines carrying the low-copy-class Rhg1, again consistent with the genomic DNA sequence data. However, a splice isoform of the Glyma18g02590 cDNA was identified in all of the tested low-copy Rhg1 lines, and this splice isoform was not found in the high-copy or single-copy Rhg1 lines (Fig. 3B; Supplemental Table S4).
A naturally occurring truncated allele encoding a predicted α-SNAP was recently implicated in SCN disease resistance derived from Peking and PI 437654 but not in PI 88788-derived resistance (Matsye et al., 2012). Our results from whole-genome sequencing, however, indicate that the sequence encoding that truncated α-SNAP is not encoded by a Glyma18g02590 gene at Rhg1 on chromosome 18 but rather by Glyma11g35820, the paralog of Glyma18g02590 on chromosome 11 (Supplemental Fig. S1; Supplemental Table S5). The SNPs at Glyma11g35820 responsible for encoding the truncated allele were also identified in the high-copy Rhg1 SCN-resistant lines Cloud and LG05-4292.
Another Rhg1 sequence polymorphism was identified in the Peking genome: a nucleotide deletion in the second exon of the Glyma18g02600 coding sequence (Table II) observed as a heterozygous deletion (see below). Translation of the resulting mRNA results in a stop codon eight codons downstream of the deletion, truncating the predicted protein by 314 amino acids (removing 58% of the wild-type protein sequence).
Table II. Amino acid polymorphisms for genes encoded within and adjacent to the Rhg1 repeat, from all analyzed soybean lines.
Position, Chromosome 18 base-pair position relative to the Williams 82 reference genome, with gene names (and putative gene product functions in parentheses) above the relevant base-pair positions. The remaining columns indicate soybean accessions. Low-Copy Lines, All five soybean lines estimated to contain three copies of Rhg1 have the same amino acid polymorphism and are represented by a single column. High-Copy Lines, All lines estimated to contain seven or more copies of Rhg1 have the same amino acid polymorphism and are represented by a single column. Only three additional soybean lines contain amino acid polymorphisms for any of the six genes analyzed and are listed in individual columns. Amino acid polymorphisms are reported as the amino acid present in Williams 82, the amino acid position, and the resulting new amino acid discovered. Not shown here is the soybean line Peking, which contains a single-nucleotide deletion in some Rhg1 repeat copies that introduces a frame shift at amino acid 214 and results in a premature stop codon after amino acid 222 of Glyma18g02600 (eliminating 58% of predicted protein).
| Position | Low-Copy Lines | High-Copy Lines | NE3001 | LG05-4317 | LG94-1128 |
|---|---|---|---|---|---|
| Glyma18g02570 (unknown function) | No polymorphisms | ||||
| Glyma18g02580 (amino acid transporter) | |||||
| 1638989 | V9I | ||||
| Glyma18g02590 (α-SNAP) | |||||
| 1643208 | Q203K | ||||
| 1643225 | D208E | ||||
| 1644965 | E285Q | ||||
| 1644968 | D286Y | D286H | |||
| 1644972 | D287EV | D287EA | |||
| 1644974 | L288I | L288I | |||
| Glyma18g02600 (PLAC-8 family) | |||||
| 1647134 | V23A | ||||
| 1647764 | 214 stop (Peking) | ||||
| 1648561 | M480L | M480L | |||
| Glyma18g02610 (unknown function) | No polymorphisms | ||||
| Glyma18g02620 (SEL-1-like) | No polymorphisms | ||||
Resistance-Conferring Rhg1 Loci Developed from a Common Source But Underwent Copy Number Expansion in Distinct Lineages
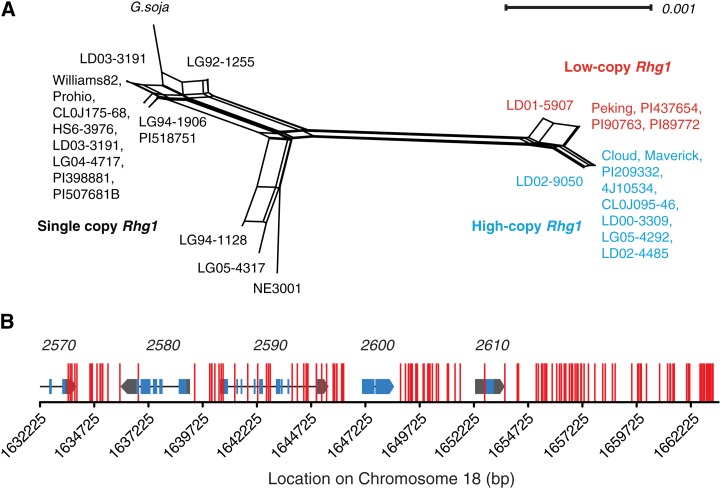
To further explore the evolutionary history of the Rhg1 locus, DNA sequence variation sites in a diverse set of soybean lines were used to construct a nonhierarchical phylogenetic network using the NeighborNet algorithm in SplitsTree (Bryant and Moulton, 2004; Huson and Bryant, 2006). The network reveals a clear split between the Rhg1 loci from SCN-resistant and SCN-susceptible lines (Fig. 4A). There is a further split in the multicopy clade, separating the low- and high-copy Rhg1 groups from each other (Fig. 4A). A common origin of the high-copy and low-copy Rhg1 repeats was suggested by the identity of their repeat-junction sequences (Cook et al., 2012) and is now further supported by the high number of DNA sequence variant sites shared by the two groups but absent in single-copy Rhg1 lines (Fig. 4B). In total, 147 DNA variant sites not detected in the single-copy Rhg1 SCN-susceptible lines are common to all of the sequenced high-copy and low-copy Hg Type Test lines. This is 75% of the 197 DNA variant sites present in at least one Hg Type Test line but not present in any of the examined SCN-susceptible lines. These data suggest that a common progenitor had accumulated the 147 DNA variant sites prior to subsequent divergence of the two copy number groups. In support of subsequent divergence of the low-copy lines from the high-copy lines, a small number of DNA variant sites not present in any tested SCN-susceptible genome were universally common within either the low-copy or the high-copy Rhg1 group: 10 sites for low copy and seven for high copy (Supplemental Fig. S2). Even more recent divergence is highlighted by the presence of a small number of DNA variants unique to a single tested genotype: Peking (six), PI 88788 (zero), PI 90763 (one), PI 437654 (zero), PI 209332 (five), PI 89772 (zero), and Cloud (one).
Figure 4.
Network analysis and shared polymorphisms support three Rhg1 locus types and a single SCN-resistant-type progenitor. A, NeighborNet analysis indicates two distinct groups based on Rhg1 sequence, separating the SCN-susceptible lines (left, single-copy Rhg1) and the SCN-resistant lines (right, multicopy Rhg1). The resistant lines further split into two groups that correspond with Rhg1 copy number (Figs. 1 and 2); lines containing three Rhg1 copies are noted in red and those containing seven or more copies are noted in blue. The four coding genes within the Rhg1 repeat, including 200 bp of sequence upstream of the start codon, were used for analysis. B, DNA variant sites present in all seven Hg Type Test soybean lines (low-copy and high-copy Rhg1, SCN resistant) but absent from all sequenced SCN-susceptible single-copy Rhg1 lines. Vertical red lines show locations of these 148 DNA variant sites, which are 75% of all of the 197 SNP or INDEL DNA variant sites present in at least one Hg Type Test line but not present in any of the examined SCN-susceptible lines. Rhg1 locus gene models are shown at correct x axis positions for reference (blue, exons; black line, introns; and gray, untranslated regions); gene names are given above the gene model (e.g. 2570 = Glyma18g02570).
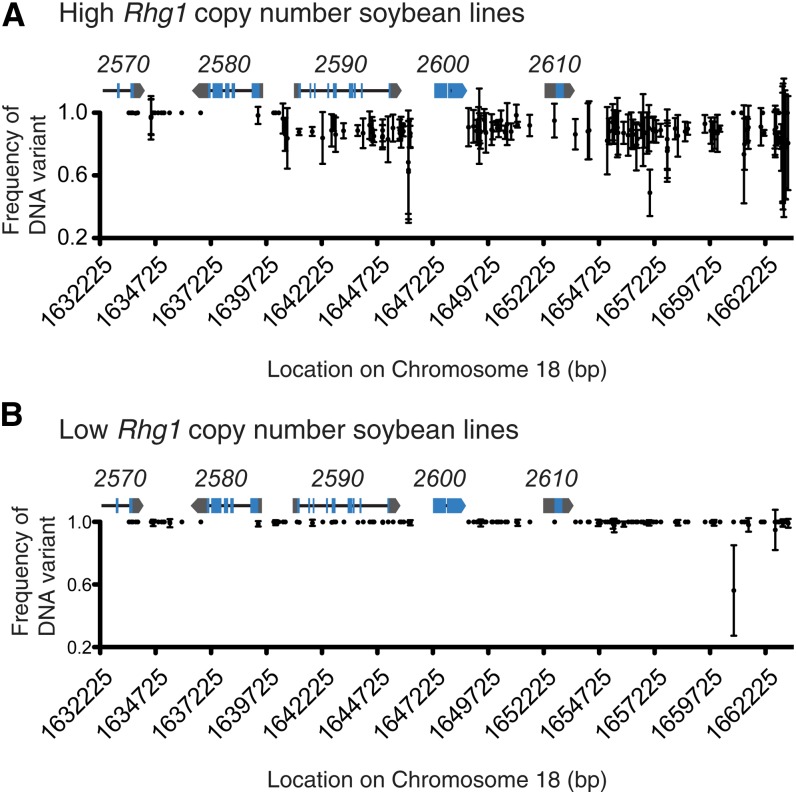
The degrees of similarity between Rhg1 repeats within any single genome or within a copy number group can be analyzed by the frequency of variant sequence relative to reference sequence from the whole-genome sequence data sets. Within the high-copy genomes of Cloud, PI 209332, and LD00-3309 (PI 88788 derivative), most of the variant sites on the right three-quarters of the interval as shown in Figure 5 have a sequence frequency of roughly 0.85 to 0.9 (Fig. 5A). The other 10% to 15% of sequence reads at these positions match the Williams 82 reference sequence, suggesting that roughly three-quarters of one of the Rhg1 repeats in the high-copy Rhg1 accessions contains Williams 82-type sequence. This is consistent with the Glyma18g02590 cDNA data described above (Supplemental Table S4). Most variant sites across the left one-quarter of the Rhg1 repeat (Fig. 5A) are invariant for the alternate sequence, indicating its presence in all copies.
Figure 5.
The frequency of DNA variant sites across Rhg1 repeats reveals heterogeneity between repeats in high-copy but not low-copy Rhg1-containing lines. A, Nearly homogenous presence of the same non-Williams 82 DNA sequence for variant sites in all copies (left one-quarter) or all but one copy (right three-quarters) of the Rhg1 repeat. The x axis shows the locations of DNA variant sites (SNP or INDEL) within the Rhg1 locus on soybean chromosome 18, and the y axis shows the proportion of all DNA sequence reads with variant (high-copy-type) sequence rather than the reference Williams 82-type (single-copy Rhg1) sequence at the designated DNA variant site. Data were combined for the three Rhg1 high-copy-class Hg Type Test soybean lines LD00-3309 (PI 88788), PI 209332, and Cloud; mean frequency and se for the three soybean lines are shown. Rhg1 locus gene models are shown at correct x axis positions for reference (blue, exons; black line, introns; and gray, untranslated regions); gene names are given above the gene model (e.g. 2570 = Glyma18g02570). B, Near identity of the three repeats in Rhg1 low-copy lines and absence of a Williams 82-type segment. Details are as in A except showing combined data for the four Rhg1 low-copy-class Hg Type Test soybean lines Peking, PI 90763, PI 437654, and PI 89772.
A small number of DNA variant sites do not follow the above trend and indicate the development and propagation of variant sequences in a smaller number of the total copies. Specifically, the DNA variant at chromosome 18 base pair position 1,657,025 is apparently found in only four of the seven copies in Cloud and in only five or six of the 10 copies in PI 209332 and LD00-3309, suggesting as one possibility the emergence of this DNA polymorphism in one repeat at an intermediate stage of copy number expansion of the locus (Supplemental Table S6). However, propagation of the repeats apparently was not symmetric between genomes, because (for example), at positions 1,657,807/1,657,816 and 1,661,264/1,661,293, Cloud and PI 209332 appear to carry only five and seven to eight copies, respectively, of the variant site while LD00-3309 appears to carry the variant site in all nine non-Williams 82 repeats. Conversely, the set of polymorphisms at positions 1,663,007 to 1,663,250 are present in only six to seven copies in LD00-3309, eight to nine copies in PI 209332, but in all six non-Williams 82 copies of Cloud (Supplemental Table S6). Inspection of raw sequence data for these nonhomogenous variant sites suggests that they are valid sequence calls rather than data-processing errors and suggests unequal propagation of specific copies during evolution of the locus. Although we cannot rule out phenotypic selection among the high-copy Rhg1 soybean lines for revertants that carry more copies of the Williams 82 reference sequence at these nonhomogenous variant sites, the sites are in intergenic regions at least 1 kb away from known transcription start sites. Hence, it may be more parsimonious to assume that they are neutral sites that reflect the source of progenitor repeats that were utilized during Rhg1 repeat expansion.
Analysis of the low-copy Rhg1 lines (Peking, PI 89772, PI 90763, and PI 437654) shows a different pattern of repeat expansion and may partially account for well-established functional differences between the high-copy (PI 88788-type) and low-copy (Peking-type) Rhg1 loci. The frequency of variant sequence to reference sequence at polymorphic sites in all Rhg1 low-copy lines is nearly 1 (i.e. mostly uniform across the 31.2-kb repeat region; Fig. 5B). This suggests that the low-copy lines experienced copy number expansion from a single shared progenitor and/or homogenization across the repeats by gene conversion or other mechanisms after at least some repeats had already formed. Loss of repeats carrying divergent copies may have also occurred. This in-depth analysis of sequencing frequencies shows that not only are the two resistance groups diverging for Rhg1 copy number but the sequence composition of the repeats is also following different evolutionary paths.
Variation in Soybean Resistance to Diverse Nematode Populations Supports the High-Copy and Low-Copy Rhg1 Groupings and Suggests a Relationship between Copy Number and Resistance
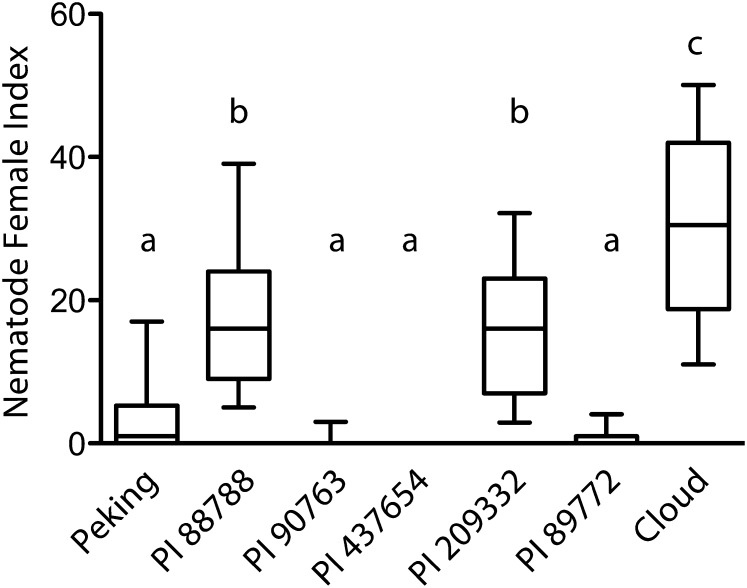
Previous research has described differences in SCN resistance between Peking-, PI 437654-, and PI 88788-derived soybean sources, measured in terms of genetics, cell biology, nematode development, and nematode race specificity or Hg type specificity, but the causes for these observations have remained elusive (Arelli and Webb, 1996; Mahalingam and Skorupska, 1996; Kim et al., 1998, 2010c; Brucker et al., 2005a; Niblack et al., 2006; Klink et al., 2011). To address this, we analyzed data for soybean resistance to SCN from greenhouse trials conducted by Alison Colgrove, Terry Niblack, and colleagues as part of the Northern Regional SCN Tests (Cary and Diers, 2010, 2011, 2012, 2013). The analysis included data from a total of 97 field populations collected from 2009 to 2012, including SCN field populations from eight to 10 north central U.S. states and/or adjacent Canadian provinces per year. The results from our analysis indicate that Cloud, which contains seven copies of Rhg1, was significantly less resistant than the other lines tested (Fig. 6). The other two lines in the high-copy Rhg1 class, PI 88788 and PI 209332, which contain nine and 10 copies, respectively, form a statistically significantly more resistant cluster than Cloud, suggesting that higher Rhg1 copy number may increase SCN resistance. Because Cloud, PI 88788, and PI 209332 lines are not isogenic at other loci, this comparison is only suggestive. The low-copy Rhg1 lines are significantly more resistant to diverse SCN populations, but since these lines carry an SCN resistance-conferring allele of Rhg4, a simple comparison of the relative contributions or efficacies of Rhg1 loci between low-copy and high-copy lines is obfuscated. Moreover, the roles of low-copy and high-copy Glyma18g02590 amino acid polymorphisms in impacting resistance to SCN are unknown.
Figure 6.
Nematode resistance data from 78 diverse SCN populations indicate similarities in resistance profiles based on copy number. Nematode development data were obtained for the seven Hg Type Test SCN-resistant lines for greenhouse assays conducted as part of the 2009 to 2012 Northern Regional SCN Tests. The data analyzed for 78 SCN populations were collected from 12 U.S. states and Canadian provinces. Female index is the percentage of SCN cysts that developed on the resistant soybean line relative to the susceptible control soybean line. Boxes show median and 25% to 75% range of data; whiskers extend to 10% and 90% of the data. For statistical analysis, variance was calculated by random replacement with 1,000 bootstrap replicates for each line within a given nematode population (see “Materials and Methods”). This calculated variance was used in a weighted ANOVA; soybean lines not sharing the same letter above the whisker had significantly different means following Bonferroni correction for multiple testing at P < 0.001.
Rhg1 Loci from Different Sources Contain Differentially Methylated Regions That Correlate with SCN Resistance
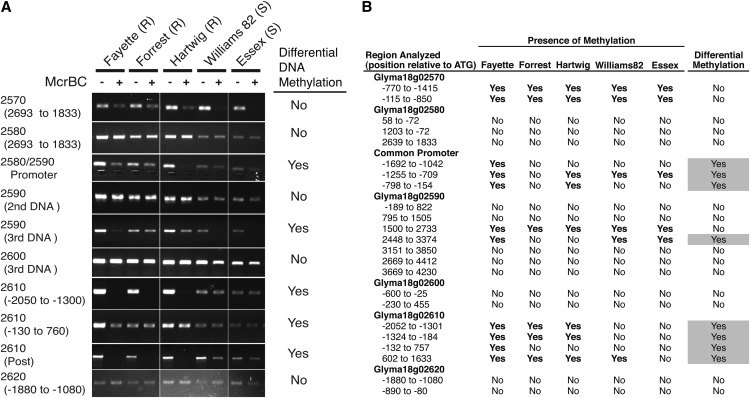
In addition to determining the genome structure and nucleic acid variation present at the Rhg1 locus from different sources, we investigated potential differences in DNA methylation states. In a broad survey of root DNA methylation patterns at Rhg1, we used DNA methylation-sensitive restriction enzymes coupled with PCR to identify differentially methylated regions (DMRs) between SCN-resistant and SCN-susceptible genotypes. The enzyme McrBC restricts DNA at sites of methylated cytosines of the sequence (G/A)mC and does not restrict unmethylated DNA (Sutherland et al., 1992). Hence, genomic DNA digestion by McrBC followed by PCR will not produce a product if the PCR product spans methylated cytosines. Using a total of 23 primer pairs, we discovered eight DMRs between SCN-susceptible genomes (carrying a single-copy Rhg1 locus) and SCN-resistant genomes (carrying low- or high-copy Rhg1 loci; Fig. 7). Hypermethylated DMRs were detected in SCN-resistant lines in the shared promoter for genes Glyma18g02580 and Glyma18g02590 and within and flanking the coding sequence of Glyma18g02610. We did not observe DMRs in the gene body of Glyma18g02580, nor did we observe substantial methylation or DMRs adjacent to or within the coding sequence of Glyma18g02600. We also used McrBC to analyze methylation at the Rhg1-adjacent but nonrepeated genes Glyma18g02570 and Glyma18g02620 and did not observe DMRs (Fig. 7).
Figure 7.
Differential Rhg1 locus DNA methylation between SCN-resistant and SCN-susceptible lines, particularly in control regions upstream of SCN resistance genes. A, Representative gel images of PCR products from a soybean root genomic DNA template treated with the restriction endonuclease McrBC (+) or buffer only (−) prior to PCR. McrBC cleaves (G/A)mC sites containing methylcytosine, preventing PCR amplification of cleaved template strands so that PCR product abundance goes down with increasing levels of methylation. Differential DNA Methylation was scored as positive if any soybean line differed from other lines in McrBC sensitivity of the PCR product in two independent tests. Soybean lines are denoted as either resistant (R) or susceptible (S) to SCN. B, Summary table for the replicated McrBC study described in A with 23 PCR primer pairs used to assess DNA methylation within the Rhg1 locus. The presence of methylation is listed as yes if both DNA samples showed reduced PCR amplification following McrBC DNA treatment. The right column reports methylation differences between different soybean lines.
During the preparation of this article, a genome-wide methylome study was published in which whole-genome bisulfite sequencing was performed for soybean lines LDX01-1-165 (referred to here as LDX), LD00-2817P (referred to here as LD), and progeny from their cross (Schmitz et al., 2013a). LD is known to have SCN resistance derived from PI 437654 (low-copy Rhg1 locus type), while LDX contains a single copy of Rhg1 (Diers et al., 2010; Kim et al., 2011). To confirm our observations and gain single-base resolution for methylation, we highlighted and reanalyzed the data of Schmitz et al. (2013a), focusing on Rhg1.
Consistent with the findings described above, our Rhg1 copy number estimate was 2.93 for LD and 1.17 for LDX, with various LD × LDX F3-derived (and hence potentially heterozygous) progeny families giving a range of Rhg1 copy number estimates between one and three (Supplemental Fig. S3A). We were also able to estimate transcript abundance for the two parents along with the two F3-derived progeny families that were subjected to RNA sequencing characterization (see “Materials and Methods”). Consistent with our present and previous findings (Fig. 1B; Cook et al., 2012), standardized RNA sequence read depth for noninfested plants, normalized to the susceptible LDX parent, showed elevated expression for the genes encoded within but not adjacent to the Rhg1 repeat in LD and progeny 11272 but not progeny 11268 (Supplemental Fig. S3B). This is consistent with elevated Rhg1 copy number as a significant cause of the elevated transcript levels.
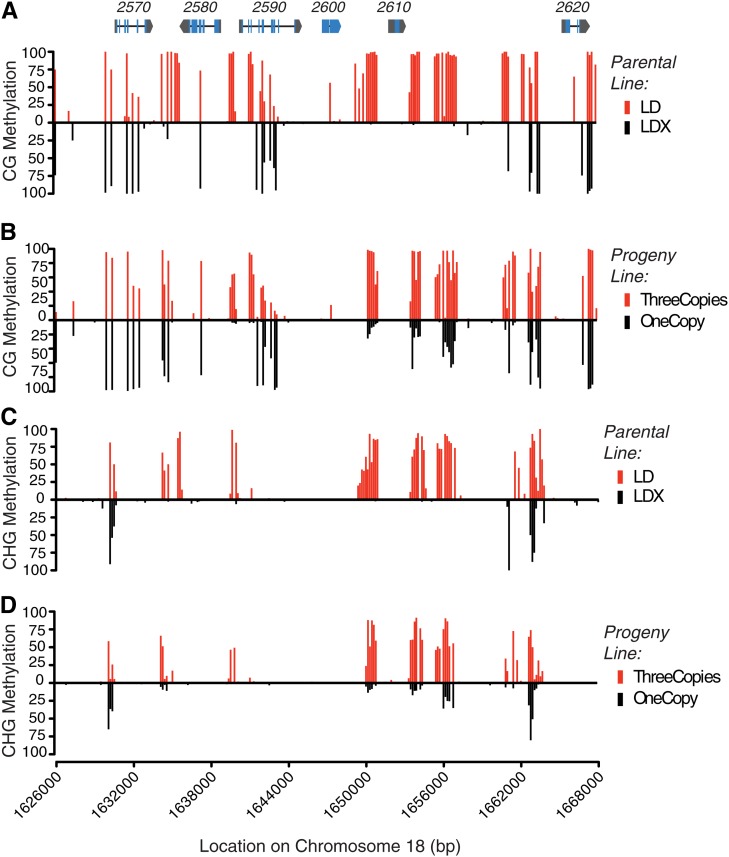
DNA methylation levels were computed from the data of Schmitz et al. (2013a) in bins of 150 bp in the CG, CHG, and CHH sequence contexts in both parents and 27 progeny lines that had at least 4× average sequencing depth. Consistent with our above findings of differential root DNA methylation in different Rhg1 copy number groups, we observed differential hypermethylated DNA in all three sequence contexts at the same regions in lines estimated to contain multiple copies of Rhg1 (Fig. 8; Supplemental Fig. S4). Data for the full set of lines can be seen in Supplemental Figure S5 (see “Materials and Methods”). Consistent with the finding that methylation patterns are largely inherited based on the parental methylation pattern (Schmitz et al., 2013a), for Rhg1 we observed high average levels of cytosine methylation (a characteristic of the LD parent that carries three Rhg1 copies) in the progeny that appeared homozygous for the three-copy Rhg1 haplotype (Fig. 8, B and D). Lower average Rhg1 methylation (a characteristic of the LDX parent that carries one Rhg1 copy) was observed in the progeny homozygous for single-copy Rhg1 haplotypes (Fig. 8, B and D). Together, our findings and the data of Schmitz et al. (2013a) describe, in detail, across tissue types and different sources of SCN resistance, stably inherited hypermethylated DNA regions at the resistance-conferring alleles of the genes shown to mediate Rhg1 resistance.
Figure 8.
DNA methylome sequence from three-copy and single-copy Rhg1 lines and their progeny further define differential methylation at Rhg1 SCN resistance genes. Levels of DNA methylation are reported as proportions of methylated cytosines detected from bisulfite sequencing. Data are for 150-bp bins represented by a single vertical line. Rhg1 locus gene models are shown at the top of A at correct x axis positions along chromosome 18, which are shown below D for reference (blue, exons; black line, introns; and gray, untranslated regions); gene names are given above the gene model (e.g. 2570 = Glyma18g02570). A, Levels of cytosine methylation for the sequence context CG, showing differential methylation of parental line LD (three copies of Rhg1; red vertical lines above the x axis) relative to parental line LDX (single copy of Rhg1; black vertical lines below the x axis). The greatest differential methylation is present upstream and downstream of the Glyma18g02580 open reading frame, in the common promoter for Glyma18g02580 and Glyma18g02590, and both upstream and downstream of the Glyma18g02610 open reading frame, with more methylation in the three-copy Rhg1 SCN-resistant line. B, Average CG methylation in F3-derived progeny families of the cross between lines LD and LDX, either for all six progeny estimated to have an Rhg1 copy number of three (red vertical lines above the x axis) or for all 16 progeny lines estimated to have an Rhg1 copy number of one (black vertical lines below the x axis). Substantial similarities to the parental CG methylation patterns are evident. C and D, Levels of cytosine methylation for the sequence context CHG, where H can be A, T, or C. C, Analysis similar to A except for the CHG sequence context. The same regions identified as differentially methylated in A are again identified as hypermethylated. D, Analysis similar to B except for the CHG sequence context, using the same progeny as in B.
DISCUSSION
SCN is the most economically limiting pathogen for soybean, causing billions of dollars of yield losses annually in the United States alone (Wrather and Koenning, 2009). Major efforts in soybean breeding and biotechnology are focused on the incorporation of desirable Rhg1 alleles and on the continued discovery of new and better sources of SCN resistance. We had previously determined that three very tightly linked genes at Rhg1 contribute to SCN resistance and that these genes reside on a 31-kb segment that is present in 10 copies in a common SCN-resistant variety along with an altered amino acid sequence for one of the genes (Cook et al., 2012). However, the extent of Rhg1 structural variation present in a broader set of soybean germplasm, the presence of alternate coding alleles and their expression levels, and the relatedness of different Rhg1 sources were not known. Here, we report the discovery of the structural, coding, and methylation differences present at Rhg1 from a diverse population of soybean lines.
The identification in different soybean lines of seven, nine, and 10 copies of an Rhg1 locus composed of highly similar sequences indicates that copy number at Rhg1 is plastic and malleable over the time scale of breeding cycles. This is evidenced by the discovery of 10 copies of Rhg1 in Fayette, a line developed by backcrossing Williams 82 (single copy) to PI 88788 (nine copies; Mikel et al., 2010). In contrast, all the sequenced SCN-resistant lines belonging to the low-copy Rhg1 group contained three copies of nearly identical Rhg1 repeats. It will be interesting to identify additional sources of SCN resistance to determine if the sequences in this Rhg1 group can persist in greater than three copies. This information, coupled with the relationship between larger numbers of Rhg1 repeats and increased resistance, suggests a new strategy to improve SCN resistance through the addition of Rhg1 copies.
There remains a need for improved assays that can inexpensively but accurately determine the copy number of Rhg1 or other high-copy-number loci that confer adaptive traits (Curtis et al., 2012; Maron et al., 2013; Stebbing et al., 2013). We initially utilized qPCR with genomic DNA templates for this purpose but found it challenging to obtain precise results for copy numbers above approximately four. Fiber-FISH provided definitive data, and whole-genome sequencing provided accurate estimates of higher copy number regions as long as the genome-wide read depth exceeded approximately 2-fold coverage. Comparative genome hybridization methods can also be used (Roberts et al., 2012). However, these relatively complex procedures are not likely to be useful, for example, in a plant breeding germplasm screen that seeks to identify rare individuals or infrequent recombination events carrying usefully elevated copy numbers.
Biochemical characterization of the wild-type (Williams 82-type), low-copy, and high-copy versions of the Glyma18g02590 α-SNAP alleles also is needed to determine what, if any, altered functions they have compared with each other and with canonical α-SNAP functions. We speculate that while the genomes containing the PI 88788-type α-SNAP have apparently benefited from an increase in Rhg1 copy number, the genomes with the Peking-type α-SNAP may have remained at three copies because of selection against an unknown negative impact of the Peking-type full-length α-SNAP. Rhg1 copy number in these genomes may also be affected by the shorter splice isoform of the Glyma18g02590 α-SNAP that was only detected in the low-copy Rhg1 lines. Alternatively, the loss of a wild-type (Williams 82-like) α-SNAP coding sequence in the three-copy genomes may have limited expansion of the locus. It is also possible that interactions with a specific Rhg4 allele may favor the Rhg1 locus configurations found in the low-copy Rhg1 haplotypes.
The identification of the different copy numbers at Rhg1 also suggests a hypothesis regarding the relatively ineffectual nature of low-copy Rhg1 in the absence of the resistance-conferring Rhg4 allele (Brucker et al., 2005a; Liu et al., 2012). In the absence of Peking-type Rhg4, the three copies of Rhg1 now known to be present in low-copy lines such as Peking have been shown to be more resistant to SCN infection than single-copy Rhg1 lines, suggesting that this Rhg1 can function independently of resistance-associated Rhg4 alleles (Brucker et al., 2005a; Liu et al., 2012). This raises the possibility that Rhg4 combined with the high-copy Rhg1 may provide a broader spectrum SCN resistance, while the Peking-type Rhg1 resistance could possibly be improved by increasing the copy number or expression level. Also, stacked deployment of both types of Rhg1 in single soybean lines could attenuate the development of virulent nematode populations. This type of research is increasingly important given the slow but ongoing erosion of the widely deployed PI 88788-derived resistance (Niblack et al., 2008; Tylka et al., 2012).
Our data help to explain the overlaps observed by many SCN-resistance specialists when comparing different soybean accessions with regard to their spectrum of resistance to a range of different SCN populations. For example, the resistance spectra of the Hg Type Test lines PI 88788, PI 209332, and Cloud (PI 548316) correlate highly, as do those of Peking (PI 548402), PI 90763, PI 89772, and PI 438489B (Colgrove and Niblack, 2008). Those two groupings match the Rhg1 DNA sequence, copy number, and α-SNAP groups discovered in this study.
PI 437654 is recognized for its particularly high levels of resistance against diverse nematode populations (Colgrove and Niblack, 2008). However, we discovered the near identity of PI 437654 Rhg1 copy number and sequence to other, less broadly resistant Rhg1 low-copy soybean lines. Although Rhg1 makes one of the strongest contributions to PI 437654-derived resistance (Webb, 2012), the present finding reemphasizes the importance of identifying and cloning additional SCN resistance quantitative trait loci from PI 437654 (Wu et al., 2009).
Current models for evolution by gene duplication are often applied to single gene duplicates. A fascinating and unusual element of Rhg1 is that gene copy number selection occurred, and research hypotheses are being tested, for an approximately 30-kb block of four genes that encode completely dissimilar proteins, three of which have been shown to contribute (Cook et al., 2012) to the phenotype that apparently has driven selection. Determining the exact course of evolution of the Rhg1 locus is difficult, but our data strongly suggest that the repeats in the low-copy and high-copy class have a common origin. It is not clear if the common Rhg1-resistant progenitor diverged from susceptible lines prior to duplication or if the divergence occurred after duplication. Either scenario could account for the highly similar sequence and the identical repeat junction found between low- and high-copy Rhg1 lines if repeat homogenization or gene conversion has played a role in the evolution of the Rhg1 locus and caused the high sequence identity between repeats within single plant lines.
Our data suggest that multiple evolutionary forces could have differentially affected the different genes in the repeat. Two of the proteins encoded at Rhg1 (Glyma18g02580 and Glyma18g02610) have identical derived amino acid sequences among the repeats and between the resistant and susceptible lines, which matches predictions for gene duplicates fixed by positive selection for increased dosage and having a low rate of nonsynonymous to synonymous substitutions (KN/KS < 1; Innan and Kondrashov, 2010). However, the presence of nonsynonymous substitutions in Glyma18g02590 in both the low- and high-copy Rhg1 lines, caused by different nucleotide polymorphisms, suggests a different evolutionary course, the duplication and divergence scenario that is applicable to many gene duplicates (Ohno, 1970). The identification of a premature stop codon in one copy of Glyma18g02600 in Peking, despite the highly similar SCN resistance between Peking and the other resistant lines in the low-copy class, is also interesting. This provides further evidence that Glyma18g02600 is not required for full Rhg1-mediated resistance and could be the first glimpse of pseudogenation (Lynch and Conery, 2000). Hence, the different genes in the Rhg1 repeat apparently represent different evolutionary trajectories.
The identification of Rhg1 DNA regions that exhibit differential methylation between SCN-resistant and SCN-susceptible accessions adds an additional layer of complexity to the control of phenotype expression at Rhg1 and probably to Rhg1 locus evolution. The observation of highly similar gene duplicates in the genomes of many organisms has led to the hypothesis that decreased expression of duplicate gene copies is a mechanism to maintain normal physiology following gene duplication (Qian et al., 2010). In recent work on mammalian gene duplicates, increased DNA methylation of promoter regions has been significantly correlated with gene duplicates and silencing, suggesting a potential mechanism for the restoration of dosage imbalance (Chang and Liao, 2012). This mechanism has also been suggested to follow whole-genome duplications, for example in soybean, where for a number of gene pairs one copy of the paralogous pair was often found to have increased repressive methylation and decreased expression (Schmitz et al., 2013a). Our observations for Rhg1 may seem to be the opposite of this, because in SCN-resistant lines with multiple Rhg1 copies, hypermethylation is observed at genes that exhibit increased transcript abundance. However, expression of the multicopy Rhg1 genes might be even greater in the SCN-resistant genomes if there were not methylation. Although beyond the scope of this study, recent identifications of dynamic methylation changes in Arabidopsis (Arabidopsis thaliana) following biotic stress (Dowen et al., 2012; Yu et al., 2013) suggest the hypothesis that the differentially methylated cytosine regions found upstream of Glyma18g02580, Glyma18g02590, and Glyma18g02610 could result in lower constitutive expression and increased expression of these genes following nematode infection. Future experiments to test this hypothesis may reveal further mechanisms that provide increased fitness and thereby impact the evolution of gene copy number variation.
MATERIALS AND METHODS
Estimating Copy Number and Transcript Abundance
To estimate the number of Rhg1 copies present in the Hg Type Test lines, we collected tissue for DNA extraction from 2-week-old plants grown in Metro Mix at 26°C. Leaf tissue was collected and flash frozen in liquid nitrogen, and DNA extraction was performed as described previously. To estimate Rhg1 copy number, qPCR was run using two separate primer pairs per sample. One set of primers described previously spanned the junction of repeated segmental Rhg1 duplicates, which failed to amplify a product in genomes with the wild-type single copy of the locus. A second primer pair used in a separate reaction amplified a product corresponding to a DNA interval from the gene Glyma18g02620, which is adjacent to but not present in the Rhg1 repeat. The ratio of the two products was used to determine the number of Rhg1 repeats.
To quantify the relative transcript abundance for the genes within and adjacent to the Rhg1 repeat interval, tissue was collected from the roots of plants 5 d after emergence. Plants were grown in a growth room in Metro Mix at 24°C and 16 h of light. The entire root-soil mass was removed from the pot, quickly immersed in water to remove excess soil, and flash frozen in liquid nitrogen. RNA was extracted using Trizol following the manufacturer’s recommended procedures. Contaminating DNA was removed from the samples using Turbo DNase following the manufacturer’s guidelines. To amplify cDNA from RNA, Bio-Rad’s iScript kit was used with 1 μg of total RNA per reaction following the manufacturer’s recommended guidelines. qPCR was performed as described previously (Cook et al., 2012). Briefly, primer pairs corresponding to transcripts of Glyma18g02570, Glyma18g02580, Glyma18g02590, Glyma18g02600, and Glyma18g02610 were used to amplify products for each sample in duplicate technical replicates. A product was also amplified from each sample corresponding to transcript of gene SKP1/ASK-interacting protein16 for use in normalizing samples across plates (Cook et al., 2012).
The soybean (Glycine max) lines previously defined to make up the Hg Type Test nematode test were chosen for analysis (Niblack et al., 2002). The lines are PI 548402 (Peking), PI 88788, PI 90763, PI 437654, PI 209332, PI 89772, and PI 548316 (Cloud). The other line used and referenced in this work is Fayette, which was developed by crossing Williams(2) with PI 88788. Progeny from this cross were backcrossed with Williams(2) while selecting for SCN resistance.
Transcript Analysis
To confirm the annotation of transcripts at Rhg1, RACE PCR was performed for 3′ analysis of Glyma18g02590 (Supplemental Table S4) using the SMARTer RACE cDNA kit per the manufacturer’s protocols (Clontech) and previously defined primers (Cook et al., 2012). Following RACE, PCR products were TA cloned into pCR8/GW/TOPO as mentioned previously. Randomly chosen colonies were sequenced to confirm the 3′ ends of individual transcripts.
Fiber-FISH
Fiber-FISH experiments were carried out using the same methods and probes as detailed previously (Cook et al., 2012), and Rhg1 repeat copy number findings are based on the maximum number of copies observed in at least 10 separate probe-hybridizing DNA fibers for a given plant genotype.
Whole-Genome Sequencing
Whole-genome sequencing was performed for lines Peking (PI 548402), PI 90763, PI 437654, PI 209332, PI 89772, and Cloud (PI 548316). Tissue was collected from at least five plants per sample totaling at least 3 g of tissue to homogenize any somatic or possible intraplant DNA variants. DNA was extracted following previously published protocols (Swaminathan et al., 2007). Two separate DNA libraries were constructed for each sample. For construction of the paired-end library, DNA was randomly sheared, separated, and enriched for DNA fragments ranging from 200 to 400 bp in length. Adapter sequence was added to the ends of each sample for bar coding following Illumina guidelines. Paired-end libraries for samples PI 209332, PI 89772, and Cloud were sequenced on a single Illumina HiSeq 2000 lane, producing reads of 101 bp sequenced from both ends of the fragment. Paired-end libraries for samples Peking, PI 90763, and PI 437654 were sequenced on Illumina’s HiSeq 2500 using the rapid sequencing run, producing sequence of 101 bp in both the forward and reverse directions. A separate library was also constructed for each sample using larger insert sizes, known as a mate-pair library. DNA for each sample was randomly sheared, separated, and collected, ranging in size from 2 to 3 kb. The mate-pair libraries were constructed using the mate-pair library preparation kit from Illumina following the manufacturer’s protocols. All six libraries were sequenced in the forward and reverse directions on a single Illumina HiSeq 2000 lane, generating sequencing lengths of 101 bp per direction. All samples were demultiplexed using their respective adapter sequences and processed following Illumina's Cassava-1.8.2 pipeline to generate data in the fastq format used for downstream applications.
An article is in preparation that will report the whole-genome sequencing data for the lines in the SoyNAM project (Q. Song, B.W. Diers, and P. Cregan, unpublished data). Briefly, each plant sample was paired-end sequenced on an Illumina HiSeq 2000, producing reads 151 bp in length in each direction. DNA insert sizes from the samples were 300 bp.
Previously sequenced Glycine soja data were downloaded from the Sequenced Read Archive section of the National Center for Biotechnology Information, stored under accession number SRA009252 (Kim et al., 2010b). Data from runs SRR020188, SRR020190, SRR020182, and SRR020184 were processed for analysis in this research.
Short-Read Genome Alignments
Rapid Genome Alignment for SoyNAM Lines
To rapidly estimate the copy number of the Rhg1 interval in the SoyNAM, reads were aligned to a limited reference using the program Bowtie2 (Langmead and Salzberg, 2012). The reference for mapping was created using the Bowtie2 build indexer function with input sequence corresponding to the Williams 82 reference genome (version 1.1, assembly 1.89) corresponding to the Rhg1 interval on chromosome 18 (1,581,000–1,714,000) and the homologous loci on chromosome 11 interval (37,361,000–37,456,000), chromosome 2 interval (47,705,000–47,855,000), chromosome 9 interval (45,995,000–46,345,000), and chromosome 14 position (4,240,265–4,340,264). Paired-end reads were mapped using default settings. Mapped reads were processed using Samtools (Li et al., 2009), and read depth was computed using the coverageBed program of BEDtools (Quinlan and Hall, 2010) over 1-kb bins ranging from 1,600,000 to 1,694,000. Read depth was estimated by summing the number of reads corresponding to the region 5′ of the Rhg1 repeat (1,600,000–1,631,999), the Rhg1 repeat (1,632,000–1,663,999), and the 3′ region (1,664,000–1,694,000). Copy number was estimated using both flanking regions, computed as the ratio of read depth corresponding to the Rhg1 interval divided by the total reads in the flanking interval. Read depth was reported as the average of these two ratios along the se.
Full Genome Alignment
Illumina sequencing reads were aligned to the full Williams 82 reference genome (build 1.89; http://www.phytozome.net/cgi-bin/gbrowse/soybean/) using the program BWA (version 0.7.1; Li and Durbin, 2009). Reads were mapped using the default settings of the aln function. Alignments were then paired using the sampe function. Alignments were further processed using the program Picard (version 1.83) to add read group information (AddOrReplaceReadGroups), mark PCR duplicates (MarkDuplicates), and merge alignments (MergeSamFiles) from separate sequencing reactions per genome. For the Hg Type Test data processing, PCR duplicates were marked at the lane level prior to merging the sequencing runs (McKenna et al., 2010).
Sequence Variant Detection
Sequence alignment files were processed for variant discovery using the GATK software package (version 2.4.9; DePristo et al., 2011). The best practices were followed as described. Insertion and deletion (INDEL) sites were identified using the RealignerTargetCreator and a set list of known INDELs. Because a known INDEL list is not publicly available for soybean, one was created following the GATK recommended guidelines. The list of known INDELs was created by selecting for concordance among high-confidence INDELs identified from the samples 4J105-34, LD00-3309, LG05-4292, and CL0J095-46 (i.e. INDELs predicted with confidence from all four genomes were used as the list of known INDELs). Following the RealignerTargetCreator, samples were realigned around INDEL sites using the IndelRealigner function with the following options: –consensusDeterminationModel USE_READS, –known INDELS, –maxConsensuses 70, –LODThresholdForCleaning 0.5, –maxReadsForConsensuses 600, –maxReadsForRealignment 100000. Following realignment, variants were called using the UnifiedGenotyper algorithm with the following options: –stand_call_conf 20, –stand_emit_conf 15, –rf BadCigar, –A VariantType, –glm BOTH. To remove false variants, a filter was applied to remove variants not sequenced at least three times and having a quality score greater than 50. Variant files were annotated with the program SnpEff as documented (Cingolani et al., 2012).
Copy Number Estimates
Read depth in the 1-kb intervals was averaged over the two flanking intervals to determine average read depth of the region per resequenced genome and used to determine the estimated copy number of the Rhg1 locus and the flanking intervals. We used average read depth over 1-kb intervals to estimate copy number from the whole-genome resequencing data. The analyzed interval was 93 kb, centered on the known 31-kb Rhg1 repeat with equally spaced flanking intervals. The average read depth in 1-kb bins was determined for the flanking Rhg1 regions and used to normalize read depth across bins. Final copy number estimates were made by averaging the normalized read depth across the three 32-kb intervals.
Network Analysis
To determine Rhg1 sequence relationships between soybean lines, we performed multiple sequence alignment using ClustalW2. The open reading frames for the genes Glyma18g02580, Glyma18g02590, Glyma18g02600, and Glyma18g02610 including 200 bp of upstream promoter sequence were concatenated and aligned. The alignment was used in SplitsTree (version 4.13.1) to construct a sequence network (Huson and Bryant, 2006). The analysis pipeline included Uncorrect P for distances and NeighborNet for network construction. Parsimony-Uninformative sites were excluded from the network.
Analysis of Nematode Resistance
To determine the relationship between nematode resistance and lines containing different copy numbers of Rhg1, we analyzed data collected as part of the Northern Regional SCN Tests (Cary and Diers, 2010, 2011, 2012, 2013). In total, we analyzed data from greenhouse nematode trials conducted on the seven Hg-type soybean lines and the susceptible control line Lee for 78 SCN field populations. Six plants per genotype were tested against the 78 different nematode populations. To more accurately estimate the variance for female index, we performed random replacement using the software R (R Development Core Team, 2009) with 1,000 bootstrap replicates per genotype-nematode combination. An ANOVA was computed using a linear mixed-effect model with bootstrap variances used to weight observations, expressed as the inverse of the variance. Residuals were checked for normality. P values were calculated using the generated test statistics, and a Bonferroni correction was applied to account for false positives resulting from multiple testing.
Methylation Analysis
Restriction Enzyme-Based Methylation Discovery
Locus-specific DNA methylation was analyzed using the methylation-specific endonuclease McrBC. McrBC digests DNA with methylated cytosines in a sequence-independent manner, while unmethylated DNA is unaffected. Restriction digestions were performed using 600 to 700 ng of DNA and manufacturer protocols. Adding the same amount of DNA to the reaction buffer with no restriction enzyme was used to set up control reactions. Samples with and without the restriction enzyme were incubated at 37°C for 90 min and heat inactivated at 65°C for 20 min. DNA was visualized on a 0.8% ethidium bromide-stained gel to ensure DNA digestion. Both digested and control DNA samples were used for subsequent PCR using GoTaq flexi DNA polymerase (Promega). For McrBC-treated DNA, PCR primers that spanned methylated DNA did not produce the intended product following PCR because the template DNA was digested by McrBC. DNA that was not methylated or not treated with the enzyme yielded a product of the expected size.
Computational Methylation Analysis
Data were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with series accession number GSE41753, deposited previously (Schmitz et al., 2013a). These data were analyzed using custom scripts written in Java or Bash to compute the data, and the results are presented in Figure 8 and Supplemental Figures S3, S4, and S5.
To estimate Rhg1 copy number, sequences from GEO accession number GSE41753 (GEO nos. GMS1024005–GMS1024008, GMS1134684, GMS1134698–GMS1134700, GMS1134705, GMS1134706, GMS1134709, GMS1134712–GMS1134714, GMS1134716, GMS1134718, GMS1134720, GMS1134722, GMS1134723, GMS1134729–GMS1134732, GMS1134734, GMS1134736, GMS1134741, GMS1134744, GMS1134749, and GMS1134756) were analyzed. The total number of cytosine sequencing reads was summed over 1-kb bins starting at position 1,600,225 and counting to the end of bin 1,696,224, for a total of 96 bins. Average sequencing coverage in the region was calculated by averaging the number of cytosine reads in the 1-kb bins over the two 32-kb intervals flanking Rhg1, which was used to normalize the read depth for each 1-kb bin. Final copy number estimates of the three 32-kb intervals were calculated as average normalized read depth over each respective 32-kb interval (Supplemental Fig. S3A).
To determine single-base cytosine methylation at the Rhg1 locus, sequences from GEO accession number GSE41753 were used for the corresponding groups: parental lines (GEO nos. GSM1024005 and GSM1024006); single-copy Rhg1 progeny (GEO nos. GSM1024007, GSM1134698–GSM1134700, GSM1134709, GSM1134712, GSM1134714, GSM1134716, GSM1134720, GSM1134723, GSM1134729–GSM1134731, GSM1134734, GSM1134741, and GSM1134749); and three-copy Rhg1 progeny (GEO nos. GSM1024008, GSM1134684, GSM1134713, GSM1134732, GSM1134744, and GSM1134756). The total number of cytosine sequencing reads and the total number of cytosine sequencing reads supporting methylation were summed over 150-bp bins starting at position 1,626,000 and counting to the end of bin 1,668,000, for a total of 280 bins. For each bin, the methylation level was computed by dividing the total number of cytosine reads supporting methylation by the total number of cytosines sequenced. Methylation levels were computed in the CG, CHG, and CHH sequence contexts. The data are represented in Figure 8 and Supplemental Figures S4 and S5.
To estimate the expression of genes within and adjacent to the Rhg1 repeat, processed RNA sequencing data were used to compare transcript levels across the four tested genotypes (GEO series GSE41753_RPKM supplementary file). To assess transcription differences, the reads per kilobase per million mapped sequence reads values from the three replicates of the single-copy Rhg1 parent LDX01-1-165 were first averaged. This number was used as a normalizer for the average of the reads per kilobase per million mapped sequence reads of the three replicates for the other three lines tested.
The whole-genome sequence data generated for this work have been deposited in the National Center for Biotechnology Information Sequence Read Archive. The samples can be accessed through the BioProject PRJNA243933 or for each BioSample: Peking (SAMN02721112), PI 90763 (SAMN02721113), PI 437654 (SAMN02721114), PI 209332 (SAMN02721115), PI 89772 (SAMN02721116), and Cloud (SAMN02721117).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleic acid sequence alignment of alpha-SNAP alleles and homolog.
Supplemental Figure S2. Positions of DNA variant sites in low- and high-copy locus types.
Supplemental Figure S3. Estimated Rhg1 copy number and expression data from a recombinant population.
Supplemental Figure S4. Differential CHH DNA methylation.
Supplemental Figure S5. Differential DNA methylation for two parents and 27 progeny at Rhg1.
Supplemental Table S1. SoyNAM sequencing summary statistics.
Supplemental Table S2. Hg-Type sequencing summary statistics.
Supplemental Table S3. Estimated Rhg1 copy number of SoyNAM lines using rapid mapping.
Supplemental Table S4. Summary of alpha-SNAP allele expression from cDNA sequencing.
Supplemental Table S5. Amino acid polymorphisms at Rhg1 paralogous locus.
Supplemental Table S6. Sequencing frequency at Rhg1 DNA variant sites in high-copy lines.
Supplementary Material
Acknowledgments
We thank Matthew Hudson and Tong Geon Lee for multiple conversations about this work and for sharing results prior to publication. We also thank Guy Plunkett, Marie Adams, John Alliet, Xiao-yu Liu, and the University of Wisconsin-Madison Biotechnology DNA Sequencing and Bioinformatics Resource Center for education and support, Nicholas Keuler for advice on statistical analysis of nematode resistance data, and the Henry Nguyen laboratory for sharing the cost of obtaining whole-genome sequence data.
Glossary
- SCN
soybean cyst nematode
- qPCR
quantitative PCR
- FISH
fluorescence in situ hybridization
- SoyNAM
soybean nested association mapping
- SNP
single-nucleotide polymorphism
- GATK
Genome Analysis Toolkit
- cDNA
complementary DNA
- DMRs
differentially methylated regions
- INDEL
insertion and deletion
- GEO
Gene Expression Omnibus
- PI
plant introduction
Footnotes
This work was supported by the United Soybean Board, the Wisconsin Soybean Marketing Board, and Wisconsin Experiment Station Hatch funds (to A.F.B.); the National Science Foundation (grant no. DBI–0922703 to J.J. and predoctoral fellowship to A.M.B.); the Pioneer Fellowship in Plant Pathology (to D.E.C.); and the American Phytopathological Society through a gift from Pioneer Hi-Bred.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. (2006) Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851–855 [DOI] [PubMed] [Google Scholar]
- Arelli APR, Webb DM. (1996) Molecular genetic diversity among soybean plant introductions with resistance to Heterodera glycines. Curr Sci 71: 230–233 [Google Scholar]
- Barnard RJ, Morgan A, Burgoyne RD. (1996) Domains of alpha-SNAP required for the stimulation of exocytosis and for N-ethylmalemide-sensitive fusion protein (NSF) binding and activation. Mol Biol Cell 7: 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard RJ, Morgan A, Burgoyne RD. (1997) Stimulation of NSF ATPase activity by alpha-SNAP is required for SNARE complex disassembly and exocytosis. J Cell Biol 139: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass C, Field LM. (2011) Gene amplification and insecticide resistance. Pest Manag Sci 67: 886–890 [DOI] [PubMed] [Google Scholar]
- Brucker E, Carlson S, Wright E, Niblack T, Diers B. (2005a) Rhg1 alleles from soybean PI 437654 and PI 88788 respond differentially to isolates of Heterodera glycines in the greenhouse. Theor Appl Genet 111: 44–49 [DOI] [PubMed] [Google Scholar]
- Brucker E, Niblack T, Kopisch-Obuch FJ, Diers BW. (2005b) The effect of rhg1 on reproduction of Heterodera glycines in the field and greenhouse and associated effects on agronomic traits. Crop Sci 45: 1721–1727 [Google Scholar]
- Bryant D, Moulton V. (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21: 255–265 [DOI] [PubMed] [Google Scholar]
- Caldwell BE, Brim CA, Ross JP. (1960) Inheritance of resistance of soybean to the cyst nematode, Heterodera glycines. Agron J 52: 635–636 [Google Scholar]
- Cary T, Diers BW (2010) 2009 Northern regional soybean cyst nematode tests. University of Illinois, Urbana. http://cropsci.illinois.edu/research/SCN-tests (May 2014) [Google Scholar]
- Cary T, Diers BW (2011) 2010 Northern regional soybean cyst nematode tests. University of Illinois, Urbana. http://cropsci.illinois.edu/research/SCN-tests (May 2014) [Google Scholar]
- Cary T, Diers BW (2012) 2011 Northern regional soybean cyst nematode tests. University of Illinois, Urbana. http://cropsci.illinois.edu/research/SCN-tests (May 2014) [Google Scholar]
- Cary T, Diers BW (2013) 2012 Northern regional soybean cyst nematode tests. University of Illinois, Urbana. http://cropsci.illinois.edu/research/SCN-tests (May 2014) [Google Scholar]
- Chang AYF, Liao BY. (2012) DNA methylation rebalances gene dosage after mammalian gene duplications. Mol Biol Evol 29: 133–144 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu XY, Ruden DM. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgrove AL, Niblack TL. (2008) Correlation of female indices from virulence assays on inbred lines and field populations of Heterodera glycines. J Nematol 40: 39–45 [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR. (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44: 1121–1131 [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209 [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S. (2010) Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol 2: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. (2009) The life and death of gene families. Bioessays 31: 29–39 [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diers BW, Cary T, Thomas D, Colgrove A, Niblack T. (2010) Registration of LD00-2817P soybean germplasm line with resistance to soybean cyst nematode from PI 437654. Journal of Plant Registrations 4: 141–144 [Google Scholar]
- Diers BW, Skorupska HT, Rao-Arelli AP, Cianzio SR. (1997) Genetic relationships among soybean plant introductions with resistance to soybean cyst nematodes. Crop Sci 37: 1966–1972 [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–E2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo BY. (1984) Ultrastructure of the esophagus of larvae of the soybean cyst nematode, Heterodera glycines. Proceedings of the Helminthological Society of Washington 51: 1–24 [Google Scholar]
- Feuk L, Carson AR, Scherer SW. (2006) Structural variation in the human genome. Nat Rev Genet 7: 85–97 [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Gohlke J, Scholz CJ, Kneitz S, Weber D, Fuchs J, Hedrich R, Deeken R. (2013) DNA methylation mediated control of gene expression is critical for development of crown gall tumors. PLoS Genet 9: e1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinberg A, Siu E, Stern C, Lawrence EA, Ferdig MT, Deitsch KW, Kirkman LA. (2013) Direct evidence for the adaptive role of copy number variation on antifolate susceptibility in Plasmodium falciparum. Mol Microbiol 88: 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Herraez I, Prado-Martinez J, Garg P, Fernandez-Callejo M, Heyn H, Hvilsom C, Navarro A, Esteller M, Sharp AJ, Marques-Bonet T. (2013) Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet 9: e1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kim DG, Riggs RD, Mauromoustakos A. (1998) Variation in resistance of soybean lines to races of Heterodera glycines. J Nematol 30: 184–191 [PMC free article] [PubMed] [Google Scholar]
- Kim M, Hyten DL, Niblack TL, Diers BW. (2011) Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci 51: 934–943 [Google Scholar]
- Kim MS, Hyten DL, Bent AF, Diers BW. (2010a) Fine mapping of the SCN resistance locus rhg1-b from PI 88788. Plant Genome 3: 81–89 [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma J, et al. (2010b) Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci USA 107: 22032–22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kim KS, Riggs RD. (2010c) Differential subcellular responses in resistance soybeans infected with soybean cyst nematode races. Plant Pathol J 26: 154–158 [Google Scholar]
- Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. (2011) Differences in gene expression amplitude overlie a conserved transcriptomic program occurring between the rapid and potent localized resistant reaction at the syncytium of the Glycine max genotype Peking (PI 548402) as compared to the prolonged and potent resistant reaction of PI 88788. Plant Mol Biol 75: 141–165 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. (2012) Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279: 5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. (2002) Selection in the evolution of gene duplications. Genome Biol 3: H0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé P, Berticat C, Berthomieu A, Unal S, Bernard C, Weill M, Lenormand T. (2007) Forty years of erratic insecticide resistance evolution in the mosquito Culex pipiens. PLoS Genet 3: e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritis JA, Rebois RV, Graney LS. (1983) Development of Heterodera glycines ichinohe on soybean, Glycine max (L) Merr., under gnotobiotic conditions. J Nematol 15: 272–281 [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Chen SY, Young ND. (2004) Effect of the rhg1 gene on penetration, development and reproduction of Heterodera glycines race 3. Nematology 6: 729–736 [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, et al. (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492: 256–260 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Skorupska HT. (1996) Cytological expression of early response to infection by Heterodera glycines Ichinohe in resistant PI 437654 soybean. Genome 39: 986–998 [DOI] [PubMed] [Google Scholar]
- Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, et al. (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci USA 110: 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Girirajan S, Eichler EE. (2009) The origins and impact of primate segmental duplications. Trends Genet 25: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson AL, Williams LF. (1965) Evidence of a fourth gene for resistance to the soybean cyst nematode. Crop Sci 5: 477 [Google Scholar]
- Matsye PD, Lawrence GW, Youssef RM, Kim KH, Lawrence KS, Matthews BF, Klink VP. (2012) The expression of a naturally occurring, truncated allele of an α-SNAP gene suppresses plant parasitic nematode infection. Plant Mol Biol 80: 131–155 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikel MA, Diers BW, Nelson RL, Smith HH. (2010) Genetic diversity and agronomic improvement of North American soybean germplasm. Crop Sci 50: 1219–1229 [Google Scholar]
- Moore RC, Purugganan MD. (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Morgan A, Dimaline R, Burgoyne RD. (1994) The ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF) is regulated by soluble NSF attachment proteins. J Biol Chem 269: 29347–29350 [PubMed] [Google Scholar]
- Niblack TL. (2005) Soybean cyst nematode management reconsidered. Plant Dis 89: 1020–1026 [DOI] [PubMed] [Google Scholar]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL. (2002) A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol 34: 279–288 [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Colgrove AL, Colgrove K, Bond JP. (2008) Shift in virulence of soybean cyst nematode is associated with use of resistance from PI 88788. Plant Health Progress. http://dx.doi.org/10.1094/PHP-2008-0118-01-RS (May 2014) [Google Scholar]
- Niblack TL, Lambert KN, Tylka GL. (2006) A model plant pathogen from the kingdom Animalia: Heterodera glycines, the soybean cyst nematode. Annu Rev Phytopathol 44: 283–303 [DOI] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Bohnert HJ, Cheeseman JM. (2012) Life at the extreme: lessons from the genome. Genome Biol 13: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer, Berlin [Google Scholar]
- Olsen KM, Wendel JF. (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64: 47–70 [DOI] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. (2008) Copy number variation and evolution in humans and chimpanzees. Genome Res 18: 1698–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian WF, Liao BY, Chang AYF, Zhang JZ. (2010) Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rice LM, Brunger AT. (1999) Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol Cell 4: 85–95 [DOI] [PubMed] [Google Scholar]
- Roberts I, Carter SA, Scarpini CG, Karagavriilidou K, Barna JC, Calleja M, Coleman N. (2012) A high-throughput computational framework for identifying significant copy number aberrations from array comparative genomic hybridisation data. Adv Bioinformatics 2012: 876976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, et al. (2010) Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, He Y, Valdés-López O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. (2013a) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res 23: 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013b) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SB (1998) The Cyst Nematodes. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Stebbing J, Filipovic A, Lit LC, Blighe K, Grothey A, Xu Y, Miki Y, Chow LW, Coombes RC, Sasano H, et al. (2013) LMTK3 is implicated in endocrine resistance via multiple signaling pathways. Oncogene 32: 3371–3380 [DOI] [PubMed] [Google Scholar]
- Steemans P, Hérissé AL, Melvin J, Miller MA, Paris F, Verniers J, Wellman CH. (2009) Origin and radiation of the earliest vascular land plants. Science 324: 353. [DOI] [PubMed] [Google Scholar]
- Sutherland E, Coe L, Raleigh EA. (1992) McrBC: a multisubunit GTP-dependent restriction endonuclease. J Mol Biol 225: 327–348 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Varala K, Hudson ME. (2007) Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Amon A. (2013) Gene copy-number alterations: a cost-benefit analysis. Cell 152: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Foote SJ, Kemp DJ, Cowman AF. (1991) Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol Cell Biol 11: 5244–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylka GL, Gebhart GD, Marrett CC, Mullaney MP (2012) Evaluation of soybean varieties resistant to soybean cyst nematode in Iowa in 2012. Iowa State University Extension and Outreach. http://www.plantpath.iastate.edu/tylkalab/files/2012%20ISU%20VT%20report_0.pdf (May 2014) [Google Scholar]
- USDA (2014) Germplasm resources information network (GRIN). National Germplasm Resources Laboratory, Beltsville, MD. http://www.ars-grin.gov/npgs/index.html (May 2014) [Google Scholar]
- Walling JG, Jiang JM (2012) DNA and chromatin fiber-based plant cytogenetics. In Plant Cytogenetics: Genome Structure and Chromosome Function, Vol 4. Springer, New York, pp 121–130 [Google Scholar]
- Webb DM (November 1, 2012) Quantitative trait loci associated with soybean cyst nematode resistance and uses thereof. US Patent No. 20120278953 A1 [Google Scholar]
- Webb DM, Baltazar BM, Rao-Arelli AP, Schupp J, Clayton K, Keim P, Beavis WD. (1995) Genetic mapping of soybean cyst nematode race-3 resistance loci in the soybean PI 437.654. Theor Appl Genet 91: 574–581 [DOI] [PubMed] [Google Scholar]
- Wrather JA, Koenning SR (2009) Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Progress. http://dx.doi.org/10.1094/PHP-2009-0401-01-RS (May 2014) [Google Scholar]
- Wu HJ, Zhang ZH, Wang JY, Oh DH, Dassanayake M, Liu BH, Huang QF, Sun HX, Xia R, Wu YR, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Blake S, Sleper DA, Shannon JG, Cregan P, Nguyen HT. (2009) QTL, additive and epistatic effects for SCN resistance in PI 437654. Theor Appl Genet 118: 1093–1105 [DOI] [PubMed] [Google Scholar]
- Young LD. (1996) Yield loss in soybean caused by Heterodera glycines. J Nematol 28: 604–607 [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lepère G, Jay F, Wang JY, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LTY, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. (2013) Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.