An open-source information system for high-throughput plant phenotyping enables large-scale image analysis for different species based on real-time imaging data obtained from different spectra.
Abstract
High-throughput phenotyping is emerging as an important technology to dissect phenotypic components in plants. Efficient image processing and feature extraction are prerequisites to quantify plant growth and performance based on phenotypic traits. Issues include data management, image analysis, and result visualization of large-scale phenotypic data sets. Here, we present Integrated Analysis Platform (IAP), an open-source framework for high-throughput plant phenotyping. IAP provides user-friendly interfaces, and its core functions are highly adaptable. Our system supports image data transfer from different acquisition environments and large-scale image analysis for different plant species based on real-time imaging data obtained from different spectra. Due to the huge amount of data to manage, we utilized a common data structure for efficient storage and organization of data for both input data and result data. We implemented a block-based method for automated image processing to extract a representative list of plant phenotypic traits. We also provide tools for build-in data plotting and result export. For validation of IAP, we performed an example experiment that contains 33 maize (Zea mays ‘Fernandez’) plants, which were grown for 9 weeks in an automated greenhouse with nondestructive imaging. Subsequently, the image data were subjected to automated analysis with the maize pipeline implemented in our system. We found that the computed digital volume and number of leaves correlate with our manually measured data in high accuracy up to 0.98 and 0.95, respectively. In summary, IAP provides a multiple set of functionalities for import/export, management, and automated analysis of high-throughput plant phenotyping data, and its analysis results are highly reliable.
Plant bioinformatics faces the challenge of integrating information from the related “omics” fields to elucidate the functional relationship between genotype and observed phenotype (Edwards and Batley, 2004), known as the genotype-phenotype map (Houle et al., 2010). One of the main obstacles is our currently limited ability of systemic depiction and quantification of plant phenotypes, representing the so-called phenotyping bottleneck phenomenon (Furbank and Tester, 2011). To get a comprehensive genotype-phenotype map, more accurate and precise phenotyping strategies are required to empower high-resolution linkage mapping and genome-wide association studies in order to uncover underlying genetic variants associated with complex phenotypic traits, which aim to improve the efficiency, effectiveness, and economy of cultivars in plant breeding (Cobb et al., 2013). In the era of phenomics, automatic high-throughput phenotyping in a noninvasive manner is applied to identify and quantify plant phenotypic traits. Plants are bred in fully automated greenhouses under predefined environmental conditions with controlled temperature, watering, and humidity. To meet the demand of data access, exchange, and sharing, several phenomics-related projects in the context of several consortia have been launched, such as the International Plant Phenotyping Network (http://www.plantphenomics.com/), the European Plant Phenotyping Network (http://www.plant-phenotyping-network.eu/), and the German Plant Phenotyping Network (http://www.dppn.de/).
Thanks to the development of new imaging and transport systems, various automated or semiautomated high-throughput plant phenotyping systems are being developed and used to examine plant function and performance under controlled conditions. PHENOPSIS (Granier et al., 2006) is one of the pioneering platforms that was developed to dissect genotype-environment effects on plant growth in Arabidopsis (Arabidopsis thaliana). GROWSCREEN (Walter et al., 2007; Biskup et al., 2009; Jansen et al., 2009; Nagel et al., 2012) was designed for rapid optical phenotyping of different plant species with respect to different biological aspects. Other systems in the context of high-throughput phenotyping include Phenodyn/Phenoarch (Sadok et al., 2007), TraitMill (Reuzeau et al., 2005; Reuzeau, 2007), Phenoscope (Tisné et al., 2013), RootReader3D (Clark et al., 2011), GROW Map (http://www.fz-juelich.de/ibg/ibg-2/EN/methods_jppc/methods_node.html), and LemnaTec Scanalyzer 3D. These developments enable the phenotyping of specific organs (e.g. leaf, root, and shoot) or of whole plants. Some of them are even used for three-dimensional plant analysis (Clark et al., 2011). Consequently, several specific software applications (a comprehensive list can be found at http://www.phenomics.cn/links.php), such as HYPOTrace (Wang et al., 2009), HTPheno (Hartmann et al., 2011), LAMINA (Bylesjö et al., 2008), PhenoPhyte (Green et al., 2012), Rosette Tracker (De Vylder et al., 2012), LeafAnalyser (Weight et al., 2008), RootNav (Pound et al., 2013), SmartGrain (Tanabata et al., 2012), and LemnaGrid, were designed to extract a wide range of measurements, such as height/length, width, shape, projected area, digital volume, compactness, relative growth rate, and colorimetric analysis.
The huge amount of generated image data from various phenotyping systems requires appropriate data management as well as an appropriate analytical framework for data interpretation (Fiorani and Schurr, 2013). However, most of the developed image-analysis tools are designed for a specific task, for specific plant species, or are not freely available to the research community. They lack flexibility in terms of needed adaptations to meet new analysis requirements. For example, it would be desirable that a system could handle imaging data from different sources (either from fully automated high-throughput phenotyping systems or from setups where images are acquired manually), different imaging modalities (fluorescence, near-infrared, and thermal imaging), and/or different species (wheat [Triticum aestivum], barley [Hordeum vulgare], maize [Zea mays], and Arabidopsis).
In this work, we present Integrated Analysis Platform (IAP), a scalable open-source framework, for high-throughput plant phenotyping data processing. IAP handles different image sources and helps to organize phenotypic data by retaining the metadata from the input in the result data set. In order to measure phenotypic traits in new or modified setups, users can easily create new analysis pipelines or modify the predefined ones. IAP provides various user-friendly interfaces at different system levels to meet the demands of users (e.g. software developers, bioinformaticians, and biologists) with different experiences in software programming.
RESULTS AND DISCUSSION
IAP as an Automated Analysis Pipeline for High-Throughput Plant Phenotyping
IAP implements an automated workflow for high-throughput phenotyping data analysis and can efficiently measure plant phenotypic traits from multiple experimental setups. It provides different user interfaces (command line and graphical interface) especially for users with different levels of background skill (Fig. 1). The underlying architecture of the core of the IAP system is shown in Supplemental Figure S1. This kind of design makes it easy to extend existing analysis pipelines or to develop new imaging analysis tools as IAP plugins. A typical run of the IAP system consists of three main steps: (1) image and metadata import as well as experiment creation; (2) parameter optimization and data analysis; and (3) report generation and result export for postprocessing. The detailed steps of the IAP workflow are diagrammed in Figure 2 and described in detail below.
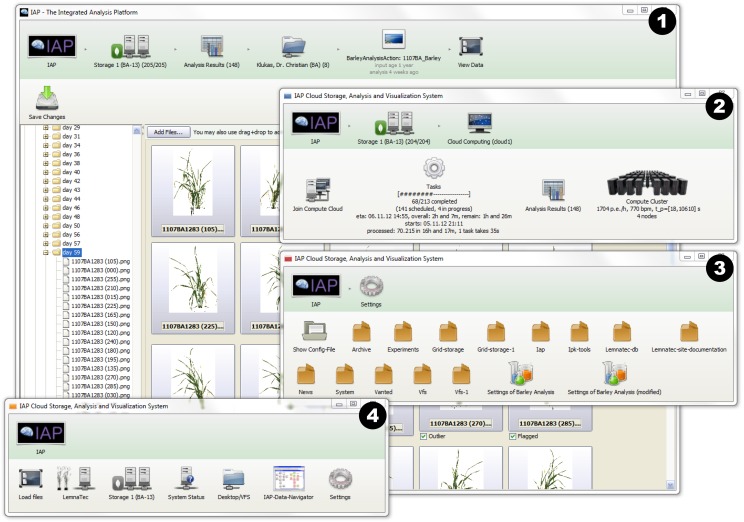
Figure 1.
The graphical user interface of the IAP system. Several windows can be opened by the user in parallel, as shown in the screenshot: (1) the main window showing the overview of experiment data (browsing and processing images); (2) the monitoring status of analysis jobs and grid-computing nodes; (3) the panel of system settings; and (4) the buttons of the main menu. [See online article for color version of this figure.]
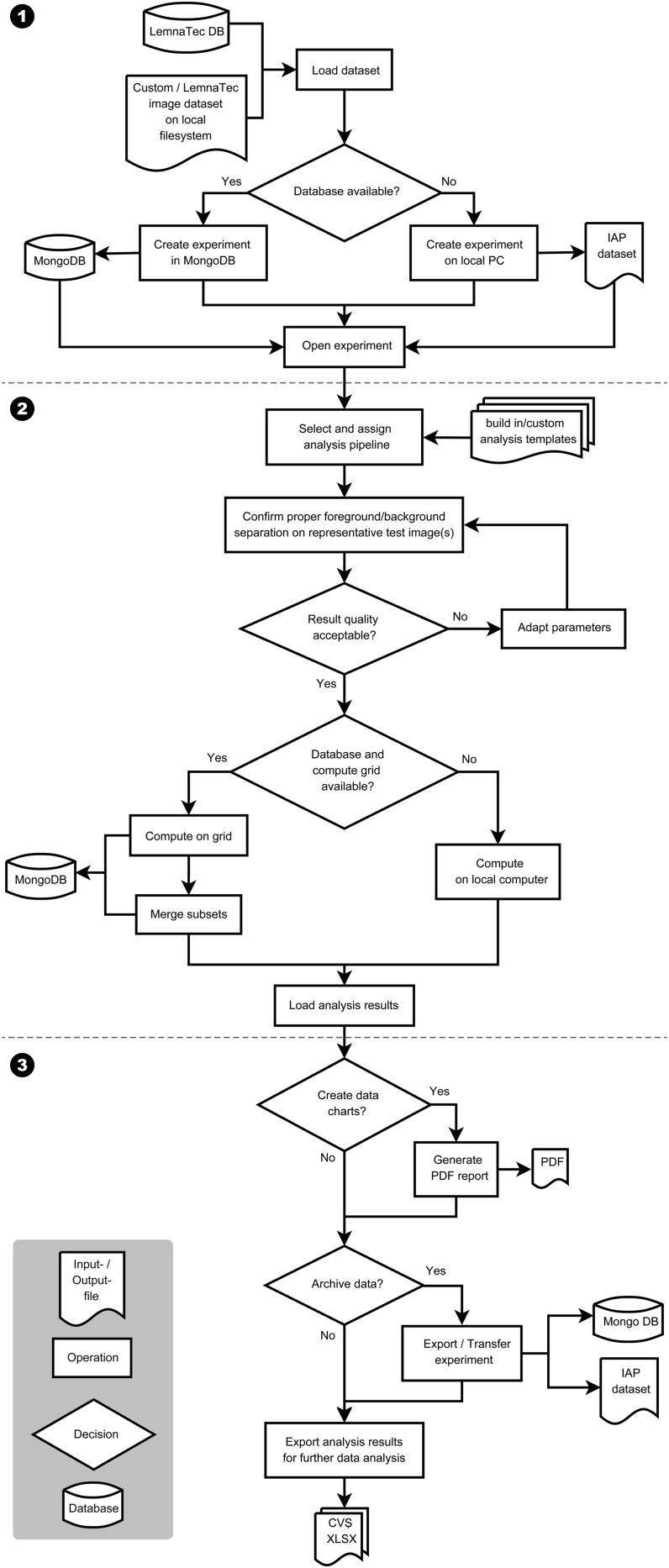
Figure 2.
A typical workflow for analyzing imaging data from high-throughput plant phenotyping experiments with IAP. Numbers indicate three main steps of the workflow: (1) loading/importing data set and experiment creation; (2) parameter optimization and image data analysis; and (3) report generation and result export.
Image data sets can be either loaded automatically from the LemnaTec system or imported from other sources, such as a manual plant image data set deposited on a local file system. A new experiment will be created based on the imported data set. If no powerful server is available, several servers and/or personal computers (PCs) could be utilized in order to speed up the analysis. In this case, the analysis is performed on a grid compute cluster. In the grid-computing mode, a MongoDB database (http://www.mongodb.org/) is required to be set up for the central storage of data and for work distribution to the computing nodes. Otherwise, the data will be saved on a single local computer or server for analysis. Depending on the type of the input data set, a specific analysis pipeline template has to be specified. The experiment templates can be built-in pipelines provided by IAP (see below) or pipelines customized by users. IAP provides tested default values for parameters of the built-in pipelines, which could be tested and further optimized for specific experiments. We also implemented a user-friendly interface to help to adjust the pipeline parameters. In general, users can choose images from a few plants at several time points to test the pipeline. Potential adjustments are automatically saved and can be exported in a separate configuration file that can be used for future analysis runs in similar situations. After completion of the quality check and potential parameter optimization in a selected pipeline, image processing can be automatically executed on grid computers or on a local PC. The system provides functionalities for generating an analysis report, supporting the user in summarizing and visualizing the calculated traits. Finally, several options are provided to export, transfer, and archive the analysis results.
Image-Processing Pipeline
The processing pipeline is implemented in a block-based manner. Each block fulfills a specific image-processing task. The block-based design allows the easy replacement of an existing block when changes are required in order to handle a new analysis task. Blocks can even be supplied by experienced end users by implementing IAP extension application program interfaces (APIs). Users are free to disable or remove existing blocks and to use a block several times within a pipeline. When creating a new pipeline to analyze image data from a new species, users need only to change the settings of some blocks instead of redefining the whole analysis pipeline. For example, if the segmentation method is needed to be changed, only the block that implements the settings of the segmentation algorithm has to be changed or replaced accordingly. The IAP system provides pipeline templates for the analysis of several plant species, such as Arabidopsis, barley, and maize. Therefore, IAP is designed as a flexible and open system for high-throughput plant phenotyping.
A detailed overview about the class definition of blocks is shown in Supplemental Figure S1. Generally, blocks can be created, modified, or extended with custom algorithms. The predefined abstract classes of blocks implement several input and output methods that can be individually extended by the user. For a specific experiment, the corresponding images and metadata will be loaded automatically into the system. Images are loaded by the block class BlLoadImages and stored in a Java object called ImageSet. Based on the fact that the experimental settings may be different at different stages of an experiment (reflected in the metadata), the implemented blocks are able to handle images with the consideration of the corresponding metadata.
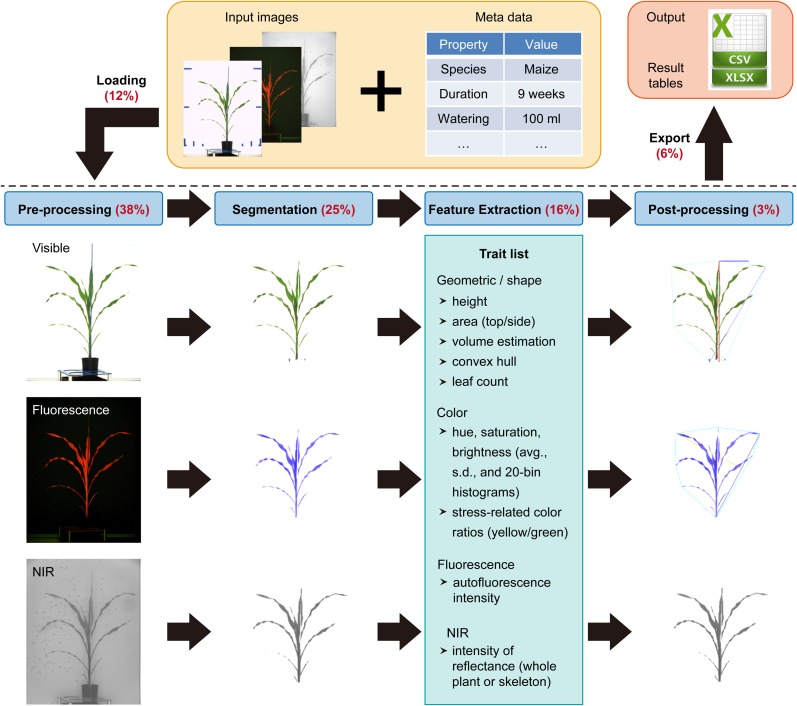
In the following sections, we elaborate on the four key components in the IAP image-processing pipeline. These key components are (1) preprocessing, (2) segmentation, (3) feature extraction, and (4) postprocessing (Fig. 3). Each part consists of several analysis blocks (Java classes) to process images from visible light and near-infrared, thermal, or fluorescence cameras. Examples of implemented blocks in each component are provided in Table I.
Figure 3.
The workflow of the proposed image-processing pipeline. Images shown are based on the example application to a maize data set (for summary of the data set, see Table I). Image data and metadata are imported via IAP functionalities (top) and subjected to image processing, including (1) preprocessing, prepare the images for segmentation; (2) segmentation, divide the image into different parts that have different meanings (foreground, plant; background, imaging chamber and machinery); (3) feature extraction, classify the segmentation result and get a trait list (examples include images from visible-light, fluorescence, and near-infrared [NIR] cameras); and (4) postprocessing, summarize calculated results for each plant. Optionally, analysis results can be marked in the images. Finally, result images are exported. Numbers in parentheses indicate the percentage of overall processing time for each analysis step.
Table I. Overview of the implemented blocks in an analysis template.
+, Processed.
| Block (Java Classes) | Description | Visible Light Image | Fluorescence Image | Near-Infrared Image |
|---|---|---|---|---|
| Preprocessing | ||||
| BlRotate | Align rotation | + | + | + |
| BlAlign | Align orientation | + | + | + |
| BlDetectBlueMarkers | Detect and delete blue markers | + | ||
| BlCutFromSide | Clear images | + | + | + |
| BlColorBalanceVerticalVis | Apply vertical color balancing on visible image | + | ||
| BlColorBalanceVerticalFluo | Apply vertical color balancing on fluorescence image | + | ||
| BlColorCorrectionNir | Removes shading from near-infrared image | + | ||
| BlColorBalanceCircularVisNir | Apply circular color balancing | + | + | |
| Segmentation | ||||
| BlRemoveBackground | Clear background by reference image | + | + | + |
| BlAdaptiveSegmentationFluo | Create intensity images for red and yellow reflectance | + | ||
| BlLabFilter | Apply filter in L*a*b* color space | + | + | |
| BlAdaptiveThresholdNir | Apply adaptive threshold on near-infrared image | + | ||
| BlMedianFilterFluo | Apply median filter | + | ||
| BlMorphologicalOperations | Apply morphological operations | + | + | + |
| BlAdaptiveRemoveSmallObjectsVisFluo | Remove artifacts | + | + | |
| BlUseFluoMaskToClearOther | Remove artifacts by image comparison with fluorescence image | + | + | |
| Feature extraction | ||||
| BlSkeletonizeVisFluo | Calculate the skeleton by thinning | + | + | |
| BlSkeletonizeNir | Calculate the skeleton by thinning | + | ||
| BlCalcWidthAndHeight | Determine plant height and width | + | ||
| BlCalcMainAxis | Calculate the plant main axis growth orientation | + | ||
| BlCalcColorHistograms | Calculate overall properties (pixel count, intensities, and plant color indices) | + | + | + |
| BlCalcConvexHull | Calculate convex hull-based shape parameters | + | + | |
| BlCalcAreas | Calculate plant area based on segmentation result | + | + | + |
| BlCalcVolumes | Estimate plant volume (digital biomass) | + | + | |
| BlDetectLeafTips | Detect leaf tips | + | + | |
| BlTrackLeafTips | Track leaf tips | + | + | |
| Postprocessing | ||||
| BlRunPostProcessors | Draw analysis results of feature extraction blocks | + | + | + |
| BlMoveMasksToImageSet | Transfer images to result image set | + | + | + |
| BlHighlightNullResults | Mark errors and outliers in result image set | + | + | + |
Preprocessing
In order to reduce influences caused by different camera systems and to further make images captured at different plant growth stages more comparable, images are first subjected to preprocessing. The following key issues are considered: (1) inconsistent orientation and alignment of images acquired from the different cameras, due to small position shifts or different zoom scales of cameras; (2) uneven brightness of images, such as reduced brightness in the image corners (vignetting), caused by the camera optics (especially for near-infrared images and images from visible light); (3) irregular illumination caused by the fluctuation of the light intensity during the imaging run or by the position of the light sources (which are located above in the imaging chamber and behind the camera, leading to shading from top to bottom); (4) noise caused by increasing temperatures of the camera sensors (with longer duration of the imaging loop, the temperature of the imaging chambers may rise by the absorption of light, leading to a higher signal-to-noise ratio of the CDD sensor); and (5) zoom change during the experiment (the zoom level of the cameras may need to change to accommodate the increasing size of the plant).
These problems can be addressed by a series of preprocessing blocks (Table I). First, it is necessary to perform image normalization and to acquire calibration values. Image normalization (removal of shading and correction of color changes) is accomplished by (1) vertical color balance, by using a linear interpolation to correct illumination and color changes in each channel of the RGB (red, green, and blue) color space from the top to the bottom of images (BlColorBalanceVertical), and (ii) circular color balancing, which is applied to images from the middle to the outside by using a spline interpolation (programming library provided by http://commons.apache.org/proper/commons-math/ and used in the block class BlColorBalanceCircular). Correction for varying zoom levels is dependent on the detection of markers in the imaging chambers, which is realized by applying an L*a*b* (L* for lightness, a* for color from green to magenta, and b* for color from blue to yellow) color filter and a cluster detection algorithm to identify blue marker points, positioned at the sides of the imaging chamber (BlDetectBlueMarkers). Next, the recognized markers are used to (1) adjust the image analysis to zoom changes, (2) get a scaling factor used to calculate absolute values of plant features (e.g. to calculate the real height of a plant in millimeters), and (3) crop images based on marker positions as used in the block BlCutFromSide.
Segmentation
Segmentation is a process of partitioning a digital image into multiple context-related segments. Every pixel in an image can be assigned to a specific label based on the result of segmentation. Thus, image segmentation is typically used to split the image into meaningful objects (e.g. plant parts and background objects, such as pots, sticks, and cages) and to recognize their boundaries. In the segmentation step, images will be divided into foreground (labeled as plant) and background parts. To this end, if available, an image of the background without the plant (reference image) is used at first for the initial background subtraction, which is a commonly used method in the object-tracking application (Yilmaz et al., 2006). During this process, the distance (computed in the L*a*b* color space) of each pixel from the same position of the currently processed image and the reference image is calculated. Objects are then identified based on the calculated distance utilizing automatic thresholding methods from ImageJ. However, based on this method, it is possible to clearly determine the foreground objects (moving objects like plant, pot, carrier, etc.).
To efficiently segment color images, we implemented an algorithm based on K-means clustering (Macqueen, 1967; Luccheseyz and Mitray, 2001), in which the color classes generally represent the plant, background, and other support objects. Further segmentation continues by analyzing the fluorescence images. We estimate a threshold by using the automated ImageJ thresholding method based on the YEN algorithm (Yen et al., 1995). The fluorescence reflection has a limited color characteristic in the yellow and red spectral channels. Processing of the fluorescence images is performed in the HSV (for hue, saturation, and brightness value) color space, which provides adequate results by using the Hue channel to characterize the fluorescence emissions from red to yellow. Very dark image parts, or parts that appear green in these images, are labeled as background pixels. Processing of comparably low-resolution near-infrared images is complicated by the fact that the plant pixels at the plant border are strongly correlated with their adjacent background pixels; therefore, segmentation of near-infrared images is still a challenge. In our pipeline, an adaptive thresholding method (Sauvola and Pietikainen, 2000) is adopted for separating the plant from the background (BlAdaptiveThresholdNir).
The segmentation result may still include small noise objects and artifacts. Therefore the images are subjected to postprocessing by image restoration methods. Filling small gaps and holes by a morphological closing operation (BlMorphologicalOperations) reconnects disconnected plant structures. A median filter (adopted from ImageJ) is applied to the fluorescence image to suppress noise (BlMedianFilterFluo). Artifacts caused by other background factors and reflections of the imaging chamber are deleted based on their size, shape, and color (BlAdaptiveRemoveSmallObjectsVisFluo). Finally, since fluorescence images provide the best segmentation results, the segmentation mask of the fluorescence image is slightly enlarged by a blur operation and then applied to the visible-light image. The result is then applied back to the fluorescence image and images from other camera types, if needed (BlUseFluoMaskToClearOther).
Feature Extraction
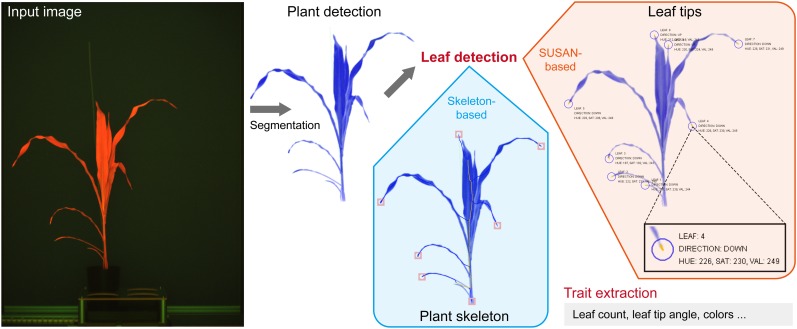
Based on the segmentation result, four classes of plant features (geometric, color, fluorescence-related, and near-infrared-related traits) are derived from the imaging data. Geometric traits like height, width, projected area, skeleton length, convex hull, and number of leaf tips (equivalent to number of leaves) are calculated in the blocks BlSkeletonize, BlCalcWidthAndHeight, BlCalcMainAxis, BlCalcConvexHull, BlCalcAreas, and BlDetectLeafTips. To calculate the number of leaf tips, we developed a new method that is based on the SUSAN approach (Smith and Brady, 1997). We modified this approach by a region-growing-based pixel-search algorithm to enhance the precision in estimating corner features. Color, position, and orientation can also be derived for each detected leaf tip (Fig. 4). The block BlTrackLeafTips uses a greedy approach to track the leaf tip progression by grouping the individual leaf tips over time based on the similarity of their positions and orientations.
Figure 4.
Detection of maize leaves with IAP. Input images (e.g. fluorescence images) were at first subjected to segmentation. Two distinct methods were performed to detect maize leaves: (1) a skeleton-based method to detect the number of leaves; and (2) the SUSAN corner detection algorithm (Smith and Brady, 1997) to detect leaf tips (marked with circles) in maize plants. By applying a region-growing algorithm, variably sized parts of the leaf tips were extracted. Furthermore, the properties of leaf tips, such as their colors and orientations, were calculated (as highlighted in the inset box).
For volume estimation, four formulas are used (BlCalcVolumes):
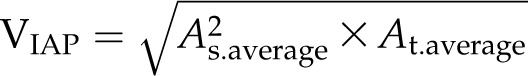 |
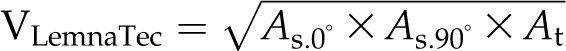 |
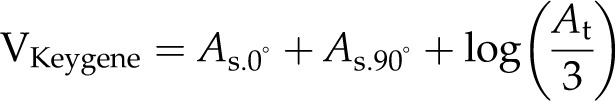 |
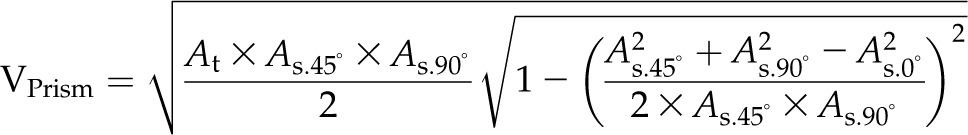 |
where As and At are the projected areas from side-view (at different angles) and top-view images, respectively. VLemnaTec is the formula used in the LemnaTec software system to estimate plant volume, considering one top-view image and two side-view images, which are taken from 0° and 90° (from the LemnaTec Scanalyzer 3D documentation), whereas KeyGene uses VKeygene for volume estimation (usually for the analysis of barley plants). The calculation formula VPrism is based on the idea for the two-side-view LemnaTec formula but extended to incorporate three side views. While the LemnaTec volume calculation is based on the assumption that the plant can be simplified to a cuboid shape, the prism formula makes the assumption that the plant shape can be generalized to a triangular prism. In our IAP system, all of these formulas are available for volume calculation, but VIAP is used by default to calculate the plant volume, as it considers and works with a flexible number of side-view images (even in cases where only one side-view image is available).
Color features are extracted in each color component in the following color spaces: RGB, HSV, and L*a*b*. A 20-bin histogram is calculated for each color channel, which provides detailed insights into the composition of the observed plant color. Partial discoloration could be seen as an increase in the number of pixels that fall into the range of a specific bin (BlCalcColorHistograms). Some of the color-dependent features could further be used to calculate derived indicators like dark green color index (Rorie et al., 2011) or the normalized differenced vegetation index (Barnes et al., 2001). The calculation of such indices also requires proper setup and calibration of the cameras and optical filters used.
Fluorescence-related features, which are related to the fluorescence activity of plants, are calculated from fluorescence images (BlSkeletonizeVisFluo). Near-infrared-related features, which are related to water content, are measured from near-infrared images and the derived skeleton (BlSkeletonizeNir). Please refer to the online document (http://iap.ipk-gatersleben.de/documentation.pdf) for a detailed description of our defined phenotypic traits.
Postprocessing
The postprocessing steps mainly involve summarizing the analysis results for each plant from different rotation angles of imaging. Several statistical results are computed, such as the average or maximal value of a plant feature (e.g. projected side area). It is also possible to visualize calculated features (e.g. the convex hull or the skeleton) in the result images (BlRunPostProcessors; Fig. 3). The result images are combined into an image set (BlMoveMasksToImageSet). Errors in imaging (e.g. missing or empty images) or failed computations are marked in the result images (BlHighlightNullResults). The final result images containing the highlighted features are saved by the block BlSaveResultImages.
Application Example
To test the performance and capability of the IAP system, 33 maize cv Fernandez plants were grown in an automated greenhouse and imaging system (LemnaTec) for noninvasive phenotyping. The summary of the experiment data set is shown in Table II. Temperature and humidity in the greenhouse were controlled during the whole experiment period. Images were acquired from the side- and top-view cameras from day 18 after sowing to the end of experiment by using the visible-light (with light spectrum of 390–750 nm), fluorescence (520–750 nm), and near-infrared (1,420–1,480 nm) cameras. In total, we daily obtained 78 images per plant from the three different camera systems. Starting from week 4 after sowing, we harvested four plants every week until the end of the experiment. To test the reliability of our system, we manually measured several traits, such as plant height, maximal width, number of leaves, maximal length/width of leaves, and plant fresh weight as well as dry weight (Supplemental Table S1). We compared these manual measurements with the corresponding image-derived traits.
Table II. Summary of the experiment data set.
| Experiment Property | Value |
|---|---|
| Duration | 64 d + 2 weeks of cultivation |
| No. of plants | 33 |
| No. of images | 33,798 |
| No. of measurements | 37,766 |
| No. of side-view angles | 25 |
| Resolution of visible image (pixels) | 2,056 × 2,454 |
| Resolution of fluorescence image (pixels) | 1,038 × 1,390 |
| Resolution of near-infrared image (pixels) | 254 × 320 |
| Size (GB; images in PNG format) | 108 |
The image and annotation (meta)data were subjected to the IAP maize pipeline for analysis. Processing images from high-throughput phenotyping experiments is time consuming and computationally demanding. IAP can process large-scale imaging data in a reasonable time on a local PC with a minimum of 6 gigabytes (GB) of system memory and using four central processing unit (CPU) cores (Table III). When analysis was performed using several computers operating in the IAP grid-computing mode, the processing time was reduced significantly. Of the overall processing time, about 12% is spent on image data loading (a block within the acquisition section of the pipeline) and 6% on result image export (a pipeline block within postprocessing). It takes 38%, 25%, 16%, and 3% of the overall time for preprocessing, segmentation, feature extraction, and postprocessing of images in the pipeline, respectively (Fig. 3). The relative image-processing times for visible (63%), fluorescence (35%), and near-infrared (2%) images vary, due to the different image resolution and number of extracted features.
Table III. Statistics of computational complexity based on different computing modes.
| Multithreading Computinga |
Grid Computingb |
||||
|---|---|---|---|---|---|
| No. of Cores | CPU Time | No. of Cores | CPU Time | No. of Nodes | CPU Time |
| h | h | h | |||
| 2 | 29.0 | 16 | 4.5 | 1 | 7.4 |
| 4 | 15.3 | 20 | 3.8 | 2 | 4.1 |
| 6 | 11.5 | 30 | 3.2 | 3 | 3.3 |
| 8 | 7.5 | 40 | 3.1 | 4 | 3.0 |
| 12 | 6.3 | 60c | 3.0 | 5 | 2.6 |
The multithreading computing tests were performed on a server with four 2.4-GHz Intel Xeon CPUs with 10 cores each (40 cores in total), with 256 GB of memory installed (60 GB was used in the test with the Java VM parameter of –Xmx60g), and used the Linux operating system.
The grid-computing tests were performed on several commonly used computers (with Windows 7 operating system installed), each with four CPU cores (Intel i7-2600 CPU, 3.4 GHz) and 16 GB of memory on average (12 GB was used for IAP tests with the Java VM parameter of –Xmx12g).
Tests were based on the utilization of the hyperthreading technique.
We extracted a representative list of phenotypic traits from the image data set. These traits are broadly categorized into four categories: geometric traits, color-related traits, fluorescence-related traits (representing plant growth conditions and health status), and near-infrared-related traits (characterizing plant water status; Fig. 3). These data could be further used to assess plant performance. For our application example, the camera systems were not calibrated; therefore, we cannot derive absolute values for water content or numeric indicators such as dark green color index and normalized differenced vegetation index. Determination of these features requires the software support for calculating them but in addition also the development of additional species-specific experimental protocols.
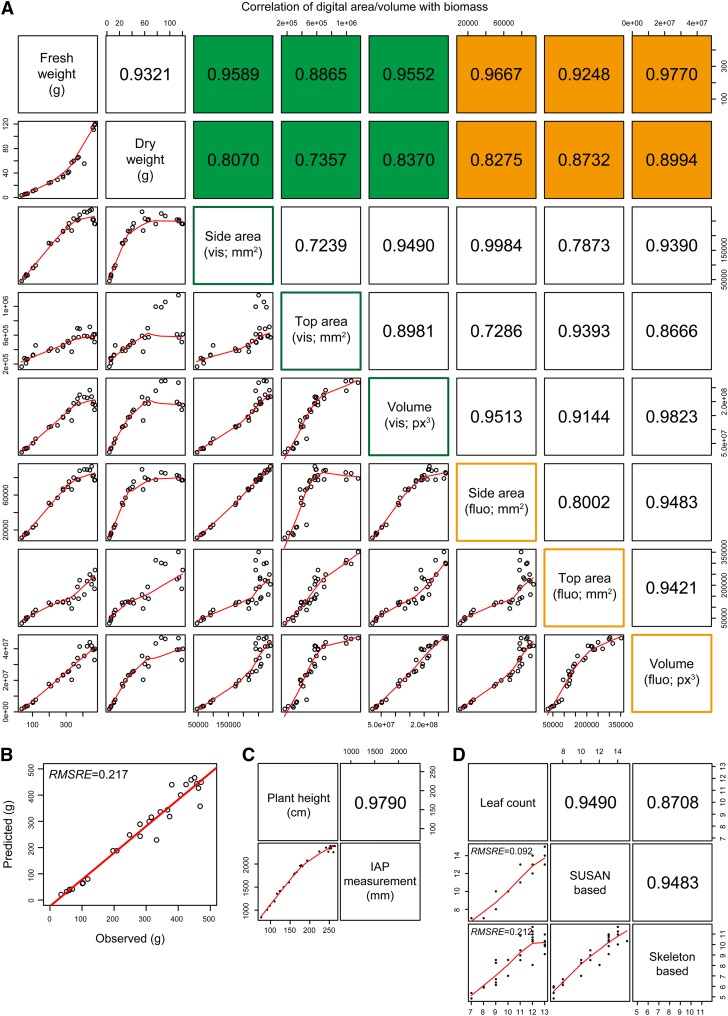
To test the reliability of traits derived from the IAP system, we correlated the calculated phenotypic data to the manual measurements (Fig. 5; Supplemental Fig. S2). We found that correlations of plant biomass (fresh or dry weight) to projected area and digital volume are sufficiently high (up to r = 0.98; Fig. 5A). The digital volume estimated from both side- and top-view images shows relatively better estimation than the projected area. In our tests, the fluorescence images showed the least noise and, therefore, provided the best information to calculate digital biomass in comparison with the visible-light images. Of the four formulas used to estimate plant volume (see “Materials and Methods”), the digital volume calculated using our defined formula (VIAP) has the best correlation with the manually acquired measurements (Supplemental Fig. S3). While the difference is very small, we prefer the newly defined formula, as it works with a flexible number of side-view images. Furthermore, when testing these formulas on other large image data sets from several different plant species, the formula VIAP performed better than the other formulas for the estimated volume, demonstrating higher correlations with manual data, indicating the robustness of our method to predict biomass. To check the accuracy of the biomass prediction based on volume traits, we developed a calibration model from an independent data set. Our calibration model is based on linear regression, in which the volume was related to plant biomass to generate a biomass prediction model. We found that the biomass data predicted based on the developed calibration model were highly heritable (root mean squared relative error = 0.22; Fig. 5B). In addition, we observed that the estimated plant height reaches a very high correlation of r = 0.98 (Fig. 5C).
Figure 5.
Correlation of IAP measurements with experimental data. A, Scatterplots showing the correlation of digital area/volume from IAP with manual measurement of biomass (plant fresh and dry weights). Area and volume were calculated from both visible (in green) and fluorescence (in orange) images. The values shown represent Pearson correlation coefficients (r). B, Precision of biomass measurements based on image-derived parameters. The accuracy of the biomass prediction is evaluated based on a calibration model using data from another independent experiment (based on the volume trait calculated from fluorescence images). RMSRE denotes the root mean squared relative error. C, Correlation of plant height from manual measurements and IAP calculations. Note that there is a bias for plants higher than 2.2 m, as they are out of the detection capability of the imaging system. D, Correlation of the number of leaves. The number of leaves was estimated by two different approaches: the SUSAN-based and skeleton-based methods. The number of leaves was calculated for each image. If a plant had multiple side-view images, the median value was used. Note that the skeleton-based approach may fail to detect small leaves, as the estimated value was smaller than expected. [See online article for color version of this figure.]
With IAP, the number of leaves can be estimated by two different methods (Fig. 4). The skeleton-based approach (Eberius and Lima-Guerra, 2009) is the most straightforward way, which shows a correlation of r = 0.87 with the manual measurement (Fig. 5D). On the other hand, the number of leaves can be derived from the detection of the leaf tips based on the SUSAN corner detection algorithm (Smith and Brady, 1997). We found that the number of leaves calculated by the SUSAN-based method achieves r = 0.95 and root mean squared relative error = 0.09 (Fig. 5D). Although the skeleton-based method is intuitive and quick, more parameters can be derived from the corner-detection-based algorithm. The region-growing pixel-search algorithm can be further used to determine the color and the direction of the identified leaf-tip regions (Fig. 4). These parameters might be used as indicators of plant health status and development. For example, the direction of leaf tips could be important indicators for light capture and the accumulation of leaf nitrogen for grain filling (Sinclair and Sheehy, 1999) and has been used as a useful trait to dissect leaf architecture through genome-wide association studies in maize (Tian et al., 2011).
Comparison with Related Tools
In the field of plant phenotyping, several software tools support high-throughput image data analysis (Cobb et al., 2013; Fiorani and Schurr, 2013). In this section, we compared our IAP system with LemnaGrid, HTPheno (Hartmann et al., 2011), and the image-analysis solution used in GROWSCREEN system collections (Walter et al., 2007; Biskup et al., 2009; Jansen et al., 2009; Nagel et al., 2012; Table IV).
Table IV. Comparison of the essential functionalities of the IAP system with other related phenotyping tools.
+, Supported feature; −, not supported feature; NA, not available or status unknown.
| Feature | IAP | HTPheno | LemnaTec | GROWSCREENa |
|---|---|---|---|---|
| License | GPLv2 | GPLv2 | Commercial | Private |
| Expandabilityb | By plugins/addons | By source code | Not possible | NA |
| Pipeline modification | On block list and pipeline settings | By source code | By predefined algorithms and visual pipeline editor | NA |
| Import of sensor data | + | — | — | NA |
| Report functionality | + | — | — | — |
| Support grid computing | + | — | + | + |
| Database support | + | — | + | + |
| Time-dependent analysis settings | + | — | — | NA |
| Documented used cases | ||||
| Maize | + | — | + | + |
| Barley | + | + | + | + |
| Arabidopsis | + | — | + | + |
| Supported camera systems | ||||
| Visible | + | + | + | + |
| Fluorescence | + | — | + | + |
| Near-infrared | + | — | + | + |
| Infrared | + | — | + | + |
Not available for testing and not exactly described in literature; comparison is based on the consideration of several GROWSCREEN systems (GROWSCREEN, GROWSCREEN-root, GROWSCREEN-fluoro, GROWSCREEN Rhizo, and GROWSCREEN 3D).
Regarding the possibility of implementing new algorithms by end users.
LemnaGrid is the software solution for high-throughput plant phenotyping provided by LemnaTec, which is designed to process imaging data from the Scanalyzer 3D system. This system was successfully used in the prediction of biomass accumulation for Arabidopsis (Arvidsson et al., 2011) and cereal plants (Golzarian et al., 2011). As a commercial solution, LemnaGrid is not intended to be further developed or significantly modified by the user (Berger et al., 2012). Thus, only predefined functionalities are available. HTPheno provides a specialized analysis pipeline for the analysis of visible-light images acquired from the LemnaTec system. Although it is published under an open-source license for usage and further development, the HTPheno image-analysis pipeline is solely tuned for barley plants and calculates a limited number of traits. It neither corrects for changing light conditions nor handles multiple zoom levels. The GROWSCREEN system is developed by the Jülich Plant Phenotyping Centre. It supports plenty of different image modalities and analysis approaches, including GROWSCREEN, GROWSCREEN-root, GROWSCREEN-fluoro, GROWSCREEN Rhizo, and GROWSCREEN 3D. However, this system seems to be closely coupled to the hardware installations, as the related software developments are not publicly available and are not reported to be used by other institutions.
Generally, IAP supports a broad set of functionalities, including both data management and data processing (Table IV). IAP implements the infrastructure supporting the analysis of other plant species and imaging modalities. Besides, IAP is an extensible system that allows users to implement new algorithms in terms of plugins and addons (see the development section on the IAP Web site).
Future Perspectives
We will continue developing the IAP software with respect to various aspects. In the near future, IAP will include more automation of image-processing steps, for example, to further improve image normalization based on reference color charts or objects and the automatic alignment of images from different camera systems, as we aim to reduce the time and effort to configure a pipeline. We are collaborating closely with other partners both within and outside the Leibniz Institute of Plant Genetics and Crop Plant Research to further develop IAP and to design new analysis pipeline templates to extract traits from roots, flowers, or stress symptoms and to support the analysis of more plant species. The development of a plant three-dimensional model could be useful to improve the accuracy and precision of plant phenotypic measurements.
With the speeded-up development in high-throughput plant phenotyping, the era of plant phenomics emerges. The traditional phenotyping bottleneck (Furbank and Tester, 2011) will be resolved in the future, but at the same time, a bottleneck of phenotypic data analysis may be created. It is important to interpret and integrate phenotypic data with other omics data domains, such as genomics, epigenomics, proteomics, transcriptomics, and metabolic data. We aim to develop IAP as a platform that is capable of performing integrative analysis of data from multiple omics data domains. As IAP includes VANTED (Junker et al., 2006) as one of its underlying modules, it will be presented as a natural multifunctional software for integrative analysis purposes.
CONCLUSION
We have presented IAP, an information system for rapid, quantitative analysis of high-throughput plant phenotyping data. IAP offers a user-friendly graphical user interface. It provides approaches for data management, image analysis, and result handling for large-scale experiments. Although we tested our system on maize plants, we already internally used the approach for other plant species. In fact, IAP now includes image-analysis pipelines to examine various traits for other species, such as Arabidopsis, wheat, and barley. The use of IAP is not limited to any image-acquisition hardware. It can be adapted to process imaging data from different monitoring systems and camera types. The provided pipeline templates can be modified or recreated for different analysis tasks by using the integrated pipeline editor and by adjusting the pipeline settings. The validation results based on the example experiment data set showed that our system is highly reliable. In summary, our IAP system is a flexible and powerful framework for high-throughput plant phenotyping and plant performance evaluation.
MATERIALS AND METHODS
Data Management
The implemented data structures for handling image data and metadata are based on HIVE (for handy integration and visualization of multimodal experimental data; Rohn et al., 2009), which was already used in our VANTED system (Junker et al., 2006). The data structures, which have been established in VANTED to support the development of the HIVE system, were further extended to handle image attributes and timing information for measurements acquired from high-throughput phenotyping applications. Details about data management and the corresponding database system have been presented previously (Klukas et al., 2012).
We developed a database access component for the LemnaTec system to automatically import imaging data and related experimental metadata into the IAP system for analysis. This component supports the import of snapshot data, plant annotation data, measurement labels, imaging time information, imaging configurations, as well as climate data from the greenhouse. Meanwhile, it is possible to import data from other sources besides the LemnaTec system, such as manually acquired images using common commercial cameras or scanners.
For grid computing, it is currently required to set up a MongoDB database (http://www.mongodb.org/) in order to make data accessible from distributed computing nodes.
Image-Analysis Workflow
An experiment data set is created after loading imaging data and metadata into the system. Afterward, the analysis is ready to be started. The core of the image processing involves several loops to handle the image data and to return results. All images from each day and each imaged plant, which are recognized by their identifiers, are processed using separate threads. The images of a specific plant are sequentially processed from the first to the last experiment day. If available, the top image is analyzed first and afterward the side images. The analysis blocks process images from a specific camera system individually or under consideration of all available camera images. For example, the mask constructed from the fluorescence image can be applied to images from the other camera systems. The order of the processing steps makes it possible to consider analysis results from previous days. Within the processing of the side images, the results of the top analysis are available; for example, the plant leaf orientation in maize (Zea mays) can be used to determine a suitable image from the side view. Finally, all analysis results for a specific plant can be postprocessed within an analysis-block function. At last, the numeric analysis results from all blocks are combined and added to the final result data set.
Data Export
For further analysis, we provide some functionality along with the IAP system to summarize and visualize the myriad numeric properties of the result data. To this end, IAP included an automated report generator based on the statistical programming language R (http://www.r-project.org/) and LaTeX (a document preparation system; http://www.latex-project.org/). The user can specify which data should be included in the result diagrams. Various experiment factors, such as species, genotype, variety, or treatment, could be considered in the report generation. The report functionality generates different types of diagrams (such as box plot, line plot, and staked bar plot) in a PDF to present the result. Finally, the numeric data can be exported in common data formats like .cvs or .xlsx for further analysis purposes.
Implementation
The IAP system is implemented in Java, taking advantage of its platform independence and the availability of numerous libraries and tools. We extended a set of analysis functionalities based on ImageJ functions (Schneider et al., 2012) and implemented various newly developed algorithms for image processing (detailed in “Results and Discussion”). Specific image-analysis tasks can be defined in independent blocks, which consist of several related, sequential image-processing operations. We provide a broad set of predefined blocks and pipeline templates for image analysis and APIs to extend IAP.
To support the development of new image-analysis procedures or novel analysis pipelines, the plugin development APIs in VANTED (GenericPlugin) have been incorporated into IAP and have been extended with a new interface, IAPplugin (Supplemental Fig. S1). By implementing this interface, it is possible to add new image-processing blocks, analysis pipelines, and experiment data manipulation commands within the user interface. The source code repository of IAP contains code for an example addon as a showcase for this functionality. The initially implemented blocks, pipelines, and commands are incorporated as individual plugins to the core system. Once a set of extensions has been developed by external users, they can be supplied to other users of IAP in the form of a single Java archive file, which bundles the newly developed plugins as a system addon.
The analysis procedures can run on a single computer by saving input and result images on the local file system. Analysis can also be performed using a parallel grid-computing approach. As high-throughput phenotyping often contains hundreds of thousands of image files, the utilization of multiple computers or servers can be desired to speed up the analysis and to calculate result data sets in a timely manner. To support the distribution of analysis tasks and to store the results in that case, the MongoDB database is needed to be installed on the PC or server. Input and output images and numeric analysis results are stored within the database using the MongoDB GridFS file storage API. As an alternative, if the database server has a limited amount of storage space, files can also be loaded and stored on a separate file server using the FTP, SCP, or other protocols. This flexibility is implemented by utilizing a technique called the virtual file system, which has its implementation in the Apache Commons Virtual File System API and library.
The number of compute threads used for analysis on an individual computer is user configurable and initially set according to the number of CPU cores in the system. The processing is performed using the so-called thread pool pattern, where a set of worker threads processes a central task queue.
Application Example: Image Acquisition
As an example shown in this study, we performed a phenotyping experiment on 33 maize (Zea mays ‘Fernandez’) plants. This energy maize cultivar was cultivated by the KWS SAAT AG. Plant images were captured using the Scanalyzer 3D (LemnaTec) at the Leibniz Institute of Plant Genetics and Crop Plant Research. Seventy-eight images were taken of every plant, of which 26 images were captured using three kinds of cameras. For each camera system, images were taken from both top view (one image) and side view (25 images). The result images were stored in PNG format. The three cameras used in the automated system are as follows: (1) visible-light camera (Basler Pilot piA2400-17gc, with resolution of 2,454 × 2,056 pixels), (2) fluorescence camera (Basler Scout scA1400-17gc, with resolution of 1,390 × 1,038 pixels), and (3) near-infrared camera (Nir 300 with resolution of 320 × 256 pixels). Images were acquired daily for 9 weeks. We obtained approximately 34,000 images in total. Image data were analyzed using IAP version 1.1 (release of November 22, 2013).
Availability and Requirements
The source code and binaries of the IAP software and the full example data set are freely available for download at http://iap.ipk-gatersleben.de/ or from SourceForge (http://sourceforge.net/projects/iapg2p/). IAP is implemented in Java and runs on Linux, Mac, and Microsoft Windows. Specific requirements are as follows: (1) run time requirements, Java JRE or JDK version 1.7 or higher; (2) operating system(s), platform independent (tested on Windows 7, Linux [Scientific Linux 6.4], and Mac OS X [10.8, Mountain Lion]); and (3) license, GPLv2 (for software) and CCA3 (for experimental data set).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. UML diagram depicting the architecture of the IAP system.
Supplemental Figure S2. Correlation analysis of phenotypic traits.
Supplemental Figure S3. Correlation of the digital volume with experimental measurements of biomass (fresh and dry weight).
Supplemental Table S1. Manual measurements and image-derived parameters for the 32 plants investigated in this study.
Supplementary Material
Acknowledgments
We thank Dr. Kerstin Neumann, Dr. Moses Mahugu Muraya, and Hea-Jung Jeon (Leibniz Institute of Plant Genetics and Crop Plant Research) as users of the imaging system and especially Thomas Altmann for valuable feedback and support during the software development; and Ming Chen (Zhejiang University) for longstanding cooperation.
Glossary
- IAP
Integrated Analysis Platform
- PC
personal computer
- API
application program interface
- CPU
central processing unit
- GB
gigabytes
Footnotes
This work was supported by the Leibniz Institute of Plant Genetics and Crop Plant Research, the National Natural Science Foundation of China (grant no. 31050110121), the Robert Bosch Stiftung (grant no. 32.5.8003.0116.0), the Federal Agency for Agriculture and Food (grant no. 15/12–13 530–06.01-BiKo CHN), the Federal Ministry of Education and Research (grant no. 0315958A), and the European Plant Phenotyping Network (grant no. 284443), funded by the FP7 Research Infrastructures Programme of the European Union. Part of this work was performed within the German Plant Phenotyping Network which is funded by the German Federal Ministry of Education and Research (grant no. 031A053B).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Arvidsson S, Pérez-Rodríguez P, Mueller-Roeber B. (2011) A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol 191: 895–907 [DOI] [PubMed] [Google Scholar]
- Barnes EM, Clarke TR, Richards SE, Colaizzi PD, Haberland J, Kostrzewski M, Waller P, Choi C, Riley E, Thompson T (2001) Coincident detection of crop water stress, nitrogen status and canopy density using ground-based multispectral data. In PC Robert, RH Rust, WE Larson, eds, Proceedings of the Fifth International Conference on Precision Agriculture. Precision Agriculture Centre, Department of Soil, Water and, Climate, University of Minnesota, Bloomington, MN, pp 1–15 [Google Scholar]
- Berger B, de Regt B, Tester M. (2012) High-throughput phenotyping of plant shoots. Methods Mol Biol 918: 9–20 [DOI] [PubMed] [Google Scholar]
- Biskup B, Scharr H, Fischbach A, Wiese-Klinkenberg A, Schurr U, Walter A. (2009) Diel growth cycle of isolated leaf discs analyzed with a novel, high-throughput three-dimensional imaging method is identical to that of intact leaves. Plant Physiol 149: 1452–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylesjö M, Segura V, Soolanayakanahally RY, Rae AM, Trygg J, Gustafsson P, Jansson S, Street NR. (2008) LAMINA: a tool for rapid quantification of leaf size and shape parameters. BMC Plant Biol 8: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV. (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JN, Declerck G, Greenberg A, Clark R, McCouch S. (2013) Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor Appl Genet 126: 867–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vylder J, Vandenbussche F, Hu Y, Philips W, Van Der Straeten D.(2012) Rosette tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiol 160: 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberius M, Lima-Guerra J (2009) High-throughput plant phenotyping: data acquisition, transformation, and analysis. In D Edwards, J Stajich, D Hansen, eds, Bioinformatics. Springer, New York, pp 259–278 [Google Scholar]
- Edwards D, Batley J. (2004) Plant bioinformatics: from genome to phenome. Trends Biotechnol 22: 232–237 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Schurr U. (2013) Future scenarios for plant phenotyping. Annu Rev Plant Biol 64: 267–291 [DOI] [PubMed] [Google Scholar]
- Furbank RT, Tester M. (2011) Phenomics: technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Golzarian MR, Frick RA, Rajendran K, Berger B, Roy S, Tester M, Lun DS. (2011) Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Green JM, Appel H, Rehrig EM, Harnsomburana J, Chang JF, Balint-Kurti P, Shyu CR. (2012) PhenoPhyte: a flexible affordable method to quantify 2D phenotypes from imagery. Plant Methods 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Czauderna T, Hoffmann R, Stein N, Schreiber F. (2011) HTPheno: an image analysis pipeline for high-throughput plant phenotyping. BMC Bioinformatics 12: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D, Govindaraju DR, Omholt S. (2010) Phenomics: the next challenge. Nat Rev Genet 11: 855–866 [DOI] [PubMed] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, Nagel K, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, Braun S, et al. (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36: 902. [DOI] [PubMed] [Google Scholar]
- Junker BH, Klukas C, Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukas C, Pape JM, Entzian A. (2012) Analysis of high-throughput plant image data with the information system IAP. J Integr Bioinform 9: 191. [DOI] [PubMed] [Google Scholar]
- Luccheseyz L, Mitray S. (2001) Color image segmentation: a state-of-the-art survey. Proceedings of the Indian National Science Academy 67: 207–221 [Google Scholar]
- Macqueen JB (1967) Some methods for classification and analysis of multivariate observations. In LM Le Cam, J Neyman, eds, Proceedings of the Fifth Berkeley Symposium on Math, Statistics, and Probability, Vol 1. University of California Press, Berkeley, CA, pp 281–297 [Google Scholar]
- Nagel KA, Putz A, Gilmer F, Heinz K, Fischbach A, Pfeifer J, Faget M, Blossfeld S, Ernst M, Dimaki C, et al. (2012) GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol 39: 891–904 [DOI] [PubMed] [Google Scholar]
- Pound MP, French AP, Atkinson JA, Wells DM, Bennett MJ, Pridmore T. (2013) RootNav: navigating images of complex root architectures. Plant Physiol 162: 1802–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuzeau C. (2007) TraitMill (TM): a high throughput functional genomics platform for the phenotypic analysis of cereals. In Vitro Cell Dev Biol Anim 43: S4 [Google Scholar]
- Reuzeau C, Pen J, Frankard V, de Wolf J, Peerbolte R, Broekaert W. (2005) TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Fenzi Zhiwu Yuzhong 3: 7534 [Google Scholar]
- Rohn H, Klukas C, Schreiber F (2009) Integration and visualisation of multimodal biological data. In I Grosse, S Neumann, S Posch, F Schreiber, PF Stadler, eds, Proceedings of the German Conference on Bioinformatics. Martin Luther University, Halle-Wittenberg, Germany, pp 105–115 [Google Scholar]
- Rorie RL, Purcell LC, Mozaffari M, Karcher DE, King CA, Marsh MC, Longer DE. (2011) Association of “greenness” in corn with yield and leaf nitrogen concentration. Agron J 103: 529–535 [Google Scholar]
- Sadok W, Naudin P, Boussuge B, Muller B, Welcker C, Tardieu F. (2007) Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ 30: 135–146 [DOI] [PubMed] [Google Scholar]
- Sauvola J, Pietikainen M. (2000) Adaptive document image binarization. Pattern Recognit 33: 225–236 [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE. (1999) Erect leaves and photosynthesis in rice. Science 283: 1455 [Google Scholar]
- Smith SM, Brady JM. (1997) SUSAN: a new approach to low level image processing. Int J Comput Vis 23: 45–78 [Google Scholar]
- Tanabata T, Shibaya T, Hori K, Ebana K, Yano M. (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol 160: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, Voisin R, Bouchez D, Durand-Tardif M, Guerche P, et al. (2013) Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J 74: 534–544 [DOI] [PubMed] [Google Scholar]
- Walter A, Scharr H, Gilmer F, Zierer R, Nagel KA, Ernst M, Wiese A, Virnich O, Christ MM, Uhlig B, et al. (2007) Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol 174: 447–455 [DOI] [PubMed] [Google Scholar]
- Wang L, Uilecan IV, Assadi AH, Kozmik CA, Spalding EP. (2009) HYPOTrace: image analysis software for measuring hypocotyl growth and shape demonstrated on Arabidopsis seedlings undergoing photomorphogenesis. Plant Physiol 149: 1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. (2008) LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J 53: 578–586 [DOI] [PubMed] [Google Scholar]
- Yen JC, Chang FJ, Chang SA. (1995) A new criterion for automatic multilevel thresholding. IEEE Trans Image Process 4: 370–378 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Javed O, Shah M. (2006) Object tracking: a survey. ACM Comput Surv 38: 13 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.