Resistance to two herbicides in Echinochloa phyllopogon is associated with overexpression of two cytochrome P450s that are simultaneously controlled by a putative single genetic element.
Abstract
Previous studies have demonstrated multiple herbicide resistance in California populations of Echinochloa phyllopogon, a noxious weed in rice (Oryza sativa) fields. It was suggested that the resistance to two classes of acetolactate synthase-inhibiting herbicides, bensulfuron-methyl (BSM) and penoxsulam (PX), may be caused by enhanced activities of herbicide-metabolizing cytochrome P450. We investigated BSM metabolism in the resistant (R) and susceptible (S) lines of E. phyllopogon, which were originally collected from different areas in California. R plants metabolized BSM through O-demethylation more rapidly than S plants. Based on available information about BSM tolerance in rice, we isolated and analyzed P450 genes of the CYP81A subfamily in E. phyllopogon. Two genes, CYP81A12 and CYP81A21, were more actively transcribed in R plants compared with S plants. Transgenic Arabidopsis (Arabidopsis thaliana) expressing either of the two genes survived in media containing BSM or PX at levels at which the wild type stopped growing. Segregation of resistances in the F2 generation from crosses of R and S plants suggested that the resistance to BSM and PX were each under the control of a single regulatory element. In F6 recombinant inbred lines, BSM and PX resistances cosegregated with increased transcript levels of CYP81A12 and CYP81A21. Heterologously produced CYP81A12 and CYP81A21 proteins in yeast (Saccharomyces cerevisiae) metabolized BSM through O-demethylation. Our results suggest that overexpression of the two P450 genes confers resistance to two classes of acetolactate synthase inhibitors to E. phyllopogon. The overexpression of the two genes could be regulated simultaneously by a single trans-acting element in the R line of E. phyllopogon.
Herbicides have become essential tools in modern agriculture for efficient weed control. Meanwhile, herbicide resistance has evolved through persistent herbicide selection exerted on huge weed populations across vast areas. The mechanisms responsible for herbicide resistance are generally grouped into two categories: target site resistance and nontarget site resistance (Yuan et al., 2007). Target site resistance, which is caused by alterations in the protein(s) that are targeted by the herbicide, accounts for a large majority of the reported cases of herbicide resistance (Powles and Yu, 2010). A single nucleotide polymorphism in a target site-encoding gene is the most frequently observed alteration, causing a structural change in the target site and reducing the ability of herbicides to interact with the target sites. Overproduction of target site proteins is another mechanism for target site resistance, as recently disclosed in glyphosate resistance (Gaines et al., 2010; Tranel et al., 2011; Salas et al., 2012). In these cases, amplification of target site-encoding genes results in overproduction of target site proteins. The molecular mechanisms of target site resistance are relatively easily analyzed, as most herbicides target specific enzymes (Yuan et al., 2007). Also, management of target site resistance can be readily achieved because resistant (R) plants can be controlled by herbicides with different modes of action.
A much less studied but more threatening mechanism is nontarget site resistance. Most nontarget site resistances are associated with enhanced metabolism and impaired translocation (Powles and Yu, 2010; Délye, 2013; Yu and Powles, 2014). In contrast to target site resistance, nontarget site resistances tend to affect multiple herbicides of different chemical classes and different modes of action (Délye, 2013), although they can be limited to several herbicides with the same mode of action (e.g. Jeffers et al., 1996; Iwakami et al., 2014b). Threatening aspects of nontarget site resistance thus are the unexpected resistances to other herbicides (Preston, 2004). However, little is known about the molecular mechanisms of nontarget site resistance. Elucidating these mechanisms, including the identification of the molecular players involved, has been the leading challenge in herbicide resistance research (Powles and Yu, 2010; Délye, 2013).
Echinochloa phyllopogon, also known as Echinochloa oryzicola, is an allotetraploid (2n = 4x = 36) and predominantly self-pollinated grass species in the Panicoideae subfamily (Yamasue, 2001). E. phyllopogon is a noxious weed in rice (Oryza sativa) fields, and severe infestations cause large decreases in rice yield. In the late 1990s, R populations were found in the Sacramento Valley of California (Fischer et al., 2000a). R plants exhibited resistance to at least nine herbicides from seven different chemical groups: sulfonylurea, triazolopyrimidine, pyrimidinyl carboxy herbicides, aryloxyphenoxy-propionate, thiocarbamate, quinclorac, and clomazone (Fischer et al., 2000b; Osuna et al., 2002; Ruiz-Santaella et al., 2006; Bakkali et al., 2007; Yasuor et al., 2009, 2010, 2012). Today, there seems to be only one herbicide left to control this weed in California (Yasuor et al., 2012). Thus, elucidating the molecular mechanism of resistance is urgently required.
The multiple herbicide-resistant E. phyllopogon exhibits resistance to bensulfuron-methyl (BSM), a sulfonylurea herbicide, and penoxsulam (PX), a triazolopyrimidine herbicide (Osuna et al., 2002; Yasuor et al., 2009), both of which are absorbed via roots (PX can also be absorbed via shoots) and inhibit acetolactate synthase (ALS), a key enzyme for the biosynthesis of the branched-chain amino acids Val, Leu, and Ile (Duggleby, 2005). Sensitivity of the ALS enzyme to BSM and PX did not significantly differ between R and susceptible (S) plants (Osuna et al., 2002; Yasuor et al., 2009), and the nucleotide sequences encoding ALS were identical in the R and S lines, lacking amino acid substitutions known to confer resistance (Iwakami et al., 2012). The BSM and PX resistances were shown to be reduced by cytochrome P450 inhibitors in R plants (Osuna et al., 2002; Yasuor et al., 2009). Also, the P450 inhibitors reduced PX metabolism in R plants to the level of that in S plants (Yasuor et al., 2009). These observations suggested that nontarget site resistance mediated by enhanced activities of P450s is involved in the mechanism of BSM and PX resistance.
P450s are a group of heme-thiolate monooxygenases that catalyze a wide variety of monooxygenation/hydroxylation reactions (Bak et al., 2011). Hundreds of P450 genes exist in plant genomes, and each P450 participates in various biochemical pathways to produce primary and secondary metabolites (Mizutani and Ohta, 2010). Several herbicide-metabolizing P450s have been identified in a number of plant species (Siminszky, 2006). In rice, BSM is mainly metabolized through O-demethylation of the methoxy group at position 4 of the pyrimidine ring (Takeda et al., 1986). Purified rice microsomes catalyzed this reaction (Deng and Hatzios, 2002), and a CYP81A6 knockout mutant of rice was susceptible to BSM (Pan et al., 2006). Therefore, P450 activity of CYP81A6 appears to catalyze the O-demethylation of BSM, although direct evidence for the reaction has yet to be reported. The CYP81A subfamily was found in E. phyllopogon as well as other species of the Panicoideae (Nelson, 2009; Iwakami et al., 2014a) but is not conserved in all plants (Nelson, 2009). Considering the putative BSM metabolizing function of the rice P450 and also the high level of BSM resistance in E. phyllopogon populations (Osuna et al., 2002; also our preliminary results), we scrutinized BSM resistance as a first step to the elucidation of multiple herbicide resistance of E. phyllopogon. This seemed a promising approach, although BSM resistance is not a significant practical problem, as BSM is not used to control E. phyllopogon because of its rather low herbicidal activity against Echinochloa spp. at the recommended dose in rice fields. In addition, we also studied PX resistance in E. phyllopogon as another type of ALS inhibitor resistance related to P450 activity; PX is frequently used to control Echinochloa spp. in rice fields. We characterized P450s of the CYP81A subfamily in E. phyllopogon and found that two CYP81A P450 genes are associated with BSM and PX resistance in E. phyllopogon. This study provides a molecular basis for the understanding of the multiple herbicide resistance in E. phyllopogon, as well as of nontarget site resistance in weeds in general.
RESULTS
BSM Metabolism in E. phyllopogon
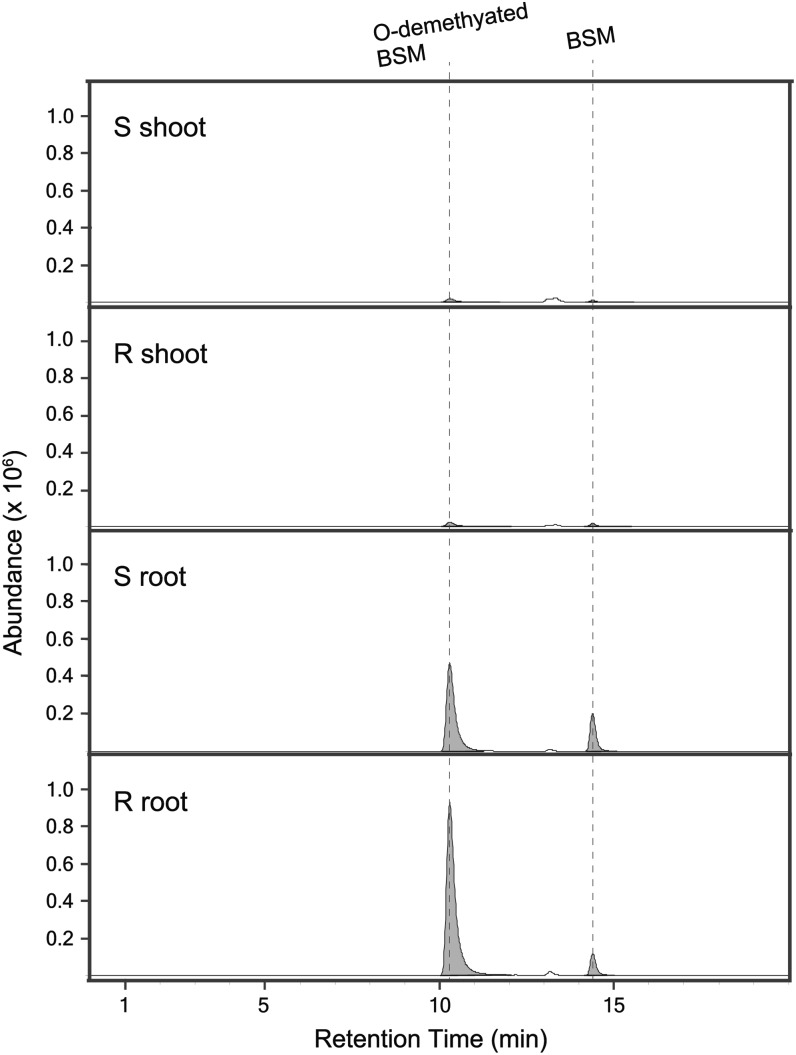
To test if BSM metabolism in the R line of E. phyllopogon is more active than in the S line and if the metabolic pathway is the same as in rice, we compared the amounts of BSM and O-demethylated BSM between the R and S lines of E. phyllopogon treated with BSM for 24 h. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) detected BSM and O-demethylated metabolites in shoots and roots of R and S lines (Fig. 1). In roots, the amount of BSM in the R line was one-half that detected in the S line, while that of O-demethylated BSM was 2-fold higher. In shoots, the amounts of BSM and O-demethylated BSM were less than one-tenth of those in roots. Together, these data suggest that BSM was metabolized primarily in the roots of E. phyllopogon and that it was metabolized more rapidly in the R line via the same metabolic pathway as in rice.
Figure 1.
LC-MS/MS analyses of a BSM metabolite formed in the shoots and roots of R and S lines of E. phyllopogon. Roots of seedlings at the second leaf stage were treated with 10 µm BSM for 24 h before analysis.
The CYP81A Subfamily in E. phyllopogon
We isolated seven CYP81A genes from the R line of E. phyllopogon, in addition to the five CYP81A genes previously described (Iwakami et al., 2014a; Supplemental Fig. S1). The seven genes were named by the Cytochrome P450 Nomenclature Committee (Dr. David Nelson, University of Tennessee Health Science Center). Three CYP81A genes, CYP81A13P, CYP81A19P, and CYP81A25P, are pseudogenes encoding truncated proteins and therefore were not analyzed further. Comparison of the other nine genes between the R and S lines revealed nucleotide polymorphisms in four genes, CYP81A12, CYP81A21, CYP81A22, and CYP81A26. In all of them except for CYP81A12, at least one nonsynonymous substitution was found (Supplemental Fig. S2). Two nonsynonymous substitutions in CYP81A22 were predicted to be within the substrate recognition site, a key region that may influence the catalytic functions of P450s (Gotoh, 1992).
Transcript Levels of CYP81A Genes in R and S E. phyllopogon
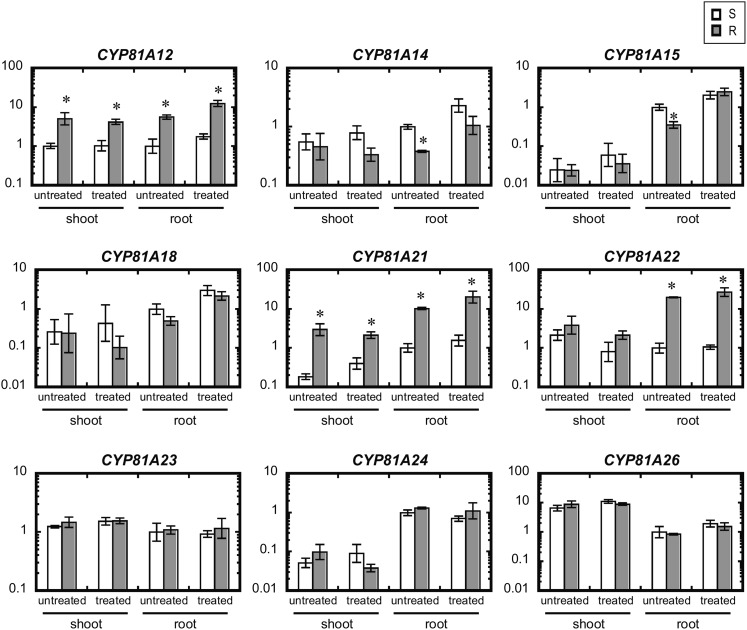
Among the nine CYP81A genes, CYP81A12 and CYP81A21 transcripts were particularly abundant in shoots and roots of R line seedlings at the second leaf stage, both in BSM-treated and untreated plants (Fig. 2). The transcript levels differed between the lines by factors of 4 or more. The expression of these genes was not influenced by BSM treatment, and they were constitutively overexpressed in the R line. CYP81A22 expression also was higher in roots of the R line than in those of the S line in BSM-treated as well as untreated plants. Transcript levels of CYP81A22 in the shoots tended to be higher in the BSM-treated condition, although the differences were not significant statistically. The other genes did not exhibit higher transcription levels in the R line; they were more actively transcribed in the S line (CYP81A14 and CYP81A15) or at similar levels in both lines.
Figure 2.
Transcript levels of P450 genes in the shoots and roots of R and S lines of E. phyllopogon. Roots of seedlings at the second-leaf stage were treated with 10 µm BSM; control plants remained untreated. Transcript levels were compared between R and S lines at 24 h after BSM treatment by real-time RT-PCR using EIF4B and RPII as internal control genes. Transcript abundance was normalized to the level in the root of the untreated S line. Data shown are means ± sd of four biological replicates (Student’s t test, *P < 0.01).
We investigated the expression profiles of the genes with higher expression levels in the R line (CYP81A12, CYP81A21, and CYP81A22) in more detail, differentiating between shoots and roots at different developmental stages, etiolated shoots, and spikelets (Supplemental Fig. S3). The transcription of the three genes was significantly higher in the R line in all organs investigated, except for CYP81A22 in shoots at the third-leaf stage, where the observed difference was not significant statistically.
Susceptibility of Transgenic Arabidopsis to BSM and PX
CYP81A12, CYP81A21, and CYP81A22, which had exhibited high transcript abundance in the R line, were introduced into Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0) under the control of the Cauliflower mosaic virus 35S promoter. For CYP81A21 and CYP81A22, R as well as S line alleles were introduced because the alleles showed an amino acid polymorphism.
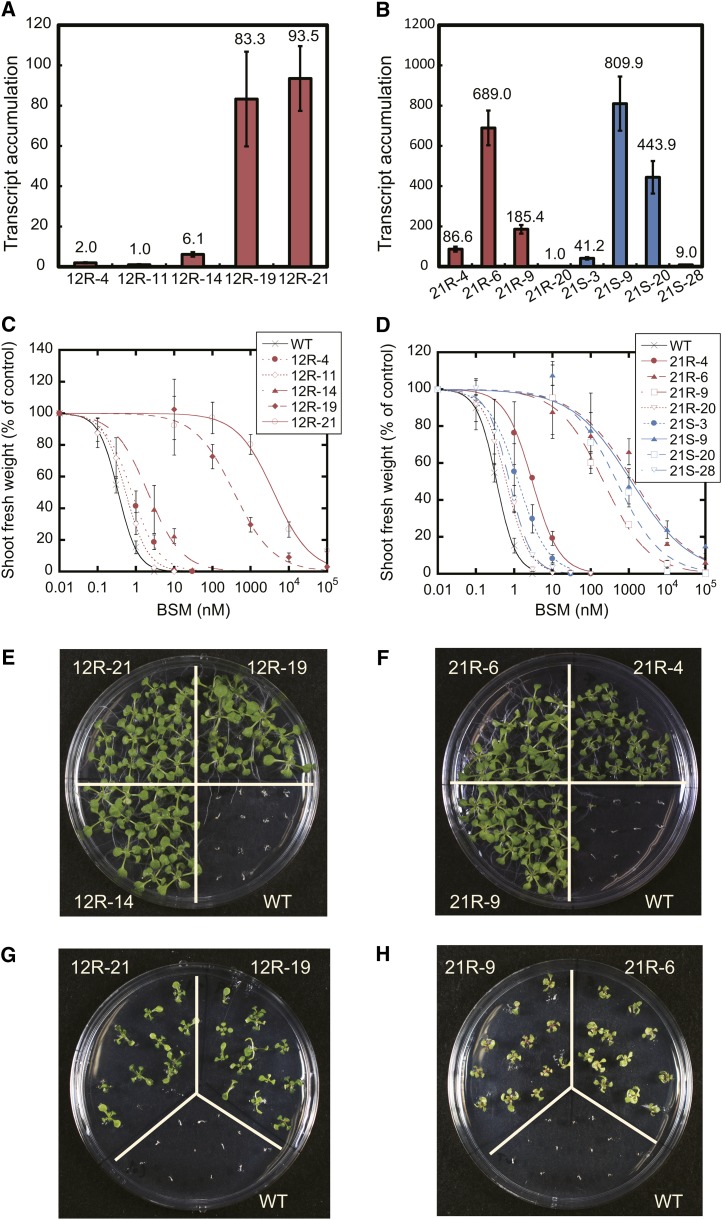
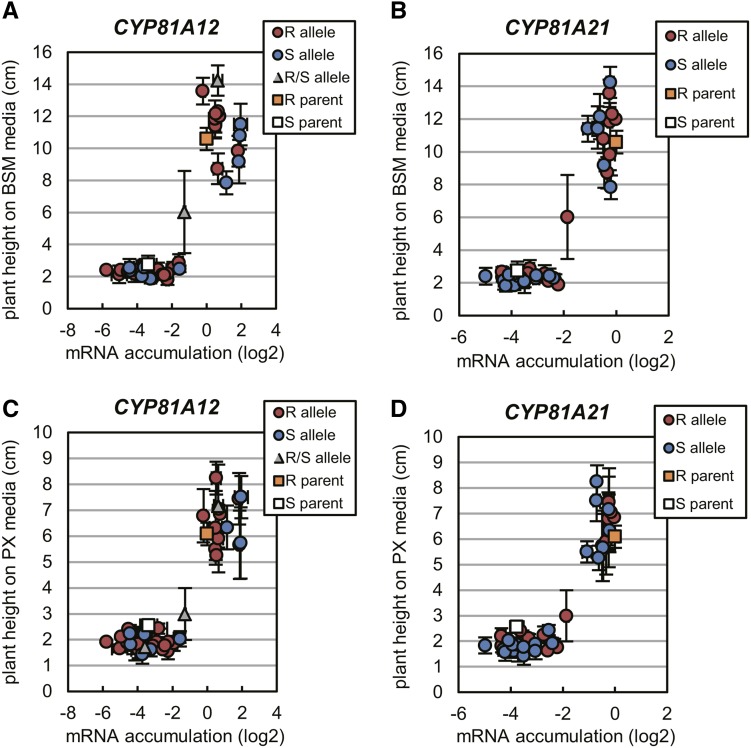
Five homozygous lines were selected for CYP81A12 expression (Fig. 3A). Transcript accumulation levels of the transgene were quite different between the five lines, and BSM susceptibility also differed (Fig. 3C). The resistance indices of 12R-19 and 12R-21, defined as the factor by which the herbicide concentration inducing 50% growth reduction (GR50) differed from the concentration required in the wild type, was more than 1,000. The BSM susceptibility was highly correlated to the transcription level of the introduced P450 gene in the CYP81A12 transgenic lines (Supplemental Fig. S4); lines with lower susceptibility corresponded to the lines with higher expression of CYP81A12. Thus, CYP81A12 conferred a decrease in BSM susceptibility to transgenic Arabidopsis depending on its transcript abundance.
Figure 3.
Transcript levels of transgenes and susceptibilities to BSM and PX in transgenic Arabidopsis expressing CYP81A12 and CYP81A21. 12R represents transgenic Arabidopsis with the CYP81A12 allele of an R line of E. phyllopogon. 21R and 21S represent transgenic Arabidopsis with the CYP81A21 alleles of an R and S line of E. phyllopogon, respectively. A and B, mRNA levels of the transgene in independent transgenic lines. The transcript levels of CYP81A12 and CYP81A21 were quantified by real-time RT-PCR using GAPDH as an internal control gene. Data are expressed as values relative to 12R-11 (CYP81A12) and 21R-20 (CYP81A21). C and D, BSM susceptibility of independent transgenic lines. Susceptibilities were evaluated by relative growth (shoot fresh weight) on Murashige and Skoog media containing BSM. Bars represent sd (n = 4). E and F, Seedlings grown for 12 d on Murashige and Skoog media with 3 nm BSM. G and H, Seedlings grown for 12 d on Murashige and Skoog media with 10 nm PX.
For CYP81A21, four homozygous lines each were selected for the R line allele and the S line allele (Fig. 3B). These lines accumulated various levels of the CYP81A21 transcript. BSM susceptibilities of each line differed greatly, and four lines (21R-6, 21R-9, 21S-9, and 21S-20) had resistance indices greater than 1,000; in other words, their GR50 values were more than 1,000-fold higher than the wild-type value (Fig. 3D). The susceptibility to BSM was highly correlated to the transgene expression, similar to the CYP81A12 transgenic lines discussed above (Fig. 3, B and D; Supplemental Fig. S4), and was independent of the inserted allele (Fig. 3, D and F; Supplemental Fig. S5). Thus, both alleles of CYP81A21 conferred a decrease in BSM susceptibility to transgenic Arabidopsis that increased with the transcript level.
Transgenic Arabidopsis expressing the R line allele of CYP81A22 did not exhibit a significant decrease in BSM susceptibility, in contrast to Arabidopsis expressing CYP81A12 or CYP81A21 (data not shown). Similarly, the S line allele of CYP81A22 did not confer significantly decreased BSM susceptibility (data not shown).
We also examined the PX susceptibilities of the transgenic lines expressing either of CYP81A12, CYP81A21, and CYP81A22. The CYP81A12- and CYP81A21-expressing lines with low BSM susceptibility also exhibited low PX susceptibility, although the effect was less pronounced (Fig. 3, G and H). For example, in line 21R-6, the resistance index was 10 for PX compared with 4,000 for BSM (Fig. 3F; Supplemental Figs. S5 and S6). A correlation between mRNA accumulation and decreasing PX susceptibility was observed, similarly as with the BSM response (Fig. 3B; Supplemental Fig. S6). The lines with low transgene expression and relatively high BSM susceptibility (e.g. lines 12R-14 and 21R-4) did not show prominent decreases in PX susceptibility (data not shown). In lines expressing CYP81A22, no significant change in PX susceptibility was observed (data not shown).
Promoter Regions and Copy Numbers of CYP81A12 and CYP81A21
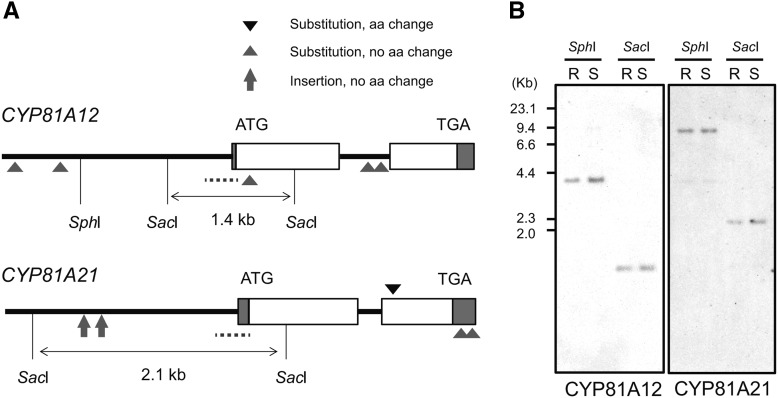
The putative promoter sequences of CYP81A12 and CYP81A21 were characterized over approximately 2 kb upstream from the transcription start site. The 300-bp region upstream from the ATG starting codon was extremely similar in the two genes. Large differences between the R and S lines were not observed in the putative promoter regions, although the promoters of the R line alleles of CYP81A12 and CYP81A21 carried two single nucleotide polymorphisms and two insertions, respectively (Fig. 4A).
Figure 4.
Molecular analysis of the CYP81A12 and CYP81A21 loci in R and S lines of E. phyllopogon. A, Schematic structure of CYP81A12 and CYP81A21 of the R and S lines. White and gray boxes represent coding regions and untranslated regions, respectively. SphI and SacI restriction sites, the translation start codon (ATG), and translation stop codon (TGA) are indicated. Dotted bars represent the positions of probes for genomic DNA-blot analysis. Triangles indicate the positions where DNA polymorphisms were observed. Arrows mark insertions observed in CYP81A21 of the R line. B, DNA-blot analysis of SphI/SacI-digested genomic DNA from R and S plants. CYP81A12 and CYP81A21 were detected using the probes depicted in Figure 4A.
The copy numbers of CYP81A12 and CYP81A21 were surveyed in a genomic DNA-blot analysis using gene-specific probes designed from the regions around the 5′-untranslated region and the promoter sequences (Fig. 4A). The band patterns were not different between the R and S lines for SacI and SphI digestions; single bands of the same size were observed in both lines (Fig. 4B). The signal intensity of the detected bands for the two genes was similar between the R and S lines. These results indicate that the copy number of CYP81A12 and CYP81A21 was one in both the R and S lines.
Segregation Analyses of Herbicide Susceptibilities in R and S E. phyllopogon
Herbicide susceptibilities of R and S plants from Californian populations of E. phyllopogon were reported to differ by at least more than 25-fold for BSM and 5- to 9-fold for PX (Osuna et al., 2002; Yasuor et al., 2009). In our evaluation of plants growing on Murashige and Skoog media, the GR50 of BSM was estimated to be 0.65 and 716 μm for the S and R lines, respectively (Supplemental Fig. S7, A and B). Thus, the resistance index for BSM was more than 1,100, which is not inconsistent with the previous report (at least more than 25-fold) that did not test high herbicide doses and therefore could not estimate GR50 accurately. On the other hand, the resistance index for PX, 6.2, was similar to published values (Supplemental Fig. S7, C and D).
Inheritance of BSM and PX resistance was assessed in the progeny of crosses between R and S plants. Resistance segregated in the F2 population on media containing 10 μm BSM or 0.3 μm PX, concentrations on which the S lines stopped growing at the first-leaf stage (Supplemental Fig. S7). The susceptibilities were assessed by measuring plant height 7 d after herbicide treatment. For BSM resistance, segregation approached a 1:2:1 ratio (R:intermediate:S = 15:25:14, χ2 = 0.3333, P = 0.8465; Supplemental Fig. S8A). The segregation of PX resistance was not as clear, likely because of a much lower resistance level of the parental R line compared with BSM. Still, the ratio of the number of plants taller than the S-parent line to plants with a similar plant height as the S-approached 3:1 (R + intermediate:S parent line = 38:16, χ2 = 0.6173, P = 0.4321; Supplemental Fig. S8B). These results suggested that the resistances to BSM and PX were each under the control of a single locus.
In the F6 population, 40 recombinant inbred lines (RILs) of the two reference lines were used to assess the linkage of the resistances to BSM and PX and the transcript levels of CYP81A12, CYP81A21, and CYP81A22. BSM and PX susceptibilities were fixed in almost all RILs and strongly linked to each other; 14 lines were resistant to BSM and PX, 25 lines were susceptible to BSM and PX, and only one line segregated for BSM and PX resistances (Fig. 5; Supplemental Table S1). Transcript levels of the three genes were determined in roots because the transcript levels differed most in the roots of the two parental lines (Supplemental Fig. S3). In the 14 R and 25 S RILs, high transcript levels of CYP81A12 and CYP81A21 perfectly cosegregated with BSM and PX resistances (Fig. 5). The transcript levels of the segregating lines were intermediate between those of the R and S groups, presumably because the levels were determined in bulk samples of seedling roots (n = 9). The transcript level of CYP81A22 was not associated with the herbicide resistances (Supplemental Fig. S9), indicating that the increased expression of CYP81A22 observed in the R parents plays no major role in herbicide resistance.
Figure 5.
Transcript levels of CYP81A12 and CYP81A21 and susceptibilities to BSM (A and B) and PX (C and D) in F6 lines derived from a cross between an R-parent and an S-parent line of E. phyllopogon. Transcript accumulations in roots of each line (n = 9) were examined by real-time RT-PCR using RPII and TUB as internal control genes. Bars for mRNA levels represent the sd of three technical replicates in real-time RT-PCR. BSM and PX susceptibilities were examined by measuring the plant height of nine plants of each line. Bars for plant height show the sd of the nine biological replicates. R and S mark homozygous lines carrying alleles of R and S parents, respectively. R/S indicates a heterozygous line having R as well as S alleles from R and S parents.
We also examined the alleles of the three genes in each RIL. The promoter sequences and coding sequences of CYP81A12 and CYP81A21 were not associated with herbicide resistance and high transcript levels (Fig. 5; Supplemental Table S1); plants that carried the S line allele of CYP81A12 and/or CYP81A21 could become resistant to both herbicides if their transcript levels were high and vice versa (Supplemental Table S1). The R and S alleles of CYP81A12 and CYP81A21 segregated independently (Supplemental Table S1), suggesting that CYP81A12 and CYP81A21 are located on different chromosomes. In contrast to CYP81A12 and CYP81A21, the transcript levels of CYP81A22 were perfectly associated with the allele (Supplemental Fig. S9). Alleles were also determined for CYP81A26 because the two lines differ by an amino acid polymorphism in the gene products. The alleles did not cosegregate with BSM and PX susceptibilities in the F6 plants (Supplemental Table S1), indicating that the amino acid polymorphism of CYP81A26 is mostly irrelevant for BSM and PX resistance.
BSM Metabolic Functions of CYP81A12 and CYP81A21
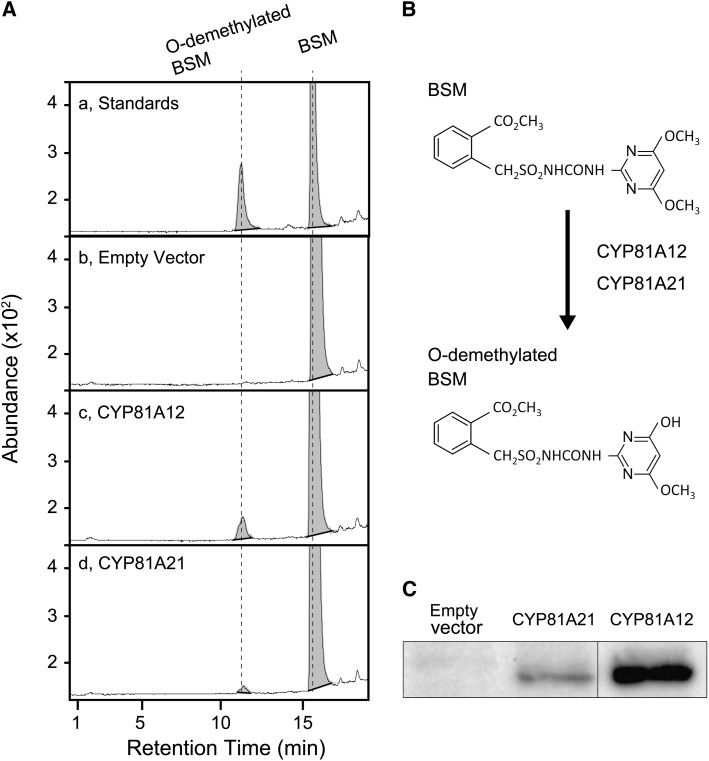
To evaluate the ability of CYP81A12 and CYP81A21 to metabolize BSM via O-demethylation, recombinant CYP81A12 and CYP81A21 proteins were expressed using a yeast (Saccharomyces cerevisiae) expression system that also carried the Arabidopsis NADPH-cytochrome P450 reductase gene ATR1 (Pompon et al., 1996). Accumulation of CYP81A12 and CYP81A21 proteins in the transgenic yeast were confirmed by immunoblotting (Fig. 6). The immunoblot showed that more CYP81A12 than CYP81A21 accumulated in the transgenic yeast.
Figure 6.
BSM metabolism catalyzed by CYP81A12 and CYP81A21. A, LC-MS/MS analyses of a BSM metabolite formed in yeast expressing CYP81A12 and CYP81A21. a, Chromatogram of standards of BSM and BSM O-demethylated at position 4 of the pyrimidine ring. b, Chromatogram of yeast harboring the empty vector (pYeDP60) as a control. BSM was detected in the culture media. c, Chromatogram of yeast expressing an R line allele of CYP81A12. A new peak, corresponding to the O-demethylated BSM, was detected. d, Chromatogram for the yeast expressing an R line allele of CYP81A21. The peak corresponding to the O-demethylated BSM was small but clearly detectable. B, Changes in the structure of BSM following O-demethylation catalyzed by CYP81A12 and CYP81A21. C, Immunoblot of microsome fractions extracted from yeast expressing CYP81A12 and CYP81A21.
For the metabolism assay, BSM was added to the yeast culture media. Fifteen hours later, the media were analyzed by LC-MS/MS. Although small, a new peak was detected in the media of yeast expressing CYP81A12 and CYP81A21, whereas no such peak occurred in the empty vector control (Fig. 6). The retention time of the new peak was 11.2 min, corresponding to the O-demethylated metabolite from BSM in the analysis of standards. The mass spectrum of the metabolite was the same as that of the O-demethylated BSM standard. These results indicated that CYP81A21 and CYP81A12 detoxify BSM via O-demethylation. The amounts of the metabolite were higher in the cultures transformed with CYP81A12 than in those expressing CYP81A21. This difference probably is caused by higher expression levels of CYP81A12 in the transformed yeast (Fig. 6), although further analysis is required to determine if BSM metabolic activities differ between the two P450s.
DISCUSSION
Our results showed that two E. phyllopogon P450 genes, CYP81A12 and CYP81A21, confer marked decreases in the susceptibilities to the two herbicides BSM and PX to Arabidopsis. The decrease in the susceptibilities was strongly correlated to the transcript levels of both genes in transgenic lines. The higher expression of the two genes in E. phyllopogon cosegregated completely with BSM and PX resistance in F6 RILs. CYP81A21 has amino acid polymorphisms that distinguish the R and S alleles, but the allelic variation had no influence on the susceptibilities to BSM and PX in transgenic Arabidopsis, and the alleles did not cosegregate with BSM and PX resistance in the F6 RILs of E. phyllopogon. Heterologously produced CYP81A12 and CYP81A21 proteins metabolized BSM into O-demethylated BSM, the highly accumulated metabolite in the R line of E. phyllopogon. These results indicate that enhanced expression of CYP81A12 and CYP81A21 plays a key role in the resistance to two different classes of ALS inhibitors in E. phyllopogon.
The resistance index observed in transformed Arabidopsis plants with high levels of CYP81A12 and CYP81A21 expression was more than 1,000 for BSM but only about 10 for PX (Fig. 3; Supplemental Fig. S6). BSM and PX responses of the R and S lines of E. phyllopogon were similar to those of the transformants; the resistance index for BSM was 1,100, and that for PX was 6.2 (Supplemental Fig. S7). The parallel herbicide responses between transformed Arabidopsis and E. phyllopogon supports the hypothesis that BSM and PX resistance in the R line of E. phyllopogon is caused by overexpression of these P450 genes.
In this study, heterologously produced CYP81A12 and CYP81A21 degraded BSM into the O-demethylated metabolite. The ALS-inhibiting activity of the O-demethylated metabolite in vitro is approximately one-three thousandth of that of BSM, and its herbicidal activity is practically abolished in plants, including E. phyllopogon (Takeda et al., 1986). The catalysis of the O-demethylation reaction by CYP81A12 and CYP81A21 can explain the low susceptibility to BSM in transgenic Arabidopsis expressing either of the two P450 genes. Importantly, the O-demethylated metabolite was the metabolite that accumulated more in the R line of E. phyllopogon. These results indicate that the two genes can confer BSM resistance to the R lines of E. phyllopogon. In rice, no direct evidence has been documented for O-demethylation of BSM by a specific P450, although P450s are known to be involved in BSM metabolism (Deng and Hatzios, 2002). Our data show that P450s in the CYP81A subfamily metabolize BSM into an O-demethylated metabolite. However, we have not investigated other metabolites that might be produced by the P450s, and further analyses using radioactive BSM might be required to fully characterize its metabolism.
We did not investigate the involvement of P450s in PX metabolism. PX is known to selectively kill wild-type Echinochloa spp. without damaging rice because of the rapid metabolic breakdown of PX in rice (Johnson et al., 2009). The major PX metabolite in rice is O-demethylated at one of the heterocyclic methoxy groups (Johnson et al., 2012). The reaction is similar to the O-demethylation of BSM. Previously, Yasuor et al. (2009) observed that PX was more rapidly metabolized into polar metabolites in the R line than in the S line of E. phyllopogon. Identification of the metabolites in E. phyllopogon and also in yeast expressing recombinant P450s will be future issues.
Our segregation study using an F2 population suggested that the resistances to BSM and PX resistances were controlled by a single genetic element each. Higher expression levels of CYP81A12 and CYP81A21 did not segregate in the F6 RILs, although the alleles of both genes including the promoter regions segregated independently (Fig. 5). The copy numbers of the two genes were not different between the R and S plants (Fig. 4B). These results suggest that a trans-element simultaneously regulates the expression of both genes in the R lines, and it appears unlikely that other mechanisms, such as gene amplification of the P450 genes or cis-acting regulatory loci, regulate expression. In contrast to CYP81A12 and CYP81A21, the magnitude of the expression levels of CYP81A22 was associated with its alleles in F6 RILs, although it was not associated with herbicide resistance. The mechanisms underlying the regulation of expression levels of CYP81A22 are unknown but likely involve cis-elements because large indels were observed in its promoter regions when comparing the R and S lines (data not shown).
The similarity of functions observed in CYP81A12 and CYP81A21 is likely attributable to their origin; these genes are putative homeologs derived from the allotetraploid nature of E. phyllopogon. The identities of the coding sequences and protein sequences of the two genes were very high, 96.2% and 94.7%, respectively. Their short genetic distance in the phylogenetic tree of coding sequences strongly suggests that the two genes are homeologs (Supplemental Fig. S1). CYP81A12 and CYP81A21 segregated independently in the F6 RILs (Supplemental Table S1). This autonomy of inheritance refutes the possibility of tandem duplication and supports the homeologous relationship of the two genes. At present, it is unknown whether increased expression of either of the two P450 genes is sufficient to acquire herbicide resistance in R plants of E. phyllopogon or whether both genes must be up-regulated.
The CYP81A subfamily belongs to the CYP81 family of P450s, a large protein family, in which lineage-specific duplication events, also known as a CYP bloom, frequently have occurred (Nelson and Werck-Reichhart, 2011). Apparently, the CYP81A subfamily is also lineage specific, because genes encoding this subfamily have only been reported in grasses (Nelson, 2009). In the CYP81A subfamily, large divergence is observed in the number and sequences of P450s in E. phyllopogon, maize (Zea mays), and Sorghum bicolor, three members of the Panicoideae subfamily (Supplemental Fig. S1). This divergence is much larger than in rice (subfamily Oryzoideae) and Brachypodium distachyon (subfamily Pooideae), indicating that lineage-specific duplication events may have occurred together with the diversification of the Panicoideae. Duplicated P450 genes could be beneficial for fine-tuning the levels of the metabolites of their gene products via up- or down-regulation of each copy (subfunctionalization; e.g. Kushiro et al., 2004; Magome et al., 2013) and may enable the acquisition of new enzymatic functions (neofunctionalization; e.g. Frey et al., 2009; Prasad et al., 2012; Weng et al., 2012). The expression of CYP81A22 in Arabidopsis failed to confer significant decreases in herbicide susceptibilities in contrast to CYP81A12 and CYP81A21, supporting the notion that members of the CYP81A subfamily may perform different enzymatic functions. The functional characterization of the diverse CYP81A genes in E. phyllopogon is an important issue in the study of herbicide resistance.
To elucidate the evolution of multiple herbicide resistance in E. phyllopogon, it will be important to identify the element responsible for the overexpression of the two CYP81A genes, a causal gene for BSM and PX resistance. Our research suggested a single causal element that contrasts, however, with well-studied cases of nontarget site-resistant weeds such as Lolium rigidum or Alopecurus myosuroides. In these species, the resistance to each herbicide is frequently regulated by multiple genes (e.g. Neve and Powles, 2005a, 2005b; Petit et al., 2010; Busi et al., 2011, 2013). This discrepancy can be explained by the mode of pollination. Outcrossing in L. rigidum and A. myosuroides can facilitate the evolution of resistance through the accumulation of minor genes, where individual genes make limited contributions to herbicide resistance but can cooperatively cause significant levels of resistance. The mode of pollination in E. phyllopogon, however, is self-crossing; outcrossing is limited in the study populations as demonstrated by Tsuji et al. (2003), and accumulations of minor resistance genes are unlikely to occur. The self-pollinating nature of this species might have favored the selection of a major gene causing a large decrease in herbicide susceptibility.
Moreover, it will be necessary to establish whether the resistance to herbicides other than BSM and PX is based on similar mechanisms. Previous studies suggested that resistances to different herbicides can be based on distinct mechanisms. For example, in the resistance to aryloxyphenoxy-propionate herbicides, significant contributions of glutathione and Cys conjugation seem to be involved (Ruiz-Santaella et al., 2006; Bakkali et al., 2007). In quinclorac resistance, insensitivity of ethylene synthesis pathways that usually respond to quinclorac and enhanced activity of β-cyanoalanine synthase were suggested (Yasuor et al., 2012). In bispyribac-sodium resistance, bispyribac-sodium-induced P450s appear involved (Yun et al., 2005), and slightly increased expression of two P450 genes (not CYP81A21 or CYP81A21) has been reported under bispyribac-sodium stress (Iwakami et al., 2014a).
Herbicides exert strong selection pressure in agroecosystems, resulting in the evolution of herbicide-resistant plant lines. P450-mediated herbicide resistance has been implicated in several weed species (Preston, 2004; Yuan et al., 2007; Powles and Yu, 2010; Beckie and Tardif, 2012). Despite their threatening impact on agriculture, the molecular mechanisms of P450-mediated resistance remained unknown for more than 25 years (Kemp and Caseley, 1987). In this study, we identified P450 genes associated with herbicide resistance in weeds causing serious problems in agricultural fields. Our findings provide a new perspective on P450-mediated herbicide resistance in weeds and will inform further studies on herbicide resistance.
MATERIALS AND METHODS
Origin of Materials and Seed Preparation
Lines 511 and 401 were used as the R and S lines of Echinochloa phyllopogon, respectively. These lines were originally collected from rice (Oryza sativa) fields in the Sacramento Valley of California in 1997 and were self-pollinated for three successive generations (Tsuji et al., 2003). BSM susceptibility of 401 was similar to those of other susceptible accessions collected from California and lacked multiple herbicide resistance, indicating that the BSM response of 401 is within the response range of wild-type E. phyllopogon. The two lines and F3 populations resulting from a crossing of 401 and 511 were provided by Dr. Yuji Yamasue (Kyoto University). The crossing procedure was described in Tsuji et al., 2003. The F3 populations were further self-pollinated for three successive generations to obtain F6 seeds. Another cross between the S and R lines was generated according to the procedure in Tsuji et al., 2003, followed by selfing to produce F2 populations. The seeds were sterilized with 0.5% (w/v) sodium hypochlorite for 15 min, further sterilized with 0.04% (w/v) sodium hypochlorite for 30 min, and then washed three times in sterile water. Seeds were germinated on wet paper for 2.5 d in a growth chamber at 25°C under constant fluorescent light (approximately 300 μE m–2 s–1) and then transferred to solid media.
Sample Preparation for BSM Metabolism and Transcript Analysis under BSM Stress
Germinated seeds of the R and S lines of E. phyllopogon were planted on solid one-half-strength Murashige and Skoog medium and grown under the same conditions used for germination. Four days after planting, the plants were transplanted onto one-half-strength Murashige and Skoog solid media with or without 10 μm BSM. After 24 h, 1 g of shoots or roots was harvested, washed in distilled water, and stored at –20°C for BSM metabolism analysis. For transcript analysis, the shoot of a plant and roots of three plants were harvested with four replications, frozen in liquid N2, and stored at –80°C. Total RNA was isolated from the frozen samples. Complementary DNA (cDNA) was synthesized from the RNA, and transcript levels were determined by real-time reverse transcription (RT)-PCR (described below).
LC-MS/MS Analysis of BSM Metabolites
For the analysis of BSM metabolism in E. phyllopogon, the stored samples were homogenized in 100 mL of acetone and filtered through glass fiber filters (Kiriyama), and the filtrate was dissolved in 200 mL of acetone. Five milliliters of water was added to 20 mL of extract, and the acetone was evaporated. The samples (10 μL) were loaded onto a solid-phase extraction cartridge (InertSep PLS-2, styrene-divinylbenzene; GL Sciences), followed by washing with 5 mL of water:acetic acid:formic acid (80:20:1). BSM and its metabolites were eluted using 10 mL of water:acetic acid:formic acid (20:80:1).
HPLC analysis was performed on an Agilent 1200 Series, and tandem mass spectrometry was performed on an Agilent 6410 Triple Quad LC/MS with a Zorbax Eclipse Plus C18 analytical column at 40°C (Agilent Technologies). As internal standards, O-demethylated BSM [methyl-α-(4-hydroxy-6-methoxypyrimidin-2-yl) carbamoylsulfamoyl-o-toluate], a gift from DuPont, and BSM (Wako) were used. Separations were performed with mobile phases consisting of methanol (solution A) and water with 0.01% (v/v) formic acid (solution B). The gradient increased linearly from 70% (v/v) solution A and 30% (v/v) solution B to 100% solution B over 15 min with a flow rate of 0.2 mL min–1. The molecules were ionized at 350°C. Collision voltages for BSM and O-demethylated BSM were 20 and 15 V, respectively. Data were analyzed using Mass Hunter software (Agilent Technologies) and quantified based on standard curves.
For the analysis of recombinant P450s in yeast (Saccharomyces cerevisiae), the stored samples were diluted 10-fold with water and filtered through 0.2-μm polytetrafluoroethylene membranes (Whatman). LC-MS/MS analysis was performed as described above.
Nucleic Acid Extraction and cDNA Synthesis
Genomic DNA was extracted from leaves of E. phyllopogon using a DNeasy Plant Mini Kit (Qiagen) or a Nucleon Phytopure extraction kit (Amersham Pharmacia Biotech). The plants were grown in a greenhouse at Tsukuba, Japan.
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) with on-column DNase I digestion (RNase-Free DNase Set, Qiagen). The RNAs were further treated with a TURBO DNA-free kit (Applied Biosystems) for complete elimination of genomic DNA. Total RNA (1 μg) was reverse transcribed using a PrimeScript II First-Strand cDNA Synthesis Kit with oligo(dT) primer (TaKaRa). The plants from which RNA was extracted are described below.
Isolation of CYP81A Genes
For the isolation of CYP81A subfamily genes from E. phyllopogon, degenerate PCR was conducted using cDNA prepared from the RNA extracted from third-leaf stage shoots of the R line. Primers used are listed in Supplemental Table S2. The plants used for RNA extractions were grown on soil in a growth chamber with the same environmental conditions as described above for seed germination. PCR amplification was carried out using TaKaRa LA Taq or PrimeSTAR GXL DNA Polymerase (TaKaRa) with the primers listed in Supplemental Table S2. The amplicons were cloned using a pGEM-T Easy Vector Systems Kit (Promega) and sequenced as described previously (Iwakami et al., 2012). The full-length CYP81A gene was obtained by RACE as described previously (Iwakami et al., 2012). The 5′-flanking sequences of CYP81A12, CYP81A21, and CYP81A22 were determined by thermal asymmetric interlaced PCR (Liu et al., 1995; Liu and Whittier, 1995).
Real-Time RT-PCR
Real-time RT-PCR of E. phyllopogon was carried out as described previously (Iwakami et al., 2012). mRNA quantitation was performed by the ΔΔCT method. Gene expression was normalized to the expression of eukaryotic translation initiation factor 4B (EIF4B) and RNA polymerase II (RPII) in the mRNA quantitation under BSM stress and to RPII and tubulin (TUB) in the analysis of F6 RILs. The stability of these genes was evaluated using geNormPLUS software (Vandesompele et al., 2002). Primers used in real-time PCR are listed in Supplemental Table S2.
Transgene expression in Arabidopsis (Arabidopsis thaliana) was quantified using the standard curve method of real-time RT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control.
Arabidopsis Transformation and Characterization of Transgenic Plants
The coding regions of CYP81A12, CYP81A21, and CYP81A22 were amplified by PCR with the primer pairs listed in Supplemental Table S2 using cDNA prepared from third-leaf stage shoots. The amplicons were subcloned into a pENTR/D-TOPO cloning vector (Invitrogen) to yield entry vectors. Binary vectors were produced in an LR Clonase-catalyzed reaction (Invitrogen) with the entry clones and the pB2GW7 vector (Karimi et al., 2002) for transformation of Arabidopsis. The binary vectors were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation. Arabidopsis (ecotype Columbia-0) was transformed with the floral dip method using the transformed A. tumefaciens strains (Clough and Bent, 1998). The transformants were selected by bialaphos (25 mg L–1) treatment.
Four to five 12-d-old seedlings of T3 or T4 homozygous lines grown on Murashige and Skoog solid medium were harvested, frozen in liquid nitrogen, and stored for RNA extraction. The cDNAs were synthesized as described above.
BSM susceptibilities of the transgenics were evaluated by growth on Murashige and Skoog solid medium containing BSM. All assays were conducted at 22°C with a 12-h photoperiod and a light intensity of about 70 μE m–2 s–1. Seeds were planted on Murashige and Skoog solid media containing BSM in petri dishes and allowed to grow for 12 d. The position of each dish in the growth chamber was changed every 1 or 2 d. The fresh weights of 10 plants were measured immediately after removing fluid (four replicates), and the GR50 value was calculated using a nonlinear computer analysis based on a log-logistic model (Seefeldt et al., 1995).
Genomic DNA-Blot Analysis
DNA (3 μg) of the R and S lines of E. phyllopogon was digested with SphI or SacI (Roche). The digested DNA samples were electrophoresed on a 0.8% (w/v) agarose gel, and DNA fragments were transferred onto a positively charged nylon membrane (Roche). Gene-specific DNA probes of approximately 300 bp were prepared using a PCR DIG Probe Synthesis Kit (Roche). For probe synthesis, cloned DNA sequences of CYP81A12 and CYP81A21 were used as templates with a forward primer (5′-CCTCTCTTCCCCTCCCTGAC-3′) and a reverse primer (5′-GCCACGTAGGCCTTATCCAT-3′). Hybridization was performed according to the DIG Application Manual (Roche). Hybridization signals were detected on an LPR-400EX chemiluminescence detection analyzer (Taitec).
Inheritance of Transcript Levels and Herbicide Resistances
Transcript levels of F6 RILs were determined by real-time RT-PCR using RNAs extracted from first leaf stage roots of nine plants. Germination was conducted as described above, and plants were grown under the same conditions as described for germination. At the first leaf stage, roots from nine plants were harvested, frozen in liquid nitrogen, and stored for RNA extraction. cDNA was synthesized as described above.
BSM and PX susceptibilities of the F6 RILs were evaluated by growth on one-half-strength Murashige and Skoog media containing 10 μm BSM and 0.3 μm PX, respectively. Nine germinated seeds were planted on one-half-strength Murashige and Skoog media containing a herbicide. Seven days after transplanting, plant height was measured. Germination was conducted as described above. Plants grew at 25°C under constant fluorescent light (approximately 300 μE m–2 s–1).
Alleles of each line were determined by sequencing PCR products amplified from DNA extracted from F5 plants. Primers used for the amplification of coding sequence and promoter regions are listed in Supplemental Table S2.
Yeast Transformation and P450 Expression
For the expression of P450s, the WAT11 yeast strain and the pYeDP60 vector system was used (Pompon et al., 1996). The WAT11 strain contains an Arabidopsis NADPH-P450 reductase gene (ATR1); pYeDP60 is an expression vector with the Gal-inducible and Glc-repressed yeast hybrid promoter GAL10-CYC1. KpnI and EcoRI sites were introduced by PCR just upstream of the ATG and downstream of the stop codon of the full-length coding sequences, respectively. In addition, the FLAG epitope sequence was fused just downstream of the ATG using the primers listed in Supplemental Table S2. The amplicons were subcloned into pGEM-T Easy (Promega). The subclones were digested with KpnI and EcoRI and then inserted into pYeDP60. The vectors were transformed into the yeast strain using a LiCl method according to the manufacturer’s instructions provided with the pYES2 vector (Invitrogen). P450 expression was induced according to the modified two-stage cultivation method using modified SLI medium (Jiang and Morgan, 2004). For construction of the expression vector, original nucleotide sequences of E. phyllopogon P450 genes were used because no significant improvements were observed by recoding the N-terminal part of the P450 genes according to the procedure described by Hehn et al. (2002).
BSM was added to the culture medium at a final concentration of 100 μm at 9 h after the induction of P450 by Gal. At 24 h after the start of Gal induction, the culture solution was centrifuged at 500g for 5 min. The supernatant was filtered through a polyvinylidene difluoride membrane (0.22-μm pore size, Merck Millipore) and stored at –20°C until LC-MS/MS analysis.
Immunoblot Analysis
Microsomes of transgenic yeast were prepared according to Pompon et al. (1996), and 30 mg of the microsomes was electrophoresed on 10% (w/v) SDS polyacrylamide gels (SDS-PAGE). The proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P, Merck Millipore) by semidry blotting. Membranes were blocked overnight at 4°C in 5% (w/v) nonfat dried milk and then incubated in 1:4,600 dilutions of anti-flag M2 monoclonal antibody for 1 h at room temperature (Sigma-Aldrich). Blots were washed in Tris-buffered saline plus Tween 20 (12.5 mm Tris–HCl, 137 mm NaCl, 0.1% [v/v] Tween 20, pH 7.5) and incubated for 1 h at room temperature in a 1:2,500 dilution of goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Thermo Fisher Scientific) in Tris-buffered saline plus Tween 20. The blot was stained using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). Signals were detected on an LPR-400EX chemiluminescence analyzer (Taitec).
The cDNA sequences of the R strain alleles are cataloged under the following accession numbers: CYP81A13P, AB733993; CYP81A14, AB733994; CYP81A15, AB733995; CYP81A18, AB733996; CYP81A19P, AB733997; CYP81A23, AB734000; CYP81A24, AB734001; CYP81A25P, AB734002; and CYP81A26, AB734003. The cDNA sequences of the S strain alleles are cataloged under the accession number CYP81A26, AB818465. The accession numbers for the genomic DNA and cDNA sequences of CYP81A12 are AB818461 for the R line allele and AB818460 for the S line allele. The accession numbers for genomic DNA and cDNA sequences of CYP81A21 are AB818462 for the R line allele and AB818463 for the S line allele. The accession numbers for the genomic DNA sequences of CYP81A22 are AB872309 for the R line allele and AB872310 for the S line allele. Genes used as reference genes for real-time RT-PCR in E. phyllopogon were EIF4B, AB720070; RPII, AB775462; and TUB, AB872311; in Arabidopsis, GAPDH, AT1G13440 was used.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenogram of CYP81A subfamily proteins from various Poaceae.
Supplemental Figure S2. Alignment of protein sequences of the CYP81A subfamily of an R line.
Supplemental Figure S3. Transcript levels of CYP81A12, CYP81A21, and CYP81A22 in several organs of an R and an S line of E. phyllopogon.
Supplemental Figure S4. Relationship of transcript level and BSM susceptibility in transgenic Arabidopsis expressing CYP81A12 or CYP81A21.
Supplemental Figure S5. BSM and PX responses of transgenic Arabidopsis expressing an S line allele of CYP81A21.
Supplemental Figure S6. PX susceptibilities in transgenic Arabidopsis expressing CYP81A12 or an R line allele of CYP81A21.
Supplemental Figure S7. BSM and PX susceptibilities of an R and an S line of E. phyllopogon.
Supplemental Figure S8. Segregation of BSM and PX resistances in an F2 population derived from a cross between an R and an S line of E. phyllopogon.
Supplemental Figure S9. Transcript levels of CYP81A22 and susceptibilities to BSM and PX in F6 lines derived from a cross between an R parent and an S parent line of E. phyllopogon.
Supplemental Table S1. Linkage between herbicide resistances and genotypes in the RILs.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. Tomonori Fujioka and Tsutomu Shimizu for technical advice on heterologous expression of P450s in yeast; Ayako Nishizawa-Yokoi for help in western-blot analysis; DuPont for providing BSM and the analytical standards of BSM metabolites; Dr. David Nelson for classifying and naming the P450s; Drs. Danièle Werck-Reichhart and Denis Pompom for providing the pYeDP60 vector and WAT11 yeast strain; Dr. Hiroyuki Shibaike for sharing experimental facilities; Sachiko Inanuma, Hiroko Kato, Kiyoko Amagai, Rieko Aoto, Chiharu Furusawa, Akemi Nagashii, Etsuko Ozawa, and Fukuko Suzuki for technical assistance; and Dr. Yuji Yamasue for providing the opportunity to conduct this research and for providing the seeds of E. phyllopogon used in the study.
Glossary
- BSM
bensulfuron-methyl
- PX
penoxsulam
- R
resistant
- S
susceptible
- ALS
acetolactate synthase
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- GR50
50% growth reduction
- RIL
recombinant inbred line
- cDNA
complementary DNA
- RT
reverse transcription
Footnotes
This work was supported by the Pesticide Science Society of Japan (grant to S.I.) and the Japan Association for Advancement of Phyto-Regulators (grant to A.U.).
The online version of this article contains Web-only data.
References
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D. (2011) Cytochromes p450. Arabidopsis Book 9: e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkali Y, Ruiz-Santaella JP, Osuna MD, Wagner J, Fischer AJ, De Prado R. (2007) Late watergrass (Echinochloa phyllopogon): mechanisms involved in the resistance to fenoxaprop-p-ethyl. J Agric Food Chem 55: 4052–4058 [DOI] [PubMed] [Google Scholar]
- Beckie HJ, Tardif FJ. (2012) Herbicide cross resistance in weeds. Crop Prot 35: 15–28 [Google Scholar]
- Busi R, Neve P, Powles S. (2013) Evolved polygenic herbicide resistance in Lolium rigidum by low-dose herbicide selection within standing genetic variation. Evol Appl 6: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi R, Vila-Aiub MM, Powles SB. (2011) Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity (Edinb) 106: 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Délye C. (2013) Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag Sci 69: 176–187 [DOI] [PubMed] [Google Scholar]
- Deng F, Hatzios KK. (2002) Characterization of cytochrome P450-mediated bensulfuron-methyl O-demethylation in rice. Pestic Biochem Physiol 74: 102–115 [Google Scholar]
- Duggleby RG. (2005) Suicide inhibition of acetohydroxyacid synthase by hydroxypyruvate. J Enzyme Inhib Med Chem 20: 1–4 [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Ateh CM, Bayer DE, Hill JE. (2000a) Herbicide-resistant Echinochloa oryzoides and E. phyllopogon in California Oryza sativa fields. Weed Sci 48: 225–230 [Google Scholar]
- Fischer AJ, Bayer DE, Carriere MD, Ateh CM, Yim KO. (2000b) Mechanisms of resistance to bispyribac-sodium in an Echinochloa phyllopogon accession. Pestic Biochem Physiol 68: 156–165 [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267: 83–90 [PubMed] [Google Scholar]
- Hehn A, Morant M, Werck-Reichhart D. (2002) Partial recoding of P450 and P450 reductase cDNAs for improved expression in yeast and plants. Methods Enzymol 357: 343–351 [DOI] [PubMed] [Google Scholar]
- Iwakami S, Uchino A, Kataoka Y, Shibaike H, Watanabe H, Inamura T. (2014a) Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag Sci 70: 549–558 [DOI] [PubMed] [Google Scholar]
- Iwakami S, Uchino A, Watanabe H, Yamasue Y, Inamura T. (2012) Isolation and expression of genes for acetolactate synthase and acetyl-CoA carboxylase in Echinochloa phyllopogon, a polyploid weed species. Pest Manag Sci 68: 1098–1106 [DOI] [PubMed] [Google Scholar]
- Iwakami S, Watanabe H, Miura T, Matsumoto H, Uchino A. (2014b) Occurrence of sulfonylurea resistance in Sagittaria trifolia L., a basal monocot species, based on target-site and non-target-site resistance. Weed Biol Manage 14: 43–49 [Google Scholar]
- Jeffers GM, O’Donovan JT, Hall LM. (1996) Wild mustard (Brassica kaber) resistance to ethametsulfuron but not to other herbicides. Weed Technol 10: 847–850 [Google Scholar]
- Jiang H, Morgan JA. (2004) Optimization of an in vivo plant P450 monooxygenase system in Saccharomyces cerevisiae. Biotechnol Bioeng 85: 130–137 [DOI] [PubMed] [Google Scholar]
- Johnson TC, Mann RK, Schmitzer PR, Gast RE, deBoer GJ (2012) Triazolopyrimidines. In W Krämer, U Schirmer, P Jeschke, W Matthias, eds, Modern Crop Protection Compounds, Ed 2, Vol 1. Wiley-VCH, Weinheim, Germany, pp 99–117 [Google Scholar]
- Johnson TC, Martin TP, Mann RK, Pobanz MA. (2009) Penoxsulam: structure-activity relationships of triazolopyrimidine sulfonamides. Bioorg Med Chem 17: 4230–4240 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kemp MS, Caseley JC (1987) Synergistic effects of 1-aminobenzotriazole on the phytotoxicity of chlorotoluron and isoproturon in a resistant population of black-grass (Aleperurus myosuroides). In 1987 British Crop Protection Conference: Weeds. BCPC Publications, Surrey, UK, pp 895–899 [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S. (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA 110: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Nelson D, Werck-Reichhart D. (2011) A P450-centric view of plant evolution. Plant J 66: 194–211 [DOI] [PubMed] [Google Scholar]
- Nelson DR. (2009) The cytochrome p450 homepage. Hum Genomics 4: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve P, Powles S. (2005a) High survival frequencies at low herbicide use rates in populations of Lolium rigidum result in rapid evolution of herbicide resistance. Heredity (Edinb) 95: 485–492 [DOI] [PubMed] [Google Scholar]
- Neve P, Powles S. (2005b) Recurrent selection with reduced herbicide rates results in the rapid evolution of herbicide resistance in Lolium rigidum. Theor Appl Genet 110: 1154–1166 [DOI] [PubMed] [Google Scholar]
- Osuna MD, Vidotto F, Fischer AJ, Bayer DE, De Prado R, Ferrero A. (2002) Cross-resistance to bispyribac-sodium and bensulfuron-methyl in Echinochloa phyllopogon and Cyperus difformis. Pestic Biochem Physiol 73: 9–17 [Google Scholar]
- Pan G, Zhang X, Liu K, Zhang J, Wu X, Zhu J, Tu J. (2006) Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol Biol 61: 933–943 [DOI] [PubMed] [Google Scholar]
- Petit C, Duhieu B, Boucansaud K, Délye C. (2010) Complex genetic control of non-target-site-based resistance to herbicides inhibiting acetyl-coenzyme A carboxylase and acetolactate-synthase in Alopecurus myosuroides Huds. Plant Sci 178: 501–509 [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Powles SB, Yu Q. (2010) Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol 61: 317–347 [DOI] [PubMed] [Google Scholar]
- Prasad KV, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, et al. (2012) A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337: 1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. (2004) Herbicide resistance in weeds endowed by enhanced detoxification: complications for management. Weed Sci 52: 448–453 [Google Scholar]
- Ruiz-Santaella JP, De Prado R, Wagner J, Fischer AJ, Gerhards R. (2006) Resistance mechanisms to cyhalofop-butyl in a biotype of Echinochloa phyllopogon (Stapf) Koss. from California. J Plant Dis Prot XX: 95–100 [Google Scholar]
- Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, Burgos NR. (2012) EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag Sci 68: 1223–1230 [DOI] [PubMed] [Google Scholar]
- Seefeldt SS, Jensen JE, Fuerst EP. (1995) Log-logistic analysis of herbicide dose-response relationships. Weed Technol 9: 218–227 [Google Scholar]
- Siminszky B. (2006) Plant cytochrome P450-mediated herbicide metabolism. Phytochem Rev 5: 445–458 [Google Scholar]
- Takeda S, Erbes DL, Sweetser PB, Hay JV, Yuyama T. (1986) Mode of herbicidal and selective action of DPX-F5384 between rice and weeds. J Weed Sci Tech 31: 157–163 [Google Scholar]
- Tranel PJ, Riggins CW, Bell MS, Hager AG. (2011) Herbicide resistances in Amaranthus tuberculatus: a call for new options. J Agric Food Chem 59: 5808–5812 [DOI] [PubMed] [Google Scholar]
- Tsuji R, Fischer AJ, Yoshino M, Roel A, Hill JE, Yamasue Y. (2003) Herbicide-resistant late watergrass (Echinochloa phyllopogon): similarity in morphological and amplified fragment length polymorphism traits. Weed Sci 51: 740–747 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034–, 0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Li Y, Mo H, Chapple C. (2012) Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337: 960–964 [DOI] [PubMed] [Google Scholar]
- Yamasue Y. (2001) Strategy of Echinochloa oryzicola Vasing. for survival in flooded rice. Weed Biol Manage 1: 28–36 [Google Scholar]
- Yasuor H, Milan M, Eckert JW, Fischer AJ. (2012) Quinclorac resistance: a concerted hormonal and enzymatic effort in Echinochloa phyllopogon. Pest Manag Sci 68: 108–115 [DOI] [PubMed] [Google Scholar]
- Yasuor H, Osuna MD, Ortiz A, Saldaín NE, Eckert JW, Fischer AJ. (2009) Mechanism of resistance to penoxsulam in late watergrass (Echinochloa phyllopogon [Stapf] Koss.). J Agric Food Chem 57: 3653–3660 [DOI] [PubMed] [Google Scholar]
- Yasuor H, Zou W, Tolstikov VV, Tjeerdema RS, Fischer AJ. (2010) Differential oxidative metabolism and 5-ketoclomazone accumulation are involved in Echinochloa phyllopogon resistance to clomazone. Plant Physiol 153: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Powles SB. (January 20, 2014) Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag Sci http//dx..org/10.1002/ps.3710 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Tranel PJ, Stewart CNJ., Jr (2007) Non-target-site herbicide resistance: a family business. Trends Plant Sci 12: 6–13 [DOI] [PubMed] [Google Scholar]
- Yun MS, Yogo Y, Miura R, Yamasue Y, Fischer AJ. (2005) Cytochrome P-450 monooxygenase activity in herbicide-resistant and -susceptible late watergrass (Echinochloa phyllopogon). Pestic Biochem Physiol 83: 107–114 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.