A maize semidominant mutant that results from a storage protein with defective signal peptide disrupts protein body assembly, triggers ER stress, and elevates programmed cell death in endosperm.
Abstract
Zeins are the major seed storage proteins in maize (Zea mays). They are synthesized on the endoplasmic reticulum (ER) and deposited into protein bodies. Failure of signal peptide cleavage from zeins can cause an opaque endosperm in the mature kernel; however, the cellular and molecular mechanisms responsible for this phenotype are not fully understood. In this study, we report the cloning and characterization of a novel, semidominant opaque mutant, floury4 (fl4). fl4 is caused by a mutated z1A 19-kD α-zein with defective signal peptide cleavage. Zein protein bodies in fl4 endosperm are misshapen and aggregated. Immunolabeling analysis indicated that fl4 participates in the assembly of zeins into protein bodies, disrupting their proper spatial distribution. ER stress is stimulated in fl4 endosperm, as illustrated by dilated rough ER and markedly up-regulated binding protein content. Further analysis confirmed that several ER stress pathways are induced in fl4 endosperm, including ER-associated degradation, the unfolded protein response, and translational suppression by the phosphorylation of eukaryotic translational initiation factor2 α-subunit. Programmed cell death is also elevated, corroborating the intensity of ER stress in fl4. These results provide new insights into cellular responses caused by storage proteins with defective signal peptides.
Maize (Zea mays) is one of the most important cereal crops in the world, the value of which is largely determined by the endosperm, the main storage tissue (Holding and Larkins, 2006). The endosperm consists of four major cell types: starchy endosperm cells, embryo-surrounding region cells, aleurone cells, and transfer cells. The starchy endosperm accumulates storage compounds, primarily starch and protein, and influences seed texture (Sabelli and Larkins, 2009). During development of the kernel, zeins are synthesized in the endosperm and deposited in protein bodies (Lending and Larkins, 1989). Newly synthesized zein proteins have a signal peptide by which they enter the endoplasmic reticulum (ER), where they undergo proper folding for packaging into protein bodies. Zein protein bodies have a peripheral layer rich in β- and γ-zein, while the α-zeins fill the center (Lopes and Larkins, 1991). Protein body formation is regulated by the transcription level of zeins and interactions between different zein proteins (Kim et al., 2002). The opaque endosperm phenotype, with an increased starchy region and decreased kernel hardness, is mostly associated with altered accumulation of zeins (Holding and Larkins, 2006; Wu and Messing, 2010). Our understandings of the mechanisms determining zein accumulation come largely from the study of opaque mutants.
A number of maize mutants with an opaque or floury kernel phenotype have been identified, including three classes: recessive mutants, semidominant mutants, and dominant mutants (Gibbon and Larkins, 2005). Four genes corresponding to recessive mutants, opaque1 (o1), o2, o5, and o7, have been cloned (Schmidt et al., 1990; Miclaus et al., 2011; Myers et al., 2011; Wang et al., 2011, 2012), the majority of which result from defects in storage protein synthesis, which affects endosperm texture. For example, O2 encodes a transcriptional factor that positively regulates the expression level of 22-kD α-zeins (Damerval and Devienne, 1993). O7 is an acyl-activating enzyme-like protein that influences amino acid and zein protein synthesis (Wang et al., 2011). O1 encodes a Myosin XI Motor Protein that affects protein body formation by disrupting ER morphology and motility (Wang et al., 2012). Four genes corresponding to semidominant or dominant mutants, floury1 (fl1), fl2, Defective endosperm-B30 (DE-B30) and Mucronate (Mc), have also been cloned (Coleman et al., 1997; Kim et al., 2004, 2006; Holding et al., 2007). Fl1 encodes an ER membrane protein involved in facilitating the localization of 22-kD α-zein in the protein bodies (Holding et al., 2007). The other semidominant and dominant mutants that have been cloned affect storage proteins themselves. fl2 encodes a 22-kD α-zein with defective signal peptide (Coleman et al., 1997). De-B30 is a 19-kD α-zein with a single amino acid replacement, resulting in a defective signal peptide (Kim et al., 2004). Mc encodes a 16-kD γ-zein with a frame shift mutation (Kim et al., 2006). These mutants manifest a general reduction in zeins, exhibit disrupted zein deposition and protein body deformation, and stimulate the ER stress response (Coleman et al., 1997; Kim et al., 2004, 2006). However, the mechanism underlying the starchy endosperm phenotype in these mutants is not fully understood.
Folding of proteins in the ER lumen includes three modifications, signal peptide cleavage, N-linked glycosylation, and disulfide bond formation, all of which require ER-resident molecular chaperones (Petersen et al., 2011). The ER stress response is a conserved mechanism that deals with misfolded or unfolded proteins. Signal peptide removal is an early step in protein folding, the failure of which can trigger ER stress responses (Braakman and Hebert, 2013). Cells are equipped with an exquisite ER quality control system (Deng et al., 2013) and a group of cell-sparing mechanisms under conditions of ER stress, including the ER-associated degradation (ERAD; Gillece et al., 1999), the unfolded protein response (UPR), and increased phosphorylation of eukaryotic translational initiation factor2 α-subunit (eIF2α), which slows protein synthesis (Harding et al., 1999). Programmed cell death (PCD) may occur if these cell-sparing mechanisms are exhausted (Liu and Howell, 2010). In fl2, De-B30, and Mc mutants, significant ER stress occurs that increases the amount of molecular chaperones, including binding protein (BIP), and decreases the amount of storage protein in the seed (Coleman et al., 1997; Kim et al., 2004, 2006; Kirst et al., 2005). But how these mutants deal with ER stress merits further investigation.
In this study, we characterized fl4, a novel semidominant opaque maize mutant that has small, misshapen, and aggregated protein bodies, along with a dilated ER. fl4, which was identified by a combination of map-based cloning and biochemical approaches, encodes a 19-kD α-zein z1A subfamily member with a defective signal peptide cleavage site. Our results indicated that fl4 disrupts the assembly of zeins into protein bodies and triggers ER stress pathways.
RESULTS
fl4 Is a Semidominant Opaque Mutant That Produces Small, Misshapen, and Aggregated Protein Bodies
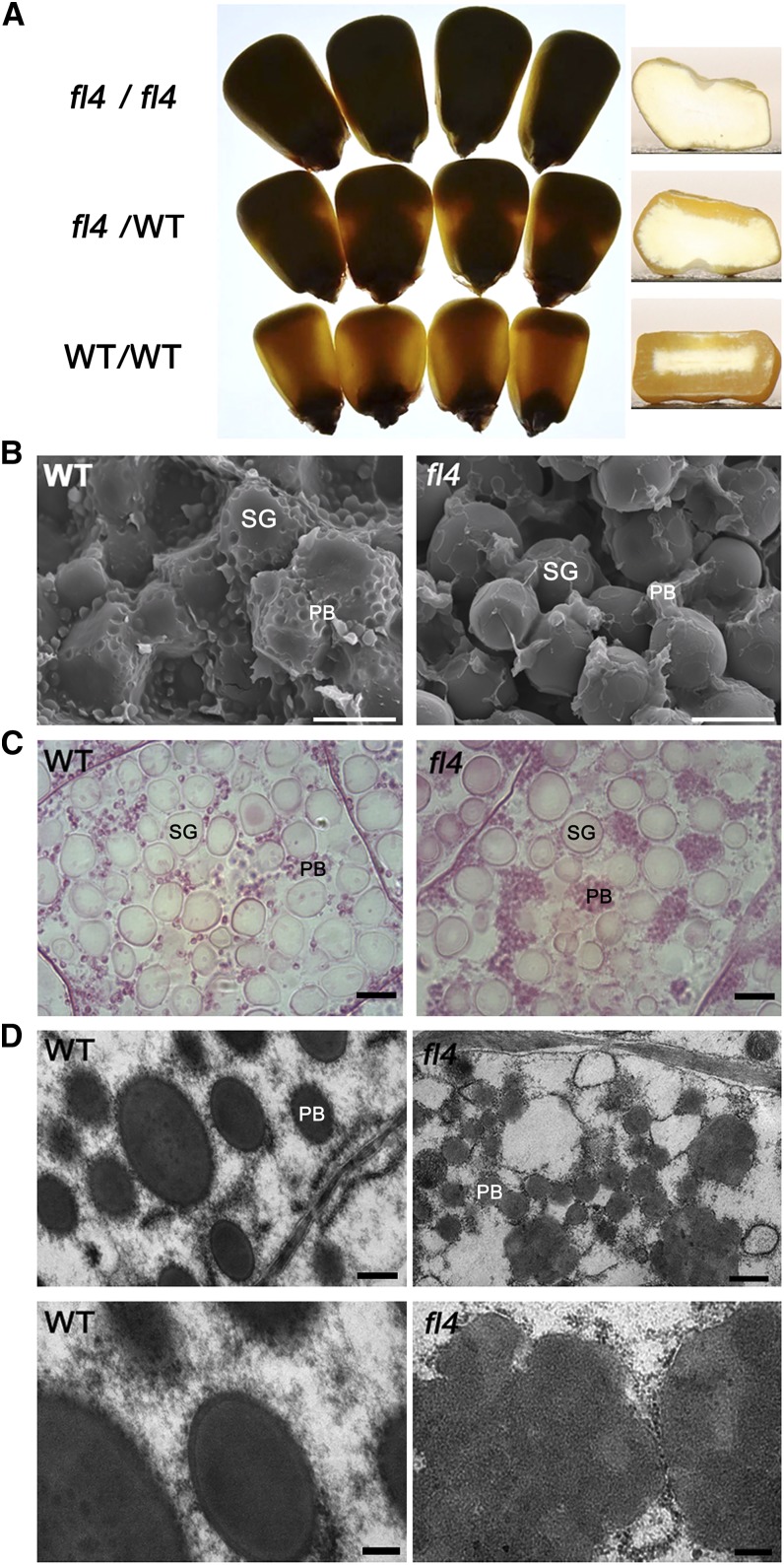
The original opaque mutant stock was obtained from the Maize Genetic Stock Center as no. 5512G. It was crossed to the W22 inbred line and an F2 population produced from the F1 progeny. The kernel phenotype in the F2 population displayed 1:2:1 segregation of fully opaque, semiopaque, and vitreous endosperm, respectively (Fig. 1A), demonstrating that the mutation in the 5512G stock is semidominant, belonging to the floury endosperm category. Gross genetic mapping placed it on the short arm of chromosome 4, which is distinct to known floury mutants, i.e. fl1 to fl3; consequently, this newly identified mutant was designated fl4.
Figure 1.
Phenotypic features of maize fl4 mutants. A, Light transmission by mature kernels. The homozygous mutant kernels (fl4/fl4), heterozygous kernels (fl4/wild type [WT]), and homozygous wild-type kernels (wild type/wild type) were randomly selected from segregating F2 population and viewed on a light box. B, Scanning electron microscopy analysis of the peripheral regions of mature wild-type and fl4 endosperm. PB, Protein body; SG, starch granules. Bars = 10 μm. C, Microstructure of developing endosperms of the wild type and fl4 (20 DAP). Protein bodies were adjoined into clumps in fl4 (right). PB, Protein body; SG, starch granules. Bars = 5 μm. D, Ultrastructure of developing endosperms of the wild type and fl4 (20 DAP). Small, misshapen, and aggregated protein bodies were observed in fl4 (right). Top, Low magnification. Bottom, High magnification. PB, Protein body; SG, starch granules. Bars = 500 nm (top) and 200 nm (bottom).
Mature fl4 and wild-type kernels were analyzed by scanning electron microscopy to reveal their endosperm texture. In the peripheral endosperm, fl4 kernels had smooth, loosely packed starch granules (Fig. 1B, right), with no marked contacts between protein bodies and starch granules. The starch granules in the same region of wild-type kernels were compact and embedded in a dense proteinaceous matrix (Fig. 1B, left). To investigate the distribution and configuration of protein bodies in fl4 and the wild type, we observed the microstructure and ultrastructure of immature endosperm cells at 20 d after pollination (DAP) using optical and transmission electron microscopy. In wild-type endosperm cells, protein bodies evenly surrounded the starch granules (Fig. 1C, left), and protein bodies were round and well separated from each other (Fig. 1D, left). In fl4 endosperm cells, protein bodies were aggregated in clumps (Fig. 1C, right) and were small, irregularly shaped, and prominently adjoined (Fig. 1D, right).
fl4 Endosperm Has Decreased Zein and Changed Soluble Amino Acid Content
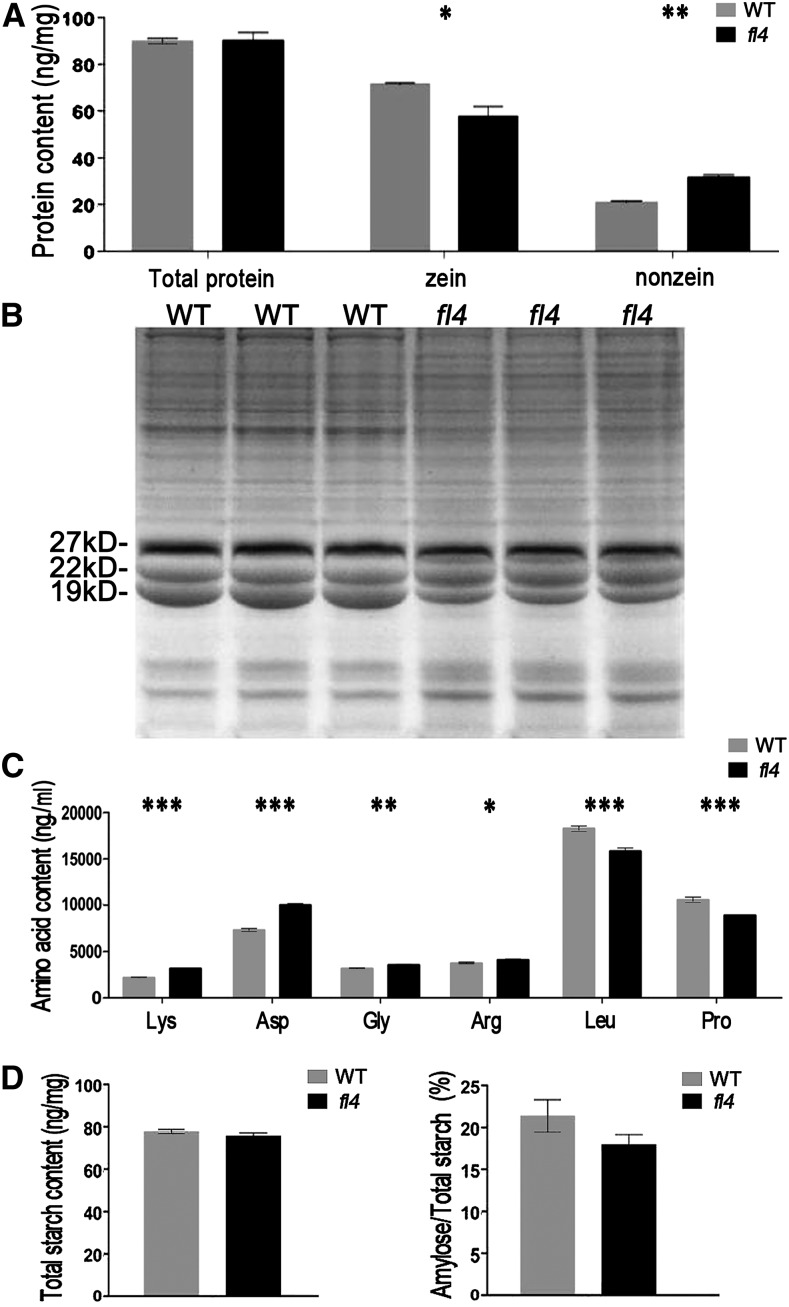
To investigate the potential biochemical reason for the opaque phenotype of fl4, we measured the major components in the mature endosperm of fl4 and wild-type kernels. We first examined the protein content to determine if the mutation caused quantitative changes in zein proteins and nonzein proteins. The results indicated that there is no significant difference in the total protein content in wild-type and opaque kernels. However, there is a general reduction in the amount of zeins, while the amount of nonzeins was found to be higher in fl4 kernels. Quantitative analysis showed that zeins were significantly decreased (19.6%) in fl4 kernels (Fig. 2, A and B). We found no obvious difference in total starch content between opaque and wild-type kernels, but there was a slight decrease in the percentage of amylose in total starch of fl4 (Fig. 2D).
Figure 2.
Biochemical analysis of wild-type and fl4 endosperm. A, Comparison of zein, nonzein, and total proteins from wild-type (WT) and fl4 mature endosperm. The measurements were done on per milligram of dried endosperm. Values are the mean values with se (n = three individuals; *P < 0.05, **P < 0.01, Student’s t test). B, SDS-PAGE analysis of total proteins from wild-type and fl4 mature endosperm. C, The soluble amino acids with different content in wild-type and fl4 mature endosperm. Values are the mean values with se (n = three individuals; *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test). D, Comparison of total starch content and the percentage of amylose in wild-type and fl4 mature endosperm. The measurements were done on per milligram of dried endosperm. Values are the mean values with se (n = three individuals).
We analyzed soluble amino acids to determine if the reduced amount of zeins in fl4 caused an alteration in Lys content. The results showed that the amount of Lys was most significantly increased (45.7%), along with a higher amount of Asn (37.2%) and a slight increase in Gly (12.5%) and Arg (9.8%) content, while the Leu and Pro content was decreased (Fig. 2C; Supplemental Table S1).
Fl4 Is Located within the 19-kD α-Zein z1A-1 Gene Cluster
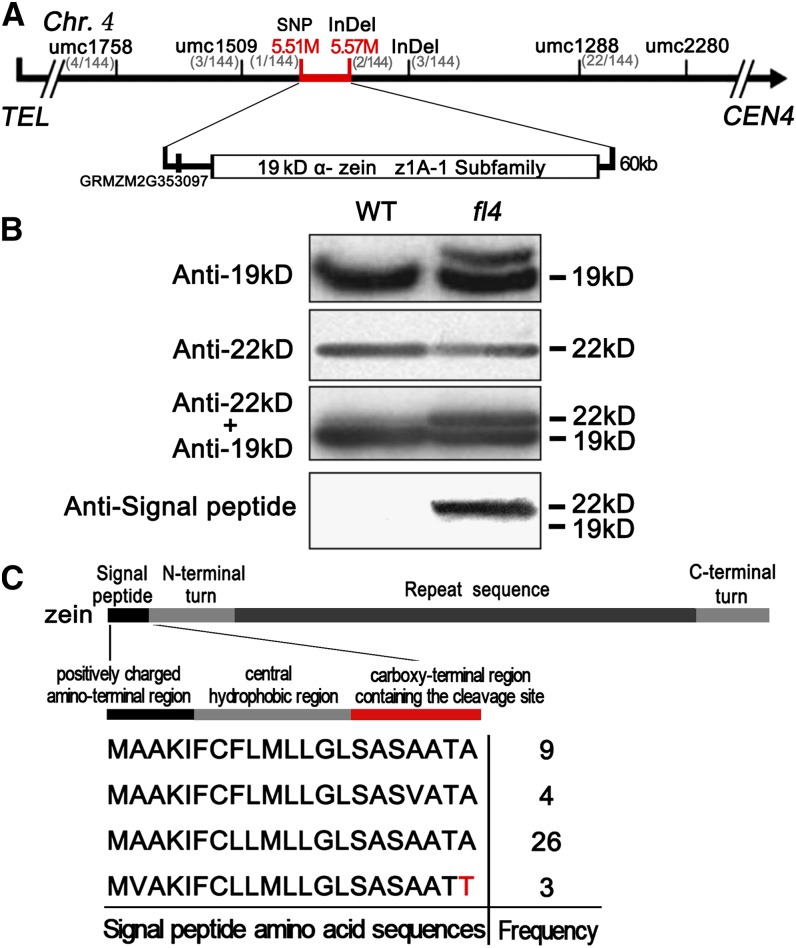
Fine mapping of Fl4 was carried out using the F2 population created with the fl4 mutant and the W22 inbred line. The Fl4 gene was mapped on the telomere side of the simple sequence repeat marker umc2280 on the short arm of chromosome 4 through an analysis of 24 individuals (Fig. 3A). After characterizing a mapping population with a total of 144 individuals, Fl4 was mapped between simple sequence repeat markers umc1758 (four recombinants) and umc1288 (22 recombinants). Additional molecular markers Insertion-deletion (InDel)5.7, InDel5.57, and Single nucleotide polymorphism (SNP)5.51 were developed, and the Fl4 gene was eventually placed between SNP5.51 (one recombinant) and InDel5.57 (two recombinants), a region encompassing a physical distance of 60 kb and containing the entire 19-kD α-zein z1A-1 gene subfamily cluster in B73 (Fig. 3A; Song and Messing, 2002). Except for the z1A-1 genes, sequence analysis identified another predicted open reading frame (GRMZM2G353097) within this region (Fig. 3A); however, its candidacy as the fl4 gene was excluded due to no sequence difference between alleles in fl4 and the wild type. Therefore, the 19-kD α-zein genes within the z1A-1 gene cluster appeared to be candidates for the Fl4 locus.
Figure 3.
Map-based cloning and identification of Fl4. A, The Fl4 locus was mapped to a 60-kb region between molecular markers SNP5.51 and InDel5.57 on chromosome 4 containing 19-kD α-zein z1A-1 subfamily. See Supplemental Table S2 for primer information. B, A novel 19-kD α-zein protein band was detected with 19-kD α-zein-specific antibody and signal peptide-specific antibody in fl4. C, Three of the 42 clones for sequencing exhibited a single amino acid replacement in the highly conserved carboxy-terminal region of the signal peptide containing the cleavage site. WT, Wild type.
The fl4 Mutation Is a Defective Signal Peptide in a 19-kD α-Zein
fl4 is a semidominant opaque mutant similar to fl2 and De-B30, which result from the alteration of zein polypeptides themselves (Coleman et al., 1997; Kim et al., 2004, 2006). The fl4 mutation is physically located within the 19-kD α-zein z1A-1 subfamily. To investigate if this mutation is associated with an alteration of a 19-kD α-zein protein, zeins were extracted from mature fl4 and wild-type kernels. After separating by SDS-PAGE, zeins were analyzed by immunoblotting with a 19-kD α-zein-specific antibody. An extra protein band was detected in fl4 that migrated more slowly than the 19-kD α-zein protein group (Fig. 3B). We determined the size of this novel band using 19-kD α-zein- and 22-kD α-zein-specific antibodies. In two zein protein samples with the same amount of 22-kD α-zeins, a 22-kD α-zein band became more prominent after a second immunoblotting with 19-kD α-zein-specific antibody. This result indicated the novel 19-kD α-zein protein band migrated approximately at the 22-kD α-zein protein group position (Fig. 3B). We used a 19-kD α-zein signal peptide-specific antibody to investigate if the slow-migrating 19-kD α-zein protein has an alteration in its signal peptide. The novel 19-kD α-zein protein showed a positive reaction with the signal peptide antibody (Fig. 3B), indicating this protein retained its signal peptide.
We amplified the 19-kD α-zein z1A-1 subfamily sequences from complementary DNAs (cDNAs) made with RNA from 18-DAP fl4 kernels. A total of 42 independent cDNA clones were sequenced. This analysis revealed the amino acid sequence of 19-kD α-zein signal peptides showed a high degree of conservation (Fig. 3C). The signal peptides are 21-amino acid sequences with a positively charged amino-terminal region, a central hydrophobic region, and a carboxy-terminal region that contains the signal peptidase cleavage site (Chou, 2001). Three of the 42 sequences exhibited a single nucleotide polymorphism in the 21st codon, which resulted in the amino acid Ala (GCG) being replaced by Thr (ACG; Fig. 3C). This mutation altered the highly conserved carboxy-terminal region containing the cleavage site, which could affect the proteolytic process of the signal peptide.
fl4 Changes Protein Body Morphology
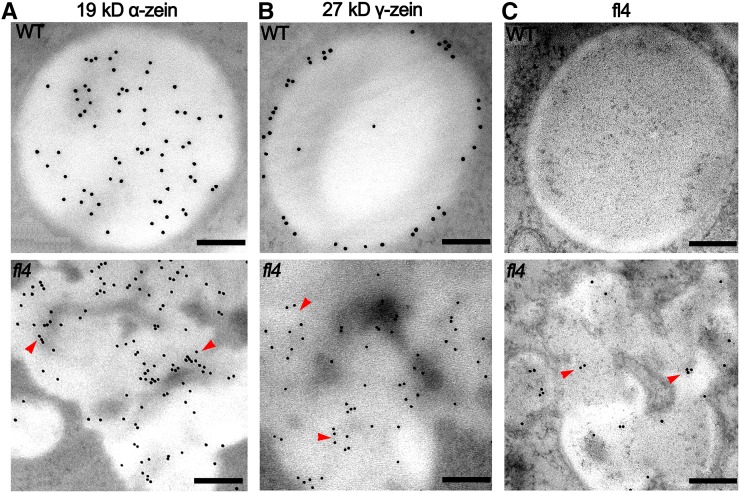
The distribution of 19-kD α-zeins within protein bodies was investigated using immunoelectron microscopy (IEM), and antibodies specifically raised against 19-kD α-zeins to determine how the structure of protein bodies is affected in fl4. Previous studies suggested that α-zeins are distributed within the protein body core but not the peripheral, γ-zein-rich region (Lending and Larkins, 1989). Similarly, our results showed that the 19-kD α-zeins are evenly distributed within the protein body core in wild-type kernels at 20 DAP (Fig. 4A, top). However, in fl4 protein bodies, the 19-kD α-zeins were redistributed to the peripheral region and were concentrated in the region connecting two aggregated misshapen protein bodies (Fig. 4A, bottom). The distribution of 27-kD γ-zeins was investigated using 27-kD γ-zein-specific antibodies. Gold particles indicating 27-kD γ-zeins localized at the periphery of wild-type protein bodies at 20 DAP (Fig. 4B, top), but in fl4, some of the 27-kD γ-zeins were redistributed into the central region (Fig. 4B, bottom).
Figure 4.
Immunolocalization of the 19-kD α-zein, 27-kD γ-zein, and fl4 protein in wild-type (WT) and fl4 endosperm cells (20 DAP). A, The distribution of gold particles indicating 19-kD α-zeins. Evenly distributed within the protein body core in the wild type (top). Inserted into the peripheral region in fl4 (bottom, red arrowhead). Bars = 200 nm. B, The distribution of gold particles indicating 27-kD γ-zeins. Located in the peripheral region in the wild type (top). Delocalized into the central region in fl4 (bottom, red arrowhead). Bars = 200 nm. C, The distribution of gold particles indicating fl4 protein with the antibodies recognizing the signal peptide. No signal in wild-type sample (top). Around the region connects two aggregated, misshapen protein bodies in fl4 (bottom, red arrowhead). Bars = 200 nm.
We investigated distribution of the fl4 protein using antibodies specifically raised against the 19-kD α-zein signal peptide to determine its accumulation in protein bodies. The 19-kD α-zein signal peptide-specific antibodies recognized unprocessed 19-kD α-zeins, which could not be detected at a low level in wild-type kernels (Fig. 4C, top). Gold particles showing the distribution of fl4 protein as unprocessed 19-kD α-zeins indicated the protein was in protein bodies, typically around the peripheral region of aggregated, misshapen protein bodies (Fig. 4C, bottom). Thus, fl4 proteins with unprocessed signal peptide are stable and affect the organization of zeins in protein bodies.
Significant ER Stress Occurs in fl4 Mutant Endosperm
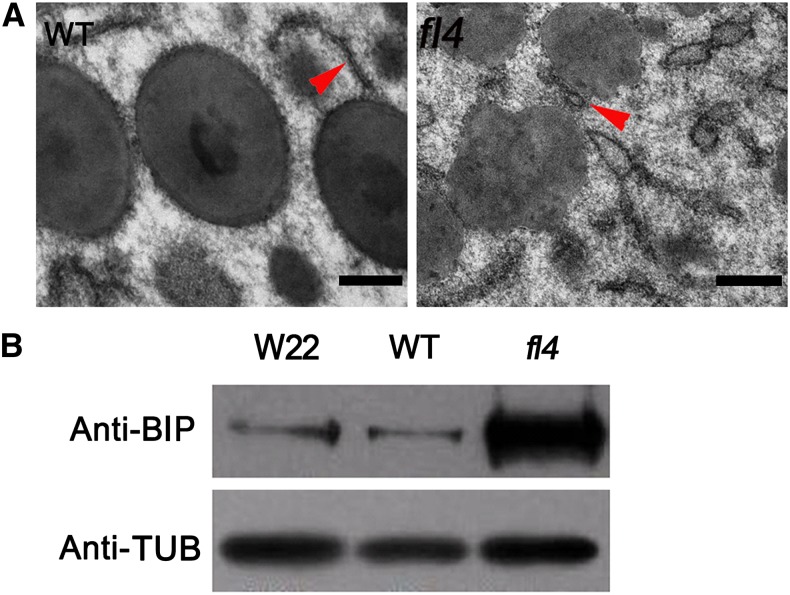
ER stress is induced in many opaque mutants, including fl2 and De-B30 (Kirst et al., 2005). ER dilation is commonly observed in previously characterized ER stress. To investigate the appearance of rough ER, we observed the ultrastructure of fl4 and wild-type endosperm cells at 20 DAP using transmission electron microscopy. Significantly dilated rough ER was observed surrounding the protein bodies in fl4 endosperm cells (Fig. 5A), indicating ER stress.
Figure 5.
ER stress evidence was observed in fl4. A, Ultrastructure of developing endosperms of the wild type (WT) and fl4 (20 DAP). Normal ER configuration in the wild type (left, red arrowhead). Dilated ER in fl4 (right, red arrowhead). Bars = 500 nm. B, Immunoblot comparing BIP accumulation in wild-type and fl4 mature kernels. Anti-Tubulin was used as a sample loading control.
Up-regulated expression of molecular chaperones, such as ER lumen BIP, was shown to be another common feature of ER stress (Gillece et al., 1999). The level of BIP was examined using a BIP-specific antibody with fl4, wild-type, and W22 mature kernels. BIP protein content was markedly increased in fl4 endosperm (Fig. 5B), suggesting ER stress is significantly triggered in the mutant.
fl4 Endosperm Exhibits Altered Expression of ER Stress-Associated Genes
We compared the gene expression profile of 19-DAP fl4 and wild-type endosperm using RNA sequencing (RNA-seq). Of the 65,536 gene transcripts detected by RNA-seq, 838 genes showed significant altered expression between fl4 and the wild type (see “Materials and Methods”). Gene Ontology (GO; http://bioinfo.cau.edu.cn/agriGO/) and the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) pathway analysis indicated genes with increased expression were mostly related to ER stress within two GO terms, GO0034976 (Response to ER stress) and GO0006983 (ER overload response; Table I), along with one Kyoto Encyclopedia of Genes and Genomes term (Protein processing in ER). These genes could be divided into four categories. The first comprised five genes encoding proteins taking part in the ERAD pathway, including a maize Endoplasmic Reticulum Oxidoreductin1 (ERO1)-like gene (Tsai and Rapoport, 2002), two Protein Disulfide Isomerase (PDI)-like genes (Tsai et al., 2001), a member of Derlins, ZmDerlin1-1 (Kirst et al., 2005), and a translocon protein SecY subunit domain container, Sec61-like gene (Plemper et al., 1999). The second category comprised four genes encoding ER-resident protein chaperones, including BIP1 (Gillece et al., 1999), precursor of BIP3, a Heat shock protein (Hsp)20-like chaperone superfamily member, and a member of the Hsp20/α crystallin protein family. The third category possessed one gene (UDP-N-acetylglucosamine transporter) encoding an enzyme involved in protein folding (Deng et al., 2013). In addition, there were five genes encoding antiapoptosis proteins as the inhibitors of ER stress-responsive PCD, which were three Bax (pro-apoptotic protein) inhibitor1 (BI1) protein family members (Ishikawa et al., 2011) and two apoptosis regulator Bcl2 (anti-apoptotic protein)-associated athanogenes (BAGs; Briknarová et al., 2001; Table I).
Table I. Expression of ER stress-associated genes in fl4 mutant.
| GO Identification | P | Category | Gene List | Description | Fold Change | P |
|---|---|---|---|---|---|---|
| GO:0034976, Response to ER stress | 2.44E-06 | ERAD | GRMZM2G108115 | ERO1 | 2.01 | 0.000112 |
| GRMZM2G389173 | PDI | 2.54 | 5.89E-05 | |||
| GRMZM2G091481 | PDI | 1.47 | 1.64E-05 | |||
| GRMZM2G117388 | Derlins; ZmDerlin1-1 | 2.00 | 0.00472 | |||
| GRMZM2G130987 | SecY subunit domain; Sec61 | 1.44 | 0.000142 | |||
| Chaperones | GRMZM2G114793 | Bip1 | 2.39 | 2.95E-10 | ||
| GRMZM2G415007 | Bip3 | 2.91 | 0.000137 | |||
| GRMZM2G331701 | HSP20-like chaperone superfamily protein | 13.48 | 0.00487 | |||
| GRMZM2G083810 | Hsp20/α crystallin family protein | 2.70 | 0.00273 | |||
| Protein folding | GRMZM2G176029 | UAA transporter | 2.18 | 1.11E-05 | ||
| GO:0006983, ER overload response | 0.00208 | Antiapoptosis | GRMZM2G465430 | BI1 family protein | 2.08 | 8.84E-05 |
| GRMZM2G029087 | BI1 family protein | 1.98 | 4.03E-06 | |||
| GRMZM2G074404 | BI1 family protein | 1.66 | 0.0260 | |||
| GRMZM2G079956 | BAG | 1.73 | 0.0239 | |||
| GRMZM2G017013 | BAG | 1.41 | 0.0429 |
Enhanced ERAD, UPR, and the Phosphorylation of eIF2α in fl4 Mutant Endosperm
After initiation of ER stress, misfolded proteins are degraded by the ERAD system. The UPR program could be activated to relieve ER stress by inducing transcriptional regulation. Protein synthesis might also be suppressed by increasing phosphorylation of eIF2α to down-regulate protein synthesis in the stressed ER (Gillece et al., 1999; Harding et al., 1999). To test this, we investigated if the mutation in Fl4 affected these ER stress-related, cell-sparing processes.
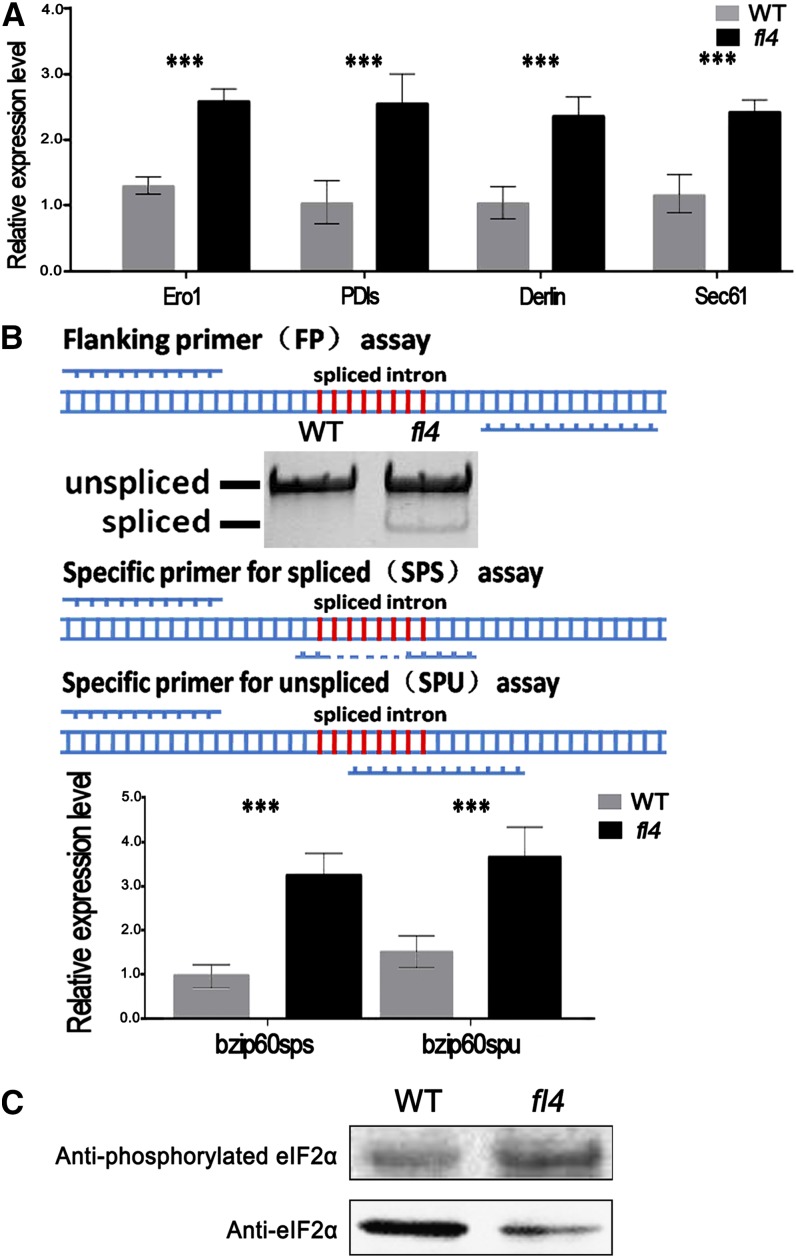
First, we used real-time PCR analysis of 19-DAP fl4 and wild-type endosperm to confirm the altered expression level of genes taking part in the ERAD system that showed the most significant differential expression in the RNA-seq data. Our results indicated that ERO1 (GRMZM2G108115) was 2.47-fold up-regulated, and PDIs (GRMZM2G389173) exhibited a 1.90-fold up-regulation. These two sharply up-regulated genes are involved in the ERAD pathway within ER lumen (Tsai et al., 2001; Tsai and Rapoport, 2002). ER membrane-localized proteins ZmDerlin1-1 (GRMZM2G117388) and Sec61 (GRMZM2G130987; Plemper et al., 1999; Kirst et al., 2005) had 2.61- and 2.02-fold up-regulation, respectively (Fig. 6A), so there was obvious up-regulation of ERAD in fl4.
Figure 6.
Effects on ERAD, UPR, and the phosphorylation of eIF2α in fl4. A, Expressions of ERAD-associated genes ERO1 (GRMZM2G108115), PDIs (GRMZM2G389173), ZmDerlin1-1 (GRMZM2G117388), and Sec61 (GRMZM2G130987) were examined in the wild type (WT) and fl4 by real-time PCR analysis (19 DAP). Values are the mean values with se (n = six individuals; *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test). B, RNA samples from wild-type and fl4 kernels (19 DAP) were analyzed for the presence of unspliced and spliced ZmbZIP60 mRNA by reverse transcription-PCR using the flanking primers (FP) assay. The level of ZmbZIP60 expression and splicing were analyzed by real-time PCR using primers for unspliced bZIP60 mRNA (SPU) or for spliced mRNA (SPS). Values are the mean values with se (n = six individuals; ***P < 0.001, Student’s t test). C, Immunoblot comparing the phosphorylated eIF2α accumulation in wild-type and fl4 kernels (18 DAP). Anti-eIF2α was used as control.
UPR involves a series of cytoplasm-localized transcriptional factors that relieve ER stress. The basic leucine zipper (bZIP) transcription factor bZIP60 is involved in the UPR in Arabidopsis (Arabidopsis thaliana; Iwata and Koizumi, 2005). This transcriptional factor resides in the ER membrane under unstressed conditions, and ER stress induces alternative splicing of the bZIP60 transcript such that the encoded protein that contains the DNA-binding domain is targeted to the nucleus (Deng et al., 2011). The increased splicing of ZmbZIP60 mRNA is an indicator of ER stress in maize as well (Li et al., 2012). We used reverse transcription-PCR to detect the existence of spliced ZmbZIP60 transcript (ZmbZIP60sps) in fl4. In contrast to the single amplified band indicating the unspliced ZmbZIP60 transcript (ZmbZIP60spu) in the wild type, there were both unspliced and spliced ZmbZIP60 transcripts in fl4 (Fig. 6B). We designed specific primers to detect unspliced and spliced ZmbZIP60 RNAs in 19-DAP fl4 and wild-type endosperm. The results indicated that in fl4, both ZmbZIP60spu and ZmbZIP60sps RNAs are up-regulated, and there is an increase in the percentage of ZmbZIP60sps (39.4% to 47.0%) among total ZmbZIP60 transcripts (Fig. 6B). Thus, the increased expression level of spliced ZmbZIP60 in fl4, together with the transcriptional increase of ER-resident chaperones in the gene expression profile (Table I), indicates up-regulation of UPR-related gene expression.
The increased phosphorylation of eIF2α is another ER stress-related, cell-sparing mechanism that was also investigated in fl4. Immunoblotting analysis of proteins from 18-DAP fl4 and wild-type kernels with both eIF2α-specific antibody and phosphorylated eIF2α-specific antibody indicated the phosphorylation level of eIF2α is elevated in fl4 endosperm (Fig. 6C), suggesting translational suppression occurred in fl4 endosperm during kernel development.
Elevated PCD in fl4 Endosperm
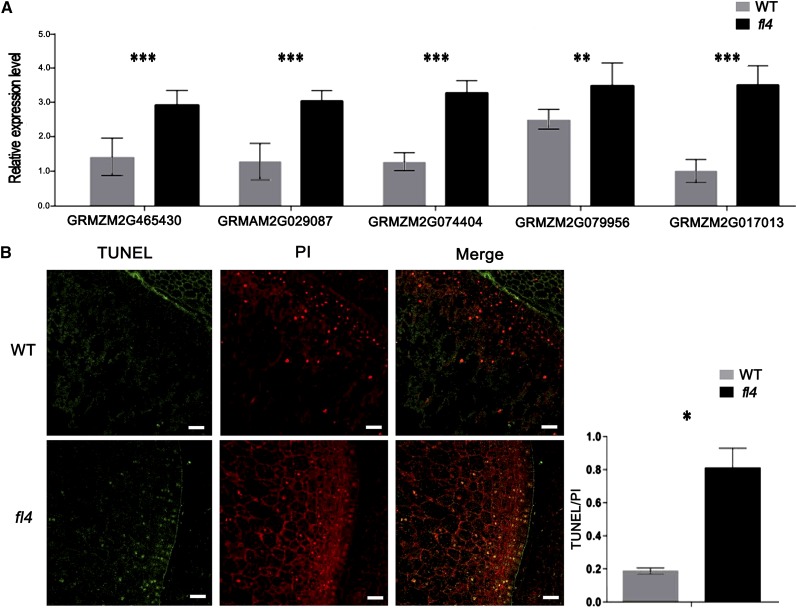
PCD may occur if ER stress-induced cell-sparing mechanisms fail (Liu and Howell, 2010), and antiapoptosis genes are also targets of UPR transcriptional regulation (Iwata et al., 2008). Five antiapoptosis genes encoding ER overloading-responsive proteins were significantly up-regulated in fl4 according to RNA-seq data (Table I). We used real-time PCR analysis of 19-DAP fl4 and wild-type kernels to confirm the expression level of these five genes. Three BI1 protein family members (GRMZM2G465430, GRMZM2G029087, and GRMZM2G074404) were 2.08-, 2.06-, and 1.98-fold up-regulated, respectively, and two apoptosis regulator BAGs (GRMZM2G079956 and GRMZM2G017013) were up-regulated 1.36- and 1.70-fold, respectively (Fig. 7A).
Figure 7.
Enhancement of PCD in fl4. A, Expressions of PCD-associated genes BI1 protein family members (GRMZM2G465430, GRMZM2G029087, and GRMZM2G074404) and BAGs (GRMZM2G079956 and GRMZM2G017013) were examined in the wild type (WT) and fl4 by real-time PCR analysis (19 DAP). Values are the mean values with se (n = six individuals; *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test). B, TUNEL assay in situ detection of DNA fragmentation in wild-type and fl4 endosperm (20 DAP). Left, TUNEL-detected nuclei (green); middle, PI-detected nuclei (red); right, merge of TUNEL-detected nuclei and PI-detected nuclei (yellow); histogram, statistical analysis of TUNEL-detected nuclei/PI-detected nuclei in different section samples. Values are the mean values with se (n = two individuals; *P < 0.05, Student’s t test). Bars = 50 μm.
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay, one of the hallmarks of PCD, was employed to detect PCD in fl4 endosperm. The 20-DAP kernel sections were TUNEL analyzed and stained with propidium iodide (PI) simultaneously to reveal all nuclei. In wild-type endosperm, few nuclei were TUNEL positive, while in fl4, the TUNEL-positive signals were much stronger (Fig. 7B). Statistical analysis of TUNEL was carried out by calculating the percentage of TUNEL-positive nuclei among PI-detected total nuclei in different section samples. The results demonstrated that DNA damage is significantly increased in fl4 endosperm.
DISCUSSION
A New Opaque Endosperm Mutant with a Defective Signal Peptide
fl4 is a newly identified semidominant opaque mutant. fl4/fl4 kernels have an obvious opaque appearance with little vitreous endosperm (Fig. 1). Using a positional cloning approach, fl4 was located in the 19-kD α-zein z1A-1 subfamily on the short arm of chromosome 4 (Fig. 3), which is different from known floury mutants, i.e. fl1 to fl3. Along with the immunoblot analysis, we identified fl4 as a 19-kD α-zein gene containing a point mutation at the signal peptide cleavage site: the 21st Ala (GCG) is replaced by Thr (ACG; Fig. 3). The high-digestibility sorghum (Sorghum bicolor) mutant that results from an identical Ala-to-Thr amino acid substitution in a storage protein at the conserved 21st amino acid of the signal peptide has a similar kernel phenotype (Wu et al., 2013). fl2 and De-B30, semidominant and dominant opaque mutants, respectively, have defective signal peptide in α-zeins as well. In fl2, Ala, the 21st amino acid, is replaced by Val in the 22-kD α-zein signal peptide (Lending and Larkins, 1992). In De-B30, Ser, the 15th amino acid, is replaced by Pro in the 19-kD α-zein signal peptide (Kim et al., 2004).
A vitreous endosperm is important for the maturation, storage, and processing of maize kernels. Considerable evidence indicates that zein protein accumulation plays a critical role in this feature (Holding and Larkins, 2006; Wu and Messing, 2010). The soft, starchy portion of the wild-type maize endosperm contains markedly less zein than the vitreous region (Dombrink-Kurtzman and Bietz, 1993). The reduced amount of zeins is reported to result in the opaque endosperm phenotype and increased amount of Lys (Mertz et al., 1964; Misra et al., 1972). In fl4, a general reduction in zeins was observed (Fig. 2), and the amount of Lys is significantly increased (Fig. 2). These features are similar to the fl2 mutant, which manifests general reductions in zeins, along with protein bodies that are small and severely deformed (Lending and Larkins, 1992). De-B30 and Mc, dominant opaque mutants, have phenotypes similar to fl2 with respect to altered protein body morphology (Kim et al., 2004, 2006).
It is unclear if the alterations in zein protein structure in fl2, De-B30, and Mc disrupt protein body morphology and vitreous endosperm formation in a way similar to fl1 (Holding et al., 2007) by being directly located in protein bodies or by indirect effects. fl4 has pleiotropic effects on endosperm development and shares many phenotypic features with fl2, De-B30, and Mc, and it offers new material for investigating the underlying mechanisms by which these semidominant and dominant mutations disrupt vitreous endosperm formation in the maize kernel.
Retention of Defective Signal Peptides Affects Protein Body Morphology
A combination of protein bodies, starch grains, and viscous cytoplasm appears to give rise to a compact matrix in the vitreous region of mature kernels (Holding and Larkins, 2006). In fl4 endosperm, small and irregularly shaped protein bodies are prominently aggregated into clumps (Fig. 1). Normally, zein protein bodies form in the ER and have a peripheral layer rich in β- and γ-zein, and the 19- and 22-kD α-zeins fill the core of the protein body as it matures (Lopes and Larkins, 1991). IEM analysis of fl2 with antibodies against α-, β-, and γ-zeins indicated the spatial distribution of zeins in protein bodies is disrupted (Lending and Larkins, 1992). In our study, a similar IEM analysis with antibodies against 19-kD α-zeins, 27-kD γ-zeins, and also the 19-kD α-zein signal peptide was carried out. Especially the signal detected with antibodies specifically raised against 19-kD α-zein signal peptide indicating the fl4 protein is stable and concentrated in misshapen protein bodies. Thus, it appears this protein disrupts the assembly of zeins into protein bodies, impairing the distribution of zeins and affecting the normal protein body morphology (Fig. 4).
Studies of proteins with unprocessed signal peptides suggest these proteins remain attached to ER membranes (Schauer et al., 1985; Shatters and Miernyk, 1991). By remaining anchored to the ER membrane, the mutated α-zeins in fl2 perturb protein body morphology (Lending and Larkins, 1992; Gillikin et al., 1997). In fl4, the 19-kD α-zein proteins are retained in the peripheral region and connect misshapen protein bodies (Fig. 4).
Previous studies indicated the essentiality of zein interactions in protein body assembly (Kim et al., 2002). It appears the temporal and spatial regulation of zein gene expression level as well as interactions between different types of zein affect protein body formation (Woo et al., 2001; Kim et al., 2002; Wu et al., 2010). Accumulated fl4 proteins remain attached to the protein body membrane, and we would postulate that fl4 proteins might interact with other normal α-zeins, keeping them in the peripheral region during protein body assembly. Small and misshapen protein bodies prominently aggregate together and perturb the formation of a rigid matrix consisting of protein bodies, starch grains, and viscous cytoplasm in fl4 endosperm. Hence, the mutation in Fl4 might disrupt vitreous endosperm formation by directly affecting protein body morphology.
Zeins with Defective Signal Peptides Stimulate ER Stress
The fl2, De-B30, and Mc mutant manifests improper zein deposition and protein body deformation and exhibits ER stress through up-regulated expression of molecular chaperones, such as BIP (Zhang and Boston, 1992; Coleman et al., 1997; Kim et al., 2004, 2006; Kirst et al., 2005). BIP is an ER-localized chaperone that functions to prevent the aggregation of protein-folding intermediates, such that terminally misfolded proteins do not occur (Kaufman, 1999). BIP is localized in the ER as well as the peripheral regions of the abnormal protein bodies in fl2, De-B30, and Mc mutants (Zhang and Boston, 1992). The mutation in Fl4 causes dilated rough ER surrounding the protein bodies, and BIP content is markedly up-regulated (Fig. 5). A series of ER stress-associated genes exhibit elevated expression levels in fl4. These differentially expressed genes are mainly involved in ERAD, protein folding, and antiapoptosis (Table I).
The ER provides a sophisticated environment for protein folding, the formation of disulfide bonds, and homeostasis in the ER lumen (Stevens and Argon, 1999; Rutkowski and Kaufman, 2004). ER stress is induced in plant cells by a variety of treatments, such as abiotic stresses, agents affecting calcium homeostasis, and inhibitors of glycosylation in the ER (van Huizen et al., 2003; Iwata et al., 2008). In response to the accumulation of unfolded and misfolded proteins in ER, UPR, as a coordinated adaptive program, is triggered, the expression of ER-resident protein chaperones and protein foldases is induced, ERAD is activated to eliminate misfolded proteins, and the rate of general translation is suppressed to reduce overload of newly synthesized proteins to stressed ER. When all these mechanisms are failing, ER stress-related apoptosis commences (Wu and Kaufman, 2006).
The fl4, fl2, De-B30, and Mc mutants provide useful materials to study ER stress in plants by producing endogenous mutated proteins that cannot achieve their native state. Earlier studies in fl2, De-B30, and Mc detected apparent ER stress in general; however, they did not characterize the nature of different ER stress pathways in detail. We dissected different ER stress pathways triggered by fl4 mutant and showed that the ERAD pathway, the UPR program, and the general reduction of translation initiation are induced, along with up-regulation of PCD (Figs. 6 and 7).
The ER lumen-resident ERO1 and PDI family proteins possibly identify ERAD substrates based on the time spent in the folding cycle (Tsai et al., 2001; Tsai and Rapoport, 2002; Martínez and Chrispeels, 2003; Deng et al., 2013). Four Derlin-like genes encoding the homologs of yeast (Saccharomyces cerevisiae) Der1p protein were identified in maize, which are involved in ERAD by escorting proteins to a retrotranslocon (Kirst et al., 2005). Another ER membrane-localized protein, sec61, is thought to serve as the retrotranslocon channel (Nakatsukasa and Brodsky, 2008). In fl4, the increased expression of genes in these families demonstrates an obvious up-regulation of ERAD (Fig. 6).
Levels of bZIP60 expression and splicing are increased by ER stress (Iwata and Koizumi, 2005; Fujita et al., 2007; Li et al., 2012). The spliced version of the transcript encodes a protein containing the DNA-binding domain, which shifts into the nucleus for activating expression of UPR genes (Iwata and Koizumi, 2005; Deng et al., 2011). In fl4, the up-regulated expression level and translocation of ZmbZIP60 provide additional evidence for the activation of the UPR (Fig. 6). The UPR program alleviates ER stress mainly through increasing transcription of protein foldases, ER-resident chaperones, and other components involved in the ER quality control system (Wu and Kaufman, 2006). The increased levels of a series of ER-resident chaperones in the fl4 gene expression profile (Table I) also indicate up-regulation in UPR transcriptional regulation.
We also observed increased phosphorylation of eIF2α during fl4 endosperm development (Fig. 6), indicating a general suppression of protein synthesis. It seems that plants do not have an ortholog of the ER stress transducer PKR-like ER kinase (PERK), which decreases general translation by inducing the phosphorylation of eIF2α in animals (Iwata and Koizumi, 2012). In addition to PERK, three other eukaryotic protein kinases are known to phosphorylate eIF2α, each of which responds to distinct signals (van Huizen et al., 2003). Plants might possess a mechanism similar to PERK to reduce the protein load in stressed ER, or the increased phosphorylation of eIF2α could be induced by other pathways.
Zeins with Defective Signal Peptides Elevate PCD in Endosperm
PCD plays an important role in endosperm development and normally begins in the central starchy-region cells around 16 DAP (Nguyen et al., 2007). An effect on PCD was poorly investigated in previous studies of opaque mutants. The increased expression of BI1 and BAG family proteins, together with the TUNEL assay showing DNA degradation, indicates an enhancement of PCD in fl4 endosperm (Fig. 7). BI1 is one of the most thoroughly characterized PCD-responsive cell death suppressors (Hückelhoven, 2004; Watanabe and Lam, 2009). Expression of AtBI1 can be up-regulated through activation of AtbZIP60 (Iwata et al., 2008). Bcl2 in mammals is an anti-apoptotic protein and has not been identified in plants. BAGs in plants are PCD-responsive cytoprotective proteins that behave as molecular cochaperones during UPR (Briknarová et al., 2001). The presence of enhanced PCD in fl4, particularly in the peripheral region, indicates that the zeins with unprocessed signal peptides trigger a strong level of ER stress responses, perhaps leading to early cell death. The central region of the endosperm is starchy in wild-type kernels, and this is where PCD begins. Some physiological process or chemical reactions might be required for the vitreous region peripheral to the central starchy endosperm cells during endosperm desiccation. In fl4, the accelerated PCD perhaps disrupts these processes or reactions because of premature cell death in the peripheral region.
An Explanation for the Phenotypic Consequences of Zeins with Uncleaved Signal Peptides in Maize Endosperm
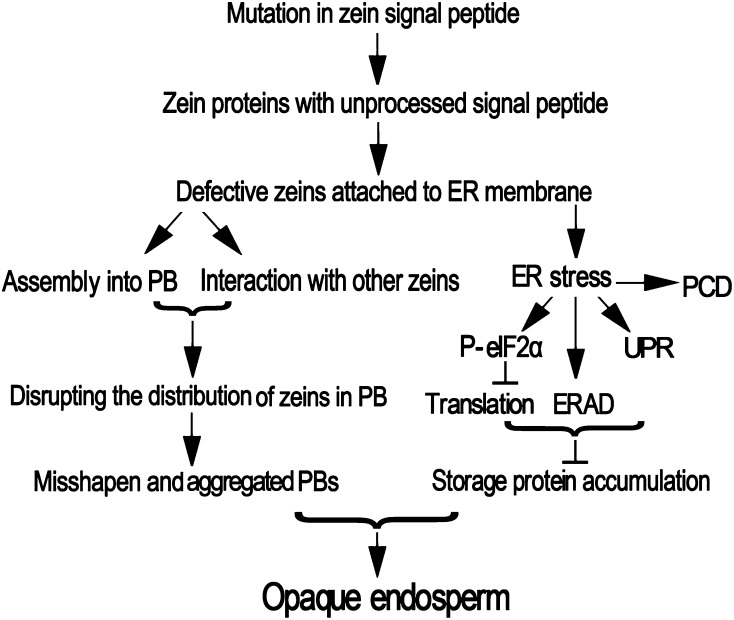
The fl4 mutant is a novel, semidominant opaque mutant with a defective signal peptide in a z1A 19-kD α-zein. This mutation disrupts normal protein body morphology and triggers ER stress responses. The fl4 mutant, together with fl2 and De-B30, provides an opportunity to characterize the cellular responses due to zeins with uncleaved signal peptides in maize endosperm. A summarized model is provided in Figure 8.
Figure 8.
An explanation for the phenotypic consequences of zeins with uncleaved signal peptides in maize endosperm.
At least some of the zeins with unprocessed signal peptides are stable and remain attached to the ER membrane (Lending and Larkins, 1992; Gillikin et al., 1997). Here, they directly participate in the packaging and organization of zeins into protein bodies, disturbing the normal spatial distribution of different zein classes and resulting in small, irregularly shaped protein bodies prominently aggregated into clumps (Figs. 1 and 4). These defective zeins that fail to achieve their native state still could be recognized by ER-resident chaperones as folding factors and trigger intense ER stress (Fig. 5). Different ER stress pathways are induced in the endosperm, especially elevated PCD process, indicating a strong intensity of ER stress in the endosperm of these mutants (Figs. 6 and 7). These cell-sparing mechanisms for releasing the ER overload condition could account for the decrease in the amount of storage proteins (Fig. 2). The presence of a defective zein signal peptide that disturbs protein body formation and decreases storage protein accumulation is responsible for the opaque endosperm phenotype.
MATERIALS AND METHODS
Plant Materials
The 5512G mutant line was obtained from the Maize Genetics Cooperation stock center. The mutation was crossed into the W22 genetic background, and a F2 was produced from the F1 progeny to generate a mapping population. Kernels were collected from F2 ears. Kernels were genotyped using DNA extracted from half of the endosperm tissue, with markers linked to the fl4 locus to determine homozygous mutant (fl4/fl4) kernels, heterozygous (fl4/+) kernels, or wild-type (+/+) kernels. All plants were cultivated in the field at the campus of Shanghai University. Immature seeds were harvested at 17, 18, 19, 20, 24, and 36 DAP.
Scanning Electron Microscopy
fl4 and wild-type kernels were prepared according to Lending and Larkins (1992): mature maize (Zea mays) kernels were rifted with a razor at the peripheral region and placed in 2.5% (w/v) glutaraldehyde. Samples were critically dried and spray coated with gold. Gold-coated samples were then observed with a scanning electron microscope (S3400N; Hitachi).
Transmission Electron Microscopy
Immature kernels of fl4 and the wild type were prepared according to Lending and Larkins (1992), with some modifications: 20-DAP kernels of fl4 and the wild type were fixed in paraformaldehyde and postfixed in osmium tetraoxide. After being dehydrated in an ethanol gradient, samples were then transferred to a propylene oxide solution and gradually embedded in acrylic resin (London Resin Company). Sections (70 nm) of samples were made with a diamond knife microtome (Reichert Ultracut E). Sample sections were stained with uranyl acetate and poststained with lead citrate. Sample sections were observed with a Hitachi H7600 transmission electron microscope.
Protein Quantification
Mature kernels of fl4 and the wild type were soaked in water, and endosperm was then separated from the embryo and pericarp. Endosperm samples were critically dried to constant weight, powdered in liquid nitrogen, and measured according to Wang et al. (2011). Proteins were extracted from 50 mg of three pooled flour endosperm samples. Extracted proteins were measured using a bicinchoninic acid protein assay kit (Pierce) according to the instructions. Measurements of all samples were replicated three times.
Measurement of Starch
Mature kernels of fl4 and the wild type were soaked in water, and endosperm was then separated from the embryo and pericarp. Endosperm samples were dried to constant weight and pulverized in liquid N2, and starch was extracted and measured using an amyloglucosidase/α-amylase method starch assay kit (Megazyme) according to the instructions and adapted as follows: 0.2 mL of aqueous ethanol (80% [v/v]) was added to 100-mg sample to aid dispersion; immediately thereafter, 3 mL of thermostable α-amylase was added. The tube was incubated in a boiling water bath for 6 min, then placed in a bath at 50°C, to which 0.1 mL of amyloglucosidase was added. The tube was stirred with a vortex mixer and incubated at 50°C for 30 min. Duplicate aliquots (0.1 mL) of the diluted solution were transferred to the bottom, 3.0 mL of GOPOD Reagent was added, and the tubes were incubated at 50°C for 20 min. The absorbance for each sample and the d-Glc control at 510 nm was read against the reagent blank.
Soluble Amino Acids Analysis
Soluble amino acids were analyzed according to Holding et al. (2010). Three milligrams of sample were refluxed for 24 h in 6 n HCl. Samples were hydrolyzed at 110°C for 24 h. Sample hydrolysates were critically dried and dissolved in 10 mL of citrate buffer. The amino acids were analyzed with a Hitachi-L8900 amino acid analyzer at Shanghai Jiaotong University. On the wild-type and opaque kernels, analyses were replicated three times.
Map-Based Cloning
Using 144 individuals from an F2 mapping population from the cross between the 5512G stock and the W22 inbred line, the Fl4 locus was mapped. For preliminary mapping, molecular markers distributed throughout maize chromosome 4 were used. SNP5.51, InDel5.57, and InDel5.7 (see Supplemental Table S2), as additional molecular markers for fine mapping, were developed to localize the Fl4 locus to a 60-kb region.
RNA Extraction and Real-Time PCR Analysis
Total RNA was extracted with TRIzol reagent (Tiangen), and DNA was removed by a treatment with RNase-Free DNase I (Takara). Using ReverTra Ace reverse transcriptase (Toyobo), RNA was reverse transcribed to cDNA. Quantitative real-time PCR was performed with SYBR Green Real-Time PCR Master Mix (Toyobo) using a Mastercycler ep realplex 2 (Eppendorf) according to the standard protocol. Specific primers were designed (see Supplemental Table S2), and the experiments were performed to two independent RNA samples sets with ubiquitin as the reference gene. From a pool of kernels collected from three individual plants, each RNA sample was extracted, for which three technical replicates were performed. A final volume of 20 mL contained 1 mL of reverse-transcribed cDNA (1–100 ng), 10 mL of 23 SYBR Green PCR buffer, and 1.8 mL of 10 mm L–1 forward and reverse primers for each sample. Relative quantifiable differences in gene expression were analyzed as described previously (Livak and Schmittgen, 2001).
Polyclonal Antibodies
For antisignal peptide antibody production, the specific polypeptide fragment of 19-kD α-zein signal peptide was synthesized. For 19-kD α-zein and 27-kD γ-zein antibody production, regions of low similarity of 19-kD α-zein and 27-kD γ-zein were selected according to previous study (Woo et al., 2001). The cDNAs responsible for selected polypeptides were cloned into pGEX-4T-1 (Amersham Biosciences), and glutathione S-transferase-tagged fusion protein was purified with the ÄKTA purification system (GE Healthcare) using a GSTrap FF column. Protein expression and purification followed established procedures. Antibodies were produced in rabbits according to standard protocols of Shanghai ImmunoGen Biological Technology.
Immunoblot Analysis
Proteins extracted from fl4 and wild-type mature kernels were separated by SDS-PAGE. Separated protein samples were then transferred to polyvinylidene difluoride membrane (0.45 mm; Millipore). The membrane with protein sample attached was incubated with primary and secondary antibodies. Using the Super Signal West Pico chemiluminescent substrate kit (Pierce), the signal was visualized according to the manufacturer’s instructions. The purified antisignal peptide, 22-kD α-zein, and 19-kD α-zein antibodies were used at 1:500, the α-tubulin antibody (Sigma-Aldrich) was used at 1:1,000, and the BIP antibody (at-95; Santa Cruz Biotechnology) was also used at 1:1,000.
Immunolabeling
fl4 and wild-type 20-DAP kernel samples were high-pressure frozen, substituted in 0.2% (v/v) uranyl acetate, secondary substituted in 0.2% (v/v) glutaraldehyde in acetone for 96 h, and rinsed several times in acetone. The samples were then treated with Lowicryl HM20 for 72 h and terminally polymerized under UV light for 48 h.
Sections of polymerized samples were placed on formvar-coated nickel grids. A 5% (w/v) solution of nonfat milk with 0.1% (v/v) Tween 20 in phosphate-buffered saline was used to block the sample. Sections were incubated with the primary antibodies (1:100 for antisignal peptide and 1:500 for 27-kD α-zein and 19-kD α-zein antibodies in phosphate-buffered saline-Tween 20) for 1 h. The secondary antibody (anti-rabbit IgG) was conjugated to 15-nm gold particles. Sections were then incubated with the secondary antibody (1:10) for 1 h. Labeled sample sections were observed with a Hitachi H7600 transmission electron microscope.
RNA-seq Analysis
Three fl4 or wild-type biological repeats were pooled together. Library construction was performed according to Illumina standard instructions. Reads were aligned to the maize B73 genome using TopHat2 (Langmead et al., 2009). Data were normalized as fragments per kilobase of exon per million fragments mapped, as the sensitivity of RNA-seq depends on the transcript length. Significant differentially expressed genes were identified as genes with at least a 1.3-fold change in expression and P < 0.05.
TUNEL Assays
TUNEL assays were performed with the DeadEnd Fluorometric TUNEL System (Roche) according to standard instructions as follows: tissues were fixed at 4°C in 4% (v/v) paraformaldehyde containing 0.1% (v/v) Triton X-100 and Tween 20 and embedded in paraffin. Ten-micrometer microtome sections were dewaxed in xylene and pretreated with 20 mg mL–1 proteinase K. Two slides, treated with DNase I in 50 mm Tris-Cl, served as positive controls. Samples were observed with a Leica microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PDI, NM_001112334, GRMZM2G389173 and NM_001112284, GRMZM2G091481; ZmDerlin1-1, NM_001112475, GRMZM2G117388; ZmbZIP60, NM_001153784, GRMZM2G025812; and ZmBIP1, NM_001112423, GRMZM2G114793. RNA-seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entry GSE54184.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Soluble amino acid content in wild-type and fl4 mature endosperm.
Supplemental Table S2. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Brian A. Larkins (University of Nebraska, Lincoln) for critical reading of the manuscript.
Glossary
- ER
endoplasmic reticulum
- BIP
binding protein
- ERAD
endoplasmic reticulum-associated degradation
- UPR
unfolded protein response
- PCD
programmed cell death
- DAP
days after pollination
- cDNA
complementary DNA
- IEM
immunoelectron microscopy
- RNA-seq
RNA sequencing
- GO
Gene Ontology
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- PI
propidium iodide
Footnotes
This work was supported by the Major Research plan of the National Natural Sciences Foundation of China (grant no. 91335208) and the Ministry of Science and Technology of China (grant nos. 2014CB138204 and 2012AA10A305).
References
- Braakman I, Hebert DN. (2013) Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 5: a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briknarová K, Takayama S, Brive L, Havert ML, Knee DA, Velasco J, Homma S, Cabezas E, Stuart J, Hoyt DW, et al. (2001) Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol 8: 349–352 [DOI] [PubMed] [Google Scholar]
- Chou KC. (2001) Using subsite coupling to predict signal peptides. Protein Eng 14: 75–79 [DOI] [PubMed] [Google Scholar]
- Coleman CE, Clore AM, Ranch JP, Higgins R, Lopes MA, Larkins BA. (1997) Expression of a mutant α-zein creates the floury2 phenotype in transgenic maize. Proc Natl Acad Sci USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, Devienne D. (1993) Quantification of dominance for proteins pleiotropically affected by opaque-2 in maize. Heredity 70: 38–51 [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH. (2013) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14: 8188–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrink-Kurtzman MA, Bietz JA. (1993) Zein composition in hard and soft endosperm of maize. Cereal Chem 70: 105–108 [Google Scholar]
- Fujita M, Mizukado S, Fujita Y, Ichikawa T, Nakazawa M, Seki M, Matsui M, Yamaguchi-Shinozaki K, Shinozaki K. (2007) Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem Biophys Res Commun 364: 250–257 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Larkins BA. (2005) Molecular genetic approaches to developing quality protein maize. Trends Genet 21: 227–233 [DOI] [PubMed] [Google Scholar]
- Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Römisch K. (1999) Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol 147: 1443–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillikin JW, Zhang F, Coleman CE, Bass HW, Larkins BA, Boston RS. (1997) A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol 114: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Holding DR, Larkins BA. (2006) The development and importance of zein protein bodies in maize endosperm. Maydica 51: 243–254 [Google Scholar]
- Holding DR, Otegui MS, Li B, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA. (2007) The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding DR, Meeley RB, Hazebroek J, Selinger D, Gruis F, Jung R, Larkins BA. (2010) Identification and characterization of the maize arogenate dehydrogenase gene family. J Exp Bot 61: 3663–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R. (2004) BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9: 299–307 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. (2011) Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ 18: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci 17: 720–727 [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13: 1211–1233 [DOI] [PubMed] [Google Scholar]
- Kim CS, Gibbon BC, Gillikin JW, Larkins BA, Boston RS, Jung R. (2006) The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response. Plant J 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Kim CS, Hunter BG, Kraft J, Boston RS, Yans S, Jung R, Larkins BA. (2004) A defective signal peptide in a 19-kD α-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol 134: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Woo YM, Clore AM, Burnett RJ, Carneiro NP, Larkins BA. (2002) Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell 14: 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS. (2005) Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol 138: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. (1989) Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. (1992) Effect of the floury-2 locus on protein body formation during maize endosperm development. Protoplasma 171: 123–133 [Google Scholar]
- Li Y, Humbert S, Howell SH. (2012) ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res Notes 5: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA. (1991) γ-Zein content is related to endosperm modification in Quality Protein Maize. Crop Sci 31: 1655–1662 [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145: 279–280 [DOI] [PubMed] [Google Scholar]
- Miclaus M, Wu Y, Xu JH, Dooner HK, Messing J. (2011) The maize high-lysine mutant opaque7 is defective in an acyl-CoA synthetase-like protein. Genetics 189: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra PS, Jambunathan R, Mertz ET, Glover DV, Barbosa HM, McWhirter KS. (1972) Endosperm protein synthesis in maize mutants with increased lysine content. Science 176: 1425–1427 [DOI] [PubMed] [Google Scholar]
- Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW. (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Sabelli PA, Larkins BA (2007) Endoreduplication and programmed cell death in the cereal endosperm. In OA Olsen, ed, Endosperm, Vol 8. Springer-Verlag, Berlin, pp 21–43 [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. (1999) Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci 112: 4123–4134 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. (2004) A trip to the ER: coping with stress. Trends Cell Biol 14: 20–28 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer I, Emr S, Gross C, Schekman R. (1985) Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol 100: 1664–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. (1990) Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA 87: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatters RG, Jr, Miernyk JA. (1991) A zein signal sequence functions as a signal-anchor when fused to maize alcohol dehydrogenase. Biochim Biophys Acta 1068: 179–188 [DOI] [PubMed] [Google Scholar]
- Song R, Messing J. (2002) Contiguous genomic DNA sequence comprising the 19-kD zein gene family from maize. Plant Physiol 130: 1626–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens FJ, Argon Y. (1999) Protein folding in the ER. Semin Cell Dev Biol 10: 443–454 [DOI] [PubMed] [Google Scholar]
- Tsai B, Rapoport TA. (2002) Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J Cell Biol 159: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104: 937–948 [DOI] [PubMed] [Google Scholar]
- van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. (2003) P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2α signaling. J Biol Chem 278: 15558–15564 [DOI] [PubMed] [Google Scholar]
- Wang G, Sun X, Wang G, Wang F, Gao Q, Sun X, Tang Y, Chang C, Lai J, Zhu L, et al. (2011) Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, et al. (2012) Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24: 3447–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. (2009) Bax Inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci 10: 3149–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R. (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. (2006) From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ 13: 374–384 [DOI] [PubMed] [Google Scholar]
- Wu Y, Holding DR, Messing J. (2010) γ-Zeins are essential for endosperm modification in quality protein maize. Proc Natl Acad Sci USA 107: 12810–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J. (2010) RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol 153: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yuan L, Guo X, Holding DR, Messing J. (2013) Mutation in the seed storage protein kafirin creates a high-value food trait in sorghum. Nat Commun 4: 2217. [DOI] [PubMed] [Google Scholar]
- Zhang F, Boston RS. (1992) Increases in binding protein (BiP) accompany changes in protein body morphology in three high lysine mutants of maize. Protoplasma 171: 142–152 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.