A diel flux balance modeling framework that integrates temporally separated metabolic networks provides realistic descriptions of light and dark metabolism in C3 and CAM leaves and suggests that energetics and nitrogen use efficiency are unlikely to have been drivers for the evolution of CAM.
Abstract
Although leaves have to accommodate markedly different metabolic flux patterns in the light and the dark, models of leaf metabolism based on flux-balance analysis (FBA) have so far been confined to consideration of the network under continuous light. An FBA framework is presented that solves the two phases of the diel cycle as a single optimization problem and, thus, provides a more representative model of leaf metabolism. The requirement to support continued export of sugar and amino acids from the leaf during the night and to meet overnight cellular maintenance costs forces the model to set aside stores of both carbon and nitrogen during the day. With only minimal constraints, the model successfully captures many of the known features of C3 leaf metabolism, including the recently discovered role of citrate synthesis and accumulation in the night as a precursor for the provision of carbon skeletons for amino acid synthesis during the day. The diel FBA model can be applied to other temporal separations, such as that which occurs in Crassulacean acid metabolism (CAM) photosynthesis, allowing a system-level analysis of the energetics of CAM. The diel model predicts that there is no overall energetic advantage to CAM, despite the potential for suppression of photorespiration through CO2 concentration. Moreover, any savings in enzyme machinery costs through suppression of photorespiration are likely to be offset by the higher flux demand of the CAM cycle. It is concluded that energetic or nitrogen use considerations are unlikely to be evolutionary drivers for CAM photosynthesis.
Photosynthetic metabolism continues to be studied intensively because of its importance for crop performance and the global carbon cycle in relation to climate change. The metabolic pathways and enzymes involved in carbon fixation and related metabolic processes, such as the synthesis of Suc and starch, have been well-characterized. However, it is apparent that full appreciation of leaf metabolism requires these metabolic processes to be placed in the context of the wider metabolic network (Szecowka et al., 2013). This is particularly important for predicting how strategies for engineering improved photosynthesis (Maurino and Weber, 2013) may affect network properties, such as redox and energy balancing (Kramer and Evans, 2011).
Flux balance analysis (FBA) has emerged as the method of choice for predicting fluxes in large metabolic network models (Sweetlove and Ratcliffe, 2011), and several flux balance models have explicitly considered photosynthetic metabolism in a variety of plants species and microorganisms, including cyanobacteria (Synechocystis sp. PCC 6803; Knoop et al., 2010, 2013; Montagud et al., 2010; Nogales et al., 2012; Saha et al., 2012), Chlamydomonas reinhardtii (Boyle and Morgan, 2009; de Oliveira Dal'Molin et al., 2011), Arabidopsis (Arabidopsis thaliana; de Oliveira Dal’Molin et al., 2010a), rapeseed (Brassica napus) embryos (Hay and Schwender, 2011), rice (Oryza sativa; Poolman et al., 2013), maize (Zea mays; Saha et al., 2011), and several C4 plants (de Oliveira Dal’Molin et al., 2010b). These models successfully predicted the metabolic routes involved in the fixation of CO2 into different biomass components in the light. However, one major feature of metabolism of photosynthetic organisms, namely the interaction between light and dark metabolism, is neglected in most of these studies. Effectively, most models assume that the organism grows in constant light, which is rarely true in natural conditions.
Apart from the obvious switch from photoautotrophic to heterotrophic metabolism between day and night, interactions between the two phases can occur through the temporal separation of storage compound synthesis and subsequent mobilization. For example, it has been shown that the carbon skeletons used for nitrogen assimilation during the day are largely provided by carboxylic acids that were synthesized and stored during the previous night (Gauthier et al., 2010). Such temporal shifts of carbon and nitrogen metabolism have substantial implications for fluxes in the central metabolic network of leaves in the light (Tcherkez et al., 2009). Interactions between temporally separated metabolic events are also a critical feature of Crassulacean acid metabolism (CAM) photosynthesis, in which CO2 is initially fixed at night by phosphoenolpyruvate carboxylase (PEPC), leading to night storage of carboxylic acids (mainly malic acid) that are decarboxylated during the day to provide CO2 for the conventional photosynthetic carbon assimilation cycle. Although this is principally an adaptation to arid environments, there are unresolved questions as to whether CAM photosynthesis is energetically more efficient than C3 photosynthesis (Winter and Smith, 1996). Such questions are becoming more important in the light of the proposed use of CAM plants as a source of biofuel (Yan et al., 2011).
One recent study used FBA to consider both light and dark metabolism in Synechocystis sp. PCC 6803 over a complete diel cycle divided into 192 time steps (Knoop et al., 2013). Time courses of metabolic flux predictions over a diel cycle were simulated by altering the constraints on metabolic outputs (biomass composition) depending on the time point and based on empirical rules. This simulation led to a highly constrained model and did not allow the range of potential interactions between the day and night phases to be fully explored. We have developed an alternative modeling framework for integrated day-night FBA, in which the metabolic fluxes in the light and dark phases were simulated simultaneously in a single optimization problem. A predefined list of storage compounds that can accumulate freely over the diurnal cycle was made available to the model. The model was then free to choose among these storage compounds, the choice being dictated by the need to satisfy the objective function within the applied constraints. This diurnal modeling framework was used to explore the interactions between light and dark metabolism and to predict the metabolic fluxes in the light in both C3 and CAM photosynthesis. We show that accounting for day-night interactions leads to an altered pattern of fluxes during the day that provides a better match with experimental observations. We were also able to simulate network flux distributions in CAM metabolism. The model successfully predicts the classic CAM cycle in the different CAM subtypes and allows a comparison of the energetic efficiency and metabolic costs between CAM and C3 photosynthetic metabolism.
RESULTS
A Day-Night Modeling Framework for Leaf Metabolism
The aim was to construct a flux balance model that accounted for the day and night phases of leaf metabolism in an integrated fashion, such that storage compounds synthesized during the day were available for use in the dark and vice versa. This was achieved by applying a specific framework of constraints to an existing genome-scale model of Arabidopsis metabolism (Cheung et al., 2013). The assumption was made that each phase (day and night) was in a pseudo-steady state, as has been done for previous flux balance models of photosynthetic metabolism. The two interdependent steady states were modeled as a single optimization problem, with photoautotrophic metabolism specified in the day by allowing a photon influx and heterotrophic metabolism specified at night by setting the photon influx to zero. For simplicity, a 12-h/12-h day-night cycle was specified. Also, for simplicity, we considered the case of a mature leaf, in which the only metabolic outputs were the synthesis of Suc and a range of amino acids for export to the phloem. It is trivial to extend the model to consider a growing leaf and account for shorter or longer days.
The relative proportions of 18 amino acids exported to the phloem (Supplemental Table S1) and the ratio of Suc:total amino acid export (2.2:1) were constrained in accordance with measurements of the composition of Arabidopsis phloem exudate (Wilkinson and Douglas, 2003). The model was required to maintain export of sugars and amino acids to the phloem during the day and night but at a ratio of 3:1 (day:night) based on measurements in Arabidopsis rosettes (Gibon et al., 2004). The sole source of nitrogen for the leaf model was nitrate import from the xylem based on the observation that nitrate represents the main form of nitrogen (80%) entering a leaf (Macduff and Bakken, 2003). The rate of nitrate import into the leaf during the day:night was constrained to be 3:2 based on measurements in various plant species (Delhon et al., 1995; Macduff and Bakken, 2003; Siebrecht et al., 2003). Other than the export of Suc and amino acids, cellular maintenance costs were accounted for by including generic ATPase and NADPH oxidase steps for maintenance and the requirement to satisfy a specified carbon conversion efficiency (Cheung et al., 2013). In this case, we assumed a carbon conversion efficiency of 50% during the night based on the day-night carbon balance calculated from measurements in Arabidopsis rosettes (Gibon et al., 2004). The ratio of ATP maintenance cost:NADPH maintenance cost was assumed to be 3:1 (Cheung et al., 2013), and maintenance costs were assumed to be the same in the light and dark phases. In addition to the above constraints, fluxes through the chloroplastic NADPH dehydrogenase and plastoquinol oxidase were set to zero because the contributions of NADPH dehydrogenase (Yamamoto et al., 2011) and plastoquinol oxidase (Josse et al., 2000) to photosynthesis are thought to be minor. The flux of Suc synthase was constrained to be irreversible in the direction of Suc degradation because it is thought that Suc synthase is not involved in Suc synthesis in leaves (Nguyen-Quoc et al., 1990).
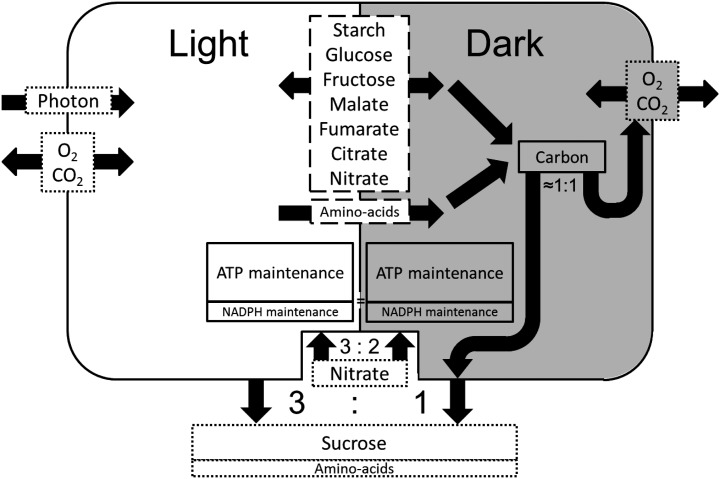
To maintain a metabolic output at night, the model must accumulate carbon and nitrogen stores during the day. To explore the metabolic interaction between the day and the night, the model included a set of sugars and carboxylic acids to be used as carbon storage molecules. Specifically, starch, Glc, Fru, malate, fumarate, and citrate were free to accumulate during either the day or night phase. No direct constraints were applied to the amount of each compound that accumulated other than the requirement for the overall model to be mass balanced. For nitrogen, nitrate and 20 common amino acids were allowed to accumulate as storage compounds, but the amino acids were constrained, such that accumulation was only permitted during the day and not at night in accordance with observations on the diel fluctuations of amino acids in leaves (Scheible et al., 2000). Again, no direct constraints were applied to the amount of each amino acid to be accumulated. Thus, the model was free to choose among a set of storage compounds to allow the constraint of export of sugars and amino acids at night to be met. The interconnections between the light and dark phases of the model as well as the input and output constraints are summarized in Figure 1. Fluxes were predicted using linear programming with an objective function to minimize the sum of fluxes within the specified constraints. The two phases were modeled simultaneously in a single optimization problem analogous to modeling interactions between two microorganisms or two tissues/cell types in multicellular organisms (de Oliveira Dal’Molin et al., 2010b). Because Arabidopsis is a C3 plant, the ratio of Rubisco carboxylation:oxygenase was set to a typical value of 3:1 to simulate photorespiration (Gutteridge and Pierce, 2006).
Figure 1.
Flux balance framework for integrated metabolic modeling of the day and night phases of a mature leaf. The light and dark phases are represented by the white and gray backgrounds of the diagram, respectively. Metabolites shown in the dashed rectangles between the two phases represent potential storage compounds. Starch, Glc, Fru, malate, fumarate, citrate, and nitrate were allowed to accumulate in the light and the dark as denoted by the arrows pointing to the light and dark states. Twenty common amino acids were allowed to accumulate in the light but not the dark as denoted by the arrow pointing from the light phase to the dark phase. Export to the phloem was set to four Suc to one amino acid, with 18 different amino acids in the proportions shown in Supplemental Table S1. The export rate was set to be 3 times greater in the light than the dark. Nitrate was set as the sole nitrogen source, with the ratio of nitrate uptake from the phloem in the light:nitrate uptake from the phloem in the dark set to 3:2. Cellular maintenance costs in the dark were set to a fixed value, where the carbon exported during the night roughly equaled the carbon released as CO2 in the dark phase. Maintenance costs were assumed to be the same in the light and the dark. The ratio of ATP maintenance cost:NADPH maintenance cost was set to 3:1.
The Integrated Day-Night Model Leads to Altered Flux Predictions Compared with a Constant Light Model
To assess the effect of the diel modeling framework, fluxes predicted in the light phase of the diel model were compared with those fluxes predicted in a constant light model. The latter model was constrained by total Suc and amino acid export and cellular maintenance costs that matched those costs over a 24-h period in the diel model.
To take account of the fact that some fluxes are not uniquely defined at the optimum, flux variability analysis was used to determine the feasible range of all fluxes. Fluxes in the light component of the diel model were considered to be significantly different from those fluxes of the same reactions in the constant light models if their flux ranges did not overlap. This procedure identified 131 reactions with nonoverlapping flux ranges in the light (Supplemental Table S2). These reactions carried different fluxes in the light, but none carried fluxes in opposite directions in the two models. Fluxes were compared using a similarity measure calculated from the fraction of the smaller flux value over the larger flux value. Reactions with a similarity measure of less than 0.25 are listed in Table I, where the smaller the value of this measure means the larger the difference in fluxes. A similarity measure of zero means that the reaction carried zero flux in one of the models. Some reactions that carried different fluxes between the diel and constant light models are to be expected. For example, starch synthesis was turned on during the day in the diel model but not the constant light model, where there was no need to store carbon because of the continuous assimilation of CO2. Similarly, transport of carboxylic acids and nitrate across the tonoplast for storage in the vacuole was activated in the diel model.
Table I. List of reactions in the light that have different fluxes in the diel model and the continuous light model.
Reactions with similarity measures less than 0.25 are listed, where a small value represents a large difference in fluxes from the two modeling approaches (Supplemental Table S2 has a complete list of reactions with nonoverlapping flux ranges). The metabolic context of the reactions is listed in the third column. PPi, Inorganic pyrophosphatase.
| Reaction Name | Similarity Measure | Metabolic Context |
|---|---|---|
| Reactions with higher flux in the diel model | ||
| Plastidic ADP Glc pyrophosphorylase | 0 | Starch synthesis |
| Plastidic starch synthase | 0 | Starch synthesis |
| Plastidic phosphoglucose isomerase | 0 | Starch synthesis |
| Plastidic phosphoglucomutase | 0 | Starch synthesis |
| Tonoplast citrate/H+ antiporter | 0 | Storage in the vacuole |
| Tonoplast malate/H+ antiporter | 0 | Storage in the vacuole |
| Tonoplast fumarate/H+ antiporter | 0 | Storage in the vacuole |
| Tonoplast nitrate transporter | 0 | Storage in the vacuole |
| Tonoplast PPiase | 0 | Storage in the vacuole |
| Plastidic alkaline pyrophosphatase | 0.015 | Related to starch synthesis |
| Reactions with higher flux in the continuous light model | ||
| Peroxisomal inorganic pyrophophatase | 0 | Related to Glu and Gln synthesis |
| Peroxisomal phosphate transporter | 0 | Related to Glu and Gln synthesis |
| Peroxisomal citrate synthase | 0 | Related to Glu and Gln synthesis |
| Peroxisomal acetyl-CoA synthetase | 0 | Related to Glu and Gln synthesis |
| Peroxisomal AMP/ATP antiporter | 0 | Related to Glu and Gln synthesis |
| Peroxisomal acetate transporter | 0 | Related to Glu and Gln synthesis |
| Peroxisomal citrate transporter | 0 | Related to Glu and Gln synthesis |
| Cytosolic Thr aldolase | 0.019 | Related to Glu and Gln synthesis |
| Cytosolic aldehyde dehydrogenase | 0.019 | Related to Glu and Gln synthesis |
| Plastidic ATP/ADP antiporter | 0.093 | Export of ATP from the chloroplast |
| Plastidic Thr transporter | 0.123 | Related to Glu and Gln synthesis |
| Plastidic Thr synthase | 0.145 | Related to Glu and Gln synthesis |
| Plastidic homo-Ser kinase | 0.153 | Related to Glu and Gln synthesis |
| Plastidic homo-Ser dehydrogenase | 0.161 | Related to Glu and Gln synthesis |
| Plastidic Asp kinase | 0.180 | Related to Glu and Gln synthesis |
| Plastidic Asp semialdehyde dehydrogenase | 0.180 | Related to Glu and Gln synthesis |
| Mitochondrial ATP/AMP antiporter | 0.202 | Related to Glu and Gln synthesis |
| Mitochondrial aldenylate kinase | 0.202 | Related to Glu and Gln synthesis |
| Plastidic Asp transporter | 0.208 | Related to Glu and Gln synthesis |
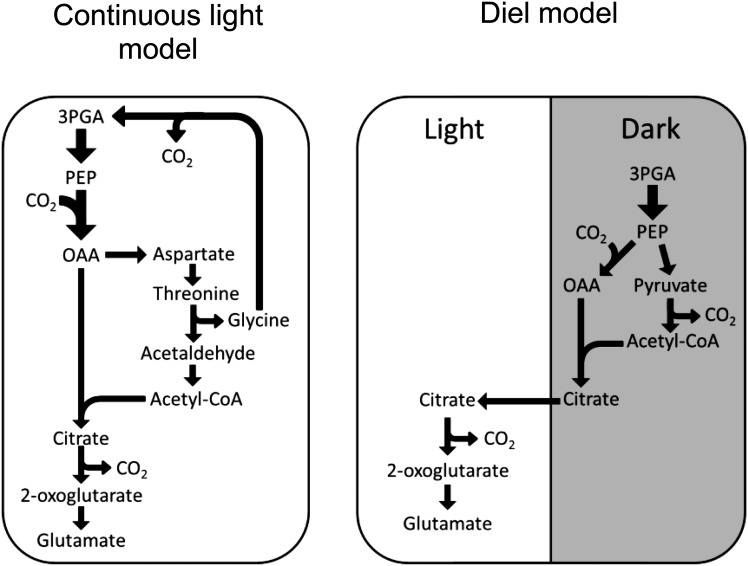
However, there were also changes in metabolic fluxes that were less directly constrained. These changes relate to the citrate used to provide carbon skeletons for Glu and Gln synthesis in the light. Table I shows that the synthesis of citrate was predicted to occur in the peroxisome through peroxisomal citrate synthase. In contrast, the diel model predicted that citrate would be synthesized through the mitochondrial tricarboxylic acid cycle during the night and stored in the vacuole. Nocturnally stored citrate was then exported from the vacuole during the day and metabolized into 2-oxoglutarate (2-OG) by aconitase and isocitrate dehydrogenase for Glu synthesis (Fig. 2). This prediction of the diel model is consistent with observations from isotopic labeling experiments (Gauthier et al., 2010), and this example shows the importance of considering the interconnections between light and dark metabolism for the prediction of realistic flux distributions.
Figure 2.
Metabolic routes for Glu biosynthesis from two modeling approaches. Left, A single steady-state model in constant light; right, the diel modeling framework. In constant light, the carbon skeletons for Gln synthesis were predicted to be supplied through a metabolic route, in which Thr is metabolized in the cytosol to acetate, transported to the peroxisome, and metabolized to citrate, which is exported to the cytosol and converted to 2-OG. Using the diel modeling framework, the model predicted the use of citrate stored in the dark to provide the carbon skeletons for Glu synthesis in the light. The thickness of the arrows is scaled to indicate relative flux magnitudes (in molar units). OAA, Oxaloacetate.
Predictions of Metabolic Network Fluxes in a Mature C3 Leaf over a Day-Night Cycle
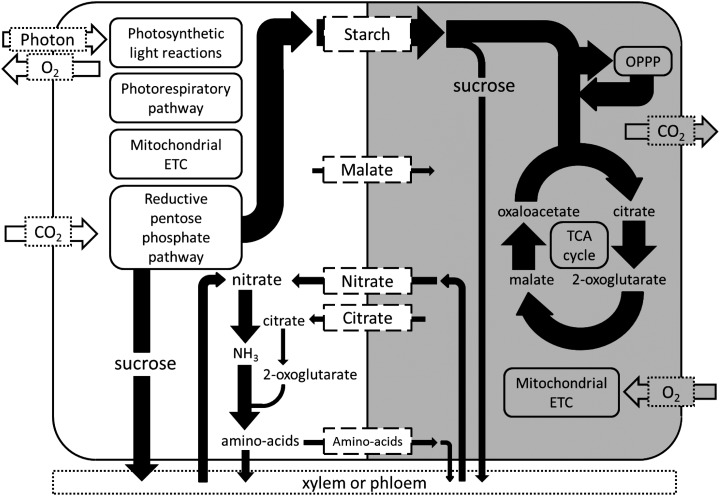
The flux solution using the day-night modeling framework can be summarized in a simple flux map (Fig. 3), and a complete list of predicted flux ranges for reactions in the light and dark is provided in Supplemental Table S3. The diurnal modeling framework successfully predicted that starch is the main carbon storage compound that accumulates in the light, although the model was free to use sugars and carboxylic acids as the carbon store. Some malate was also predicted to accumulate in the light, which then fed into the tricarboxylic acid cycle in the dark. Interestingly, the model predicted that both citrate and nitrate were stored at night to support nitrogen assimilation, which was predicted to occur primarily during the day. The nitrate store was generated by transporting imported nitrate from the xylem into the vacuole during the night and releasing the vacuolar store to the cytosol during the day. The model also predicted the storage during the day of a proportion of amino acids synthesized in the light, which were then exported into the phloem in the dark.
Figure 3.
Predicted flux map of metabolism for a mature C3 leaf over a day-night cycle. The light and dark phases are represented by the white and gray backgrounds, respectively. Metabolites shown in the dashed rectangles between the two phases represent storage compounds. The thickness of the arrows is proportional to the metabolic flux through the reactions (in molar units). Metabolic processes listed within rounded rectangles carry fluxes too large to be represented on the flux map. TCA, Tricarboxylic acid.
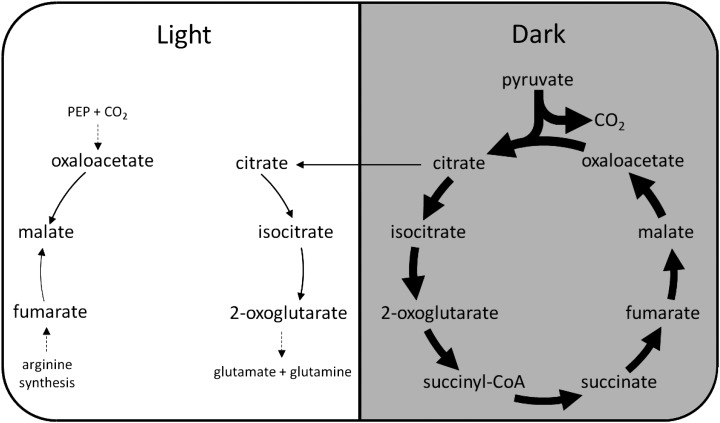
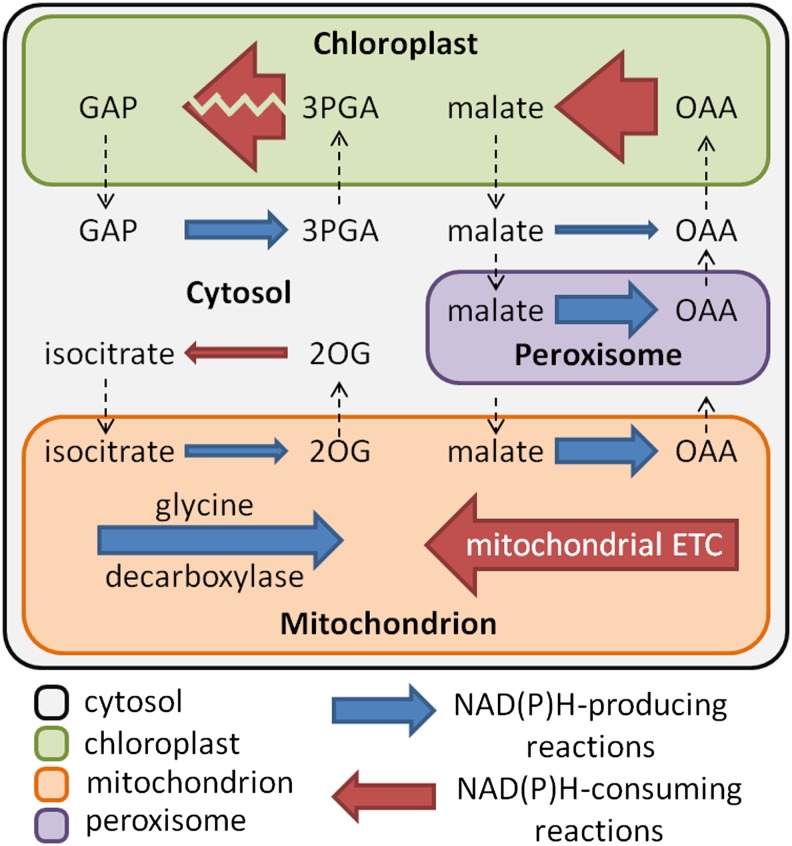
In the light phase, the model predicted large fluxes through the linear photosynthetic electron transport pathway and the Calvin-Benson pathway to support Suc and amino acid synthesis and starch accumulation. The classical photorespiratory pathway for the oxidation of 2-phosphoglycolate was also predicted to be active because of a constraint in the model that set the Rubisco carboxylation:oxygenase ratio to 3:1. Note that, if this constraint is removed, the Rubisco oxygenase reaction becomes inactive. The tricarboxylic acid cycle was predicted to operate in a noncyclic mode in the light with two separate branches (Fig. 4), consistent with the understanding based on isotope labeling experiments and the predictions from other flux balance models (Sweetlove et al., 2010). Oxaloacetate produced from PEPC was converted into malate, which accumulated in the light. The model predicted that the mitochondrial electron transport chain (ETC) is active in the light, carrying a flux equivalent to 10% of the flux through photosynthetic linear electron transport. The mitochondrial ATP synthase was predicted to contribute to 18% of total ATP synthesis in the light, with the rest produced by the chloroplast thylakoid ATP synthase. The two sources of NADH feeding into the mitochondrial ETC are Gly decarboxylase and the malate-oxaloacetate shuttle (Fig. 5), with contributions of 53% and 47%, respectively (the mitochondrial isocitrate dehydrogenase reaction is predicted to exclusively generate NADPH, which is used for maintenance reactions in the mitochondrion). The ultimate source of reductant in a photosynthetic leaf is from photosynthetic linear electron transport. Although most of the reducing power is used for photosynthetic CO2 fixation in the chloroplast, a small proportion of reductant was shuttled out of the chloroplast by the malate-oxaloacetate shuttle (11%) and the 3-phosphoglycerate (3PGA)-glyceraldehyde 3-phosphate (GAP) shuttle (4%; Fig. 5). Malate-oxaloacetate exchanges were responsible for transfer of reductant from the chloroplast into the cytosol, the mitochondrion, and the peroxisome. The 3PGA-GAP shuttle transported reductant from the chloroplast to the cytosol to produce NADPH by the nonphosphorylating NADP-glyceraldehyde 3-phosphate dehydrogenase. A proportion of NADPH produced in the cytosol was shuttled into the mitochondrion by the isocitrate-2-OG shuttle (Fig. 5).
Figure 4.
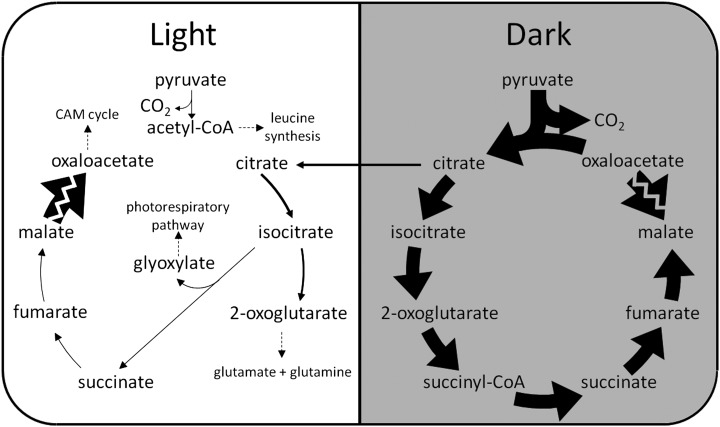
Flux predictions through the tricarboxylic acid cycle and pyruvate dehydrogenase in a C3 leaf in the light and the dark. A noncyclic mode with two separate branches, citrate to 2-OG and oxaloacetate and fumarate to malate, was predicted to operate in the light. A cyclic mode of the tricarboxylic acid cycle was predicted to operate in the dark, mainly to produce ATP through oxidative phosphorylation. Fluxes illustrated are net conversion between metabolites over all subcellular compartments. The thickness of the arrows is proportional to the metabolic fluxes through the reactions (in molar units). Citrate stored in the dark is represented by an arrow pointing from the dark phase to the light phase. Malate storage is not illustrated in this diagram.
Figure 5.
Reductant shuttling between subcellular compartments and the production and consumption of NADH in mitochondria in the light. Left-pointing arrows represent reductant-consuming reactions; right-pointing arrows represent reductant-producing reactions. The thickness of the arrows is proportional to the metabolic flux through the reactions (in molar units), except for the conversion between 3PGA and GAP in the chloroplast, where the zigzag line across the arrow indicates that the flux is too large to be illustrated to scale in the diagram. Reductant shuttles among four subcellular compartments (cytosol, chloroplast, mitochondria, and peroxisome) are shown with dashed arrows representing transfer of metabolites between compartments. OAA, Oxaloacetate; 2OG, 2-OG. [See online article for color version of this figure.]
In the dark phase, the largest fluxes are related to starch degradation, Suc synthesis, glycolysis, the oxidative pentose phosphate pathway (OPPP), the tricarboxylic acid cycle, and the mitochondrial ETC (Fig. 3). The breakdown of starch was predicted to be carried out by the hydrolytic pathway generating maltose for transport from the chloroplast to the cytosol. This finding is the result of constraining the chloroplastic hexose phosphate transporter to carry zero flux, which is in line with experimental observations (Niewiadomski et al., 2005), thus limiting the possible metabolic routes for carbon export from the chloroplast. When this constraint was removed, starch breakdown occurred through starch phosphorylase. Glycolysis and the OPPP were predicted to occur in both the cytosol and the chloroplast, with 72% of the total flux through glycolytic glyceraldehyde 3-phosphate dehydrogenase and 55% of that through the OPPP Glc-6-P dehydrogenase steps occurring in the cytosol. The tricarboxylic acid cycle operates in a cyclic mode in the dark (Fig. 4), consuming pyruvate produced from stored starch by glycolysis. A small proportion of the citrate produced by citrate synthase (15%) is stored in the dark to provide carbon skeletons for Glu and Gln synthesis in the light.
Predictions of Metabolic Fluxes in a Mature CAM Leaf over a Diurnal Cycle
From a structural metabolic modeling perspective, the Arabidopsis genome-scale metabolic model contains all the metabolic reactions required to carry out the various subtypes of CAM photosynthesis. However, because the metabolic cycle that constitutes CAM photosynthesis is temporally segregated, metabolism in CAM leaves cannot be modeled with a single steady-state FBA model. Therefore, we applied the diel modeling framework to predict metabolic fluxes in a mature CAM leaf over a 24-h cycle. As for the mature C3 leaf, the model was constrained by Suc and amino acid export and cellular maintenance costs. The main additional constraint for simulating CAM photosynthesis was that CO2 exchange with the environment was set to have zero flux in the light to simulate the closure of stomata in CAM plants during the day. Other differences between the C3 and CAM models arose from the treatment of the Rubisco oxygenase activity and the export of carbon from the plastids. First, Rubisco oxygenase activity is likely to be suppressed in CAM leaves in the light because of the increase in the internal partial pressure of CO2, but because there is also an increase in internal oxygen concentration, it is unclear to what extent photorespiration is prevented in CAM plants (Lüttge, 2011). Accordingly, the Rubisco carboxylase-oxygenase ratio was left unconstrained in the light phase of the CAM model. Second, in the ice plant Mesembryanthemum crystallinum, which is a facultative CAM plant, chloroplasts of CAM-induced plants mainly export Glc-6-phosphate (Neuhaus and Schulte, 1996), and this finding is coincident with a 71-fold increase in transcript level for the chloroplastic Glc-6-P transporter (Cushman et al., 2008). On this basis, the chloroplastic Glc-6-P transporter was not constrained in the CAM model. Finally, the various subtypes of CAM photosynthesis were simulated by setting the reactions specific to other subtypes to carry zero flux. The constraints applied for modeling the different modes of photosynthesis are summarized in Table II.
Table II. Constraints applied for modeling leaf metabolism in C3 and various subtypes of CAM.
Pi, Inorganic pyrophosphatase.
| Constraints for Different Modes of Photosynthesis | C3 |
CAM |
||||
|---|---|---|---|---|---|---|
| Generic | Starch PEPCK | Starch ME | Sugars PEPCK | Sugars ME | ||
| CO2 exchange | ||||||
| CO2 exchange in light | Free | 0 | 0 | 0 | 0 | 0 |
| Photorespiration | ||||||
| Rubisco CO2-O2 | 3:1 | Unconstrained in light; 3:1 in dark | Unconstrained in light; 3:1 in dark | Unconstrained in light; 3:1 in dark | Unconstrained in light; 3:1 in dark | Unconstrained in light; 3:1 in dark |
| Transporter | ||||||
| Chloroplast Glc 6-P inorganic phosphate | 0 | Free | Free | Free | Free | Free |
| Metabolic reactions | ||||||
| PEPCK | Free | Free | Free | 0 | Free | 0 |
| ME | Free | Free | 0 | Free | 0 | Free |
| Pyruvate, Pi dikinase | Free | Free | 0 | Free | 0 | Free |
| Storage compounds | ||||||
| Starch | Free | Free | Free | Free | 0 | 0 |
| Soluble sugars | Free | Free | 0 | 0 | Free | Free |
Using the diel modeling framework with CAM-specific constraints, FBA successfully predicted metabolic fluxes consistent with the well-known CAM cycle (Supplemental Table S3). In the light, nocturnally stored malate was decarboxylated by malic enzyme or PEPCK (phosphoenolpyruvate carboxykinase) depending on the subtype. In the generic CAM model (no subtype constraints), 89% of malate decarboxylation was predicted to be carried out by PEPCK, with the remaining 11% carried out by the cytosolic malic enzyme. In addition to the photosynthetic linear electron transport and reductive pentose phosphate pathway, which were expected to carry high fluxes in the light, the gluconeogenesis pathway also carried a high predicted flux in the light to convert phosphoenolpyruvate (PEP) from malate decarboxylation to starch or soluble sugars for storage as part of the CAM cycle. In the generic CAM model, only starch was stored during the day, not soluble sugars. In our initial models of the sugar-storing subtypes, Fru was produced in the cytosol by Suc synthase and subsequently transported into the vacuole by the vacuolar hexose transporter. However, it is thought that tonoplast hexose transport activity is restricted to the night (in an efflux capacity) in CAM plants to avoid futile cycling (Holtum et al., 2005; Antony et al., 2008). When the vacuolar hexose transporter activity was subsequently restricted to the dark period, the model predicted that Suc is imported into the vacuole and catabolized into Glc and Fru by vacuolar invertase, which is in line with postulated models in the literature (Smith and Bryce, 1992; McRae et al., 2002; Holtum et al., 2005).
The Rubisco carboxylase-oxygenase ratio was unconstrained in modeling CAM, and the model predicted no Rubisco oxygenase flux, suggesting that Rubisco oxygenase is not necessary for the functioning of a mature CAM leaf under the assumption of minimizing total flux. The mitochondrial ETC and ATP synthase carried high fluxes in the light, with the mitochondrial ETC carrying a flux equivalent to 29% of the flux through photosynthetic linear electron transport and the mitochondrial ATP synthase contributing to 34% of the total ATP produced in the light in the generic CAM model. Similar values were predicted for the various CAM subtypes. Similar to C3 leaves, an incomplete tricarboxylic acid cycle was predicted in the light phase with two separate branches: citrate to 2-OG and succinate to oxaloacetate (Fig. 6). Interestingly, the oxaloacetate branch differed from that found for the C3 model in that oxaloacetate was produced (not consumed), and succinate was the starting point of the branch (not fumarate; Figs. 4 and 6). In the CAM tricarboxylic acid cycle, the two branches are joined by the activity of isocitrate lyase in the peroxisome to produce succinate (Fig. 6).
Figure 6.
Flux predictions through the tricarboxylic acid cycle and related reactions in a CAM leaf. A noncyclic mode with two distinct branches, citrate to 2-OG and succinate to oxaloacetate, was predicted to operate in the light. The two branches of the tricarboxylic acid cycle are connected by isocitrate lyase, which converts isocitrate into succinate and glyoxylate. A cyclic mode of the tricarboxylic acid cycle was predicted to operate in the dark, mainly to contribute to ATP production through oxidative phosphorylation. Fluxes illustrated are net conversion between metabolites over all subcellular compartments. The thickness of the arrows is proportional to the metabolic flux through the reactions (in molar units), except for the conversion between malate and oxaloacetate, where the zigzag line across the arrow indicates that the flux is too large to be illustrated to scale in the diagram. Citrate stored in the dark is represented by an arrow pointing from the dark phase to the light phase. Malate storage is not illustrated in this diagram.
In the dark phase, the stored carbon (starch or soluble sugars) was predicted to be catabolized to produce PEP, the substrate for carboxylation, through glycolysis. PEP was carboxylated by PEPC to produce oxaloacetate, which was subsequently converted into malate for storage in the vacuole as malic acid during the night. With respect to starch degradation, the phosphorolytic route through starch phosphorylase was predicted in CAM because it is more energetically efficient than the hydrolytic route through maltose, which is consistent with the current view of the CAM cycle (Weise et al., 2011). Although most of the carbon store was used to provide the substrate for carboxylation in the CAM cycle, some went into the OPPP to produce NADPH for maintenance processes and the tricarboxylic acid cycle to produce NADH for ATP synthesis through the mitochondrial ETC and ATP synthase. As in a mature C3 leaf, the model predicted a complete tricarboxylic acid cycle in the dark in a mature CAM leaf (Fig. 6).
Quantitative Comparisons of Metabolism between C3 and CAM Leaves
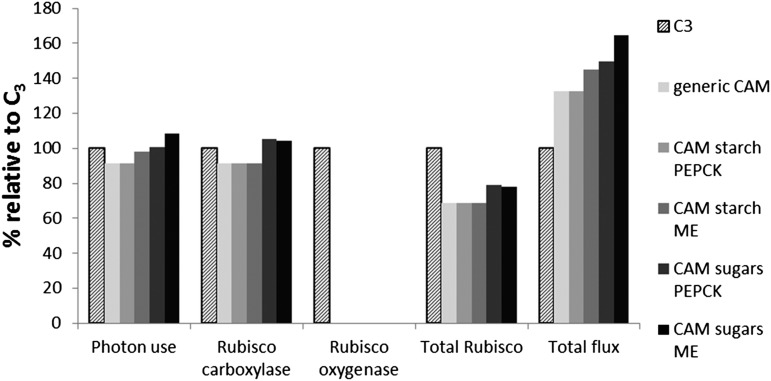
The CO2-concentrating mechanism in CAM is an energy-requiring process, and the energetic cost of supporting Suc and amino acid export from mature leaves and cell maintenance processes was compared between C3 and CAM in terms of photon use (Fig. 7). Photon use in a mature CAM leaf is similar to that in a C3 leaf, being ±10% of C3 depending on the CAM subtype.
Figure 7.
Comparison of model predictions between C3 and the various subtypes of CAM defined in Table II. The model predictions for photon use, Rubisco carboxylase flux, Rubisco oxygenase flux, total flux through Rubisco, and total flux in the metabolic model are shown with values scaled as a percentage of the value in the C3 leaf model.
Rubisco is the most abundant protein on earth, contributing up to 50% of the soluble protein and 20% to 30% of the total nitrogen in a C3 leaf (Feller et al., 2008). The investment in Rubisco, in terms of energy and nitrogen, is likely to contribute significantly to the evolutionary fitness tradeoff in plants. By comparing the model predictions, it was apparent that total flux through Rubisco (carboxylase + oxygenase flux) was lower in CAM than C3, mainly as a consequence of the prediction of no oxygenase activity in the CAM model (Fig. 7). Although this finding might suggest a cost saving for a CAM plant, it is offset by the fact that the CO2-concentrating mechanism requires high fluxes through the CAM cycle. As a consequence, the total flux through the entire metabolic network is 32% to 64% higher in CAM than in C3 (Fig. 7).
DISCUSSION
Integration of Light and Dark Metabolism Is Required to Capture Known Features of Leaf Metabolism by FBA
The usefulness of constraint-based modeling tools, such as FBA, depends on the extent to which they capture known features of the metabolic network. Good agreement has been reported between the predicted and measured fluxes for a heterotrophic Arabidopsis cell culture, leading to insights into cell maintenance costs under stress conditions (Cheung et al., 2013). There is an expectation that a similar modeling approach will be useful in analyzing photosynthetic metabolism, but most applications to date have been based on the analysis of the network under conditions of constant light. Here, analysis of the Arabidopsis model revealed that there are several features of leaf metabolism that can only be captured by FBA by considering the interaction between the temporally separated phases of light and dark metabolism. For example, the diel model accumulated citrate in the vacuole during the night, and it was released during the day to provide carbon skeletons for nitrogen assimilation. This finding agrees with evidence from isotope labeling experiments (Gauthier et al., 2010) and the current view of carboxylic acid metabolism in leaves in the light (Tcherkez et al., 2009; Sweetlove et al., 2010). In contrast, a single steady-state, constant light FBA leaf model generated citrate through an unusual metabolic route involving Thr aldolase and peroxisomal citrate synthase. This route is most likely chosen by the model because of its efficiency in terms of carbon use: The route leads to the fixation of 1 CO2 by PEPC and releases 0.5 CO2 from the recycling of Gly through the photorepiratory pathway. In another Arabidopsis genome-scale metabolic model, the Thr aldolase reaction was absent, and the model predicted that citrate would be synthesized from pyruvate by pyruvate dehydrogenase and mitochondrial citrate synthase (de Oliveira Dal’Molin et al., 2010a), contradicting well-established evidence that the mitochondrial pyruvate dehydrogenase is inhibited transcriptionally and posttranscriptionally in leaves in the light (Tovar-Méndez et al., 2003).
A possible explanation for the accumulation of citrate in the dark over the synthesis of citrate from primary photosynthate in the light could be that the degradation of starch to produce citrate (through glycolysis, PEPC, pyruvate dehydrogenase, and citrate synthase) provides a carbon-neutral route for transferring ATP and reducing power from the light phase to the dark phase. This conserves the net carbon required to support dark metabolism, where citrate in the dark would otherwise be metabolized through the tricarboxylic acid cycle and the carbon would be released as CO2. Confirming this result, if the model was configured with citrate accumulation prevented, the total CO2 release in the dark was increased by 2%.
The diel model was free to choose between a range of carbon (starch, sugars, and carboxylic acids) and nitrogen storage compounds (nitrate and amino acids) to support the export of sugars and amino acids during the night and to meet the overnight maintenance costs. The fact that the model solution selected starch and amino acids as the main daytime storage carbon and nitrogen compounds, respectively, consistent with the known biochemistry of C3 leaves, reflects the success of the objective function (minimization of sum of fluxes) in capturing the drivers that shape the metabolic system. Essentially, the choices of starch over sugars and amino acids over nitrate storage lead to a more efficient metabolic network. For example, it is energetically less costly to mobilize a plastidic starch store than a vacuolar Suc store. Leaf starch breakdown initially occurs hydrolytically in the plastid to generate maltose, which is then metabolized to hexose phosphate by a combination of the cytosolic enzymes glucosyltransferase, hexokinase, and α-glucan phosphorylase (Zeeman et al., 2004). The latter enzyme uses inorganic phosphate rather than ATP as the phosphoryl donor and hence, saves an ATP compared with the breakdown of vacuolar Suc by invertase and phosphorylation by hexokinases. The choice of starch over carboxylic acids is again driven by the efficiency driver of the objective function: carboxylic acids are more oxidized than starch; hence, the energy stored per carbon is less than for starch. The model also correctly predicted that nitrate assimilation would occur only during the day because it is overall more efficient (lower sum of fluxes) to store nitrate that is imported into the leaf at night and release it from the vacuole (along with appropriate quantities of citrate) during the day for assimilation into amino acids, consistent with experimental measurements (Stitt et al., 2002).
However, it should be noted that the behavior of all these models is highly dependent on the accuracy of the reaction list and the nature of the constraints applied. The choice of objective function will also affect the flux solution, although we have previously shown that the choice of objective function is less important than the constraints applied to an FBA model of heterotrophic metabolism (Cheung et al., 2013). Similarly, for this model, the use of other objective functions (e.g. minimization of photon use) did not change the main conclusions about the operation of the network through the diel cycle. Having established that the diel modeling framework can capture realistic aspects of leaf metabolism through the day-night cycle, it can be used in the future to examine how the predicted flux distribution changes in response to different metabolic scenarios, such as variations in available light for photosynthesis and the transition from sink to source leaves.
The Diel Metabolic Modeling Framework Allows Simulation and Analysis of CAM
The diel framework for FBA also allows CAM photosynthesis to be tackled for the first time to our knowledge. Only minimal changes to the constraints of the diel C3 model were required to capture the classical CAM cycle, the major change being to constrain CO2 exchange with the environment to zero during the light. This constraint forced the model to carry out net carbon fixation during the night. Even though the model was set up with free choices for the use of carboxylating enzymes and the nocturnal carbon store (from malate, fumarate, citrate, starch, and soluble sugars), the conventional CAM cycle was predicted with PEPC as the carboxylating enzyme and malate as the main temporary storage of fixed carbon in the dark. In addition to malate, a small amount of citrate was predicted to accumulate during the night, and it was used during the day in part to support nitrate assimilation and in part for conversion to malate by isocitrate lyase. In fact, some CAM plants accumulate a large amount of citrate during the night (Lüttge, 1988; Borland and Griffiths, 1989), and it has been speculated that either ATP citrate lyase or the tricarboxylic acid cycle is used for catabolism of this stored citrate (Holtum et al., 2005). ATP citrate lyase produces oxaloacetate and acetyl-CoA from citrate, but because there is no major sink for acetyl-CoA in the output constraints, this route was not chosen by the model. One possible reason for preferring isocitrate lyase over the tricarboxylic acid cycle is that the conversion of citrate to malate by the former route releases less CO2 per molecule of citrate (glyoxylate produced from isocitrate lyase is metabolized through the photorespiratory pathway, producing 0.5 molecules of 3PGA and 0.5 molecules of CO2 per molecule of citrate catabolized) than the latter route (two molecules of CO2 released per molecule of citrate catabolized by aconitase, isocitrate dehydrogenase, and 2-OG dehydrogenase). In the absence of measurements of ATP citrate lyase and isocitrate lyase in CAM plants, it is not clear whether these two enzymes are involved in the catabolism of citrate in vivo. Transcripts for both enzymes were identified in a recent sequencing study of two agave species (Gross et al., 2013), but at present, there are no quantitative data on the expression or activity levels that could be used to discriminate between the possible routes. Ultimately, detailed metabolic flux analysis would be required to examine this issue.
Analysis of the Energetics of CAM Metabolism
In the model of CAM leaf metabolism, where the model was free to choose between different decarboxylating enzymes and storage compounds, malate decarboxylation was predicted to be mainly carried out by PEPCK. This finding can be explained by the observation that the PEPCK subtypes of CAM require fewer photons, which leads to a lower total flux through the metabolic network (the objective function that is optimized in the model), than the malic enzyme subtypes (Fig. 7). In other words, considering the whole network, the PEPCK subtype is more energetically efficient. This conclusion contrasts with previous manual calculations of CAM subtype energetics that considered the stoichiometry of a much smaller metabolic network (Winter and Smith, 1996) and emphasized that the net balance of ATP and NAD(P)H consumption is a property of the whole network.
Although CAM undoubtedly represents an adaptation to arid conditions (Cushman, 2001; Silvera et al., 2010), the CAM cycle also acts as a CO2-concentrating mechanism, and reduced CO2 availability may have been the common original selection pressure that led to the evolution of CAM in both terrestrial and aquatic environments (Keeley and Rundel, 2003). The CO2 concentrating mechanism may also suppress the carbon- and energy-consuming processes of photorespiration, although it is open to debate and depends critically on the internal CO2:O2 concentration ratio. However, there is a tradeoff between the energetic investment in the CO2-concentrating mechanism in the form of the CAM cycle and the benefit of suppressing photorespiration. The modeling results suggest that the photon use of C3 and CAM leaves is similar (Fig. 7), meaning that there is little to be gained in terms of energetics in running the CAM cycle. However, as well as the running costs of the network, there is also the cost of the enzyme machinery. A consequence of suppression of Rubisco oxygenase activity is that less Rubisco protein is required to fix a given amount of carbon. Given that Rubisco contributes up to 50% of the soluble proteins and 20% to 30% of the total nitrogen in a C3 leaf (Feller et al., 2008), the predicted reduction of 22% to 32% of total Rubisco flux in CAM compared with C3 could be a significant benefit in terms of nitrogen use efficiency. However, at the same time, there are additional enzyme machinery costs for operating the CAM cycle. There are contradicting experimental observations relating to nitrogen efficiency in CAM plants (Lüttge, 2004). From the model predictions, CAM requires 12% to 43% more total flux through the metabolic network than C3. Although enzyme machinery cost per metabolic flux for each reaction will vary depending on factors, such as the size and the turnover rate of the enzyme, the total network flux can be used as a proxy for the overall enzyme machinery cost by averaging out the variation between reactions, although the extent to which this assumption is distorted by very abundant enzymes with low Kcat values, such as Rubisco, has not been tested. By balancing the benefit from the reduced Rubisco requirement with the increase in enzyme machinery costs in the rest of the metabolic network, the total nitrogen invested in enzyme machinery is likely to be similar in C3 and CAM. Thus, the modeling results suggest that energetics and nitrogen use efficiency are unlikely to have been contributory drivers for the evolution of CAM photosynthesis.
CONCLUSION
The diel FBA modeling framework not only predicted known features of leaf metabolism more accurately than a continuous light model, but it also allowed CAM photosynthesis to be modeled at a network scale. This has allowed an accurate accounting of the energetics of CAM metabolism, showing that there are unlikely to be substantial energetic benefits in CAM photosynthesis over C3, despite the potential for suppression of photorespiration because of the CO2-concentrating effect of the CAM cycle. In this diel FBA framework, it is assumed that the light and dark phases each represent pseudo-steady states, which ignores known differences in metabolic behavior that occur within each phase, particularly at the light-dark transition points, in both C3 and CAM leaves. The framework can easily be extended to account for such transitions by further subdividing each phase. However, this requires a more detailed knowledge of the input-output constraints at each time step and will lead to a more tightly constrained model, where each time step is solved as an independent or concatenated FBA problem. For this initial exploration, it was preferable to apply minimal constraints to establish whether a metabolic network optimized for efficiency (lowest overall flux) matched known configurations of C3 and CAM metabolism over a diel cycle.
MATERIALS AND METHODS
Construction of a Diel Metabolic Model
The genome-scale metabolic model of Arabidopsis (Arabidopsis thaliana) used for the analysis of heterotrophic metabolism (Cheung et al., 2013) was adapted to model leaf metabolism over a day-night cycle. The diel modeling framework was developed by dividing the day-night cycle into two phases, light and dark, with metabolism in each phase assumed to be at steady state. The model was constructed by duplicating the Arabidopsis genome-scale metabolic model, with the reactions and metabolites in each duplicate labeled _Light or _Dark before the compartmentation suffix. In addition, dummy reactions for transporting storage metabolites from one phase to another were manually added with the suffix _LightDark. The diel model is available in SBML (for systems biology markup language) format (Supplemental Data S1). FBA and flux variability analysis were implemented as previously described (Cheung et al., 2013).
Quantitative Comparison between Flux Ranges Calculated from the Diel Model and the Continuous Light Model
Flux ranges for reactions in the light were calculated using flux variability analysis for the diel model (vdielmin and vdielmax) and the continuous light model (vsinglemin and vsinglemax). Reversible reactions carrying fluxes in opposite directions in the two sets of flux ranges were defined as reactions where either vdielmin ≥ 0 and vsinglemax ≤ 0 or vsinglemin ≥ 0 and vdielmax ≤ 0. Reactions with nonoverlapping flux ranges were defined as reactions where either vdielmin > vsinglemax or vsinglemin > vdielmax. For reactions with nonoverlapping flux ranges that did not carry fluxes in opposite directions, a similarity measure was calculated as vlowmax/vhighmin, where vlowmax is the maximum flux of the reaction with the lower flux range of the pair, and vhighmin is the minimum flux of the reaction with the higher flux range. This measure varies from 0 to 1, where a value close to zero represents a large difference between the flux ranges.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Relative amino acid composition in the phloem of Arabidopsis.
Supplemental Table S2. List of reactions with nonoverlapping flux ranges between the diel model and the single steady-state model and their similarity measures.
Supplemental Table S3. Predicted fluxes for reactions in the light and the dark in a leaf with either C3 or CAM photosynthesis.
Supplemental Data S1. Diel model of Arabidopsis leaf metabolism in SBML format.
Supplementary Material
Acknowledgments
We thank J.A.C. Smith (University of Oxford) for discussions on the energetics of CAM metabolism.
Glossary
- CAM
Crassulacean acid metabolism
- ETC
electron transport chain
- FBA
flux balance analysis
- GAP
3-phosphoglycerate
- 2-OG
2-oxoglutarate
- OPPP
oxidative pentose phosphate pathway
- PEP
phosphoenolpyruvate
- PEPC
phosphoenolpyruvate carboxylase
- 3PGA
glyceraldehyde 3-P
Footnotes
This work was supported by the University of Oxford Systems Biology Doctoral Training Centre funded by the Clarendon Fund (studentship to C.Y.M.C.) and by Keble College (Sloane-Robinson award to C.Y.M.C.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Antony E, Taybi T, Courbot M, Mugford ST, Smith JAC, Borland AM. (2008) Cloning, localization and expression analysis of vacuolar sugar transporters in the CAM plant Ananas comosus (pineapple). J Exp Bot 59: 1895–1908 [DOI] [PubMed] [Google Scholar]
- Borland AM, Griffiths H. (1989) The regulation of citric acid accumulation and carbon recycling during CAM in Ananas comosus. J Exp Bot 40: 53–60 [Google Scholar]
- Boyle NR, Morgan JA. (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst Biol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CYM, Williams TCR, Poolman MG, Fell DA, Ratcliffe RG, Sweetlove LJ. (2013) A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J 75: 1050–1061 [DOI] [PubMed] [Google Scholar]
- Cushman JC. (2001) Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiol 127: 1439–1448 [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Tillett RL, Wood JA, Branco JM, Schlauch KA. (2008) Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). J Exp Bot 59: 1875–1894 [DOI] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L. (1995) Diurnal regulation of NO3− uptake in soybean plants: I. Changes in NO3− influx, efflux, and N utilization in the plant during the day/night cycle. J Exp Bot 46: 1585–1594 [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. (2010a) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. (2010b) C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol 154: 1871–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Nielsen LK. (2011) AlgaGEM: a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genomics (Suppl 4) 12: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller U, Anders I, Mae T. (2008) Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot 59: 1615–1624 [DOI] [PubMed] [Google Scholar]
- Gauthier PPG, Bligny R, Gout E, Mahé A, Nogués S, Hodges M, Tcherkez GGB. (2010) In folio isotopic tracing demonstrates that nitrogen assimilation into glutamate is mostly independent from current CO2 assimilation in illuminated leaves of Brassica napus. New Phytol 185: 988–999 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. (2004) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gross SM, Martin JA, Simpson J, Abraham-Juarez MJ, Wang Z, Visel A. (2013) De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics 14: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S, Pierce J. (2006) A unified theory for the basis of the limitations of the primary reaction of photosynthetic CO2 fixation: was Dr. Pangloss right? Proc Natl Acad Sci USA 103: 7203–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J, Schwender J. (2011) Computational analysis of storage synthesis in developing Brassica napus L. (oilseed rape) embryos: flux variability analysis in relation to ¹³C metabolic flux analysis. Plant J 67: 513–525 [DOI] [PubMed] [Google Scholar]
- Holtum JAM, Smith JAC, Neuhaus HE. (2005) Intracellular transport and pathways of carbon flow in plants with crassulacean acid metabolism. Funct Plant Biol 32: 429–449 [DOI] [PubMed] [Google Scholar]
- Josse EM, Simkin AJ, Gaffé J, Labouré AM, Kuntz M, Carol P. (2000) A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley JE, Rundel PW. (2003) Evolution of CAM and C4 carbon-concentrating mechanisms. Int J Plant Sci 164: S55–S77 [Google Scholar]
- Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R. (2013) Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol 9: e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H, Zilliges Y, Lockau W, Steuer R. (2010) The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol 154: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Evans JR. (2011) The importance of energy balance in improving photosynthetic productivity. Plant Physiol 155: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U. (1988) Day-night changes of citric-acid levels in crassulacean acid metabolism: phenomenon and ecophysiological significance. Plant Cell Environ 11: 445–451 [Google Scholar]
- Lüttge U. (2004) Ecophysiology of crassulacean acid metabolism (CAM). Ann Bot (Lond) 93: 629–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U. (2011) Photorespiration in phase III of crassulacean acid metabolism: evolutionary and ecophysiological implications. Prog Bot 72: 371–384 [Google Scholar]
- Macduff JH, Bakken AK. (2003) Diurnal variation in uptake and xylem contents of inorganic and assimilated N under continuous and interrupted N supply to Phleum pratense and Festuca pratensis. J Exp Bot 54: 431–444 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Weber AP. (2013) Engineering photosynthesis in plants and synthetic microorganisms. J Exp Bot 64: 743–751 [DOI] [PubMed] [Google Scholar]
- McRae SR, Christopher JT, Smith JAC, Holtum JAM. (2002) Sucrose transport across the vacuolar membrane of Ananas comosus. Funct Plant Biol 29: 717–724 [DOI] [PubMed] [Google Scholar]
- Montagud A, Navarro E, Fernández de Córdoba P, Urchueguía JF, Patil KR. (2010) Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium. BMC Syst Biol 4: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Schulte N. (1996) Starch degradation in chloroplasts isolated from C3 or CAM (crassulacean acid metabolism)-induced Mesembryanthemum crystallinum L. Biochem J 318: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Krivitzky M, Huber SC, Lecharny A. (1990) Sucrose synthase in developing maize leaves: regulation of activity by protein level during the import to export transition. Plant Physiol 94: 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. (2012) Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA 109: 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman MG, Kundu S, Shaw R, Fell DA. (2013) Responses to light intensity in a genome-scale model of rice metabolism. Plant Physiol 162: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Suthers PF, Maranas CD. (2011) Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Verseput AT, Berla BM, Mueller TJ, Pakrasi HB, Maranas CD. (2012) Reconstruction and comparison of the metabolic potential of cyanobacteria Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. PLoS ONE 7: e48285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Krapp A, Stitt M. (2000) Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ 23: 1155–1167 [Google Scholar]
- Siebrecht S, Herdel K, Schurr U, Tischner R. (2003) Nutrient translocation in the xylem of poplar: diurnal variations and spatial distribution along the shoot axis. Planta 217: 783–793 [DOI] [PubMed] [Google Scholar]
- Silvera K, Neubig KM, Whitten WM, Williams NH, Winter K, Cushman JC. (2010) Evolution along the crassulacean acid metabolism continuum. Funct Plant Biol 37: 995–1010 [Google Scholar]
- Smith JAC, Bryce JH. (1992) Metabolite compartmentation and transport in CAM plants. In Tobin AK, ed, Plant Organelles. Cambridge University Press, Cambridge, UK, pp 141–167 [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A. (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Ratcliffe RG. (2011) Flux-balance modeling of plant metabolism. Front Plant Sci 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecowka M, Heise R, Tohge T, Nunes-Nesi A, Vosloh D, Huege J, Feil R, Lunn J, Nikoloski Z, Stitt M, et al. (2013) Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25: 694–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G, Mahé A, Gauthier P, Mauve C, Gout E, Bligny R, Cornic G, Hodges M. (2009) In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol 151: 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Méndez A, Miernyk JA, Randall DD. (2003) Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem 270: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Weise SE, van Wijk KJ, Sharkey TD. (2011) The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot 62: 3109–3118 [DOI] [PubMed] [Google Scholar]
- Wilkinson TL, Douglas AE. (2003) Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol Exp Appl 106: 103–113 [Google Scholar]
- Winter K, Smith JACS. (1996) Crassulacean acid metabolism: current status and perspectives. In Winter K, Smith JACS, eds, Crassulacean Acid Metabolism: Biochemistry, Ecophysiology and Evolution. Springer-Verlag, Berlin, pp 389–426 [Google Scholar]
- Yamamoto H, Peng LW, Fukao Y, Shikanai T. (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XY, Tan DKY, Inderwildi OR, Smith JAC, King DA. (2011) Life cycle energy and greenhouse gas analysis for agave-derived bioethanol. Energy Environ Sci 4: 3110–3121 [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. (2004) The breakdown of starch in leaves. New Phytol 163: 247–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.