Proteins accumulating in the apoplast of Arabidopsis during the emission of systemic immune signals include an aspartyl protease that dampens systemic acquired resistance and a legume lectin-like protein that promotes it.
Abstract
Systemic acquired resistance (SAR) is an inducible immune response that depends on ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1). Here, we show that Arabidopsis (Arabidopsis thaliana) EDS1 is required for both SAR signal generation in primary infected leaves and SAR signal perception in systemic uninfected tissues. In contrast to SAR signal generation, local resistance remains intact in eds1 mutant plants in response to Pseudomonas syringae delivering the effector protein AvrRpm1. We utilized the SAR-specific phenotype of the eds1 mutant to identify new SAR regulatory proteins in plants conditionally expressing AvrRpm1. Comparative proteomic analysis of apoplast-enriched extracts from AvrRpm1-expressing wild-type and eds1 mutant plants led to the identification of 12 APOPLASTIC, EDS1-DEPENDENT (AED) proteins. The genes encoding AED1, a predicted aspartyl protease, and another AED, LEGUME LECTIN-LIKE PROTEIN1 (LLP1), were induced locally and systemically during SAR signaling and locally by salicylic acid (SA) or its functional analog, benzo 1,2,3-thiadiazole-7-carbothioic acid S-methyl ester. Because conditional overaccumulation of AED1-hemagglutinin inhibited SA-induced resistance and SAR but not local resistance, the data suggest that AED1 is part of a homeostatic feedback mechanism regulating systemic immunity. In llp1 mutant plants, SAR was compromised, whereas the local resistance that is normally associated with EDS1 and SA as well as responses to exogenous SA appeared largely unaffected. Together, these data indicate that LLP1 promotes systemic rather than local immunity, possibly in parallel with SA. Our analysis reveals new positive and negative components of SAR and reinforces the notion that SAR represents a distinct phase of plant immunity beyond local resistance.
Systemic acquired resistance (SAR) is a form of induced resistance that is triggered in the systemic uninfected parts of plants after a local pathogen infection (Fu and Dong, 2013). The establishment of the systemic defense response is dependent on salicylic acid (SA), although SA itself is not the mobile signal responsible for SAR (Vernooij et al., 1994; Vlot et al., 2009). Over the past 10 years, a number of long-distance signals have been proposed to mediate systemic immune signaling. These signals include methyl salicylate, azelaic acid, glycerol-3-phosphate, dehydroabietinal, and pipecolic acid (Park et al., 2007; Jung et al., 2009; Chanda et al., 2011; Chaturvedi et al., 2012; Návarová et al., 2012). SAR signal generation has also been associated with actions of an extracellular aspartyl protease (Xia et al., 2004; Prasad et al., 2009), whereas SAR signal perception requires an intact cuticle in the systemic uninfected tissue (Xia et al., 2009, 2010). Together with the effects of environmental cues, such as light, on SAR (Zeier et al., 2004; Griebel and Zeier, 2008; Liu et al., 2011a), it is becoming clear that multiple signaling pathways converge to regulate SAR.
Recent findings provide further insight into defense-associated long-distance signaling, including the cooperation between signaling molecules mediating SAR (for review, see Dempsey and Klessig, 2012; Spoel and Dong, 2012; Kachroo and Robin, 2013; Shah and Zeier, 2013). Upon infection, SA is induced and converted to methyl salicylate (MeSA), which is believed to move to the systemic tissue, where its conversion back to SA is essential for SAR (Park et al., 2007). The putative lipid transfer protein DEFECTIVE IN INDUCED RESISTANCE1 (DIR1) promotes SAR, possibly in part by directing the MeSA-SA equilibrium toward SA in systemic uninfected tissues (Maldonado et al., 2002; Liu et al., 2011b). Additionally, DIR1 contributes to a positive feedback loop promoting SAR together with another lipid transfer protein, AZELAIC ACID INDUCED1 (AZI1), and glycerol-3-phosphate (Yu et al., 2013). Azelaic acid acts upstream of DIR1/AZI1/glycerol-3-phosphate to activate SAR (Yu et al., 2013) and acts partly redundantly with dehydroabietinal, a mobile SAR signal that is active at picomolar concentrations (Chaturvedi et al., 2012). The SAR defects of Arabidopsis (Arabidopsis thaliana) mutants, in which the accumulation of MeSA or glycerol-3-phosphate is compromised, appear to be conditional and dependent on the length of light exposure of the plants after a primary SAR-inducing infection (Liu et al., 2011a). A similar effect of light was observed on the SAR-defective phenotype of dir1 mutant plants, suggesting that MeSA, glycerol-3-phosphate, and DIR1 are essential for SAR only if plants receive a limited light exposure after infection (Liu et al., 2011a). Alternatively, SAR in the dir1 mutant can be supported by the DIR1 homolog DIR1-like (Champigny et al., 2013). Finally, accumulation of the nonprotein amino acid pipecolic acid in systemic uninfected tissues is essential for SAR (Návarová et al., 2012). Pipecolic acid potentiates systemic SA-mediated defenses via FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1), which is also required for signaling downstream of azelaic acid and dehydroabietinal and essential for SAR irrespective of the length of light exposure of the plants after infection (Mishina and Zeier, 2006; Liu et al., 2011a; Chaturvedi et al., 2012; Shah and Zeier, 2013).
SAR is induced in response to local infections that trigger SA signaling and is effective in systemic tissues at protecting against attack by a broad range of pathogens that are normally sensitive to SA-mediated defense (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013; Fu and Dong, 2013; Henry et al., 2013; Kachroo and Robin, 2013). A central regulator of SA signaling is encoded by ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1; Falk et al., 1999; Wiermer et al., 2005; Straus et al., 2010). EDS1 contains a noncatalytic N-terminal lipase-like domain with a classical α/β-hydrolase fold connected to an α-helical bundle C-terminal domain, and both domains are critical for EDS1 immune signaling functions (Wagner et al., 2013). These functions include promoting SA-mediated basal resistance, a low-level postinfection response that impedes the growth of virulent pathogens, and SAR (Falk et al., 1999; Wiermer et al., 2005; Truman et al., 2007; Vlot et al., 2009; Rietz et al., 2011). Additionally, EDS1 regulates SA-independent defense signaling in one or more pathways acting redundantly with SA (Bartsch et al., 2006; Venugopal et al., 2009; Roberts et al., 2013). FMO1, for example, acts upstream of SA in systemic uninfected tissues (Mishina and Zeier, 2006; Shah and Zeier, 2013) and is associated with EDS1-dependent local responses that are genetically separable from SA (Bartsch et al., 2006). Thus, EDS1 directs both the SA-dependent and SA-independent branches of basal resistance and SAR.
EDS1 also promotes effector-triggered immunity (ETI) mediated by a subclass of plant nucleotide-binding/leucine-rich repeat (NLR) receptors that have N-terminal Toll-Interleukin1 Receptor-like homology (referred to as TIR-NLRs) and at least two NLRs that contain an N-terminal coiled-coil domain (referred to as CC-NLRs; Aarts et al., 1998; Zhu et al., 2011; Roberts et al., 2013). ETI is induced by the direct or indirect recognition of pathogen effectors by NLR receptors and is a robust defense response often culminating in localized programmed cell death at infection foci known as a hypersensitive response (HR; Jones and Dangl, 2006; Maekawa et al., 2011; Bonardi and Dangl, 2012; Spoel and Dong, 2012). Arabidopsis EDS1 signals with two sequence-related proteins, PHYTOALEXIN DEFICIENT4 (PAD4) and SENESCENCE-ASSOCIATED GENE101 (SAG101), by forming separate nucleocytoplasmic and nuclear complexes, respectively, with each partner (Feys et al., 2001, 2005; Rietz et al., 2011; Wagner et al., 2013). Also, Arabidopsis EDS1 was found to reside in nuclear complexes with the TIR-NLR receptors RESISTANT TO PSEUDOMONAS SYRINGAE4 (RSP4), RPS6, and VARIATION IN COMPOUND TRIGGERED ROOT growth response, the RPS4- and RPS6-recognized effectors AvrRps4 and HopA1, as well as the CC-NLR HYPERSENSITIVE RESPONSE TO Turnip crinkle virus (HRT), recognizing turnip crinkle virus coat protein (Bhattacharjee et al., 2011; Heidrich et al., 2011; Zhu et al., 2011; Kim et al., 2012). EDS1 shuttles between the cytoplasm and nucleus through the nuclear pore machinery, and the accumulation of EDS1 inside nuclei is necessary for both TIR-NLR-mediated ETI and basal resistance and their associated transcriptional reprogramming of defense pathways (García et al., 2010; Heidrich et al., 2011). Therefore, direct or indirect interactions between EDS1 and NLR receptors have been proposed to connect effector recognition to nuclear transcriptional outputs and pathogen resistance (Bhattacharjee et al., 2011; Heidrich et al., 2011; Sohn et al., 2012).
In Arabidopsis, ETI mediated by the CC-NLR RESISTANCE TO PSEUDOMONAS SYRINGAE pathovar MACULICOLA1 (RPM1) is triggered by Pseudomonas syringae delivering the effector protein AvrRpm1 (Dangl et al., 1992). Although ETI conferred by RPM1 and another CC-NLR (RPS2) does not genetically require EDS1 (Aarts et al., 1998), SAR induced by both CC-NLRs was abolished in the eds1 mutant (Truman et al., 2007; Rietz et al., 2011). Similarly, pad4 mutant plants were SAR defective in response to a local activation of RPM1 or RPS2 (Jing et al., 2011; Rietz et al., 2011). In this work, we show that EDS1 contributes to Arabidopsis SAR signaling in both the primary infected and systemic uninfected tissues. We utilize the SAR defect in eds1 mutant plants in response to the P. syringae effector AvrRpm1 to investigate systemic rather than local defense responses, focusing on EDS1-dependent changes in the leaf apoplast. Previous studies have shown that Arabidopsis protein secretion is highly regulated during P. syringae-induced defense responses, including SAR, with effectors such as AvrRpm1 likely directly affecting protein secretion (Hauck et al., 2003; Wang et al., 2005; Kaffarnik et al., 2009). Arabidopsis is an apoplastic phloem loader (Gottwald et al., 2000), and the phloem is believed to be the main conduit for SAR signal transmission (Guedes et al., 1980; Gaupels and Vlot, 2013). Therefore, we relate SAR to proteins differentially accumulating in the apoplast of wild-type and eds1 leaves in response to AvrRpm1. Among 12 SAR-associated apoplastic proteins, we identify contrasting roles of a predicted aspartyl protease that appears to suppress SAR and a legume lectin-like protein that promotes SAR without affecting local resistance to virulent or avirulent P. syringae. In this work, we show that there are mechanistically distinct phases in local and systemic immunity.
RESULTS
EDS1 Is Necessary for SAR Signal Generation and Perception
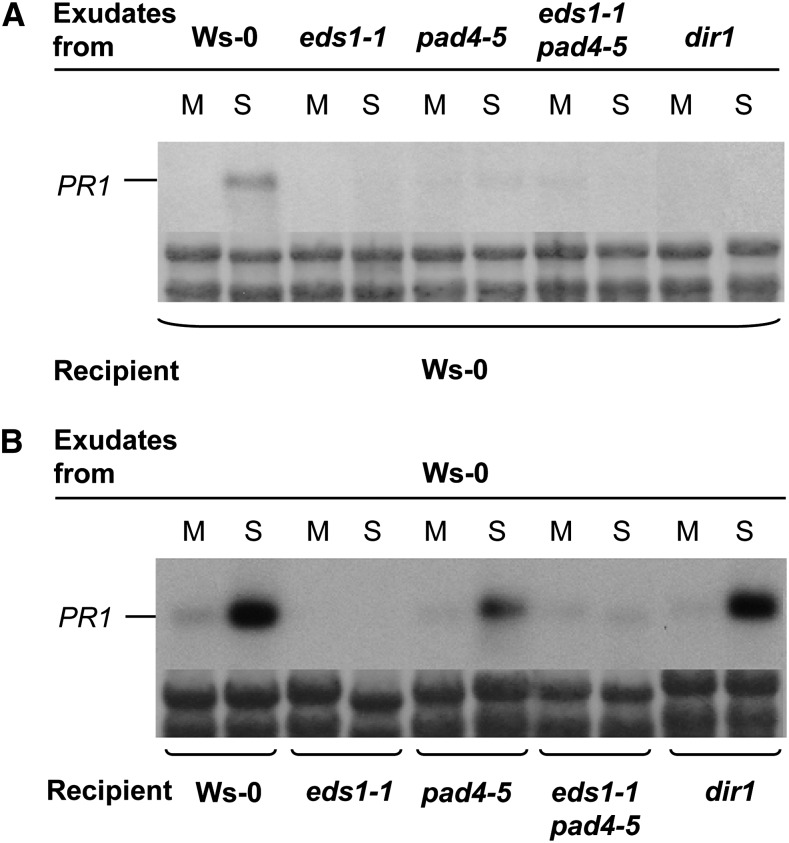
The SAR-specific defect in Arabidopsis eds1 null mutant plants in response to a primary infection with P. syringae pv tomato strain DC3000 (Pst) expressing AvrRpm1 (Pst/AvrRpm1; Aarts et al., 1998; Truman et al., 2007) may be due to compromised responses in the locally infected tissue, the systemic untreated tissue, or both. To distinguish between these possibilities, we tested for the presence of defense-inducing signals in petiole exudates from leaves infected with Pst/AvrRpm1. In accordance with the terminology of previous studies (Chaturvedi et al., 2008), these petiole exudates will be referred to as AvrPEX. AvrPEX were collected from Wassilewskija-0 wild-type, eds1-1 and pad4-5 single mutant, and eds1-1pad4-5 double mutant plants. As a control, AvrPEX from the dir1 mutant were included (Maldonado et al., 2002). The presence of defense-inducing signals in the AvrPEX was investigated upon infiltration of the AvrPEX into the leaves of previously untreated plants using the induction of the SAR marker gene PATHOGENESIS RELATED1 (PR1) as a marker for defense induction (Fig. 1).
Figure 1.
EDS1 is required for SAR signal generation (A) and perception (B). PR1 transcript accumulation is shown 24 h after infiltration of leaves with petiole exudates collected from mock-treated (M; 10 mm MgCl2) or Pst/AvrRpm1-infected (S) leaves. The exudates were collected from the genotypes indicated above each RNA blot and infiltrated into the recipient plants indicated below each RNA blot. PR1 transcript accumulation was analyzed on northern blots, and equal loading was controlled by on-blot Methylene Blue staining of ribosomal RNA (shown below each RNA blot). This experiment was repeated three times with similar results.
AvrPEX from wild-type plants induced PR1 expression in wild-type recipient plants (Fig. 1). As observed previously (Maldonado et al., 2002), AvrPEX from dir1 failed to induce PR1 expression in wild-type recipient plants (Fig. 1A). Similarly, AvrPEX from eds1-1, pad4-5, or eds1-1pad4-5 mutants did not induce PR1 expression in wild-type recipients. Thus, similar to DIR1, EDS1 and its signaling partner PAD4 are required for SAR signal generation in primary infected leaves. Reciprocally, AvrPEX from wild-type plants induced PR1 expression in the pad4-5 mutant and, as expected, the dir1 mutant control (Fig. 1B; Maldonado et al., 2002). In contrast, wild-type AvrPEX did not induce PR1 expression in eds1-1 recipient plants or in the eds1-1pad4-5 double mutant (Fig. 1B). Thus, unlike DIR1, whose activity is required only in the primary infected tissue, EDS1 acts in both the primary infected and systemic uninfected tissues during SAR signal propagation.
Conditional Expression of AvrRpm1 Triggers EDS1-Dependent Systemic Immunity
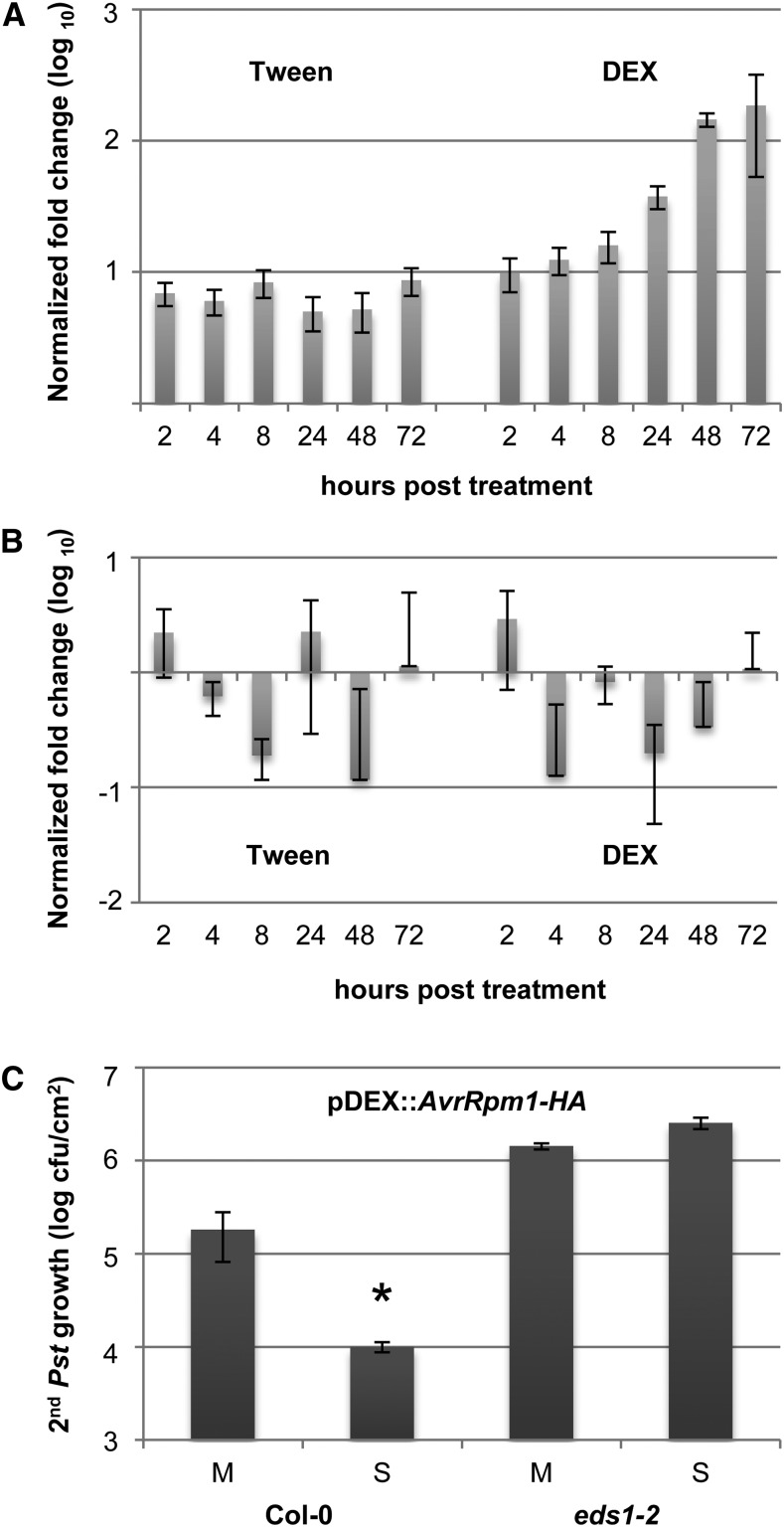
In screening for plant-derived SAR components, we sought to synchronize SAR induction as much as possible and to limit the presence of pathogen-derived proteins in the plants. Therefore, we used transgenic plants carrying the dexamethasone (DEX)-inducible transgene pDEX:AvrRpm1-HA that encodes C-terminally hemagglutinin (HA)-tagged AvrRpm1 (Mackey et al., 2002). Before employing these transgenic plants for the identification of SAR-associated proteins, we assessed whether local DEX application, which induces the expression of AvrRpm1-HA, also effectively triggers SAR. The application of 30 µm DEX with a paintbrush to the first two true leaves of pDEX:AvrRpm1-HA plants resulted in the appearance of HR lesions on the systemic untreated leaves, presumably caused by DEX traveling from the treated tissues. Therefore, we monitored the expression of the transgene in local treated and systemic untreated leaves after the application of a much lower concentration of 1 µm DEX in 0.01% (v/v) Tween 20 using 0.01% (v/v) Tween 20 as a negative control. The Tween 20-treated leaves reproducibly displayed very low AvrRpm1-HA expression, as measured by quantitative reverse transcription (qRT)-PCR (Fig. 2A; Supplemental Fig. S1A). In contrast, the DEX application induced local AvrRpm1-HA expression to more than 100-fold over 48 to 72 h (Fig. 2A; Supplemental Fig. S1A), which is 2- to 8-fold higher than bacterial AvrRpm1 transcript accumulation in Columbia-0 (Col-0) leaves 3 d after infiltration with 106 colony-forming units (cfu) mL−1 Pst/AvrRpm1 (Supplemental Fig. S1B). Induced AvrRpm1-HA expression was not observed in systemic untreated leaves of the DEX-treated pDEX:AvrRpm1-HA plants (Fig. 2B; Supplemental Fig. S1C), suggesting that the 1 µm DEX application remained local.
Figure 2.
Pathogen-free, effector-triggered systemic immunity. A and B, AvrRpm1-HA transcript accumulation was normalized to TUBULIN in Col-0 pDEX:AvrRpm1-HA plants treated with 0.01% (v/v) Tween 20 or 1 µm DEX. Transcript accumulation is shown in the treated (A) and systemic untreated (B) leaves relative to the transcript levels in the same leaves from untreated plants. C, Pst titers are shown 4 d after a secondary infection of pDEX:AvrRpm1-HA plants that was systemic to primary treatments with 0.01% (v/v) Tween 20 (mock; M) or 1 µm DEX (SAR; S). The asterisk indicates a significant difference from the mock-treated plants (P < 0.05, Student’s t test). This experiment was repeated three times with similar results.
We subsequently tested if the local application of 1 µm DEX in 0.01% (v/v) Tween 20 triggers systemic resistance in Col-0 pDEX:AvrRpm1-HA or eds1-2 pDEX:AvrRpm1-HA plants. The first two true leaves per pDEX:AvrRpm1-HA plant were treated with DEX or Tween 20, and the systemic untreated leaves were inoculated 3 d later with virulent Pst. The Pst titers in the secondary infected leaves were determined 4 d later, showing that the primary DEX treatment of Col-0 pDEX:AvrRpm1-HA but not eds1-2 pDEX:AvrRpm1-HA plants induced systemic resistance, resulting in reduced Pst growth in DEX-treated compared with Tween-treated plants (Fig. 2C). Because local DEX treatment of nontransgenic Col-0 plants did not trigger a systemic resistance response to Pst (Supplemental Fig. S2), the systemic resistance in the DEX-treated pDEX:AvrRpm1-HA plants was a consequence of local AvrRpm1-HA expression. Together, these data show that AvrRpm1-HA expression triggers EDS1-dependent systemic immunity.
Identification of Potential SAR Regulatory Proteins
Because EDS1 is required for SAR signal generation (Fig. 1) but not local resistance in response to Pst/AvrRpm1 (Aarts et al., 1998), we hypothesized that any proteins accumulating in an EDS1-dependent manner during AvrRpm1-HA-triggered responses may be related specifically to SAR. Proteomics experiments are often disturbed by highly abundant cellular proteins, such as Rubisco, that mask low-abundance proteins of interest. Therefore, we enriched the plant extracts for apoplastic proteins prior to proteomic analysis to both reduce the Rubisco content of the extracts and focus on the secreted proteins potentially related to SAR (see introduction; Wang et al., 2005). For protein isolation, we sprayed lawns of 3- to 4-week-old pDEX:AvrRpm1-HA plants with 30 µm DEX in 0.01% (v/v) Tween 20, which did not confine the expression of the transgene to a specific location within the plant but instead maximized the leaf response to AvrRpm1-HA. Pilot experiments showed that the leaves treated with 1 µm DEX did not wilt and needed to remain on the plant for 4 to 6 h before removal without loss of systemic resistance to virulent Pst bacteria (Supplemental Fig. S3). Because the leaves treated with 30 µm DEX became severely wilted at 6 h after treatment, we harvested the aboveground tissue for protein isolation 4.5 to 5 h after spraying the plants with 30 µm DEX.
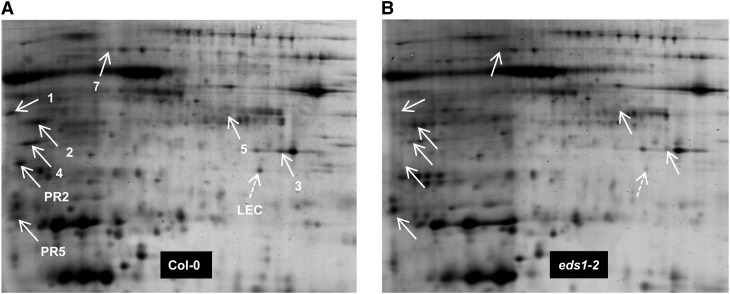
The protein accumulation in the apoplast-enriched extracts from AvrRpm1-HA-expressing wild-type and eds1 mutant plants was compared using two-dimensional (2D) PAGE (Fig. 3; Supplemental Fig. S4A). Infection of Arabidopsis cultured cells with Pst/AvrRpm1 induces the secretion of host proteins that do not carry an N-terminal secretion signal peptide (Kaffarnik et al., 2009). Nevertheless, proteins were selected as apoplastic only if they possessed a predicted signal peptide (determined by SignalP 4.1; Petersen et al., 2011), because the apoplast-enriched extracts from Arabidopsis contained a high proportion of cytosolic proteins due to cell membrane leakage during the extraction (Supplemental Table S1). Of the EDS1-dependent protein spots that were detected on the 2D gels, eight apoplastic proteins containing N-terminal signal peptides were identified after in-gel trypsin digestion of the spots followed by matrix-assisted laser desorption/ionization (MALDI)-mass spectrometry (MS; Fig. 3; Table I; Supplemental Fig. S4). This experiment was repeated four times with four biologically independent sample sets. Because not all of the differentially accumulating protein spots were trypsin digested and sequenced by MALDI-MS in every repetition, the reproducibility with which AED proteins were found is summarized in Supplemental Figure S4B. Four of the AED proteins were previously characterized as ASPARTIC PROTEASE IN GUARD CELL1 (ASPG1; Yao et al., 2012), β-d-XYLOSIDASE4 (XYL4; Minic et al., 2004), and PR2 and PR5 (van Loon et al., 2006). Similar but not identical to ASPG1, AED1 and AED3 are predicted aspartyl proteases (Simões and Faro, 2004; Faro and Gal, 2005). AED4 and AED5 are GDSL-motif lipases (Akoh et al., 2004).
Figure 3.
2D gel analysis of apoplast-enriched extracts from AvrRpm1-HA-expressing Col-0 (A) and eds1-2 mutant (B) plants. Isoelectric focusing was performed along the horizontal axis across a pI range from 4 to 7, and proteins were resolved according to their mass along the vertical axis on 12% (w/v) polyacrylamide gels. White arrows indicate the positions of AED proteins. A putative lectin encoded by At3g16530 was identified on this gel set only and is marked with the dashed arrows (LEC). Numbers correspond to the number of each AED protein in Table I. This experiment was repeated multiple times (for reproducibility per spot, see Supplemental Fig. S4).
Table I. Summary of AED proteins identified by 2D gel or ICPL analyses with their ICPL quantitation and significance as well as possible biological functions (Gene Ontology analysis; www.arabidopsis.org)ND, Not determined.
| Gene | Locus | Protein (Predicted Size) | ICPL Ratio, Wild Type to eds1-2 | P (ICPL)a | Biological Process |
|---|---|---|---|---|---|
| AED1 | At5g10760 | Eukaryotic aspartyl protease family protein (49.4 kD) | ND | ND | Response to stress/response to abiotic or biotic stimulus/protein metabolism/signal transduction/other cellular processes |
| ASPG1, AED2 | At3g18490 | Aspartic protease in guard cell1 (53.2 kD) | 1.39 | 0.20 | Response to stress/response to abiotic or biotic stimulus/protein metabolism |
| AED3 | At1g09750 | Eukaryotic aspartyl protease family protein (47.6 kD) | 1.22 | 0.45 | Protein metabolism |
| AED4 | At1g29660 | GDSL-motif lipase (40.1 kD) | 1.45 | 0.15 | Response to stress/response to abiotic or biotic stimulus/transport |
| AED5 | At3g05180 | GDSL-motif lipase (42.3 kD) | ND | ND | Other metabolic processes |
| XYL4, AED7 | At5g64570 | β-Xylosidase (84.3 kD) | 1.15 | 0.61 | Other metabolic and cellular processes |
| PR2 | At3g57260 | Pathogenesis-related protein2 (37.3 kD) | 5.62 | 1.5E-11 | Response to stress/response to abiotic or biotic stimulus/signal transduction/other cellular and metabolic processes |
| PR5 | At1g75040 | Pathogenesis-related protein5 (25.2 kD) | 2.9 | 3.2E-05 | Response to stress/response to abiotic or biotic stimulus/signal transduction/other cellular and metabolic processes |
| LLP1, AED9 | At5g03350 | Legume lectin family protein (30.1 kD) | 2.0 | 0.007 | Unknown |
| PNP-A, AED14 | At2g18660 | Plant natriuretic peptide A (14.5 kD) | 2.65 | 0.0001 | Response to stress/response to abiotic or biotic stimulus/electron transport or energy pathways/other metabolic and cellular processes |
| AED15 | At2g43570 | Putative chitinase (29.7 kD) | 2.34 | 0.0009 | Other metabolic and cellular processes |
| EP1, AED19 | At4g23170 | Cys-rich receptor-like kinase (29.7 kD) | 1.96 | 0.009 | Other cellular processes |
According to the Perseus statistical tool (www.perseus-framework.org; Cox and Mann, 2011).
To quantify the relative differences in protein accumulation between the AvrRpm1-HA-induced wild-type and eds1 mutant plants, proteins in the different extracts were analyzed by isotope-coded protein labeling (ICPL). To this end, the proteins were chemically labeled with mass tags containing different isotopic substitutions and mixed prior to liquid chromatography-tandem mass spectrometry (MS/MS)-based protein identification, thus allowing the immediate and relative quantification of the proteins in extracts from wild-type and eds1 plants. Differentially labeled standards were added as internal controls (the quantitation results are shown in Supplemental Fig. S5A). Across three biologically independent replicates, 758 proteins were identified in the apoplast-enriched extracts, 609 of which could be quantified (Supplemental Table S1). As expected, the log normal distribution of quantitation followed a Gaussian distribution (Supplemental Fig. S5B). In the ICPL experiment, ASPG1 and AED4 each displayed trends toward an approximately 1.4-fold enhanced accumulation in the extracts from the wild type compared with those from eds1 mutant plants (Table I; Supplemental Table S1). The accumulation of AED3 and XYL4 was not different between wild-type and eds1 extracts (Supplemental Table S1). However, each of these proteins was found in multiple spots on the 2D gels. Notably, only a single spot per protein (i.e. one isoform or posttranslationally modified variant) displayed differential accumulation between the extracts from the wild-type and eds1 mutant plants (Supplemental Fig. S4). Finally, AED1 and AED5 were not identified in the ICPL analyses. For AED5, this result might be due to its relatively low abundance (Fig. 3A). AED1, however, appears to be a quite abundant protein in the wild-type extracts (Fig. 3A). The amino acid sequence and the theoretical peptide masses after an in silico trypsin digestion of AED1 did not indicate why this protein was not identified by liquid chromatography-MS/MS.
In the ICPL experiment, six AED proteins displaying a statistically significant EDS1-dependent accumulation were detected in all three independent biological replicates and contained predicted N-terminal secretion signal peptides (Table I; Supplemental Table S1). These AEDs included PR2, PR5, the putative PR protein PLANT NATRIURETIC PEPTIDE A (PNP-A; Meier et al., 2008), the predicted chitinase AED15, a receptor-like kinase that is encoded by the SA-responsive EP1 gene (Blanco et al., 2005), and the SA-inducible LEGUME LECTIN-LIKE PROTEIN SAi-LLP1 (LLP1; Krinke et al., 2007; Blanco et al., 2009; Armijo et al., 2013). LLP1 is 63% identical to a protein that is encoded by locus At3g16530 and was detected on one 2D gel set (Fig. 3). The LLP1-like protein showed a slight trend of 1.3-fold enhanced accumulation in the extracts from the wild type compared with those from eds1 mutant plants (Supplemental Table S1). Another sequence-related protein, AtLEC (for Arabidopsis thaliana LECTIN; Lyou et al., 2009), which is 60% identical to LLP1 and 89% identical to the LLP1-like protein, did not differentially accumulate between the wild-type and eds1 extracts (Supplemental Table S1).
In summary, we assigned 12 AED proteins that (besides PR2, PR5, and PNP-A) have not previously been associated with SAR. A Gene Ontology term analysis (www.arabidopsis.org) linked six AED proteins to responses to biotic and/or abiotic stresses (Table I).
Infection Induces Local and Systemic AED Gene Expression Changes
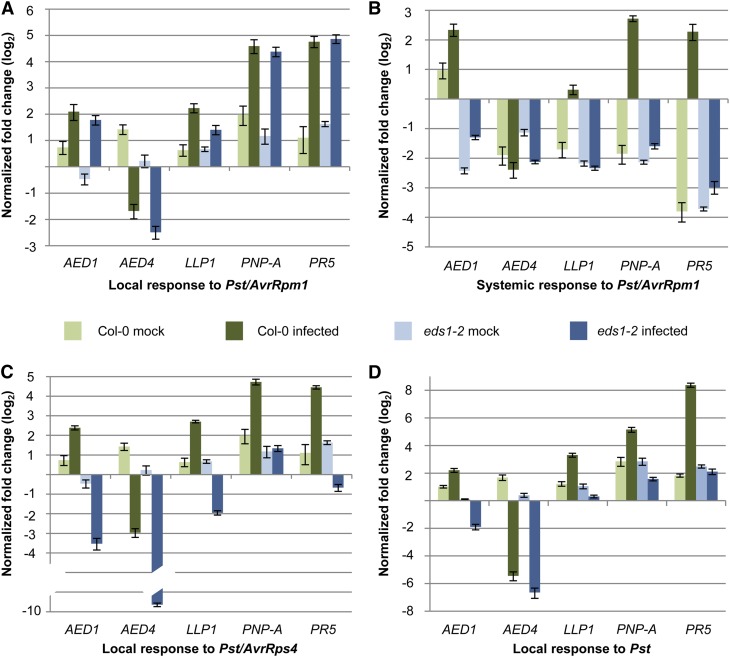
To gain further insight into the regulation of the AED genes in locally infected and systemic uninfected leaves during SAR establishment, we analyzed AED transcript accumulation using qRT-PCR in the local infected and systemic uninfected leaves of Pst/AvrRpm1-inoculated plants. The transcript accumulation of AED1, LLP1, PNP-A, and PR5 was induced by Pst/AvrRpm1 relative to the mock-treated controls both locally and systemically (Fig. 4, A and B). Their local induction was independent of EDS1 (Fig. 4A). Similarly, AED4 transcript accumulation was locally repressed by Pst/AvrRpm1 compared with the mock-treated controls, and this repression was independent of EDS1 (Fig. 4A). Systemically, a local mock treatment led to a slight induction of AED1 transcript accumulation and an apparent repression in these tissues of the other genes tested, shown in Figure 4B. Although local Pst/AvrRpm1 infection enhanced systemic AED1 transcript accumulation approximately 2-fold compared with the mock-treated controls in both the wild-type and eds1 mutant plants, the relative level of AED1 transcripts remained much lower in eds1 compared with the wild type (Fig. 4B). The systemic LLP1 transcript accumulation was 4-fold higher in the locally Pst/AvrRpm1-infected compared with the mock-treated plants, and this induction was dependent on EDS1 (Fig. 4B). Similarly, the transcript accumulation of the putative PR gene PNP-A and PR5 was induced systemically in an EDS1-dependent manner by Pst/AvrRpm1. In contrast, the systemic regulation of AED4 was not different between the local mock and Pst/AvrRpm1 treatments and did not require EDS1. Taken together, we established that the systemic but not local transcript accumulation of AED1 and the transcriptional regulation of LLP1, PNP-A, and PR5 in response to Pst/AvrRpm1 are dependent on EDS1 and thus likely related to SAR.
Figure 4.
AED transcript accumulation in response to avirulent and virulent Pst. Col-0 wild-type (green bars) and eds1-2 mutant (blue bars) plants were treated with 10 mm MgCl2 (mock; light colored bars) or infected (dark colored bars). Three days later, the transcript accumulation of AED1, AED4, LLP1, PNP-A, and PR5 was analyzed by qRT-PCR and normalized to UBIQUITIN in leaves infected with Pst/AvrRpm1 (A), in systemic untreated leaves of Pst/AvrRpm1-infected plants (B), in leaves infected with Pst/AvrRps4 (C), and in leaves infected with Pst (D). The transcript accumulation is shown relative to the corresponding transcript levels in leaves of untreated plants of the same age. These experiments were repeated three (AED4, LLP1, and PNP-A) or more (AED1 and PR5) times with similar results.
Similar to Pst/AvrRpm1, two other Pst strains, Pst expressing the effector AvrRps4 (Pst/AvrRps4) and virulent Pst, locally repressed AED4 transcript accumulation (Fig. 4, C and D). This repression occurred independently of EDS1 or was enhanced in the infected eds1-2 compared with wild-type plants (Fig. 4C). In contrast to an EDS1-independent induction of AED1, LLP1, PNP-A, and PR5 in leaves that were infected with Pst/AvrRpm1, these genes were locally induced by Pst/AvrRps4 or Pst in an EDS1-dependent manner (Fig. 4, C and D). Because ETI triggered by Pst/AvrRpm1 is EDS1 independent but ETI triggered by Pst/AvrRps4 and basal resistance to Pst genetically require EDS1 (Aarts et al., 1998; Falk et al., 1999; Rietz et al., 2011), the transcriptional up-regulation of the AED1, LLP1, PNP-A, and PR5 genes in infected leaves appears to correlate with the local resistance response rather than SAR.
AED1 Likely Suppresses SAR
CONSTITUTIVE DISEASE RESISTANCE1 (CDR1) encodes an apoplastic aspartyl protease that is 30% identical to AED1 at the amino acid level and is reported to contribute to SAR (Xia et al., 2004). Here, we studied SAR in two transfer DNA (T-DNA) insertion mutants of AED1. Whereas AED1 transcript accumulation was virtually undetectable in the aed1-1 knockout mutant, AED1 expression was reduced to approximately 15% compared with wild-type levels in the aed1-2 knockdown mutant, which carried a T-DNA insertion in the promoter region of AED1 (Fig. 5A). SAR was induced in these plants by infection of the first two true leaves per plant with Pst/AvrRpm1. Three days later, the next two systemic leaves were infected with virulent Pst, and the bacterial titers were determined at 4 d post infection (dpi). Wild-type plants and both aed1-1 and aed1-2 mutants preinfected with Pst/AvrRpm1 supported significantly lower Pst growth than did the mock-pretreated plants, which was indicative of an SAR response (Fig. 5B). Because the transcripts of the neighboring locus At5g10770, which is 70% homologous at the nucleotide level with AED1, were elevated in the aed1-1 and aed1-2 mutant plants compared with the wild-type (Fig. 5A), we cannot exclude overcompensation for the loss of functional AED1 in, for example, aed1-1 by At5g10770.
Figure 5.
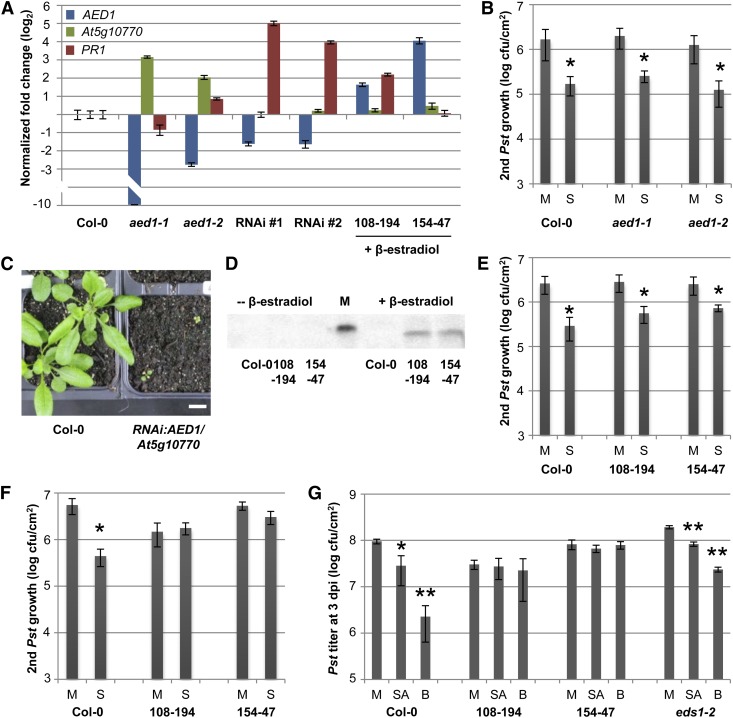
AED1 likely suppresses SAR. A, Transcript levels of AED1 (blue bars), its neighboring locus At5g10770 (green bars), and PR1 (red bars) were normalized to UBIQUITIN in the aed1-1 and aed1-2 mutant plants, in two aerial tissue pools that each contained three RNAi:AED1/At5g10770 transgenic plants (T1), and in XVE:AED1-HA lines 108-194 and 154-47 at 24 h after treatment with 30 µm β-estradiol. For each set of mutant or transgenic plants, the transcript levels are shown relative to those in Col-0 plants of the same age (one representative Col-0 sample is shown). B, SAR in aed1-1 and aed1-2. Pst titers are shown 4 d after a secondary infection that was systemic to primary treatments with 10 mm MgCl2 (mock; M) or Pst/AvrRpm1 (SAR; S). C, Five-week-old RNAi:AED1/At5g10770 transgenic plants (T1) compared with Col-0. Both genotypes were grown for the first 2 weeks on Murashige and Skoog medium without (Col-0) or with (transgenic plants) antibiotics, transferred to soil, and propagated for another 3 weeks. D, Anti-HA western blot of whole protein extracts before (left) or 24 h after (right) treatment of Col-0 plants and XVE:AED1-HA lines 108-194 and 154-47 with 30 µm β-estradiol. M, Molecular size marker; the visible band corresponds to 50 kD. E and F, SAR in XVE:AED1-HA plants. The plants were either untreated (E) or sprayed with 30 µm β-estradiol 24 h before SAR induction (F). SAR was analyzed as in B. G, SA-induced resistance in XVE:AED1-HA plants. At 24 h after the treatment of Col-0, XVE:AED1-HA lines 108-194 and 154-47, and eds1-2 plants with 30 µm β-estradiol, all of the plants were sprayed with 0.01% (v/v) Tween 20 (mock; M) or with 1 mm SA or 1 mm BTH (B) in 0.01% (v/v) Tween 20. After another 24 h, the treated leaves were infected with Pst. The in planta Pst titers are shown at 4 dpi. Asterisks (B and E–G) indicate significant differences compared with the mock-treated controls (*P < 0.05, **P < 0.005, Student’s t test). These experiments were repeated two (aed1-2 [A and B] and RNAi lines [B]) to at least three times with similar results.
To exclude any effects of possible functional redundancy between AED1 and its neighboring locus, we attempted to silence the expression of both AED1 and At5g10770 using double-stranded RNA interference (RNAi). The RNAi construct included two consecutive 400-nucleotide stretches with full complementarity to the respective transcripts and was used to transform Col-0 plants. T1 plants were cultivated on sterile medium for 2 weeks as described in “Materials and Methods” and subsequently transferred to soil. Compared with similarly treated Col-0 plants, the RNAi plants were severely stunted (Fig. 5C). With time, these plants produced a few flowers and seeds, but we were unable to retrieve viable T2 transgenic plants. Therefore, we pooled the aboveground tissue of three 5-week-old T1 plants to produce sufficient material to allow for gene expression analysis. In two biologically independent samples, the AED1 transcript accumulation was 3-fold lower in RNAi:AED1/At5g10770 plants compared with wild-type plants, while At5g10770 transcript accumulation was not reduced. In contrast to the aed1-1 and aed1-2 mutants, the RNAi plants accumulated wild-type but not elevated levels of At5g10770 transcripts (Fig. 5A). Therefore, the stunted phenotype was most likely related to the reduced AED1 expression. Notably, the transcript accumulation of the SAR marker gene PR1 was elevated in both of the RNAi samples (Fig. 5A), suggesting the hyperactivation of SAR in these plants. These data show that AED1 likely restricts SA or SAR signaling, although it is possible that the stunted phenotype of the RNAi:AED1/At5g10770 plants is due to another function of AED1.
To gain further insight into the role of AED1 during SAR, we constructed transgenic lines that conditionally overaccumulate AED1 with a C-terminal HA tag. The expression of the transgene was controlled by the β-estradiol-inducible XVE promoter, containing the DNA-binding domain of the bacterial repressor LexA (X), the acidic transactivating domain of VP16 (V) and the regulatory region of the human estrogen receptor (E; Zuo et al., 2000). In the absence of β-estradiol, two independent homozygous XVE:AED1-HA transgenic lines did not display AED1-HA protein accumulation, as detected by immunoblot analysis using anti-HA antibodies (Fig. 5D). After spraying the same plants with 100 µm β-estradiol in 0.01% Tween 20 (v/v), we detected the accumulation of a protein band corresponding to the predicted size of AED1-HA (49 kD; Fig. 5D). Compared with untreated Col-0 plants, AED1 transcript accumulation was induced 3- to 16-fold in XVE:AED1-HA plants that were treated with 30 µm β-estradiol (Fig. 5A), the concentration used to induce AED1-HA accumulation in subsequent experiments. Thus, compared with the approximately 4-fold induction of AED1 expression locally or systemically by infection of wild-type plants (Fig. 4), the treatment of the XVE:AED1-HA plants with 30 µm β-estradiol caused either a comparable (transgenic line 108-194) or an approximately 4-fold higher (transgenic line 154-47) induction of AED1 transcript accumulation, suggesting that the overaccumulation of AED1-HA likely remains within or close to physiologically relevant levels.
Without β-estradiol, both of the XVE:AED1-HA transgenic lines mounted normal SAR responses, resulting in the reduced growth of virulent Pst in the systemic leaves of preinfected compared with mock-pretreated plants (Fig. 5E). β-Estradiol is normally dissolved in ethanol. Because we observed that the spray treatment of wild-type plants with a low concentration of ethanol repressed SAR (Supplemental Fig. S6), we dissolved β-estradiol in methanol for the following experiments. A control spray treatment of plants with a low concentration of methanol did not affect SAR (Supplemental Fig. S6). Spraying the XVE:AED1-HA plants with 30 µm β-estradiol 1 d prior to the start of the experiment suppressed Pst/AvrRpm1-induced SAR against Pst (Fig. 5F; Supplemental Fig. S6), although local resistance to Pst/AvrRpm1 and Pst did not change (Supplemental Fig. S7). Because the same β-estradiol treatment did not alter SAR in Col-0 wild-type plants (Fig. 5F; Supplemental Fig. S6), we concluded that the overaccumulation of the AED1 protein leads to a suppression of SAR.
Because SAR is highly correlated with SA, we investigated the SA-induced resistance response in plants overaccumulating AED1-HA. For this, we treated plants first with 30 µm β-estradiol and 24 h later with SA or the SA functional analog benzo 1,2,3-thiadiazole-7-carbothioic acid S-methyl ester (BTH) in 0.01% (v/v) Tween 20 or with 0.01% (v/v) Tween 20 as a mock control. Twenty-four hours later, the treated leaves were inoculated with Pst, and the in planta Pst titers were determined at 4 dpi. Both SA and BTH locally enhanced resistance in Col-0 plants, resulting in reduced Pst growth in treated plants compared with the mock-treated controls (Fig. 5G). As expected, the eds1-2 plants responded to both SA and BTH with enhanced resistance to Pst (Fig. 5G; Falk et al., 1999). Notably, overaccumulation of AED1-HA rendered the XVE:AED1-HA plants nonresponsive to SA and BTH, with both independent transgenic lines supporting Pst growth to similar levels in the mock-treated, SA-treated, and BTH-treated plants (Fig. 5G). Taken together, these data suggest that AED1 acts downstream of SA to suppress systemic immunity.
LLP1 Is Essential for SAR
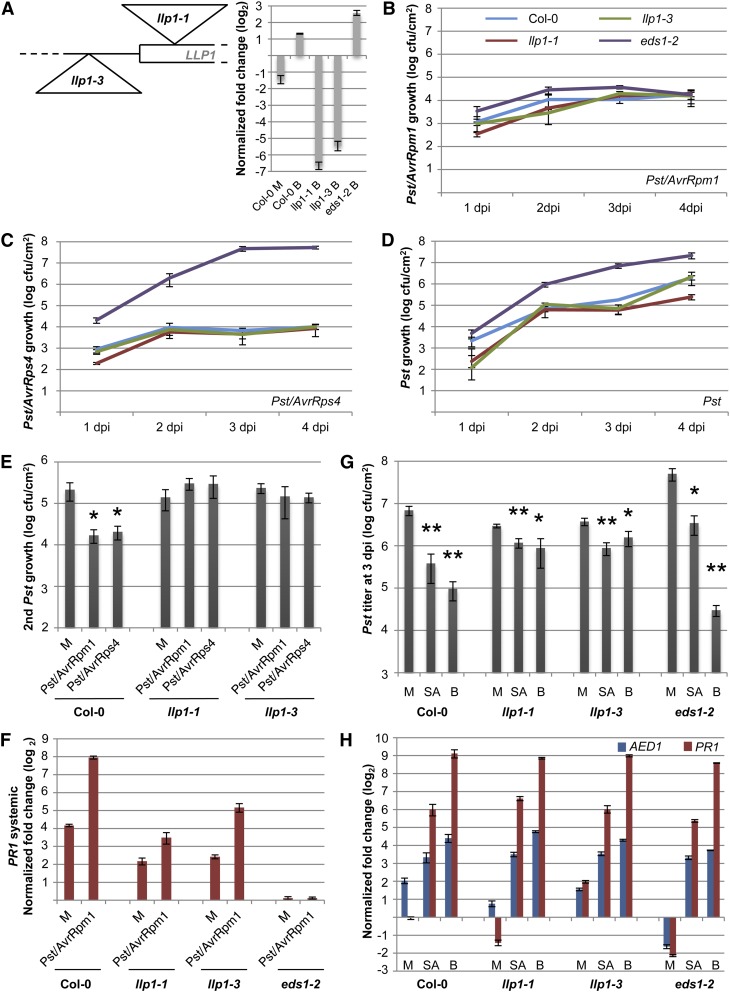
To investigate a role of the LLP1 gene in SAR, we examined the local and systemic defense responses of two Col-0 mutant lines carrying T-DNA insertions upstream or in the vicinity of the start codon of LLP1 (Fig. 6A). Both insertions strongly reduced LLP1 transcript accumulation. Because LLP1 transcript accumulation is enhanced by SA (Krinke et al., 2007; Blanco et al., 2009) and BTH (Fig. 6A), we measured LLP1 expression in BTH-treated leaves of the llp1 insertion lines. In llp1-1 and llp1-3 mutant plants, BTH elevated the LLP1 transcript accumulation to 1% and 2%, respectively, of the BTH-induced levels in wild-type plants, confirming that both are knockdown mutants (Fig. 6A). To study local resistance responses, we monitored the growth of three Pst strains in the llp1 mutants compared with the wild-type and eds1-2 control plants. The llp1-1 and llp1-3 mutants supported the growth of Pst/AvrRpm1 to comparable levels as the Col-0 and eds1-2 plants (Fig. 6B), indicating that local resistance to Pst/AvrRpm1 was not affected by mutations in LLP1. Supporting this conclusion, free SA was induced to comparable levels in the Pst/AvrRpm1-infected leaves of wild-type and llp1-1, llp1-3, and eds1-2 mutant plants (Supplemental Fig. S8). In contrast to eds1-2, which displayed enhanced susceptibility to Pst/AvrRps4 and virulent Pst, both llp1-1 and llp1-3 supported growth of these Pst strains that was comparable to that in the Col-0 wild type (Fig. 6, C and D), indicating that local EDS1-dependent immunity was not altered in the llp1 mutants. However, SAR triggered by Pst/AvrRpm1 was abolished in both the llp1-1 and llp1-3 mutant plants (Fig. 6E). Because plants overexpressing LLP1 showed marginally higher resistance than wild-type plants against Pst/AvrRpm1 (Armijo et al., 2013), we reasoned that the llp1 SAR-defective phenotype might arise from weak local effects of llp1 mutations specifically to Pst/AvrRpm1. Therefore, we used Pst/AvrRps4, whose growth is not affected in plants overexpressing LLP1 (Armijo et al., 2013), as a primary inoculum to trigger SAR. The pretreatment of Col-0 plants with Pst/AvrRps4 triggered SAR, leading to reduced Pst growth in systemic leaves compared with the mock pretreatment (Fig. 6E). Similar to Pst/AvrRpm1, Pst/AvrRps4 did not trigger SAR in the llp1-1 and llp1-3 mutant plants (Fig. 6E). Together, these data suggest an SAR-specific function for LLP1 in plant immunity.
Figure 6.
LLP1 promotes SAR. A, Schematic drawing of the positions of the llp1-1 and llp1-3 T-DNA insertions near the start codon of LLP1 and transcript levels of LLP1 normalized to UBIQUITIN in the Col-0, llp1-1, llp1-3, and eds1-2 plants 24 h after infiltration of the leaves with 10 mm MgCl2 (mock; M) or with 100 µm BTH (B; inset). Transcript accumulation is shown relative to that in untreated Col-0 plants. B to D, Growth curves of Pst/AvrRpm1 (B), Pst/AvrRps4 (C), and Pst (D) in the wild-type (blue), llp1-1 (red), llp1-3 (green), and eds1-2 (purple) plants. The bacterial titers in the infected leaves were determined at 1, 2, 3, and 4 dpi. E, SAR in Col-0, llp1-1, and llp1-3 plants. Pst titers are shown 4 d after a secondary infection that was systemic to primary treatments with 10 mm MgCl2 (M) or with Pst/AvrRpm1 or Pst/AvrRps4. F, Systemic PR1 induction. PR1 transcript levels were normalized to UBIQUITIN in the systemic untreated leaves 3 d after a primary treatment of Col-0, llp1-1, llp1-3, and eds1-2 plants with 10 mm MgCl2 (M) or Pst/AvrRpm1. G and H, SA-induced resistance in the llp1 mutants. Col-0 and llp1-1, llp1-3, and eds1-2 mutant plants were sprayed with 0.01% (v/v) Tween 20 (mock; M) or with 1 mm SA or 1 mm BTH (B) in 0.01% (v/v) Tween 20. After 24 h, leaves of the treated plants were either infected with Pst (G) or harvested for qRT-PCR analysis (H). In G, the in planta Pst titers are shown at 4 dpi; in H, the transcript levels of AED1 and PR1 were normalized to UBIQUITIN and are shown relative to those in untreated Col-0 plants of the same age. Asterisks indicate significant differences from the mock-treated controls (*P < 0.05, **P < 0.005, Student’s t test). These experiments were repeated two times (E [Pst/AvrRps4], F, and H) to at least three times (A–D, E [Pst/AvrRpm1], and G) with similar results.
SAR induced by Pst/AvrRpm1 was accompanied by enhanced PR1 transcript accumulation in the systemic uninfected leaves of the preinfected compared with the mock-pretreated wild-type plants (Fig. 6F). In contrast to the SAR-defective eds1-2 mutant (Truman et al., 2007; Rietz et al., 2011), which did not support the systemic induction of PR1 (Fig. 6F), the llp1 mutant plants responded to the local infection with modestly induced PR1 expression in the systemic uninfected tissue that was 8- to 20-fold lower compared with that in the wild-type plants (Fig. 6F). These data indicate that immune signaling was compromised but not abolished in the systemic uninfected tissue of locally infected llp1 mutant plants. To gain further insight into a possible role of LLP1 in SA signaling, we investigated the responses of the llp1 mutants to the application of exogenous SA or BTH. In the wild-type and eds1-2 plants, both SA and BTH induced resistance against Pst (Fig. 6G), which was accompanied by enhanced PR1 transcript accumulation in the treated leaves (Fig. 6H). Similarly, endogenous AED1 transcript accumulation was induced by both SA and BTH in the wild-type and eds1-2 plants to a comparable degree as by infection of the wild-type plants (Figs. 4 and 6H). These results confirmed the role of EDS1 upstream of SA in SA-induced immunity (Falk et al., 1999). Additionally, LLP1 transcript accumulation was induced in BTH-treated wild-type and eds1-2 plants (Fig. 6A), indicating that BTH induced LLP1 expression independently of EDS1. Both the llp1-1 and llp1-3 mutants responded to SA and BTH with enhanced resistance to Pst, which was not significantly different from that induced in the wild-type plants (Fig. 6G). Additionally, SA and BTH induced comparable levels of AED1 and PR1 transcript accumulation in the llp1-1 and llp1-3 mutants compared with those in the wild-type and eds1-2 plants (Fig. 6H). These data show that LLP1 is not necessary for immune signaling downstream of SA. Altogether, the data suggest that LLP1 is an important promoter of systemic resistance, possibly acting in parallel with SA.
DISCUSSION
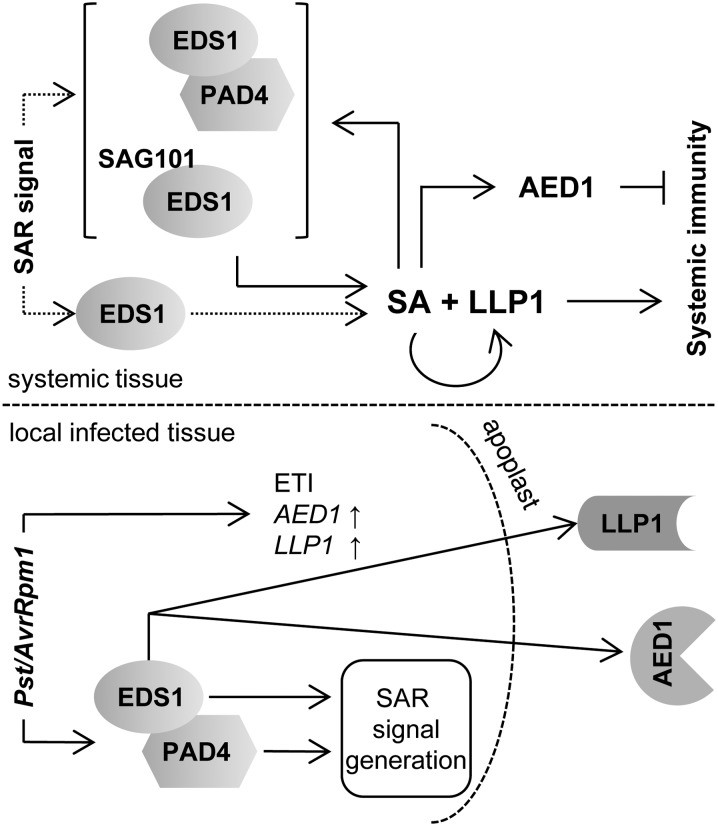
Here, we have utilized the SAR-specific defect of eds1 mutant plants in response to the Pst effector AvrRpm1 to identify new SAR components using a comparative proteomics approach to identify potential proteinaceous signaling components in the apoplast. We showed that EDS1 is required for both SAR signal generation in the primary infected leaves and SAR signal perception in the systemic tissues. Therefore, the identified proteins might operate during one or more phases of SAR. Of the 12 identified AED proteins, nine have not previously been associated with SAR. Of these, the predicted aspartyl protease AED1 appears to suppress systemic immunity as part of a homeostatic control mechanism regulating SAR (Figs. 5 and 7), whereas the legume lectin-like protein LLP1 is essential for promoting SAR, possibly in parallel with SA (Figs. 6 and 7). Systemic but not local AED1 and LLP1 transcript accumulation was correlated with EDS1-dependent Pst/AvrRpm1-induced SAR (Fig. 4). Moreover, both AED1 and LLP1 have pronounced effects on SAR but only mild or no influence on local defenses (Figs. 5 and 6; Supplemental Fig. S7; Armijo et al., 2013). Thus, together with documented differences between local and systemic transcriptional and metabolic profiles (Gruner et al., 2013), our data support a mechanistic separation between the local and systemic phases of SAR. Both AED1 and LLP1 predominantly operate in the systemic phase.
Figure 7.
Model integrating EDS1, PAD4, AED1, and LLP1 in SAR signaling. Both EDS1 and PAD4 are essential for SAR signal generation in Pst/AvrRpm1-infected tissue. In addition, signaling downstream of EDS1 leads to the accumulation of AED1 and LLP1 in the apoplast. Systemically, EDS1 either alone or together with PAD4 or SAG101 (both options are shown in dotted lines) mediates SAR signal perception upstream of SA. SA signaling is most likely fortified by the positive feedback loop of SA with EDS1/PAD4/SAG101. The local accumulation of AED1 and LLP1 transcripts in response to Pst/AvrRpm1 is EDS1 independent and appears to be related to ETI, whereas systemic AED1 and LLP1 expression is regulated by EDS1 and SA and may be associated with SAR. The predicted aspartyl protease AED1 likely suppresses systemic immunity, and the legume lectin-like protein LLP1 appears to promote systemic immunity via actions in parallel with SA.
Similar to mitogen-activated protein kinases 3 and 6 (Tsuda et al., 2013), EDS1/PAD4 signaling can compensate for the loss of SA signaling during local resistance responses activated via CC-NLRs similar to RPM1 responding to AvrRpm1 (Venugopal et al., 2009). For SAR, however, EDS1 and SA do not appear to act redundantly (Fig. 1; Truman et al., 2007; Rietz et al., 2011). Although we cannot rule out that EDS1 and PAD4, as a nucleocytoplasmic complex functioning in basal resistance (Feys et al., 2005; García et al., 2010; Rietz et al., 2011), function in the synthesis of one or more SAR signaling molecules, a more upstream transcriptional reprogramming role seems more likely between AvrRpm1 recognition and SAR signal generation (Bartsch et al., 2006; García et al., 2010). Whereas both EDS1 and PAD4 are necessary for SAR signal generation, EDS1 but not PAD4 is essential for SAR signal perception (Fig. 1). Because the eds1 mutant responds normally to SA (Figs. 5 and 6; Falk et al., 1999), the data place EDS1 upstream of SA, also during SAR signal perception. Arabidopsis EDS1, PAD4, and SAG101 have separable functions in TIR-NLR and basal resistance responses (Rietz et al., 2011; Wagner et al., 2013) and in viral resistance mediated by the CC-NLR HRT (Chandra-Shekara et al., 2004; Zhu et al., 2011). Also, it is known that PAD4 has a unique action, without EDS1 or SAG101, in phloem-based defenses against green peach aphid infestation (Pegadaraju et al., 2007). Similarly, EDS1 might individually contribute to SAR signal perception without its signaling partner PAD4 (Figs. 1 and 7). Because the EDS1/PAD4 complex is essential for basal resistance and the full extent of SAR (Rietz et al., 2011), loss of PAD4 is likely partially compensated for by SAG101 for at least a subset of SAR defense outputs (Feys et al., 2005; Wagner et al., 2013; Fig. 7). Thus, EDS1, together with its signaling partner PAD4 or SAG101, might support SAR signal perception by fortifying SA-mediated defenses, including SA-associated transcriptional reprogramming (Fig. 7; Feys et al., 2005; Wiermer et al., 2005; Vlot et al., 2009; García et al., 2010; Rietz et al., 2011).
AvrRpm1 transcript accumulation upon treatment of pDEX:AvrRpm1-HA plants with 1 µm DEX reaches higher levels than during an SAR-inducing infection of Col-0 plants with Pst/AvrRpm1 (Supplemental Fig. S1). Because it is unclear how bacterial translation and subsequent protein transfer into plant cells compares with the translation of the foreign transcript by the Arabidopsis translational machinery, it is difficult to draw conclusions on the relative potency of each AvrRpm1 delivery. However, the macroscopic cell death symptoms that are induced locally by both treatments are comparable (Supplemental Fig. S1, D and E). Prior studies have shown that ETI-inducing pathogens trigger SAR independently of HR cell death (Cameron et al., 1994; Mishina and Zeier, 2007; Liu et al., 2010). Altogether, the systemic resistance induced by the local expression of AvrRpm1-HA (Fig. 2) is likely caused by an AvrRpm1-dependent signaling event and not, for example, by nonspecific processes following tissue damage in DEX-treated pDEX:AvrRpm1-HA tissues. In the absence of effectors, systemic immunity can be induced by the local application of purified pathogen-associated molecular patterns (PAMPs), such as flagellin or lipopolysaccharides (Mishina and Zeier, 2007), eliciting basal PAMP-triggered immunity (PTI), which is not normally associated with HR cell death (Jones and Dangl, 2006; Henry et al., 2013). The fact that either PAMPs (Mishina and Zeier, 2007) or the effector AvrRpm1 (Fig. 2) can trigger systemic immunity indicates that local defense signaling in the form of PTI or ETI is possibly sufficient for SAR.
Using the eds1 mutant with an SAR-specific phenotype in response to Pst/AvrRpm1 (Aarts et al., 1998; Truman et al., 2007), we expected to identify proteins that are more predominantly associated with systemic than local defenses. In support of this hypothesis, there was limited overlap between the AED proteins and proteins that were identified in proteomic studies of local responses of Arabidopsis plants or cultured cells to either SA or virulent and avirulent Pst (Oh et al., 2005; Jones et al., 2006; Kaffarnik et al., 2009). However, these studies identified proteins with predicted enzymatic activities that are related to some of the AEDs found here. For example, GDSL LIPASE-LIKE1 shares 25% identity with AED4 at the amino acid level, is secreted upon the SA treatment of Arabidopsis cultured cells (Oh et al., 2005), and is involved in the local and systemic defense responses that are associated with ethylene but not SA (Oh et al., 2005; Kwon et al., 2009). Our data implicate the GDSL-motif lipase AED4 in EDS1-dependent SAR (Fig. 3). The protein was found in petiole exudates from 6-week-old, long-day-grown Arabidopsis plants, indicating that AED4 may be mobile through the phloem (Benning et al., 2012; Guelette et al., 2012). Because the expression of AED4 is repressed by the infection of plants with virulent or avirulent Pst (Fig. 4) or by drought (Huang et al., 2008), it is tempting to speculate that AED4 is involved in phloem-mediated long-distance signaling regulating responses to biotic and abiotic stress.
The expression of AED1, AED4, LLP1, PNP-A, and PR5 in leaves infected with virulent or avirulent Pst was predominantly associated with local defenses (Figs. 4 and 7). In contrast, the accumulation of the corresponding proteins in apoplast-enriched extracts from the AvrRpm1-HA-expressing plants was EDS1 dependent and thus was correlated with SAR (Figs. 3 and 7; Supplemental Table S1). The accumulation or modification of AED proteins is possibly regulated posttranscriptionally or posttranslationally independently of or prior to transcriptional changes. Such a scenario was previously suggested to explain early changes in intracellular protein accumulation in Arabidopsis plants infected with different Pst strains, including Pst/AvrRpm1 (Jones et al., 2006), and is supported by the identification of two AED proteins in multiple 2D gel spots (Supplemental Fig. S4). Also, for differentially apoplast-enriched protein spots, we could not distinguish between differences in overall protein accumulation or their secretion. In the systemic uninfected leaves of Pst/AvrRpm1-infected plants, the transcript accumulation of AED1, LLP1, PNP-A, and PR5 was tightly correlated with the extent of SAR (Figs. 4 and 7). For PNP-A, the data provide further experimental support for a previous bioinformatics study linking PNP-A to SA-mediated defense signaling and SAR (Meier et al., 2008). Natriuretic peptides occur in animals and plants and have similarities with cell wall-loosening expansins (Gehring and Irving, 2003). PNP-A localizes to the apoplast and is associated with plant homeostasis regulating ion fluxes and cellular water uptake (Gehring and Irving, 2003; Wang et al., 2011). Because PNP-A was suggested to regulate dark respiration via long-distance, possibly phloem-mediated signaling (Ruzvidzo et al., 2011), we will investigate whether the PNP-A-triggered local and/or systemic changes in plant homeostasis affect SAR.
The aspartyl protease CDR1 accumulates in the apoplast upon infection of Arabidopsis plants with Pst/AvrRpm1 (Xia et al., 2004). Similar to its rice (Oryza sativa) homolog OsCDR1 (Prasad et al., 2009), AtCDR1 is believed to release a peptide between 3 and 10 kD in size that triggers PR2 transcription both locally and systemically (Xia et al., 2004). Similar to CDR1, AED1 is an atypical aspartyl protease lacking a plant-specific insertion that is found in canonical plant aspartyl proteases (Faro and Gal, 2005). However, in contrast to CDR1, AED1 appears to repress SAR by restricting systemic immunity downstream of SA (Fig. 5). Previous studies have firmly linked the expression of AED1 in Arabidopsis with EDS1 or SA-mediated defense responses (Chini et al., 2004; Eulgem et al., 2004; Mosher et al., 2006; van Damme et al., 2008), but its biological role has remained elusive. We showed that AED1 transcript accumulation is induced locally and systemically by infection and locally by SA (Figs. 4 and 6). Because the conditional overexpression of AED1-HA repressed both SAR and SA-induced resistance without affecting the growth of Pst in healthy plants (Fig. 5; Supplemental Fig. S7), AED1 might be part of a homeostatic mechanism to limit SAR signaling (Fig. 7) and thus regulate the resource allocation in the tradeoff between defense and plant growth (Heidel and Dong, 2006; van Hulten et al., 2006; Traw et al., 2007; Pajerowska-Mukhtar et al., 2012). In support of this, we found that reduced AED1 transcript levels in Arabidopsis RNAi:AED1/At5g10770 plants caused severe stunting, a phenotype that is often observed in constitutive defense mutants (Shirano et al., 2002; Yoshioka et al., 2006; Bi et al., 2010; Zhu et al., 2010). The stunted growth of the RNAi:AED1/At5g10770 plants was accompanied by an elevated transcript accumulation of the SAR marker gene PR1, underscoring a negative regulatory role of AED1 during SAR.
The primary structure of AED1 resembles that of its tobacco (Nicotiana tabacum) homolog, CHLOROPLASTIC NUCLEOID DNA-BINDING41 (CND41), which degrades the chloroplast protein Rubisco during the initial steps of senescence (Nakano et al., 1997; Kato et al., 2004, 2005). Consequently, CND41 antisense tobacco displayed delayed senescence in older leaves, whereas the plants were dwarfed, similar to the RNAi:AED1/At5g10770 Arabidopsis plants (Fig. 5; Nakano et al., 2003; Kato et al., 2004). Therefore, we speculate that CND41 may also play a role in the defense against pathogens, given the proposed connections between senescence and innate immunity, with autophagy-associated genes antagonizing SA-mediated immune signaling (for review, see Dickman and Fluhr, 2013). Elucidating the exact mode of AED1 action during SAR, including its apparent recruitment in the systemic rather than the local phase of SA-mediated immunity, requires further investigation. Similar to tobacco and tomato (Solanum lycopersicum) apoplastic aspartyl proteases that degrade PR proteins (Rodrigo et al., 1989, 1991), AED1 may suppress SAR by degrading one or more proteins in the apoplast of SAR-induced leaves.
LLP1 is one of 226 lectin genes encoding carbohydrate-binding proteins in the Arabidopsis genome (Peumans and Van Damme, 1995; Sharon and Lis, 2004; Armijo et al., 2013). The plant lectins are divided into 12 families, and LLP1, along with its two closest homologs At3g16530 and AtLEC, belongs to the lectin-legB family of legume lectin-like proteins (Jiang et al., 2010; Armijo et al., 2013). In contrast to LLP1, most Arabidopsis legume lectin-like proteins contain kinase domains (Armijo et al., 2013). Notably, several lectin receptor kinases with other types of lectin domains promote SA-associated immunity, including PTI (for review, see Singh and Zimmerli, 2013). In contrast to the LLP1 homolog AtLEC, whose expression is associated with jasmonic acid- and ethylene-mediated signaling (Lyou et al., 2009), the expression of LLP1 is induced early after the SA treatment of Arabidopsis plants in a NONEXPRESSOR OF PR GENES1 (NPR1)-dependent manner (Krinke et al., 2007; Blanco et al., 2009) and by virulent and avirulent Pst in an EDS1-dependent manner (Fig. 4). The LLP1 expression that was induced by BTH was independent of EDS1 (Fig. 6), suggesting that the induction of LLP1 by SA occurs downstream of EDS1 (Fig. 7). Similar to EDS1 (Falk et al., 1999; Truman et al., 2007; Heidrich et al., 2011), LLP1 is necessary for systemic immunity triggered by the local inoculation of plants with Pst/AvrRpm1 or Pst/AvrRps4 but not for the local immunity triggered by the application of exogenous SA (Fig. 6). The latter result indicates that LLP1 probably does not act downstream of SA. However, a function of LLP1 upstream of SA seems equally unlikely, because LLP1 does not appear to affect the local resistance responses to virulent and avirulent Pst that are normally associated with EDS1 and SA (Fig. 6; Armijo et al., 2013). Additionally, LLP1 did not affect EDS1-independent free SA accumulation in Arabidopsis in response to Pst/AvrRpm1. Altogether, the data link LLP1 most closely with systemic rather than local immunity and suggest that LLP1 may act in parallel with SA. Because the constitutive overaccumulation of LLP1 did not trigger a significant resistance response to, for example, Pst (Armijo et al., 2013), LLP1 likely cooperates with one or more additional components, possibly including SA. Further investigation is required to clarify how LLP1 acts and which signaling components, in addition to SA, may cooperate with LLP1 to promote the systemic phase of SAR.
Mammalian lectins perform essential functions in animal innate immunity (summarized in Rabinovich et al., 2012). These lectins are believed to operate in the recognition of self, nonself, or altered-self molecules or associated entities (e.g. tumor cells) and to mediate cellular trafficking, cell-cell communication, and immune signaling, among other activities (Sharon and Lis, 2004; Rabinovich et al., 2012). Similar to plant lectin receptor kinases (Singh and Zimmerli, 2013), a subset of lectins functions as PAMP receptors in mammalian immunity (Rabinovich et al., 2012). Analogous to animal systems, LLP1 may regulate SAR in plants by mediating the recognition of, for example, altered-self glycan structures accumulating in locally infected or systemically responding tissues. Interestingly, LLP1 is localized to the plasma membrane facing the apoplast (Armijo et al., 2013) and might sense changes to the glycan composition of the cell wall, possibly caused by another AED, XYL4, which functions as an extracellular β-d-xylosidase (Fig. 3; Table I; Minic et al., 2004). Alternatively, LLP1 might recognize components of the cuticle, which contributes to SAR signal perception or propagation in systemic uninfected tissues via a currently unknown mechanism (Xia et al., 2009, 2010).
In conclusion, we uncovered two new components that regulate different aspects of SAR. An aspartyl protease, AED1, represses SAR likely as part of a feedback regulatory mechanism. In contrast, the legume lectin-like protein LLP1 promotes SAR, possibly in parallel with SA. Future research will be directed at elucidating whether LLP1-mediated extracellular glycan sensing acts on systemic immunity.
MATERIALS AND METHODS
Plants and Cultivation Conditions
The Arabidopsis (Arabidopsis thaliana) cv Wassilewskija-0 (petiole exudate analyses) and cv Col-0 (all other experiments) were used. The mutants eds1-1, pad4-5, eds1-1pad4-5, dir1, and eds1-2 have been described previously (Falk et al., 1999; Feys et al., 2001; Maldonado et al., 2002; Bartsch et al., 2006). Col-0 pDEX:AvrRpm1-HA has been described previously (Mackey et al., 2002) and was crossed with eds1-2 to yield eds1-2 pDEX:AvrRpm1-HA. The T-DNA insertion lines SALK_111104 (aed1-1), SALK_091655 (aed1-2), SALK_036814 (llp1-1), and SALK_074760 (llp1-3) were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). Homozygous plants were selected for seed stocks and experiments. The plants were grown on normal potting soil mixed with silica sand at a ratio of 5:1 and kept at 20°C/22°C (night/day), 70% relative humidity, and 100 µE m−2 s−1 light for 10-h days.
DNA Constructs and Plant Transformation
The RNAi construct targeting AED1 and At5g10770 was made in pHANNIBAL (Wesley et al., 2001). From each target gene, a 400-nucleotide fragment of the coding sequence was amplified by reverse transcription-PCR with the primer sets pRNAi-1-5′cl/pRNAi-1-3′cl and pRNAi-70-5′cl/pRNAi-70-3′cl (Supplemental Table S2), respectively, using RNA that was isolated from Pseudomonas syringae pv tomato strain DC3000 expressing AvrRpm1-infected Col-0 plants as a template. Subsequently, the two fragments were annealed and amplified by PCR using the primers pRNAi-1-5′cl and pRNAi-70-3′cl. The resulting 800-nucleotide fragment was cloned into pENTR/dTOPO (Invitrogen), sequenced, and cloned into pHANNIBAL in the sense and antisense orientations using EcoRI/XhoI and HindIII/XbaI, respectively. Finally, the AED1/At5g10770 RNAi cassette from pHANNIBAL was transferred to the binary vector pART27 using NotI.
The AED1 overexpression construct was made in pER8-GW-C-term-3XHA-Strep. To generate pER8-GW-C-term-3XHA-StrepII, the 3XHA-StrepII fragment from pXCSG-HAStrep (Stuttmann et al., 2009) was cut with XhoI and XbaI and transferred to Gateway-compatible pER8 (Curtis and Grossniklaus, 2003) that was cut with XhoI and SpeI. The coding sequence of AED1 (At5g10760) lacking the stop codon was amplified by reverse transcription-PCR with the primers pAED1-5′cl and pAED1-3′cl (Supplemental Table S2) using RNA isolated from Col-0 plants infected with Pst/AvrRpm1 as a template. The resulting DNA fragment was cloned into pENTR/dTOPO (Invitrogen) and sequenced. Subsequently, AED1 complementary DNA was transferred into the destination vector pER8-GW-C-term-3XHA-StrepII by standard Gateway technology (Invitrogen).
Both pART27 containing the AED1/At5g10770 RNAi cassette and pER8-GW-C-term-3XHA-StrepII carrying AED1 were transferred into Agrobacterium tumefaciens strain GV3101. Subsequently, Col-0 plants were transformed with each construct by floral dipping according to Logemann et al. (2006). Transgenic T1 plants were selected on Murashige and Skoog medium containing 50 µg mL−1 kanamycin (RNAi lines) or 50 µg mL−1 hygromycin (AED1 overexpression lines). The surviving RNAi plants were transferred to soil 2 weeks after germination and used for experiments 3 weeks later. Homozygous AED1 overexpression lines of the third (T3; line 108-194) or fourth (T4; line 154-47) generation were used for all of the experiments.
Pathogens, Infection Methods, and SAR Assays
Four- to 5-week-old plants were used for all of the infection experiments. Bacterial propagation and the infection of plants with virulent or avirulent Pst were carried out as described previously (Aarts et al., 1998; Vlot et al., 2008). For bacterial growth curves or gene expression analysis in infected tissues, rosette leaves 3 and 4 were infiltrated with 105 cfu mL−1 of the appropriate bacterium. SAR or systemic gene expression changes were induced by infiltrating the first two true leaves per plant with 106 cfu mL−1 Pst/AvrRpm1 or Pst/AvrRps4 or 10 mm MgCl2 as a control. For DEX-induced SAR, the first two true leaves of the pDEX:AvrRpm1-HA plants were surface treated (painted) with 1 µm DEX (Sigma-Aldrich) in 0.01% (v/v) Tween 20 or 0.01% (v/v) Tween 20 as a control. For systemic gene expression or SAR analyses, two systemic leaves (leaves 3 and 4) were either harvested or infected with 105 cfu mL−1 Pst 3 d after the primary treatment. The in planta Pst titers were determined 4 d later as described previously (Vlot et al., 2008). For experiments with the AED1-HA-overexpressing plants, the XVE:AED1-HA plants were sprayed with 30 µm β-estradiol in 0.01% (v/v) Tween 20 at 24 h prior to the start of the experiment. β-Estradiol was freshly dissolved in methanol and diluted to the appropriate concentration prior to every experiment for the optimal induction of AED1-HA accumulation.
Chemically Induced Resistance
BTH (purchased commercially under the trade name Bion; Ciba-Geigy) and SA (Sigma-Aldrich) were each dissolved in water to produce stock solutions of 100 mm. The local induction of LLP1 transcript accumulation was triggered via syringe infiltration of the first two true leaves of 4- to 5-week-old plants with 100 µm BTH. To induce local resistance with SA or BTH, 4- to 5-week-old plants were sprayed until dropoff with 1 mm of either compound in 0.01% (v/v) Tween 20 or with 0.01% (v/v) Tween 20 as a control. After 24 h, leaves 3 and 4 of the treated plants were either harvested for gene expression analysis or syringe infiltrated with 105 cfu mL−1 Pst. The in planta Pst titers were determined at 4 dpi as described above.
Petiole Exudates
The petiole exudates were collected as described previously (Maldonado et al., 2002), except that leaves were treated with 106 cfu mL−1 Pst/AvrRpm1 or 10 mm MgCl2. The exudates from five to 10 leaves in 1.5 mL of 1 mm EDTA were diluted 2-fold with water and infiltrated into untreated healthy plants. The exudate-infiltrated leaves were collected 24 h later, and PR1 expression was analyzed on northern blots as described previously (Maldonado et al., 2002). In parallel, 100 µL of exudate per petiole was plated on sterile medium and cultivated for Pst/AvrRpm1 growth as described (Aarts et al., 1998); experiments were evaluated and considered for this work only if the petiole exudates did not contain bacteria.
Isolation of Apoplast-Enriched Protein Extracts
The Col-0 DEX::AvrRpm1-HA and eds1-2 DEX::AvrRpm1-HA plants were sown as a lawn. Three- to 4-week-old plants were sprayed until dropoff with 30 µm DEX in 0.01% (v/v) Tween 20. Four to 5 h later, all of the aboveground tissue was harvested. The apoplast-enriched protein extracts were isolated in either APO buffer I (2.5 mm Tris, pH 7.4, 1 mm EDTA, and 30 mm mannitol; used for 2D gel analysis) or APO buffer II (2.5 mm HEPES, pH 7.4, 1 mm EDTA, and 30 mm MgCl2; used for ICPL analysis). The plant tissue in the appropriate buffer was exposed to a mild vacuum for up to 10 min in a normal vacuum chamber. Afterward, the vacuum was slowly released. This procedure was repeated two to three times until the tissue was completely infiltrated. The infiltrated plants were transferred to 20- to 30-mL syringes that were hung in 50-mL tubes and centrifuged at 2,250 rpm at 4°C for 20 min. The flow through was collected as the apoplast-enriched protein extract.
Proteomics
2D Gel Analysis
A total of 200 µg of protein was loaded onto 17-cm (two biological replicates, including Fig. 3 and Supplemental Fig. S4A) or 7-cm (two additional replicates in Supplemental Fig. S4B) nonlinear isoelectric focusing strips, pI 4 to 7 (Bio-Rad), and run according to the manufacturer’s instructions. The second dimension with the 17-cm strips was performed using 12% (w/v) polyacrylamide gels in the Laemmli buffer system and run in a Bio-Rad Protean II cell. The second dimension using the 7-cm strips was performed using NuPAGE precast 4% to 12% (w/v) polyacrylamide gradient gels (Invitrogen) in the MES buffer system. Both large gels and one small gel were then stained using SYPRO Ruby (Invitrogen); the other small gel was stained using Coomassie PageBlue (Fermentas). For the software-assisted comparison of 2D gels containing proteins from the wild-type and eds1 mutant plants, gel images were overlapped, combined, and analyzed using the Delta2D gel-analysis system (DECODON). The spots selected for MALDI analysis were robotically excised, digested, and spotted using the Proteineer SP + DP systems (Bruker Daltonik), and the data were collected on an Ultraflex III (Bruker Daltonik) using two-stage data collection as described previously (March et al., 2012). The peptide mass fingerprints and peptide fragmentation fingerprints were searched against the National Center for Biotechnology Information nonredundant database using Mascot (Matrix Science; www.matrixscience.com) with the parameters described in Supplemental Protocol S1.
ICPL
The proteins in the apoplast-enriched extracts were concentrated on 3-kD size-exclusion columns (Amicon Ultra; Millipore) according to the manufacturer’s instructions. Subsequently, 70 µg of protein per sample (at a concentration of 3.5 µg µL−1) was labeled with d0-N-nicotinoyloxy-succinimide (ICPL-0 or light tag; wild-type extracts) or d4-N-nicotinoyloxy-succinimide (ICPL-4 or heavy tag; eds1-2 extracts) according to the ICPL duplex kit instructions (Serva Electrophoresis). The differentially labeled wild-type and mutant extracts were then pooled per biologically independent repetition, and the proteins were separated on one-dimensional polyacrylamide gels (12% [w/v]). After Coomassie Blue staining and washing of the gels, three slices with visible protein bands were excised per sample and subjected to in-gel trypsin digestion as described previously (Sarioglu et al., 2006). The digested peptides were analyzed according to Gaupels et al. (2012) by nano-HPLC using the Ultimate 3000 (Dionex) device coupled to a linear quadrupole ion-trap Orbitrap (LTQ Orbitrap XL) mass spectrometer (Thermo Fisher) equipped with a nanoelectrospray ionization source. All of the MS/MS spectra were analyzed using Mascot (version 2.2.06; Matrix Science). Mascot was set up to search the National Center for Biotechnology Information Arabidopsis database (version of April 19, 2012 [64,961 sequences]), assuming the digestion enzyme trypsin and with a fragment ion mass tolerance (MS/MS) of 0.6 D and parent ion tolerance of 10 ppm (MS). The iodoacetamide derivatization of Cys was specified in Mascot as a stable modification. The oxidations of Met and ICPL-0 and ICPL-4 for Lys were specified as variable modifications. The data processing for the ICPL analysis was performed according to Gaupels et al. (2012) with one exception: proteins that were identified by at least one unique peptide but were present in all three biological replicates were considered, of which proteins displaying a significant differential accumulation between the extracts from the wild-type and eds1 mutant plants were considered AEDs (P < 0.05, Perseus statistical tool; http://www.perseus-framework.org/; Cox and Mann, 2011).
SA Measurement
Free SA was isolated from pooled leaves from six individual plants per sample and measured essentially as described previously (von Saint Paul et al., 2011), except that the extraction was performed with a mixture of 98:2 (v/v) methanol:formic acid. For the solid phase extraction, the mixtures were adjusted to 30:69.4:0.6 (v/v) methanol:water:formic acid, and the quantification was based on SA fluorescence (excitation, 305 nm; emission, 438 nm), with 3,4-dihydroxy benzoic acid added as an internal control during the extraction.
RNA Isolation and qRT-PCR
Total RNA was isolated using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions. The complementary DNA was generated using oligo(dT) (20-mer) and SuperScript II Reverse Transcriptase (Invitrogen), and PCR was performed using the primers outlined in Supplemental Table S2. Quantitative PCR was performed with the SensiMix SYBR Low-ROX Kit (Bioline) on a 7500 real-time PCR system (Applied Biosystems). The transcript accumulation of target genes was normalized to TUBULIN or UBIQUITIN using relative quantification with the 7500 Fast System Software 1.3.1. The presented quantitative PCR results are averages of three technical repetitions per sample ± sd.
Protein Immunoblots
Four- to 5-week-old XVE:AED1-HA plants (or Col-0 as a control) were sprayed until dropoff with 100 µm β-estradiol in 0.01% (v/v) Tween 20. After 24 h, 100 mg of leaf tissue was ground in 200 µL of 2× Laemmli sample buffer and boiled for 5 min. Subsequently, 15 µL per sample was analyzed by western blotting (Towbin et al., 1979) using Hybond-P polyvinylidene difluoride membranes (GE Healthcare) and monoclonal anti-HA antibodies conjugated to horseradish peroxidase (HRP; Sigma-Aldrich). HRP was visualized using the chemiluminescence kit Immun-Star WesternC (Bio-Rad) and a Typhoon Trio+ (GE Healthcare). The protein size was compared with the Precision Plus Protein Size Marker (Bio-Rad) tagged on the blot with Precision Protein Strep Tactin-HRP conjugate (Bio-Rad) according to the manufacturer’s instructions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AvrRpm1-HA transcript accumulation in infected Col-0 plants and in eds1-2 pDEX:AvrRpm1-HA plants.
Supplemental Figure S2. DEX does not induce SAR in Col-0 plants.
Supplemental Figure S3. SAR signals are emitted from the DEX-treated leaves of pDEX:AvrRpm1-HA plants between 4 and 6 h after DEX treatment.
Supplemental Figure S4. Summary of 2D gel analyses of apoplast-enriched extracts from AvrRpm1-HA-expressing plants.
Supplemental Figure S5. ICPL controls.
Supplemental Figure S6. Effects of organic solvents on SAR.
Supplemental Figure S7. Pathogen growth in infected leaves of plants overaccumulating AED1-HA.
Supplemental Figure S8. Local accumulation of free SA before and after infection of llp1 mutants.
Supplemental Table S1. Summary of ICPL data.
Supplemental Table S2. Primers used in this study.
Supplemental Protocol S1. MALDI-MS parameters for protein identification in 2D gel spots.
Supplementary Material
Acknowledgments
We thank Dr. Laurent Noël for pXCSG-HAStrep, Jaqueline Bautor and Anne Harzen for technical assistance, Dr. Juergen Schmidt for technical advice, and Drs. Anton Schaeffner and Joerg Durner for critically reading the article.
Glossary
- SAR
systemic acquired resistance
- SA
salicylic acid
- MeSA
methyl salicylate
- ETI
effector-triggered immunity
- NLR
nucleotide-binding/leucine-rich repeat
- HR
hypersensitive response
- TIR-NLR
nucleotide-binding/leucine-rich repeat receptors with N-terminal Toll-Interleukin1 receptor-like homology
- CC-NLR
nucleotide-binding/leucine-rich repeat receptors with an N-terminal coiled-coil domain
- Pst
Pseudomonas syringae pv tomato strain DC3000
- Pst/AvrRpm1
Pseudomonas syringae pv tomato strain DC3000 expressing AvrRpm1
- DEX
dexamethasone
- HA
hemagglutinin
- qRT
quantitative reverse transcription
- cfu
colony-forming units
- Col-0
Columbia-0
- 2D
two-dimensional
- MALDI
matrix-assisted laser desorption/ionization
- MS/MS
tandem mass spectrometry
- MS
mass spectrometry
- ICPL
isotope-coded protein labeling
- Pst/AvrRps4
Pseudomonas syringae pv tomato strain DC3000 expressing the effector AvrRps4
- T-DNA
transfer DNA
- dpi
days post infection
- RNAi
double-stranded RNA interference
- BTH
benzo 1,2,3-thiadiazole-7-carbothioic acid S-methyl ester
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- HRP
horseradish peroxidase
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. VL66/1–1 and SFB924 project B6 to A.C.V.), by Marie Curie Intraeuropean fellowships (grant nos. HPMF–CT–2001–01197 to L.J. and MEIF–CT–2006–040357 to A.C.V.), by the European Molecular Biology Organization (long-term fellowship no. ALTF 137–2006 to A.C.V.), by the Max-Planck Society, and by an Alexander von Humboldt Sofja Kovalevskaja Award (to J.E.P.).
This article is dedicated to the memory of Chris Lamb (1950–2009).
The online version of this article contains Web-only data.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534–552 [DOI] [PubMed] [Google Scholar]
- Armijo G, Salinas P, Monteoliva MI, Seguel A, García C, Villarroel-Candia E, Song W, van der Krol AR, Álvarez ME, Holuigue L. (2013) A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Mol Plant Microbe Interact 26: 1395–1406 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning UF, Tamot B, Guelette BS, Hoffmann-Benning S. (2012) New aspects of phloem-mediated long-distance lipid signaling in plants. Front Plant Sci 3: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W. (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Bi D, Cheng YT, Li X, Zhang Y. (2010) Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol 153: 1771–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F, Garretón V, Frey N, Dominguez C, Pérez-Acle T, Van der Straeten D, Jordana X, Holuigue L. (2005) Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59: 927–944 [DOI] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K. (2013) Salicylic acid: an old hormone up to new tricks. Mol Plant Pathol 14: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Dangl JL. (2012) How complex are intracellular immune receptor signaling complexes? Front Plant Sci 3: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ. (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5: 715–725 [Google Scholar]
- Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK. (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front Plant Sci 4: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, Yu K, Sekine KT, Gao QM, Selote D, Hu Y, Stromberg A, Navarre D, et al. (2011) Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat Genet 43: 421–427 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Navarre D, Kachroo A, Kang HG, Klessig D, Kachroo P. (2004) Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J 40: 647–659 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J. (2008) Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Venables B, Petros RA, Nalam V, Li M, Wang X, Takemoto LJ, Shah J. (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J 71: 161–172 [DOI] [PubMed] [Google Scholar]
- Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J 38: 810–822 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 80: 273–299 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Ritter C, Gibbon MJ, Mur LAJ, Wood JR, Goss S, Mansfield J, Taylor JD, Vivian A. (1992) Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell 4: 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF. (2012) SOS: too many signals for systemic acquired resistance? Trends Plant Sci 17: 538–545 [DOI] [PubMed] [Google Scholar]
- Dickman MB, Fluhr R. (2013) Centrality of host cell death in plant-microbe interactions. Annu Rev Phytopathol 51: 543–570 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Weigman VJ, Chang HS, McDowell JM, Holub EB, Glazebrook J, Zhu T, Dangl JL. (2004) Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol 135: 1129–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro C, Gal S. (2005) Aspartic proteinase content of the Arabidopsis genome. Curr Protein Pept Sci 6: 493–500 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels F, Sarioglu H, Beckmann M, Hause B, Spannagl M, Draper J, Lindermayr C, Durner J. (2012) Deciphering systemic wound responses of the pumpkin extrafascicular phloem by metabolomics and stable isotope-coded protein labeling. Plant Physiol 160: 2285–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels F, Vlot AC (2013) Plant defense and long-distance signaling in the phloem. In GA Thompson, AJE van Bel, eds, Phloem, Molecular Cell Biology, Systemic Communication, Biotic Interactions. Wiley-Blackwell, Ames, IA, pp 227–247 [Google Scholar]
- Gehring CA, Irving HR. (2003) Natriuretic peptides: a class of heterologous molecules in plants. Int J Biochem Cell Biol 35: 1318–1322 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T, Zeier J. (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J. (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes MEM, Richmond S, Kuc J. (1980) Induced systemic resistance to anthracnose in cucumber as influenced by the location of the inducer inoculation with Colletotrichum lagenarium and the onset of flowering and fruiting. Physiol Plant Pathol 17: 229–233 [Google Scholar]
- Guelette BS, Benning UF, Hoffmann-Benning S. (2012) Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J Exp Bot 63: 3603–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Dong X. (2006) Fitness benefits of systemic acquired resistance during Hyaloperonospora parasitica infection in Arabidopsis thaliana. Genetics 173: 1621–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Henry E, Yadeta KA, Coaker G. (2013) Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol 199: 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Ma Z, Ramachandran S. (2010) Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol Biol 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing B, Xu S, Xu M, Li Y, Li S, Ding J, Zhang Y. (2011) Brush and spray: a high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol 157: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AME, Thomas V, Bennett MH, Mansfield J, Grant M. (2006) Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol 142: 1603–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Robin GP. (2013) Systemic signaling during plant defense. Curr Opin Plant Biol 16: 527–533 [DOI] [PubMed] [Google Scholar]
- Kaffarnik FAR, Jones AME, Rathjen JP, Peck SC. (2009) Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics 8: 145–156 [DOI] [PubMed] [Google Scholar]
- Kato Y, Murakami S, Yamamoto Y, Chatani H, Kondo Y, Nakano T, Yokota A, Sato F. (2004) The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 220: 97–104 [DOI] [PubMed] [Google Scholar]
- Kato Y, Yamamoto Y, Murakami S, Sato F. (2005) Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta 222: 643–651 [DOI] [PubMed] [Google Scholar]
- Kim TH, Kunz HH, Bhattacharjee S, Hauser F, Park J, Engineer C, Liu A, Ha T, Parker JE, Gassmann W, et al. (2012) Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 24: 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke O, Ruelland E, Valentová O, Vergnolle C, Renou JP, Taconnat L, Flemr M, Burketová L, Zachowski A. (2007) Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiol 144: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Jin HC, Lee S, Nam MH, Chung JH, Kwon SI, Ryu CM, Park OK. (2009) GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J 58: 235–245 [DOI] [PubMed] [Google Scholar]
- Liu PP, Bhattacharjee S, Klessig DF, Moffett P. (2010) Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Mol Plant Pathol 11: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, von Dahl CC, Klessig DF. (2011a) The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol 157: 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, von Dahl CC, Park SW, Klessig DF. (2011b) Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and tobacco. Plant Physiol 155: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ülker B, Somssich IE. (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyou SH, Park HJ, Jung C, Sohn HB, Lee G, Kim CH, Kim M, Choi YD, Cheong JJ. (2009) The Arabidopsis AtLEC gene encoding a lectin-like protein is up-regulated by multiple stimuli including developmental signal, wounding, jasmonate, ethylene, and chitin elicitor. Mol Cells 27: 75–81 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- March TJ, Richter D, Colby T, Harzen A, Schmidt J, Pillen K. (2012) Identification of proteins associated with malting quality in a subset of wild barley introgression lines. Proteomics 12: 2843–2851 [DOI] [PubMed] [Google Scholar]
- Meier S, Bastian R, Donaldson L, Murray S, Bajic V, Gehring C. (2008) Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, Rihouey C, Do CT, Lerouge P, Jouanin L. (2004) Purification and characterization of enzymes exhibiting β-d-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol 135: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Murakami S, Shoji T, Yoshida S, Yamada Y, Sato F. (1997) A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell 9: 1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Nagata N, Kimura T, Sekimoto M, Kawaide H, Murakami S, Kaneko Y, Matsushima H, Kamiya Y, Sato F, et al. (2003) CND41, a chloroplast nucleoid protein that regulates plastid development, causes reduced gibberellin content and dwarfism in tobacco. Physiol Plant 117: 130–136 [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J. (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK. (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17: 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong X. (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Pegadaraju V, Louis J, Singh V, Reese JC, Bautor J, Feys BJ, Cook G, Parker JE, Shah J. (2007) Phloem-based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. Plant J 52: 332–341 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJM. (1995) Lectins as plant defense proteins. Plant Physiol 109: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BD, Creissen G, Lamb C, Chattoo BB. (2009) Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol Plant Microbe Interact 22: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Rabinovich GA, van Kooyk Y, Cobb BA. (2012) Glycobiology of immune responses. Ann N Y Acad Sci 1253: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Vlot AC, Feys BJ, Niefind K, Parker JE. (2011) Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V. (2013) Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet 9: e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo I, Vera P, Conejero V. (1989) Degradation of tomato pathogenesis-related proteins by an endogenous 37-kDa aspartyl endoproteinase. Eur J Biochem 184: 663–669 [DOI] [PubMed] [Google Scholar]
- Rodrigo I, Vera P, Van Loon LC, Conejero V. (1991) Degradation of tobacco pathogenesis-related proteins: evidence for conserved mechanisms of degradation of pathogenesis-related proteins in plants. Plant Physiol 95: 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzvidzo O, Donaldson L, Valentine A, Gehring C. (2011) The Arabidopsis thaliana natriuretic peptide AtPNP-A is a systemic regulator of leaf dark respiration and signals via the phloem. J Plant Physiol 168: 1710–1714 [DOI] [PubMed] [Google Scholar]
- Sarioglu H, Brandner S, Jacobsen C, Meindl T, Schmidt A, Kellermann J, Lottspeich F, Andrae U. (2006) Quantitative analysis of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced proteome alterations in 5L rat hepatoma cells using isotope-coded protein labels. Proteomics 6: 2407–2421 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Zeier J. (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Lis H. (2004) History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology 14: 53R–62R [DOI] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF. (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões I, Faro C. (2004) Structure and function of plant aspartic proteinases. Eur J Biochem 271: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Singh P, Zimmerli L. (2013) Lectin receptor kinases in plant innate immunity. Front Plant Sci 4: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, Jones JDG, Banfield MJ. (2012) Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci USA 109: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE. (2010) Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 62: 628–640 [DOI] [PubMed] [Google Scholar]
- Stuttmann J, Lechner E, Guérois R, Parker JE, Nussaume L, Genschik P, Noël LD. (2009) COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J Biol Chem 284: 7920–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traw MB, Kniskern JM, Bergelson J. (2007) SAR increases fitness of Arabidopsis thaliana in the presence of natural bacterial pathogens. Evolution 61: 2444–2449 [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme M, Huibers RP, Elberse J, Van den Ackerveken G. (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J 54: 785–793 [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J. (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103: 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Venugopal SC, Jeong RD, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y, Hersh M, Stromberg AJ, Navarre D, Kachroo A, et al. (2009) Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, Uknes S, Kessmann H, Ryals J. (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu PP, Cameron RK, Park SW, Yang Y, Kumar D, Zhou F, Padukkavidana T, Gustafsson C, Pichersky E, et al. (2008) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J 56: 445–456 [DOI] [PubMed] [Google Scholar]
- von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Kessler T, Schmitt-Kopplin P, Schäffner AR. (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23: 4124–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE. (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630 [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang YH, Gehring C, Irving HR. (2011) Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol 52: 837–850 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Xia Y, Gao QM, Yu K, Lapchyk L, Navarre D, Hildebrand D, Kachroo A, Kachroo P. (2009) An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5: 151–165 [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C. (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23: 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yu K, Navarre D, Seebold K, Kachroo A, Kachroo P. (2010) The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol 154: 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Xiong W, Ye TT, Wu Y. (2012) Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J Exp Bot 63: 2579–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. (2006) The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Soares JM, Mandal MK, Wang C, Chanda B, Gifford AN, Fowler JS, Navarre D, Kachroo A, Kachroo P. (2013) A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep 3: 1266–1278 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S. (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. (2011) SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog 7: e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.