Abstract
Printable multi-marker biochips that enable simultaneous quantitative detection of multiple target biomarkers in point-of-care and resource-limited settings are a holy grail in the field of biodiagnostics. However, preserving the functionality of biomolecules, which are routinely employed as recognition elements, during conventional printing approaches remains challenging. In this article, we introduce a simple yet powerful approach, namely plasmonic calligraphy, for realizing multiplexed label-free bioassays. Plasmonic calligraphy involves a regular ballpoint pen filled with biofunctionalized gold nanorods as plasmonic ink for creating isolated test domains on paper substrates. Biofriendly plasmonic calligraphy approach serves as a facile method to miniaturize the test domain size to few mm2, which significantly improves the sensitivity of the plasmonic biosensor compared to bioplasmonic paper fabricated using immersion approach. Furthermore, plasmonic calligraphy also serves as a simple and efficient means to isolate multiple test domains on a single test strip, which facilitates multiplexed biodetection and multi-marker biochips. Plasmonic calligraphy, which can be potentially automated by implementing with a robotic arm, serves as an alternate path forward to overcome the limitations of conventional ink-jet printing.
Keywords: Localized surface plasmon resonance, Calligraphy, Gold nanorods, Plasmonic ink
1. Introduction
Owing to numerous advantages such as high specific surface area, excellent wicking properties, compatibility with conventional printing approaches, significant cost reduction and easy disposability, paper substrates are gaining increased attention in biodiagnostics, food quality testing, environmental monitoring, flexible energy and electronic devices (Chen et al., 2008; Cheng et al., 2010; Huang et al., 2013; Lee et al., 2010, 2011; Li et al., 2010, 2012; Martinez et al., 2007, 2009; Nergiz et al., 2013; Parolo and Merkoci, 2013; Tian et al., 2012c). Recent surge in the activity related to paper-based diagnostic devices is primarily focused on realizing microfluidic paper-based analytical devices (µPADs) for point-of-care assays and inexpensive diagnostic tools for resource-limited environments (Lewis et al., 2012; Martinez et al., 2009). Most of these developments rely on labor-, time- and/or resource-intensive patterning techniques such as photolithography, wax printing, ink-jet printing of polydimethylsiloxane (PDMS), to create fluidic pathways and/or different functional regions for site-selective adsorption of the biochemical reagents (Abe et al., 2008; Bruzewicz et al., 2008; Carrilho et al., 2009; Martinez et al., 2007; Noh and Phillips, 2010; Olkkonen et al., 2010; Osborn et al., 2010; Qu et al., 2012; Yu and White, 2013). Moreover, implementing ink-jet printing with biomolecules can result in loss of recognition functionality due to the inherent temperature variations associated with ink-jet printing process. These considerations clearly highlight the need for a simple and biofriendly technique that enables multi-marker biochips for point-of-care and resource-limited settings.
The refractive index sensitivity of localized surface plasmon resonance (LSPR) of plasmonic nanostructures renders it an attractive transduction platform for chemical and biological sensing (Abbas et al., 2013b; Anker et al., 2008; Englebienne, 1998; Haes et al., 2005; Haes and Van Duyne, 2002; Kattumenu et al., 2011; Maier and Atwater, 2005; Mayer and Hafner, 2011; Riboh et al., 2003; Rosi and Mirkin, 2005; Sepúlveda et al., 2009; Svedendahl et al., 2009; Yonzon et al., 2004). We have recently demonstrated plasmonic paper comprised of biofunctionalized gold nanorods (AuNRs) uniformly adsorbed on paper substrates (Tian et al., 2012c). The bioplasmonic paper enabled the detection of aquaporin-1, a kidney cancer biomarker in artificial urine down to a concentration of 10 ng/ml (Morrissey et al., 2010). Bioplasmonic paper, fabricated by immersing a paper substrate into biofunctionalized AuNRs solution, facilitates the detection of one specific target protein in the analyte solution (e.g., urine). Perceivably, this immersion approach hinders spatial multiplexing (i.e., realizing multiple test domains for the detection of more than one target biomolecule on the same substrate) as it results in uniform adsorption of the bioconjugated nanorods over the entire paper surface.
Here, we demonstrate a simple yet powerful plasmonic calligraphy approach for realizing multiplexed label-free bioassays using a regular ballpoint pen filled with gold nanorods or biofunctionalized gold nanorods as (bio)plasmonic ink. Plasmonic calligraphy offers two distinct advantages over plasmonic paper substrates obtained by immersion method as reported previously. Firstly, plasmonic calligraphy serves as a facile method to miniaturize the test domain size to few mm2, which significantly improves the sensitivity of the plasmonic biosensor compared to bioplasmonic paper fabricated using immersion approach (Tian et al., 2012c). Secondly, bioplasmonic calligraphy enables simple and efficient multiplexed biodetection on paper substrates thus leading to multi-marker biochips. In this study, we demonstrate these two aspects using gold nanorods as plasmonic nano-transducers.
2. Experimental
2.1. Materials
Cetyltrimethylammonium bromide (CTAB), chloroauric acid, ascorbic acid, sodium borohydride, poly(styrene sulfonate) (PSS) (Mw = 70,000 g/mol), and poly(allyl amine hydrochloride) (PAH) (Mw = 56,000 g/mol) were purchased from Sigma–Aldrich. Silver nitrate and filter paper (Whatman #1) was purchased from VWR international. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxy succinimide (NHS), Rabbit IgG, Goat anti-Rabbit IgG, Human IgG, Goat anti-human IgG, Mouse IgG, and Goat anti-mouse IgG were purchased from Thermo scientific. SH–PEG–COOH (Mw = 5000 g/mol) was purchased from Jenkem Technology. All the chemicals have been used as received with no further purification. Paper mate profile retractable ballpoint pens were bought from Amazon.
2.2. Synthesis of gold nanorods (AuNRs)
Gold nanorods were synthesized using a seed-mediated approach (Huang et al., 2009; Orendorff and Murphy, 2006). Seed solution was prepared by adding 0.6 ml of an ice-cold sodium borohydride solution (10 mM) into 10 ml of 0.1 M cetyltrimethylammonium bromide (CTAB) and 2.5 × 10−4 M chloroauric acid (HAuCl4) solution under vigorous stirring at room temperature. The color of the seed solution changed from yellow to brown. Growth solution was prepared by mixing 95 ml of CTAB (0.1 M), 0.5 ml of silver nitrate (10 mM), 4.5 ml of HAuCl4 (10 mM), and 0.55 ml of ascorbic acid (0.1 M) consecutively. The solution was homogenized by gentle stirring. To the resulting colorless solution, 0.12 ml of freshly prepared seed solution was added and set aside in the dark for 14 h. Prior to use, the AuNRs solution was centrifuged twice at 10,000 rpm for 10 min to remove excess CTAB and redispersed in nanopure water (18.2 MΩ cm).
2.3. Preparation of polyelectrolytes coated gold nanorods (AuNRs)
AuNRs were modified with polyelectrolytes as previously reported (Pastoriza-Santos et al., 2006). Briefly, 1 ml of twice centrifuged AuNRs solution was added drop-wise to 0.5 ml of PSS solution (0.2%, w/v) in 6 mM NaCl aqueous solution under vigorous stirring, and left undisturbed for 3 h. To remove excess PSS, the above solution was centrifuged at 10,000 rpm for 10 min, and the pellet was dispersed in nanopure water after removing the supernatant. To modify AuNRs with PAH, 1 ml of PSS coated AuNRs solution was added drop-wise to 0.5 ml of PAH (0.2%, w/v) solution in 6 mM NaCl, stirred for 3 h. The resultant 1 ml of PAH coated AuNRs solution was centrifuged and concentrated to 10 µl and employed as ink to write on paper substrates. The surface charge of CTAB stabilized AuNRs, PSS and PAH coated AuNRs were estimated by measuring the zeta potential of corresponding solution (Fig. S3).
2.4. AuNRs–IgG conjugates preparation
To a 37.5 µl solution of heterobifunctional polyethylene glycol (SH–PEG–COOH) in water (20 µM, Mw = 5000 g/mol), EDC and NHS with the same molar ratio as SH–PEG–COOH were added followed by shaking for 1 h. The pH of the above reaction mixture was adjusted to 7.4 by adding 10x concentrated phosphate buffered saline (PBS), followed by the addition of 10 µl of rabbit immunoglobulin G (IgG) solution (75 µM, Mw = 150 kDa). The reaction mixture was incubated for an additional 2 h, and then filtered to remove any byproduct during the reaction by centrifugation using a centrifuge tube with 50 kDa filter. The final SH–PEG–IgG conjugates solution (0.75 µM) was obtained after washing with PBS buffer (pH 7.4) twice. AuNRs–IgG conjugates’ solution was prepared by adding 50 µl of SH–PEG–IgG conjugates solution to 1 ml of twice centrifuged AuNRs solution with incubation for 1 h. The amount of SH–PEG–IgG was optimized to obtain maximum coverage of IgG on AuNRs surface (Fig. S4). Using SDS–PAGE, we have confirmed that the affinity of SH–PEG–IgG remains essentially the same as that of pristine IgG (see Fig. S5 and supporting information for details).
2.5. Bioplasmonic paper substrates preparation
A regular laboratory filter paper (Whatman™ #1) was immersed into a 1% (w/v) BSA in PBS buffer (pH 7.5) for 1 h as a pretreatment step to prevent nonspecific binding (Fig. S6A). We noted ~30% improvement in plasmonic biosensor response (i.e., longitudinal LSPR shift of AuNRs) for BSA-blocked paper compared to pristine paper (Fig. S6B and C). Plasmonic ink was prepared by concentrating 1 ml of twice centrifuged as synthesized AuNRs to 10 µl after centrifugation. Bioplasmonic ink was concentrated from 1 ml of NR–IgG conjugates solution by centrifugation at 3000 rpm for 20 min. The plasmonic ink was injected into an empty ballpoint pen refill cleaned with ethanol and nanopure water by sonication. The adsorption of AuNRs–IgG conjugates on paper was achieved by direct writing with bioplasmonic ink filled pen, or exposing written AuNRs paper in SH–PEG–IgG conjugates solution for 30 min, followed by thorough rinsing with buffer and nanopure water. Bioplasmonic paper was exposed to various concentrations of anti-IgG in PBS for 1 h, followed by thorough rinsing with PBS and water and drying with a stream of nitrogen.
2.6. Extinction spectra measurements
Extinction spectra from paper substrates were collected using a CRAIC microspectrophotometer (QDI 302) coupled to a Leica optical microscope (DM 4000 M) with 20 × objective in the range of 450–800 nm with 10 accumulations and 0.1 s exposure time in reflection mode. The spectral resolution of the spectrophotometer is 0.2 nm. Several UV–vis extinction spectra (~ 10) were collected for each substrate before and after anti-IgG exposure. Each spectrum represented a different spot within the same substrate. Shimadzu UV-1800 spectrophotometer was employed for collecting UV–vis extinction spectra from solution.
2.7. Characterization
Transmission electron microscopy (TEM) micrographs were recorded on a JEM-2100F (JEOL) field emission instrument. Samples were prepared by drying a drop of the solution on a carboncoated grid, which had been previously made hydrophilic by glow discharge. Scanning electron microscope (SEM) images were obtained using a FEI Nova 2300 Field Emission SEM at an accelerating voltage of 10 kV. Plasmonic paper was gold sputtered for 60 s before SEM imaging.
3. Results and discussion
3.1. Characterization of plasmonic calligraphed paper
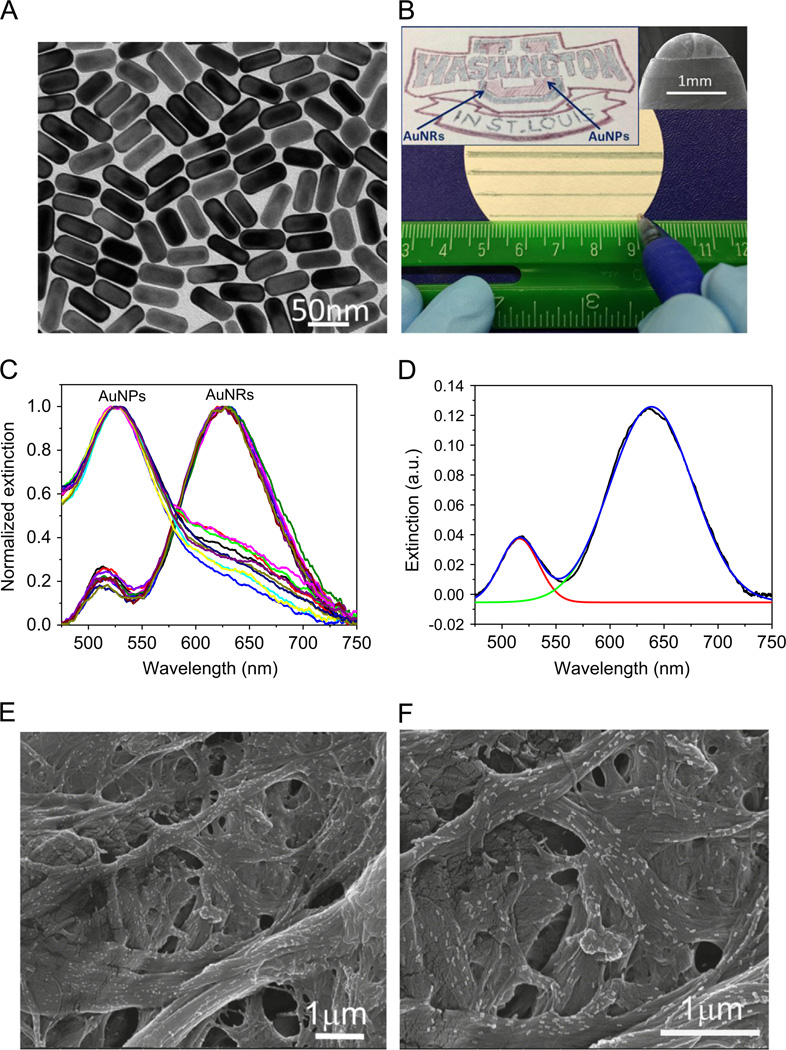
Plasmonic calligraphy using a ballpoint pen to form sensing islands on paper offers a unique advantage in that the volume of ink deposited can be well-controlled by altering the viscosity of the ink and ‘finesse’ of the ball used for writing. On the other hand, a more conventional approach of micropipette-based deposition of sensing elements (i.e., biofunctionalized AuNR) on paper surface results in fuzzy boundaries and non-uniform drying patterns due to uncontrolled evaporation on heterogeneous paper surface. Gold nanorods are particularly attractive as plasmonic transducers considering the high refractive index sensitivity of longitudinal LSPR, facile and large tunability of the LSPR wavelength with aspect ratio and the electromagnetic (EM) hot-spots at the tips (Nusz et al., 2009; Tian et al., 2012a,b). AuNRs, synthesized using a seed-mediated approach, are positively charged with a length of 56.373.7 nm and a diameter of 22.471.8 nm (TEM image in Fig. 1A) (Huang et al., 2009; Orendorff and Murphy, 2006). Fig. 1B depicts plasmonic calligraphy on a laboratory filter paper (Whatman #1) using a regular ballpoint pen with AuNRs ink, which yields continuous and clearly defined lines visible to even unaided eye. Ball pens are particularly well suited for dispensing nanoparticle inks due to their compatibility with liquid and gels (Gostony and Schneider, 1998). The viscosity of AuNRs ink was measured to be ~ 1.25 Pa s, which is close to the optimal viscosity for silver nanoparticle ink reported previously (Russo et al., 2011). The left inset image of Fig. 1B shows the logo of Washington University in St. Louis, a complex pattern, drawn on a laboratory filter paper using cetyltrimethylammonium chloride (CTAC) stabilized gold nanospheres (AuNPs, red region) and cetyltrimethylammonium bromide (CTAB) stabilized gold nanorods (AuNRs, green region). The right inset image of Fig. 1B depicts the SEM image of the tip of a ballpoint pen with a ball diameter of ~ 1.5 mm, showing the residue of AuNRs ink left on the ball surface. Extinction spectra collected from several locations of red and green region of the university logo drawn with AuNPs and AuNRs ink revealed excellent optical uniformity of the plasmonic paper substrate (Fig. 1C). UV–vis extinction spectrum obtained from AuNRs region is characterized by two distinct bands corresponding to the transverse (lower wavelength) and longitudinal (higher wavelength) oscillation of electrons with the incident EM field (Fig. 1C).
Fig. 1.
(A) TEM image of AuNRs. (B) Demonstration of writing AuNRs on paper substrate (left inset: the logo of Washington University in St. Louis drawn using gold nanospheres (AuNPs, red region) and gold nanorods (AuNRs, green region). Right inset: SEM image of the tip of a ball pen with AuNRs ink residue on the surface). (C) Extinction spectra measured from ten spots of the red and green region of the university logo, showing excellent spectral homogeneity. (D) A representative LSPR spectrum of AuNRs deconvoluted using two Gaussian peaks. (E and F) SEM images of AuNRs adsorbed on paper substrates by plasmonic calligraphy approach. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
The extinction spectrum of AuNRs was deconvoluted by fitting the extinction spectrum with two Gaussian peaks to obtain the longitudinal LSPR wavelength of AuNRs, which was used to monitor the binding of target proteins to AuNRs (Fig. 1D). It is known that longitudinal LSPR of AuNRs is more sensitive to the refractive index change of the surrounding medium compared to its transverse band and LSPR of AuNPs (Chen et al., 2008; Tian et al., 2012c). Longitudinal LSPR wavelength measured from 10 different spots of the green region of the university logo exhibited a small standard deviation of ~ 1 nm (Fig. 1C). The excellent spectral homogeneity is due to the uniform adsorption of AuNRs on paper substrates as evidenced by the SEM images (Fig. 1E and F). The spectral homogeneity observed here is quite remarkable considering the simplicity of the writing process and the inherent heterogeneity of the paper substrates (large surface roughness and hierarchical nature of the fibrous mat). The density of the nanostructures on the paper substrate can be controlled by the number of strokes. The density of the AuNRs adsorbed on the paper substrate for a single stroke was found to be 31 ± 9/µm2 determined from SEM micrographs. Notably, the adsorption of AuNRs on paper is sufficiently strong to resist desorption from paper surface even after extensive rinsing with water or buffer as confirmed by little change in the intensity and shape of extinction spectra collected before and after rinsing. In addition to AuNRs, various shape-controlled nanostructures stabilized with different ligands, including gold nanospheres stabilized with citrate ions, gold nanoshells capped with poly(vinyl pyrrolidone) (PVP), can be written on paper with no sign of aggregation or patchiness (Fig. S1).
3.2. Significant improvement on sensitivity of bioassays
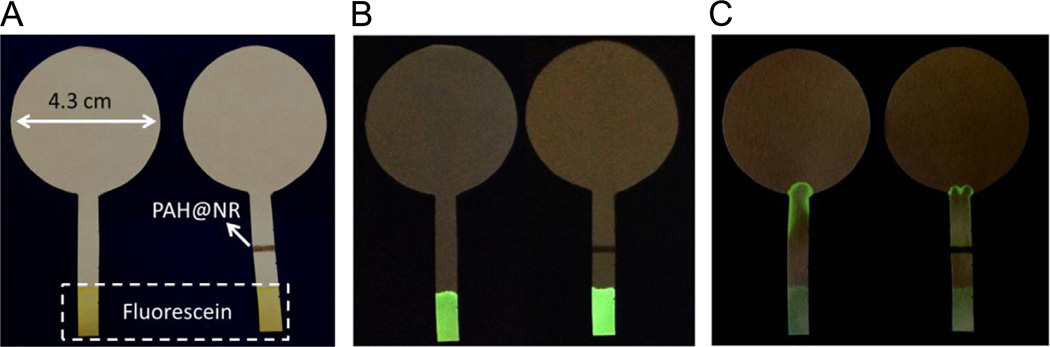
First, we set out to demonstrate that the plasmonic calligraphy approach serves as a simple and powerful tool to miniaturize the test domain size, which leads to dramatic improvement in plasmonic paper-based biosensor performance compared to previous immersion method. Capillary-driven flow of the analyte solution across the test domain written on paper is employed to maximize the target analyte interaction with the recognition elements on the plasmonic nanostructures (Abbas et al., 2013a). To visually demonstrate the concept of capillary-driven flow-based sensing, AuNRs modified with positively charged poly(allylamine hydrochloride) (PAH@AuNRs) were written on the stem portion of a paper substrate cut in the shape of a badminton racket with a head of 4.3 cm diameter and a stem of 4 × 0.6 cm2 (Fig. 2A). The head portion serves as a wicking pad or collection reservoir and the bottom end of the stem is immersed in the analyte solution of a predefined volume. The model analyte solution comprised of negatively charged fluorescein molecules was deposited at the lower end of the stem (Fig. 2B). The capillary-driven flow results in the transport of fluorescein from the tip of the stem to the wicking pad. In the case of paper substrate without PAH@AuNRs line, most of the fluorescein is collected at the neck of the substrate as indicated by the strong green fluorescence from the neck region under UV illumination (Fig. 2C). On the other hand, reduced fluorescence was observed at the neck region of the substrate with PAH@AuNRs line as most of the negatively charged fluorescein was bound to the positively charged PAH@AuNRs line (Fig. 2C). Absence of strong fluorescence from the PAH@AuNRs line is possibly due to the non-radiative quenching of fluorescence by the plasmonic nanostructures (Fig. 2C) (Dulkeith et al., 2002; Kang et al., 2011).
Fig. 2.
(A) Optical and (B) fluorescence images of paper strips with 10 µl of 10mM fluorescein molecules adsorbed at the bottom of strip under visible light and UV light. (C) Transport of the fluorescein molecules toward wicking pad with 50 µl of water, showing the adsorption of negatively charged fluorescein molecules on positively charged PAH @AuNRs band.
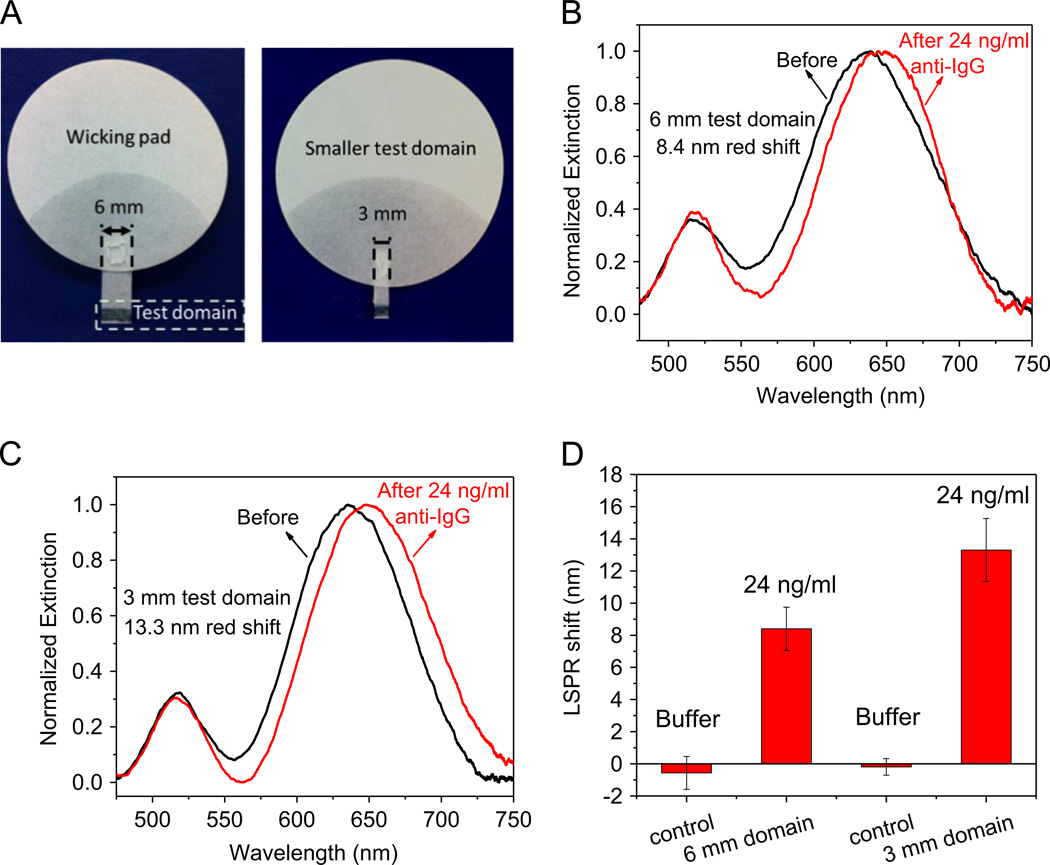
In most of sensing systems, miniaturization of the test domain size results in improved sensitivity and lower limit of detection while adversely affecting the dynamic range. In the case of plasmonic sensors, individual nanostructures and even specific parts of individual nanostructures have been employed for chemical and biological detection, which exhibit remarkable sensitivities but limited dynamic range (Mayer et al., 2010; Rycenga et al., 2012) Most of these demonstrations involve complex and tedious fabrication methods (e.g., e-beam lithography) and/or signal collection and processing methods (e.g., dark-field scattering spectroscopy). Plasmonic calligraphy approach serves as a facile tool to optimize the test domain size for achieving a balance between sensitivity and dynamic range (e.g., covering physiological and pathological concentration of a protein biomarker). The test domain size can be controlled by simply cutting the paper substrates to vary the feature size written on the paper substrate using plasmonic ink. Fig. 3A shows AuNRs line was written at the bottom end of the stem portion of a test strip followed by functionalization of AuNR with rabbit immunoglobulin G (IgG) (see experimental section for details). A predefined volume of the target protein solution (100 µl of 24 ng/ml anti-rabbit IgG) was transported from the bottom of the stem to wicking pad across test domains of different sizes using capillary force. The approach adapted here ensures the analyte to pass through test domain, overcoming one of the drawbacks of miniaturizing the test domain i.e., low probability for the analyte molecules to ‘find and bind’ to the test domain. The LSPR wavelength shift was observed to be 13.3 nm when the domain size was reduced to 3 × 1.5 mm2 compared to 8.4 nm for a test domain of 6 × 1.5 mm2 upon exposure to 24 ng/ml of anti-IgG (Fig. 4B and C). The increase in LSPR shift by about 58%, indicates an improvement in sensitivity by reducing the test domain size (Fig. 3D). Plasmonic calligraphy in combination with ‘paper cutting’ forms a powerful tool to dial in the required sensitivity or dynamic range of a paper-based biosensor.
Fig. 3.
(A) Images of paper strips with different size of test domains. (B and C) Extinction spectra of AuNRs paper upon 24 ng/ml of anti-IgG binding corresponding to different test domain size. (D) Comparison of LSPR shifts for different test domain size of bioassays.
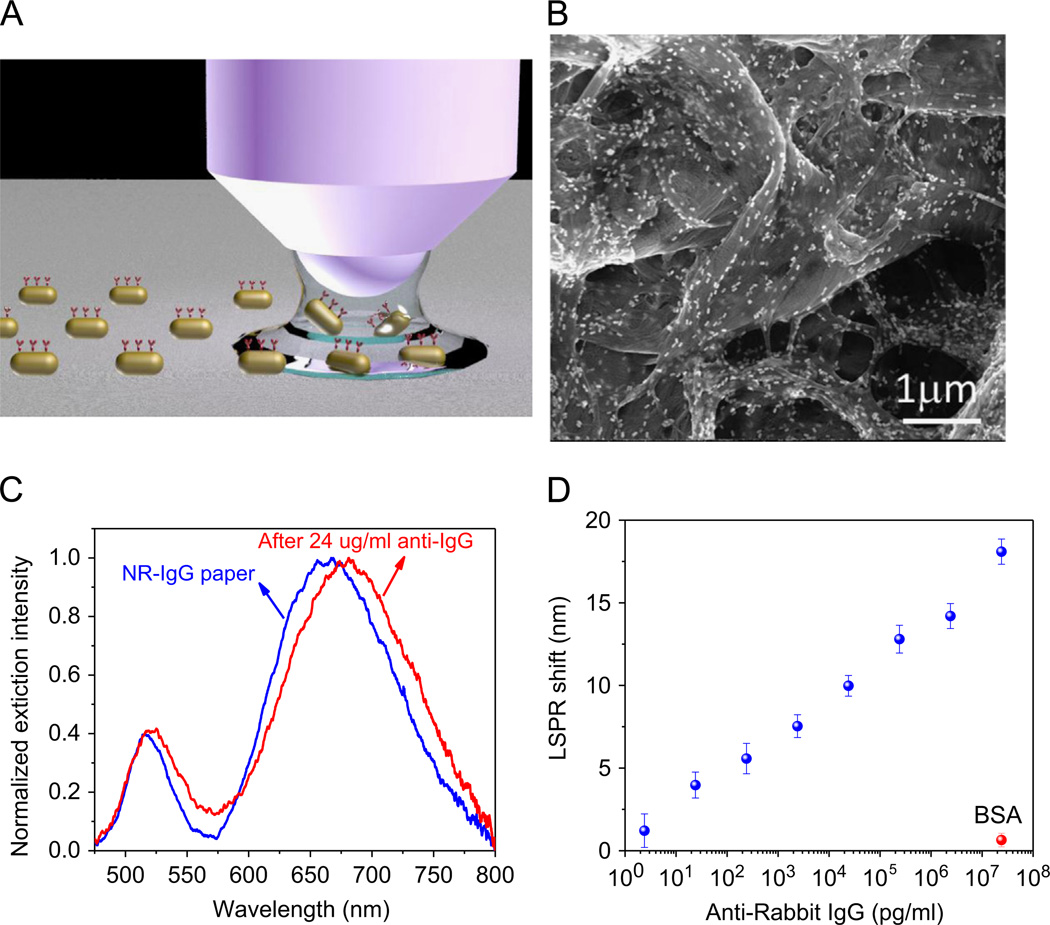
Fig. 4.
(A) Schematic illustration of the concept of bioplasmonic calligraphy. (B) SEM images of NR–IgG conjugates adsorbed on paper substrates by bioplasmonic calligraphy approach. (C) Extinction spectra of AuNRs–IgG conjugates on the paper substrate before (blue) and after binding of anti-IgG (red). (D) Plot showing the LSPR peak shift of bioplasmonic paper for various concentrations of anti-IgG and BSA. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
3.3. Multiplexed biosensing based on bioplasmonic calligraphy
Multi-marker plasmonic biochips using paper substrates that enable multiplexed biosensing will be an extremely powerful tool to facilitate the detection and quantification of multiple prognostic biomarkers using the same substrate. To achieve such a multi-marker biochip, individual test domains should be comprised of plasmonic nanostructures with differential functionalization specific to target biomarkers. To realize the differential functionalization of test domains on paper substrates, we employ biofunctionalized nanostructures as ink (called bioplasmonic ink henceforth) rather than biofunctionalization after creating the test domains as described above (Fig. 3A). Such bioplasmonic ink facilitates one to write with distinct biofunctionalized nanostructures on paper substrates adjacent to each other without cross-contaminating the test domains based on the concept of bioplasmonic calligraphy as illustrated in Fig. 4A. SEM images revealed highly uniform distribution of gold nanorods modified with rabbit IgG (NR-rabbit IgG) conjugates on paper surface with no signs of aggregation or patchiness on the substrate (Fig. 4B). Higher magnification image reveals the preferential alignment of AuNRs– rabbit IgG conjugates along the cellulose fibers (Fig. S2). Extinction spectra were obtained from paper substrates calligraphed with NRrabbit IgG and subsequently exposed to 24 µg/ml of anti-rabbit IgG (Fig. 4C). LSPR wavelength exhibited a red shift of ~ 17 nm upon specific binding of anti-rabbit IgG to rabbit IgG appended on the AuNRs. A semi-log plot of the longitudinal LSPR wavelength shift for different concentrations of anti-rabbit IgG revealed that LSPR shift monotonically increases with increase in the concentration of antirabbit IgG. An extremely small LSPR shift (~ 1 nm) was noted for relatively high concentration of BSA (24 µg/ml) due to nonspecific binding (Fig. 4D). Detection limit was determined to be 24 pg/ml (~0.16 pM), which is on par with that observed in the case of other rigid substrates (Mayer and Hafner, 2011). It is worth noting that the biomolecules appended to the nanostructure preserve their recognition capabilities confirming that the simple bioplasmonic calligraphy approach suggested here is ‘biofriendly’ and can be potentially employed for multiplexed biodetection as demonstrated below.
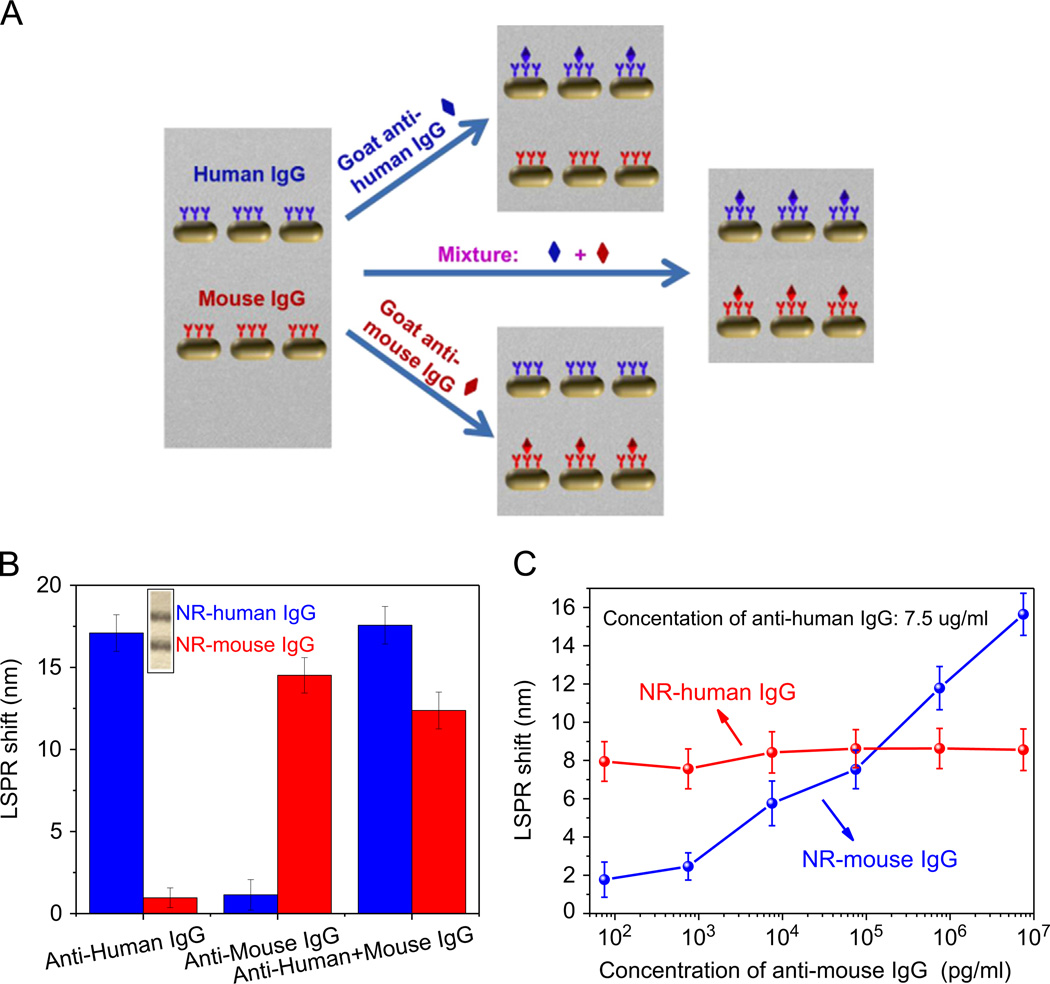
To test capability of multiplexed detection, we wrote two distinct test domains comprised of AuNRs with human IgG and mouse IgG, and then obtained the LSPR shift upon exposure to the different combination of target proteins (goat anti-human IgG, and goat anti-mouse IgG) (Fig. 5A and inset of Fig. 5B). Goat anti-human IgG and goat anti-mouse IgG are affinity-purified secondary antibodies with well-characterized specificity for human IgG and mouse IgG, respectively, which have been tested by ELISA and/ or solid-phase adsorbed to ensure minimal cross-reaction with each other. Extinction spectra of AuNRs functionalized with human IgG (NR–human IgG) showed LSPR shift of ~ 17.1 nm and AuNRs functionalized with mouse IgG (NR–mouse IgG) showed extremely small LSPR shift (~1.0 nm) upon exposure to 24 µg/ml of anti-human IgG (Fig. 5B). On the other hand, upon exposure to 24 µg/ml of anti-mouse IgG, NR–human IgG line showed extremely small shift (~ 1.1 nm) while LSPR shift of NR–mouse IgG was measured to be ~ 14.5 nm (Fig. 5B). Upon exposure to a mixture of anti-human IgG and anti-mouse IgG (24 µg/ml each), NR–human IgG showed ~ 17.6 nm of LSPR shift and NR–mouse IgG showed ~ 12.3 nm. The spectral response of the two lines upon exposure to the mixture closely corresponds to the LSPR shift measured for exposure to individual target biomolecules. This multiplexed bioassay was also challenged with exposure to a mixture of anti-mouse IgG of different concentrations and anti-human IgG of a fixed concentration (Fig. 5C). A monotonic increase in the LSPR shift of NR–mouse IgG band was observed with increasing the concentration of anti-mouse IgG while NR–human IgG band exhibited a stable ~ 8 nm LSPR shift corresponding to the fixed concentration of anti-human IgG (7.5 µg/ml) in the mixture. A detection limit of 750 pg/ml of anti-mouse IgG was noted even in the presence of a constant interfering 7.5 µg/ml of anti-human IgG. These results clearly show the capability of multiplexed biosensing based on bioplasmonic calligraphy approach. The approach suggested here obviates the need for any complex multi-step process such as formation of hydrophilic test domains and hydrophobic barriers to achieve label-free multiplexed biodetection.
Fig. 5.
(A) Schematic illustration of multiplexed detection based on bioplasmonic calligraphy. (B) Longitudinal LSPR wavelength shift of AuNRs functionalized with human IgG and mouse IgG corresponding to exposure of the calligraphed paper to goat anti-human IgG, goat anti-mouse IgG, and the mixture of both (inset: image of NR–human IgG and NR–mouse IgG written on paper). (C) Longitudinal LSPR wavelength shift of NR–human IgG and NR–mouse IgG corresponding to exposure of the calligraphed paper to the mixture of different concentrations of goat anti-mouse IgG and the constant concentration of goat anti-human IgG.
4. Conclusion
Plasmonic calligraphy approach serves as a simple and powerful tool to miniaturize test domain size by controlling the calligraphed feature size and simply cutting the paper to desired dimensions, which results in dramatic improvement in sensitivity and lowering limit of detection. We introduced a low-cost novel approach for fabricating multiplexed label-free biosensing on paper substrates in the form of bioplasmonic calligraphy. The calligraphy approach allows one to create well-isolated test domains on paper substrates using biofunctionalized plasmonic nanostructures as ink. We have demonstrated the feasibility of such an approach for multiplexed biosensing using two target proteins. Bioplasmonic calligraphy can serve as a powerful tool enabling the synergism of paper-based microfluidics and plasmonic biosensing, which is expected to be truly transformative by opening up novel possibilities to realize the fabrication of multimarker paper-based biochips.
Supplementary Material
Acknowledgments
We would like to thank Ms. Marilee Fisher for technical help in performing antibody conjugates affinity study. We acknowledge financial support from National Science Foundation under award number CBET-1254399 (CAREER), NCI R01CA141521, and Air Force Research Laboratories. We would like thank Nano Research Facility (NRF), a member of the National Nanotechnology Infrastructure Network (NNIN), for providing access to electron microscopy facilities.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bios.2014.03.043.
References
- Abbas A, Brimer A, Slocik JM, Tian L, Naik RR, Singamaneni S. Anal. Chem. 2013a;85(8):3977–3983. doi: 10.1021/ac303567g. [DOI] [PubMed] [Google Scholar]
- Abbas A, Tian L, Morrissey JJ, Kharasch ED, Singamaneni S. Adv. Funct. Mater. 2013b;23(14):1789–1797. doi: 10.1002/adfm.201202370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Suzuki K, Citterio D. Anal. Chem. 2008;80(18):6928–6934. doi: 10.1021/ac800604v. [DOI] [PubMed] [Google Scholar]
- Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat. Mater. 2008;7(6):11. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- Bruzewicz DA, Reches M, Whitesides GM. Anal. Chem. 2008;80(9):3387–3392. doi: 10.1021/ac702605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho E, Martinez AW, Whitesides GM. Anal. Chem. 2009;81(16):7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Chen H, Kou X, Yang Z, Ni W, Wang J. Langmuir. 2008;24(10):5233–5237. doi: 10.1021/la800305j. [DOI] [PubMed] [Google Scholar]
- Cheng C-M, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM. Angew. Chem. Int. Ed. 2010;49(28):4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel FCJM, Reinhoudt DN, Moller M, Gittins DI. Phys. Rev. Lett. 2002;89(20):203002. doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]
- Englebienne P. Analyst. 1998;123(7):1599–1603. doi: 10.1039/a804010i. [DOI] [PubMed] [Google Scholar]
- Gostony H, Schneider SL. USA: Schiffer Publishing, PA; 1998. [Google Scholar]
- Haes AJ, Chang L, Klein WL, Van Duyne RP. J. Am. Chem. Soc. 2005;127(7):2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- Haes AJ, Van Duyne RP. J. Am. Chem. Soc. 2002;124(35):10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhu H, Chen Y, Preston C, Rohrbach K, Cumings J, Hu L. ACS Nano. 2013;7(3):2106–2113. doi: 10.1021/nn304407r. [DOI] [PubMed] [Google Scholar]
- Huang X, Neretina S, El-Sayed MA. Adv. Mater. 2009;21(48):4880–4910. doi: 10.1002/adma.200802789. [DOI] [PubMed] [Google Scholar]
- Kang K, Wang J, Jasinski J, Achilefu S. J. Nanobiotechnol. 2011;9(1):16. doi: 10.1186/1477-3155-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattumenu R, Lee CH, Tian L, McConney ME, Singamaneni S. J. Mater. Chem. 2011;21(39):15218–15223. [Google Scholar]
- Lee CH, Hankus ME, Tian L, Pellegrino PM, Singamaneni S. Analyt. Chem. 2011;83(23):8953–8958. doi: 10.1021/ac2016882. [DOI] [PubMed] [Google Scholar]
- Lee CH, Tian L, Singamaneni S. ACS Appl. Mater. Interfaces. 2010;2(12):3429–3435. doi: 10.1021/am1009875. [DOI] [PubMed] [Google Scholar]
- Lewis GG, DiTucci MJ, Phillips ST. Angew. Chem. Int. Ed. 2012;51(51):12707–12710. doi: 10.1002/anie.201207239. [DOI] [PubMed] [Google Scholar]
- Li X, Ballerini DR, Shen W. Biomicrofluidics. 2012;6(1) doi: 10.1063/1.3687398. 011301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tian J, Shen W. Cellulose. 2010;17(3):649–659. [Google Scholar]
- Maier SA, Atwater HA. J. Appl. Phys. 2005;98(1) 011101. [Google Scholar]
- Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angew. Chem. Int. Ed. 2007;46(8):1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal. Chem. 2009;82(1):3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- Mayer KM, Hafner JH. Chem. Rev. 2011;111(6):3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- Mayer KM, Hao F, Lee S, Nordlander P, Hafner JH. Nanotechnology. 2010;21:25. doi: 10.1088/0957-4484/21/25/255503. [DOI] [PubMed] [Google Scholar]
- Morrissey JJ, London AN, Luo J, Kharasch ED. Mayo. Clin. Proc. 2010;85(5):413–421. doi: 10.4065/mcp.2009.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergiz SZ, Gandra N, Farrell ME, Tian L, Pellegrino PM, Singamaneni S. J. Mater. Chem. A. 2013;1(22):6543–6549. [Google Scholar]
- Noh H, Phillips ST. Anal. Chem. 2010;82(10):4181–4187. doi: 10.1021/ac100431y. [DOI] [PubMed] [Google Scholar]
- Nusz GJ, Curry AC, Marinakos SM, Wax A, Chilkoti A. ACS Nano. 2009;3(4):795–806. doi: 10.1021/nn8006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen J, Lehtinen K, Erho T. Anal. Chem. 2010;82(24):10246–10250. doi: 10.1021/ac1027066. [DOI] [PubMed] [Google Scholar]
- Orendorff CJ, Murphy CJ. J. Phys. Chem. B. 2006;110(9):3990–3994. doi: 10.1021/jp0570972. [DOI] [PubMed] [Google Scholar]
- Osborn JL, Lutz B, Fu E, Kauffman P, Stevens DY, Yager P. Lab Chip. 2010;10(20):2659–2665. doi: 10.1039/c004821f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolo C, Merkoci A. Chem. Soc. Rev. 2013;42(2):450–457. doi: 10.1039/c2cs35255a. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Santos I, Pérez-Juste J, Liz-Marzán LM. Chem. Mater. 2006;18(10):2465–2467. [Google Scholar]
- Qu L-L, Li D-W, Xue J-Q, Zhai W-L, Fossey JS, Long Y-T. Lab Chip. 2012;12(5):876–881. doi: 10.1039/c2lc20926h. [DOI] [PubMed] [Google Scholar]
- Riboh JC, Haes AJ, McFarland AD, Ranjit Yonzon C, Van Duyne RP. J. Phys. Chem. B. 2003;107(8):1772–1780. [Google Scholar]
- Rosi NL, Mirkin CA. Chem. Rev. 2005;105(4):1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Russo A, Ahn BY, Adams JJ, Duoss EB, Bernhard JT, Lewis JA. Adv. Mater. 2011;23(30):3426–3430. doi: 10.1002/adma.201101328. [DOI] [PubMed] [Google Scholar]
- Rycenga M, Langille MR, Personick ML, Ozel T, Mirkin CA. Nano Lett. 2012;12(12):6218–6222. doi: 10.1021/nl3032235. [DOI] [PubMed] [Google Scholar]
- Sepúlveda B, Angelomé PC, Lechuga LM, Liz-Marzán LM. Nano Today. 2009;4(3):244–251. [Google Scholar]
- Svedendahl M, Chen S, Dmitriev A, Käll M. Nano Lett. 2009;9(12):4428–4433. doi: 10.1021/nl902721z. [DOI] [PubMed] [Google Scholar]
- Tian L, Chen E, Gandra N, Abbas A, Singamaneni S. Langmuir. 2012a;28(50):17435–17442. doi: 10.1021/la3034534. [DOI] [PubMed] [Google Scholar]
- Tian L, Fei M, Kattumenu R, Abbas A, Singamaneni S. Nanotechnology. 2012b;23(25):255502. doi: 10.1088/0957-4484/23/25/255502. [DOI] [PubMed] [Google Scholar]
- Tian L, Morrissey JJ, Kattumenu R, Gandra N, Kharasch ED, Singamaneni S. Anal. Chem. 2012c;84(22):9928–9934. doi: 10.1021/ac302332g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonzon CR, Jeoung E, Zou S, Schatz GC, Mrksich M, Van Duyne RP. J. Am. Chem. Soc. 2004;126(39):12669–12676. doi: 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- Yu WW, White IM. Analyst. 2013;138(4):1020–1025. doi: 10.1039/c2an36116g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.