Abstract
Analysis of cerebrospinal fluid (CSF) offers key insight into the status of the central nervous system. Characterization of murine CSF proteomes can provide a valuable resource for studying central nervous system injury and disease in animal models. However, the small volume of CSF in mice has thus far limited individual mouse proteome characterization. Through non-terminal CSF extractions in C57Bl/6 mice and high-resolution 2D-LC MS/MS analysis of individual murine samples, we report the most comprehensive proteome characterization of individual murine CSF to date. We identified a total of 566 unique proteins, including 128 proteins from three individual CSF samples that have been previously identified in brain tissue. Our methods and analysis provide a mechanism for individual murine CSF proteome analysis. The data are available in the ProteomeXchange with identifier PXD000248.
Keywords: Cerebrospinal fluid, CSF, mouse, proteomics, proteome, mass spectrometry
MS-based proteomics is providing increasingly detailed views of human CSF proteomes in both healthy and diseased states [1–4]. Because of its contact with brain tissue and relative isolation from other tissues, in part due to the blood brain barrier [5], CSF can provide insights into the pathophysiology of central nervous system (CNS) disease, such as Alzheimer’s, meningitis, multiple sclerosis, and CNS tumors [6]. While the mouse blood/plasma proteome has been studied in increasing detail [7,8], little is known about the mouse CSF proteome despite its use for controlled testing of hypotheses of human disease states. Protein concentration in CSF is typically ~0.2%–0.5% that of blood, presenting analysis challenges [9]. Studies of the murine CSF proteome have been limited by sample collection, small volumes, sample preparation (e.g. centrifuge samples to remove cellular contaminants), and measurement sensitivity [10–12]. The small CSF volume of ~36 microliters [13] is perhaps the most significant challenge. Mouse CSF replenishes rapidly at ~3.3×10−4 ml/min [17].
To collect samples, three Isoflurane-anesthetized wild-type male C57BL/6 mice (Charles River) were mounted on a stereotaxic system (David Kopf Instruments). After drilling through the skull, a 5-microliter syringe (Hamilton) was lowered in the lateral ventricle (1.0mm lateral, 0.4–0.5mm posterior, 2.5–3.0mm depth from bregma). CSF was withdrawn at a rate of 400nL/minute. Approximately 4uL of CSF was collected per mouse, comparable to previous studies [12,14]. Samples were extracted using a non-terminal procedure and analyzed individually (not pooled). Protocols were approved by the University of Washington Institutional Animal Care and Use Committee. After extraction, CSF was transferred to a sterile vial, snap frozen in liquid nitrogen, and stored at −80C° until analysis preparation. CSF proteins were denatured following addition of trifluoroethanol (final concentration 50% v/v) and sonicated for 1-min in a bath sonicator at RT, then incubated for 2-hrs at 60C°. The CSF proteins were reduced by DTT to a final concentration of 2mM, sonicated for 1-min at RT, and incubated at 37C° for 1-hr. Protein concentration was estimated at 0.3mg/ml. Samples were diluted 5-fold with 50mM NH4HCO3 and proteins were digested following the addition of trypsin at an estimated 1:50 (w/w) trypsin-to-protein ratio and incubated overnight at 37C°.
Samples were subjected to 2D-LC (i.e. LC/LC)-MS/MS analysis. The 2D-LC system was custom built using two Agilent 1200 nanoflow pumps and one 1200 capillary pump (Agilent Technologies). Use of dual trapping and reversed-phase columns allowed for parallel event coordination, allowing for fraction trapping and washing offline while analytical separation occurred on the other reversed-phase column. Columns were manufactured in-house by slurry packing media into fused silica (Polymicro Technologies Inc) using a 1cm sol-gel frit for media retention. First dimension SCX column; 5-Nm PolySULFOETHYL-A (PolyLC Inc.), 15cm × 360um o.d. × 150um i.d. Trapping columns; 5-Nm Jupiter C18 (Phenomenex), 4cm × 360um o.d. × 150um i.d. Second dimension reversed-phase columns; 3-um Jupiter C18 (Phenomenex), 35cm × 360um o.d. × 75um i.d. Mobile phases consisted of 0.1mM NaH2PO4 (A) and 0.3M NaH2PO4 (B) for the first dimension and 0.1% formic acid in water (A) and 0.1% formic acid acetonitrile (B) for the second dimension. The SCX separation provided 10 fractions for the second dimension reversed phase gradient nanoLC; 20uL for each fraction having an estimated concentration of ~10ng/uL, totaling ~200ng. MS analysis used a LTQ Orbitrap Velos mass spectrometer (ThermoScientific) using 150um o.d. × 20um i.d. chemically etched fused silica electrospray emitters [15]. The SCX separation for 10 fractions was performed stepwise, each step taking 20-min. Each fraction was trapped on C18 material and washed for 50-min using the second dimension mobile phase A. The gradient was then started and 15-min later acquisition was started and continued for 100-min, with overlapping steps this resulted in 1085-min of total analysis time per sample. Electrospray emitters were chemically etched fused silica 150μm o.d. × 20μm i.d. Orbitrap spectra (AGC 1×106) were collected from 400–2000 m/z at a resolution of 100k followed by data dependent ion trap CID (collision energy 35%, AGC 3×104) and second-stage MS analysis of the ten most abundant ions and a dynamic exclusion time of 180-sec.
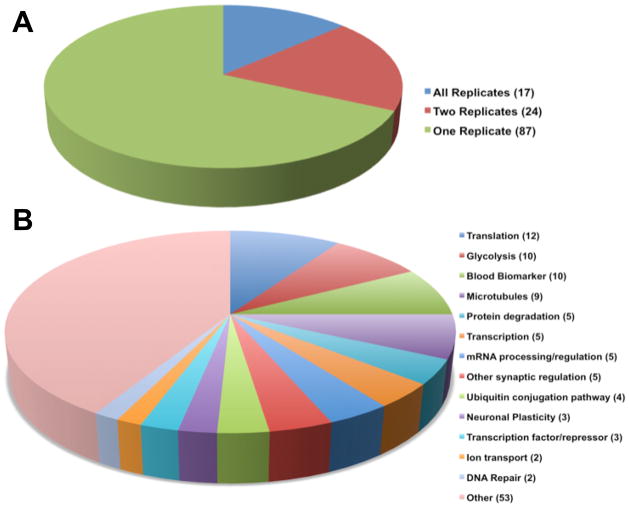
In three samples, 566 unique proteins were identified at a false discovery rate (FDR) of 0.5% at the spectrum level (~1% at the unique peptide level, and ~3% at the protein level). To further reduce false positives, we excluded proteins not identified by ≥2 unique peptides. Of 566 total proteins identified, 261 (46%) met this ≥2 unique peptide criteria. 128 of the 261 were found previously in mouse brain (49%). A similar number of unique proteins were in each of the three samples, although the number of brain-specific proteins varied due to factors including inherent under-sampling of shotgun measurements [16]. We identified 102 unique proteins that met our criteria from mouse 1; 30 previously identified in brain tissue (29%). In mouse 2, we identified 214 unique proteins; 128 previously found in brain tissue (60%). In mouse 3, we identified 74 unique proteins; 20 previously identified in brain tissue (27%). All proteins identified in the first and third CSF samples were also identified in the second. Seventeen of the 128 total proteins found in brain tissue were identified across all three CSF samples (Figure 1A). UNIPROT database was used to determine protein functionality (Figure 1B) [18], with proteomics data uploaded to The Proteomics Identifications (PRIDE) database [19].
Figure 1. Distribution and function of proteins identified by at least two unique peptides and 0.5% FDR across biological replicates.
A) An overview of the distribution of unique proteins found in brain tissue identified across the three samples. 17 such proteins were identified in all three CSF replicates, 24 proteins in two of the three CSF replicates, and 87 proteins in one replicate. B) Proteins grouped by similar biological function.
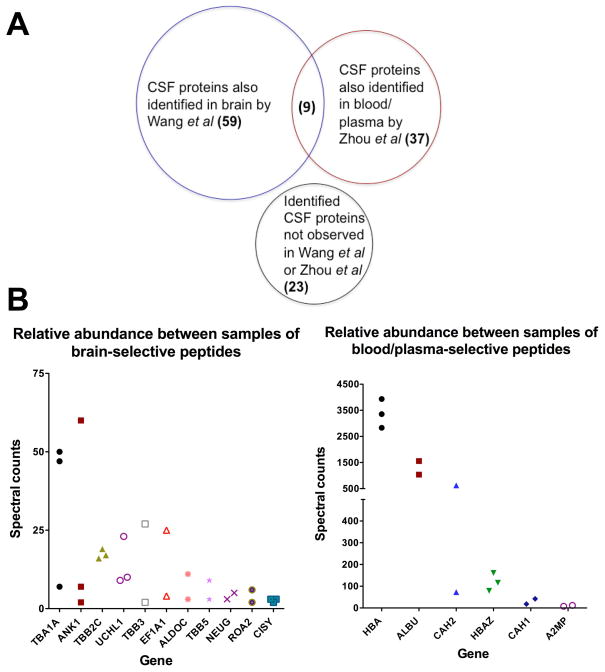
Supplemental Table 1 provides a list of proteins identified by our criteria. The most abundant proteins, including hemoglobin subunits, albumin, carbonic anhydrase, can be attributed to blood contamination. Nevertheless, our multidimensional analysis enabled the confident identification of CSF proteins, including synapsin-1 and synapsin-2, tubulin alpha 1-a chain, alpha-synuclein, neurogranin, calcium/calmodulin-dependent protein kinase type II subunit alpha, and microtubule-associated protein 6. We compared proteins identified in CSF to proteins previously identified in mouse brain tissue [17] and plasma [8]. We expected that the mouse CSF proteome would more closely align with the mouse brain tissue proteome than the plasma proteome if blood/plasma contamination of the CSF were minimal. Conversely, if blood/plasma contamination of mouse CSF were considerable, we expected to identity few proteins exclusive to brain tissue. Of the 128 proteins, 59 of the proteins (46%) were shown by Wang et al. to be found in brain tissue, but not found by Zhou et al. in blood plasma. Thirty-seven proteins (29%) were identified in both brain tissue and blood/plasma. Nine of the proteins (7%) were identified in both the UNIPROT database as expressed in brain tissue and found in mouse blood/plasma in Zhou et al. [8,17]. However, these nine proteins were not identified in brain tissue by Wang et al. [17]. Twenty-three proteins (18%) identified in the UNIPROT database as expressed in brain tissue and were neither identified by Wang et al. nor Zhou et al. (Figure 2A). Relative abundances (as spectral counts) of unique proteins previously observed in murine brain [8] or murine blood/plasma [17] and identified in at least two of the three mouse CSF samples were compared (Figure 2B).
Figure 2. Literature comparison of murine brain and plasma proteins.
A) Comparison of unique proteins identified in murine CSF to previously reported unique proteins in the brain [17] and plasma [8]. The remaining proteins (“Other”) have been identified in murine brain tissue by other studies [18]. B) Relative abundance, measured by spectral counts, of unique proteins detected across at least two of the three CSF samples previously identified in Wang et al. in murine brain tissue or Zhou et al. in plasma. HBA, ALBU, CAH2, and HBAZ were categorized as “blood/plasma-selective,” although Wang et al. identified these proteins in brain tissue as well.
Of the 37 proteins identified by Wang et al. and Zhou et al. in both brain tissue and blood/plasma, many are critical to general functionality in heterogeneous cell types. Proteins essential for glycolysis (Triosephosphate isomerase, Pyruvate kinase isozymes M1/M2, Fructose-bisphosphate aldolase A, Phosphoglycerate kinase 1, L-lactate dehydrogenase A-chain, L-lactate dehydrogenase B-chain, Phosphoglycerate mutase 1) were detected in brain tissue and blood/plasma [8,17]. The histone protein H4, as well as ubiquitously expressed 14-3-3 proteins critical for regulation of intracellular signaling, were identified in both brain tissue and blood/plasma.
While the four most abundant proteins identified in mouse CSF were almost certainly due to blood contamination, the high relative abundance of blood components did not preclude identification of brain-derived proteins. We also note that because the blood-brain barrier is not impermeable [20], it is possible that our brain tissue protein identification criteria excluded proteins normally found in mouse CSF, but that are not found in brain tissue. Mouse 2 CSF analysis yielded more brain-derived proteins than mouse 1 and 3, likely because of reduced blood contamination. Consistent with this explanation, CSF from mouse 2 had the lowest spectral counts of hemoglobin subunit alpha. In our analysis, we also observed proteins associated with cytoplasmic functions, such as ribosomal subunit proteins. These findings are consistent with human CSF proteome, where ribosomal proteins were also found [4]. Many factors could account for the presence of cytoplasmic proteins in human and mouse CSF samples, including glial cell turn over, neural pruning, or potentially cell lysis during extraction.
Comparisons of the mouse CSF proteome to the brain tissue and plasma proteomes revealed the greatest overlap with proteins identified solely in brain tissue, supporting the effectiveness of our extraction and analytical methodology for reducing blood/plasma contamination. A sizeable number of proteins discovered in CSF also overlapped with both the brain tissue and blood/plasma proteomes. Most of these overlapping proteins are relatively abundant and with annotated “housekeeping” functions, allowing heterogeneous cell types to perform tasks vital for survival. However, other proteins identified in mouse CSF found in both mouse brain tissue and blood/plasma have critical roles within the CNS. Proteins such as neurogranin and calcium/calmodulin dependent protein kinase are mediators of intracellular calcium concentrations, which are critical to neurotransmitter release and synaptic plasticity [21]. Multiple proteins (or protein isomers) found in mouse CSF including Synapsin-1, Tubulin beta-2A chain, Ubiquitin carboxyl-terminal hydrolase isozyme-L1, and Alpha-adducin, were also identified in the human CSF proteome, but not in human plasma [4]. Proteins previously identified in the supernatant of mouse choroid plexus cell cultures including plasminogen activator inhibitor, complement factor H, and Apolipoprotein-E [22], were also identified.
A recent study identified 289 proteins in mouse CSF through pooling 10 collections from 10 different mice [12]. Our study builds upon these results by providing the most detailed view to date of individual mice CSF proteomes, and demonstrates a method for detailed proteomic analysis suitable for the associated small volume and low protein concentration CSF samples. Comparisons with previous murine samples and normal human CSF further validate extraction techniques and support utilizing individual mouse CSF sample analysis. Increases in analytical sensitivity provided by MS-based approaches are dramatically enhancing biomarker development opportunities [23]. The present data provide a foundation for utilizing mouse models for CSF biomarker development, CNS disease progression, and a basis for selection of a more limited set of peptide targets for quantitative measurements using ultra-sensitive targeted MS measurements [24].
Supplementary Material
Supplemental Table 1. Summary of the unique proteins identified across three individual mice by independent CSF samples. Column 1 contains UNIPROT Gene ID (for mouse). Column 2 contains the protein name corresponding to the gene. Column 3 briefly summarizes the function attributed to the protein. Columns 4–6 contain the sum of spectral counts (SC) for peptides for the listed protein identified in mouse 1, 2, and 3 CSF samples, respectively. Columns 7–9 catalog the number of unique peptide identified for the listed protein in mouse 1, 2, and 3 CSF samples, respectively. Column 10 gives the sum of total spectrum counts for each protein in samples 1–3, which provides an estimate of protein relative abundance.
Acknowledgments
Portions of this work were supported by the National Institute of General Medical Sciences (P41GM103493). Research was performed at the University of Washington and at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
References
- 1.Hu Y, Malone JP, Fagan AM, Townsend RR, et al. Comparative proteomic analysis of intra- and inter-individual variation in human cerebrospinal fluid. Mol Cell Proteomics. 2005 Dec;4(12):2000–9. doi: 10.1074/mcp.M500207-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Spudich SS, Nilsson AC, Lollo ND, et al. Cerebrospinal fluid HIV infection and pleocytosis: relation to systemic infection and antiretroviral treatment. BMC Infect Dis. 2005;5:98. doi: 10.1186/1471-2334-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schutzer SE, Liu T, Natelson BH, Angel TE, et al. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010 Jun 11;5(6):e10980. doi: 10.1371/journal.pone.0010980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angel TE, Jacobs JM, Smith RP, Pasternack MS, et al. Cerebrospinal fluid proteome of patients with acute Lyme disease. J Proteome Res. 2012 Oct 5;11(10):4814–22. doi: 10.1021/pr300577p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelhardt B, Sorokin L. The blood–brain and the blood-cerebrospinal fluid barriers: function and dysfunction. Semin Immunopathol. 2009;31:497–511. doi: 10.1007/s00281-009-0177-0. [DOI] [PubMed] [Google Scholar]
- 6.Krizanac-Bengez L, Mayberg MR, Janigro D. The cerebral vasculature as a therapeutic target for neurological disorders and the role of shear stress in vascular homeostatis and pathophysiology. Neurol Res. 2004 Dec;26(8):846–53. doi: 10.1179/016164104X3789. [DOI] [PubMed] [Google Scholar]
- 7.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, et al. In depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 8.Zhou JY, Petritis BO, Petritis K, Norbeck AD, et al. Mouse-specific tandem IgY7-SuperMix immunoaffinity separations for improved LC-MS/MS coverage of the plasma proteome. J Proteome Res. 2009 Nov;8(11):5387–95. doi: 10.1021/pr900564f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You JS, Gelfanova V, Knierman MD, Witzmann FA, et al. The impact of blood contamination on the proteome of cerebrospinal fluid. Proteomics. 2005 Jan;5(1):290–6. doi: 10.1002/pmic.200400889. [DOI] [PubMed] [Google Scholar]
- 10.Lafon-Cazal M, Adjali O, Galotti N, Poncet J, et al. Proteomic analysis of astrocytic secretion in the mouse. Comparison with the cerebrospinal fluid proteome. Biol Chem. 2003;278(27):24438–48. doi: 10.1074/jbc.M211980200. [DOI] [PubMed] [Google Scholar]
- 11.Swan EE, Peppi M, Chen Z, Green KMx, et al. Proteomics analysis of perilymph and cerebrospinal fluid in mouse. Laryngoscope. 2009 May;119(5):953–8. doi: 10.1002/lary.20209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham R, Jany P, Messing A, Li L. Protein changes in immunodepleted cerebrospinal fluid from transgenic mouse models of Alexander disease detected using mass spectrometry. J Proteome Res. 2013 Feb 1;12(2):719–28. doi: 10.1021/pr300785h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudick RA, Zirretta DK, Herndon RM. Clearance of albumin from mouse subarachnoid space: a measure of CSF bulk flow. J Neurosci Methods. 1982 Sep;6(3):253–9. doi: 10.1016/0165-0270(82)90088-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Duff K. A Technique for Serial Collection of Cerebrospinal Fluid from the Cisterna Magna in Mouse. J Vis Exp. 2008 Nov 10;(21) doi: 10.3791/960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly RT, Page JS, Luo Q, Moore RJ, et al. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem. 2006 Nov 15;78(22):7796–801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001 Dec 1;73(23):5683–90. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Qian WJ, Mottaz HM, Clauss TR, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005 Nov-Dec;4(6):2397–403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vizcaino JA, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013 Jan 1;41(D1):D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quaegebeur A, Lange C, Carmeliet P. The neurovascular link in health and disease: molecular mechanisms and therapeutic implications. Neuron. 2011 Aug 11;71(3):406–24. doi: 10.1016/j.neuron.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhong L, Gerges NZ. Neurogranin targets calmodulin and lowers the threshold for the induction of long-term potentiation. PLoS One. 2012;7(7):e41275. doi: 10.1371/journal.pone.0041275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thouvenot E, Lafon-Cazal M, Demettre E, Jouin P, et al. The proteomic analysis of mouse choroid plexus secretome reveals a high protein secretion capacity of choroidal epithelial cells. Proteomics. 2006 Nov;6(22):5941–52. doi: 10.1002/pmic.200600096. [DOI] [PubMed] [Google Scholar]
- 23.Shi T, Su D, Liu T, Tang K, et al. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012 Apr;12(8):1074–92. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson PL, Smith RD. Proteome analysis by mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:399–424. doi: 10.1146/annurev.biophys.32.110601.141854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Summary of the unique proteins identified across three individual mice by independent CSF samples. Column 1 contains UNIPROT Gene ID (for mouse). Column 2 contains the protein name corresponding to the gene. Column 3 briefly summarizes the function attributed to the protein. Columns 4–6 contain the sum of spectral counts (SC) for peptides for the listed protein identified in mouse 1, 2, and 3 CSF samples, respectively. Columns 7–9 catalog the number of unique peptide identified for the listed protein in mouse 1, 2, and 3 CSF samples, respectively. Column 10 gives the sum of total spectrum counts for each protein in samples 1–3, which provides an estimate of protein relative abundance.