Abstract
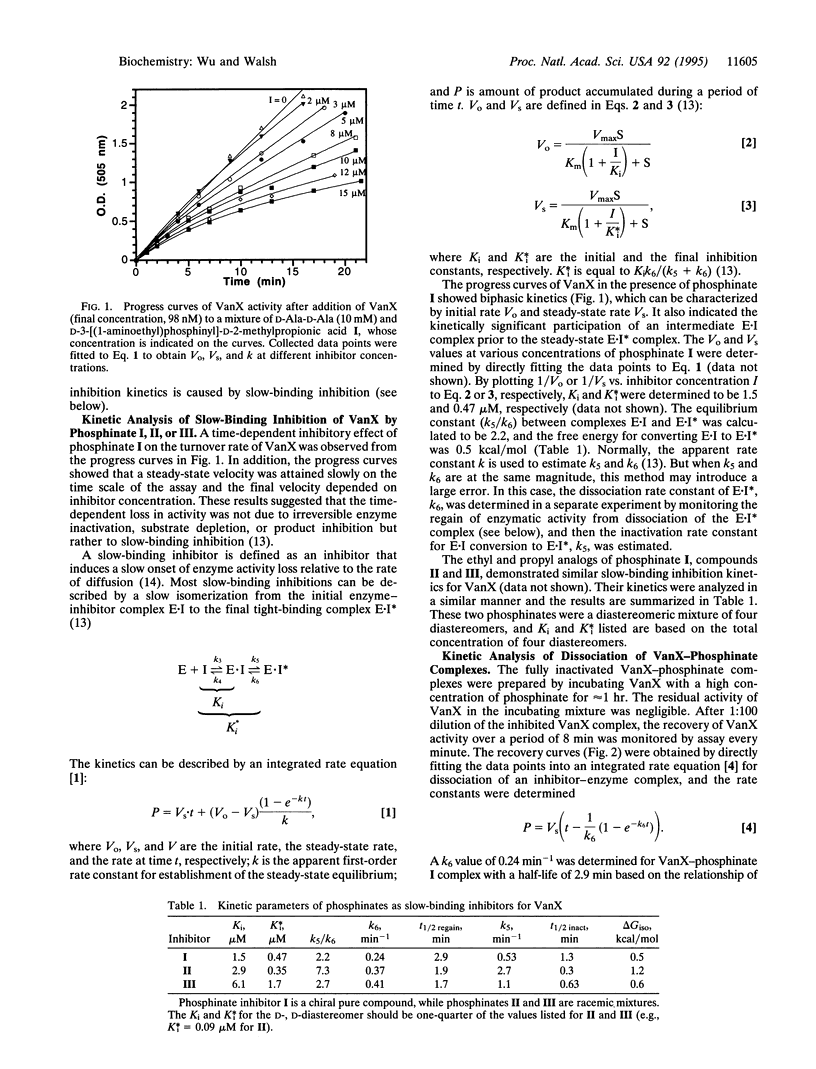
VanX is a D-Ala-D-Ala dipeptidase that is essential for vancomycin resistance in Enterococcus faecium. Contrary to most proteases and peptidases, it prefers to hydrolyze the amino substrate but not the related kinetically and thermodynamically more favorable ester substrate D-Ala-D-lactate. The enzymatic activity of VanX was previously found to be inhibited by the phosphinate analogs of the proposed tetrahedral intermediate for hydrolysis of D-Ala-D-Ala. Here we report that such phosphinates are slow-binding inhibitors. D-3-[(1-Aminoethyl)phosphinyl]-D-2-methylpropionic acid I showed a time-dependent onset of inhibition of VanX and a time-dependent return to uninhibited steady-state rates upon dilution of the enzyme/inhibitor mixture. The initial inhibition constant Ki after immediate addition of VanX to phosphinate I to form the E-I complex is 1.5 microM but is then lowered by a relatively slow isomerization step to a second complex, E-I*, with a final K*i of 0.47 microM. This slow-binding inhibition reflects a Km/K*i ratio of 2900:1. The rate constant for the slow dissociation of complex E-I* is 0.24 min-1. A phosphinate analog with an ethyl group replacing what would be the side chain of the second D-alanyl residue in the normal tetrahedral adduct gives a K*i value of 90 nM. Partial proteolysis of VanX reveals two protease-sensitive loop regions that are protected by the intermediate analog phosphinate, indicating that they may be part of the VanX active site.
Full text
PDF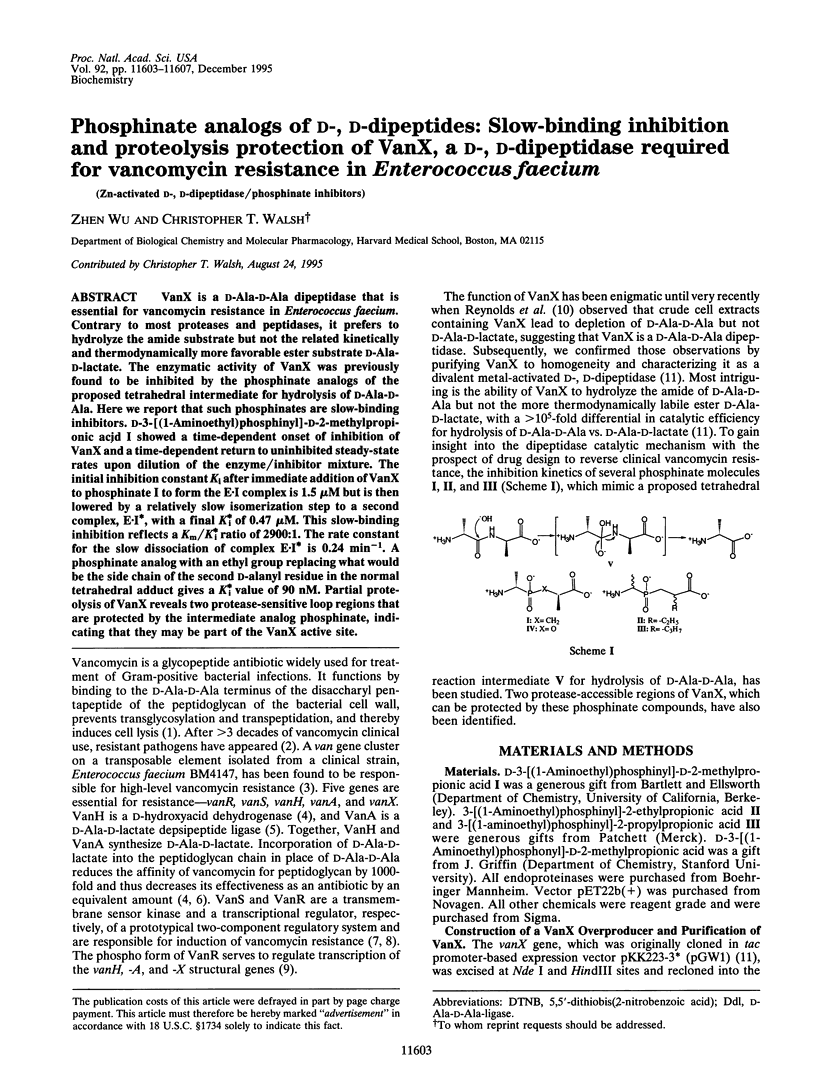


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur M., Molinas C., Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992 Apr;174(8):2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Depardieu F., Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993 Jan;175(1):117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna J. C., Williams D. H. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- Bugg T. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. Identification of vancomycin resistance protein VanA as a D-alanine:D-alanine ligase of altered substrate specificity. Biochemistry. 1991 Feb 26;30(8):2017–2021. doi: 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- Bugg T. D., Wright G. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991 Oct 29;30(43):10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Duncan K., Walsh C. T. ATP-dependent inactivation and slow binding inhibition of Salmonella typhimurium D-alanine:D-alanine ligase (ADP) by (aminoalkyl)phosphinate and aminophosphonate analogues of D-alanine. Biochemistry. 1988 May 17;27(10):3709–3714. doi: 10.1021/bi00410a028. [DOI] [PubMed] [Google Scholar]
- Erion M. D., Walsh C. T. 1-Aminocyclopropanephosphonate: time-dependent inactivation of 1-aminocyclopropanecarboxylate deaminase and Bacillus stearothermophilus alanine racemase by slow dissociation behavior. Biochemistry. 1987 Jun 16;26(12):3417–3425. doi: 10.1021/bi00386a025. [DOI] [PubMed] [Google Scholar]
- Fan C., Moews P. C., Walsh C. T., Knox J. R. Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 A resolution. Science. 1994 Oct 21;266(5184):439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- Holman T. R., Wu Z., Wanner B. L., Walsh C. T. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994 Apr 19;33(15):4625–4631. doi: 10.1021/bi00181a024. [DOI] [PubMed] [Google Scholar]
- Keck W., van Leeuwen A. M., Huber M., Goodell E. W. Cloning and characterization of mepA, the structural gene of the penicillin-insensitive murein endopeptidase from Escherichia coli. Mol Microbiol. 1990 Feb;4(2):209–219. doi: 10.1111/j.1365-2958.1990.tb00588.x. [DOI] [PubMed] [Google Scholar]
- Leclercq R., Derlot E., Duval J., Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988 Jul 21;319(3):157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- Messer J., Reynolds P. E. Modified peptidoglycan precursors produced by glycopeptide-resistant enterococci. FEMS Microbiol Lett. 1992 Jul 1;73(1-2):195–200. doi: 10.1016/0378-1097(92)90608-q. [DOI] [PubMed] [Google Scholar]
- Parsons W. H., Patchett A. A., Bull H. G., Schoen W. R., Taub D., Davidson J., Combs P. L., Springer J. P., Gadebusch H., Weissberger B. Phosphinic acid inhibitors of D-alanyl-D-alanine ligase. J Med Chem. 1988 Sep;31(9):1772–1778. doi: 10.1021/jm00117a017. [DOI] [PubMed] [Google Scholar]
- Reynolds P. E., Depardieu F., Dutka-Malen S., Arthur M., Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol Microbiol. 1994 Sep;13(6):1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Shi Y., Walsh C. T. Active site mapping of Escherichia coli D-Ala-D-Ala ligase by structure-based mutagenesis. Biochemistry. 1995 Mar 7;34(9):2768–2776. doi: 10.1021/bi00009a005. [DOI] [PubMed] [Google Scholar]
- Wright G. D., Holman T. R., Walsh C. T. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993 May 18;32(19):5057–5063. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- Wright G. D., Walsh C. T. Identification of a common protease-sensitive region in D-alanyl-D-alanine and D-alanyl-D-lactate ligases and photoaffinity labeling with 8-azido ATP. Protein Sci. 1993 Oct;2(10):1765–1769. doi: 10.1002/pro.5560021020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Wright G. D., Walsh C. T. Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1995 Feb 28;34(8):2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]



