ABSTRACT
Acinetobacter baumannii is recognized as an emerging bacterial pathogen because of traits such as prolonged survival in a desiccated state, effective nosocomial transmission, and an inherent ability to acquire antibiotic resistance genes. A pressing need in the field of A. baumannii research is a suitable model strain that is representative of current clinical isolates, is highly virulent in established animal models, and can be genetically manipulated. To identify a suitable strain, a genetically diverse set of recent U.S. military clinical isolates was assessed. Pulsed-field gel electrophoresis and multiplex PCR determined the genetic diversity of 33 A. baumannii isolates. Subsequently, five representative isolates were tested in murine pulmonary and Galleria mellonella models of infection. Infections with one strain, AB5075, were considerably more severe in both animal models than those with other isolates, as there was a significant decrease in survival rates. AB5075 also caused osteomyelitis in a rat open fracture model, while another isolate did not. Additionally, a Tn5 transposon library was successfully generated in AB5075, and the insertion of exogenous genes into the AB5075 chromosome via Tn7 was completed, suggesting that this isolate may be genetically amenable for research purposes. Finally, proof-of-concept experiments with the antibiotic rifampin showed that this strain can be used in animal models to assess therapies under numerous parameters, including survival rates and lung bacterial burden. We propose that AB5075 can serve as a model strain for A. baumannii pathogenesis due to its relatively recent isolation, multidrug resistance, reproducible virulence in animal models, and genetic tractability.
IMPORTANCE
The incidence of A. baumannii infections has increased over the last decade, and unfortunately, so has antibiotic resistance in this bacterial species. A. baumannii is now responsible for more than 10% of all hospital-acquired infections in the United States and has a >50% mortality rate in patients with sepsis and pneumonia. Most research on the pathogenicity of A. baumannii focused on isolates that are not truly representative of current multidrug-resistant strains isolated from patients. After screening of a panel of isolates in different in vitro and in vivo assays, the strain AB5075 was selected as more suitable for research because of its antibiotic resistance profile and increased virulence in animal models. Moreover, AB5075 is susceptible to tetracycline and hygromycin, which makes it amenable to genetic manipulation. Taken together, these traits make AB5075 a good candidate for use in studying virulence and pathogenicity of this species and testing novel antimicrobials.
INTRODUCTION
Acinetobacter baumannii is an opportunistic, Gram-negative pathogen that thrives in clinical settings and is often multidrug resistant (MDR), factors which earn it a place among the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens of clinical importance (1). Some recent isolates are resistant to all typically used antibiotics except colistin and tigecycline and thus are called extensively or extremely drug-resistant (XDR) A. baumannii (2). MDR/XDR A. baumannii strains are a worldwide problem for clinicians and caregivers in the hospital setting, particularly in the intensive care unit (ICU) (3). A. baumannii is also often isolated from infections of severe wounds sustained in military combat. These infections are responsible for increased morbidity, with prolonged wound healing and amputations of extremities when limbs cannot be salvaged (4, 5). A. baumannii was a predominant isolate from wounded soldiers serving in Iraq (4, 5) and was associated with wartime polytrauma injuries in the past (6). Additionally, there may be a link between A. baumannii and crush injuries, as A. baumannii infections were also prevalent after the recent large earthquakes in Haiti (7) and China (8).
Another disturbing development that has increased the clinical importance of A. baumannii infections is that many strains have become highly antibiotic resistant. For example, in previous decades, A. baumannii isolates obtained from both military and civilian settings were often carbapenem sensitive. Now, the majority of U.S. military isolates are carbapenem resistant (9). This trend has also been mirrored in civilian hospitals around the world (10). Recently, even colistin-resistant strains have emerged in the military health care system (11). The latter development is deeply troubling, as colistin is considered the last line of defense against these MDR isolates. Exacerbating this problem is the lack of new treatments in the pharmaceutical pipeline (12); therefore, research on A. baumannii virulence factors is urgently needed, as they could constitute potentially novel targets for future antimicrobials.
While previous studies attempted to examine the virulence of different clinical A. baumannii strains utilizing in vivo model systems (13, 14), the majority of A. baumannii researchers still use two American Type Culture Collection (ATCC) strains, ATCC 19606T and ATCC 17978, which were isolated more than 50 years ago and are not significantly antibiotic resistant. These strains are certainly more amenable to genetic manipulation than most clinical isolates (15, 16) and share considerable genome homology (>90%) to current A. baumannii isolates (17), but they are not representative of contemporary isolates of this rapidly evolving pathogen. Some researchers, recognizing that the ATCC isolates are dated, have performed studies with more recent clinical isolates; however, genetic manipulation of such isolates has depended on susceptibility to aminoglycosides (18–20), which is often not found in clinical strains (21). Therefore, our goal was to carry out a systematic study of our own contemporary clinical strains isolated from patients in the U.S. military health care system to identify a strain that is more representative of current clinical isolates, that is highly virulent in established model infections, and that can be genetically manipulated without a potential sacrifice with respect to virulence and antibiotic resistance. Not only does identifying such a strain account for more recent clinical outcomes, but the increased virulence in animal models allows greater statistical power in screening new therapeutics. Moreover, the ability to manipulate the genome allows the study of virulence factors, some of which may be responsible for the emergence of this pathogen in more recent years.
RESULTS
Defining genetic characteristics of A. baumannii isolates.
In order to identify potential reference strains, a diverse set of 33 A. baumannii isolates was chosen based on genetic, isolation site, and antibiotic resistance differences from more than 200 A. baumannii strains isolated between 2004 and 2010 from patients in the U.S. military health care system. AB0057, first isolated in 2004 at Walter Reed Army Medical Center, was also included as a comparator because this strain is well characterized, and its genome was previously sequenced (22).
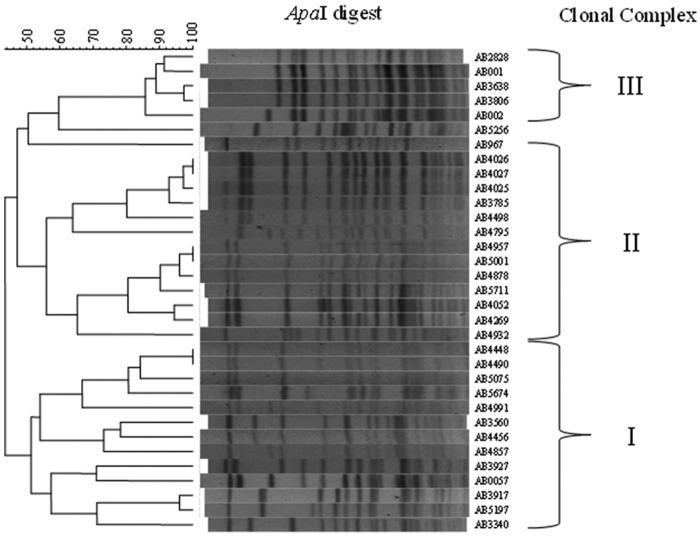
The diversity set of A. baumannii isolates was determined via pulsed-field gel electrophoresis (PFGE) analysis and a multiplex PCR assay previously developed to identify the international clonal complexes (ICC). Separately, antibiograms were determined using two different automated bacterial identification systems. The majority of strains were found to be multidrug resistant, typical of current clinical strains (see Table S1 in the supplemental material). As shown in Fig. 1, the genetic similarity of the strains, as determined by PFGE, ranged from 45 to 100%. PFGE types were considered to represent the same clones when their genetic similarity was >80% (23); based on this cutoff, the 33 strains represent 19 unique clones. When the genetic relatedness of these 19 clones was compared, it was found that the majority of them clustered into three groups, which generally aligned with the ICC designations determined by multiplex PCR (Fig. 1 and Table 1) (24). Exceptions were the isolates AB3560, AB4456, and AB4857, which were determined to be ICC III by the multiplex assay but appeared to be ICC I via PFGE. In this case, we relied on the multiplex data (Table 1) to be definitive. These data were used to select four representative strains for genome sequencing and evaluation in animal models.
FIG 1 .
Pulsed-field gel electrophoresis of A. baumannii strains. Genomic DNA was isolated from 33 A. baumannii clinical isolates, digested with ApaI, and separated by pulsed-field gel electrophoresis (PFGE). Patterns of electrophoresis were compared using BioNumerics 6.0 software. The ICC was determined by multiplex PCR analysis, and brackets delineate the approximate grouping of each strain.
TABLE 1 .
A. baumannii strains used in this studya
| Strain | MRSN | Isolation site | Yr isolated | Clonal complexb | Source |
|---|---|---|---|---|---|
| AB001 | 1332 | ND | ND | ND | C. Murray |
| AB002 | 1333 | ND | ND | ND | C. Murray |
| AB0057 | 1311 | Blood/sepsis | 2004 | I | This study |
| AB967 | 1308 | Blood/sepsis | 2003 | III | This study |
| AB2828 | 846 | Blood/sepsis | 2006 | III | This study |
| AB3340 | 847 | Blood/sepsis | 2006 | I | This study |
| AB3560 | 848 | Blood/sepsis | 2006 | III | This study |
| AB3638 | 849 | Posterior wound | 2007 | III | This study |
| AB3785 | 853 | Blood/sepsis | 2007 | II | This study |
| AB3806 | 854 | Leg wound | 2007 | III | This study |
| AB3917 | 1309 | Blood/sepsis | 2007 | ND | This study |
| AB3927 | 856 | Tibia/osteomyelitis | 2007 | I | This study |
| AB4025 | 858 | Femur/osteomyelitis | 2007 | II | This study |
| AB4026 | 859 | Fibula/osteomyelitis | 2007 | II | This study |
| AB4027 | 860 | Femur/osteomyelitis | 2007 | II | This study |
| AB4052 | 863 | War wound | 2007 | II | This study |
| AB4269 | 877 | War wound | 2007 | II | This study |
| AB4448 | 899 | War wound | 2007 | I | This study |
| AB4456 | 903 | Tracheal aspirate | 2007 | III | This study |
| AB4490 | 906 | War wound | 2008 | I | This study |
| AB4498 | 907 | Blood | 2008 | II | This study |
| AB4795 | 930 | Bone/osteomyelitis | 2008 | II | This study |
| AB4857 | 939 | Ischial/osteomyelitis | 2008 | III | This study |
| AB4878 | 941 | War wound | 2008 | II | This study |
| AB4932 | 949 | Sputum | 2008 | II | This study |
| AB4957 | 951 | Sacral/osteomyelitis | 2008 | II | This study |
| AB4991 | 953 | War wound | 2008 | I | This study |
| AB5001 | 954 | Blood/sepsis | 2008 | II | This study |
| AB5075 | 959 | Tibia/osteomyelitis | 2008 | I | This study |
| AB5197 | 960 | STS/tissue | 2008 | I | This study |
| AB5256 | 961 | Blood/sepsis | 2009 | NA | This study |
| AB5674 | 963 | Blood/sepsis | 2009 | I | This study |
| AB5711 | 1310 | Blood/sepsis | 2009 | II | This study |
| ATCC 19606T | NA | Urine | 1948 | ND | ATCC |
| ATCC 17978 | NA | Spinal meningitis | 1951 | ND | ATCC |
| RUH134 | NA | Urine | 1982 | II | L. Dijkshoorn |
| RUH875 | NA | Urine | 1984 | I | L. Dijkshoorn |
| RUH5875 | NA | Unknown, Netherlands | 1997 | III | L. Dijkshoorn |
| ACICU | NA | Outbreak isolate, Rome, Italy | 2005 | II | M. Tolmasky |
MRSN, The Multidrug-resistant Organism Repository and Surveillance Network; ND, no data; NA, not applicable; STS, sterile swab site (most likely from an infected wound).
As determined by multiplex assay performed in this study. AB5256 was considered NA because only the OXA-51 amplicon was amplified from group 1 primer set (31).
Three of the strains chosen each represented one of the three ICC groups, AB5075 (ICC I), AB5711 (ICC II), and AB4857 (ICC III). The fourth strain, AB5256 was an outlier, as the OXA-51 allele from this strain was amplified with group 1 primers (24), while the csuE allele was not. The isolates were sequenced (25) and compared to previously sequenced A. baumannii genomes using the BLAST score ratio (BSR) approach (26). This method compares putative peptides encoded in each genome based on the ratio of BLAST scores to determine if they are conserved (BSR value ≥ 0.8), divergent (0.8 > BSR > 0.4), or unique (BSR < 0.4). The majority of the proteomes were similar among strains, meaning they had a BSR of >0.4; however, each isolate also had a set of unique proteins (see Table S2 in the supplemental material). These results are similar to what has been found previously with MDR A. baumannii clinical isolates (17), suggesting that the strains used in this study are not genetic outliers.
Virulence assessed in the Galleria mellonella model.
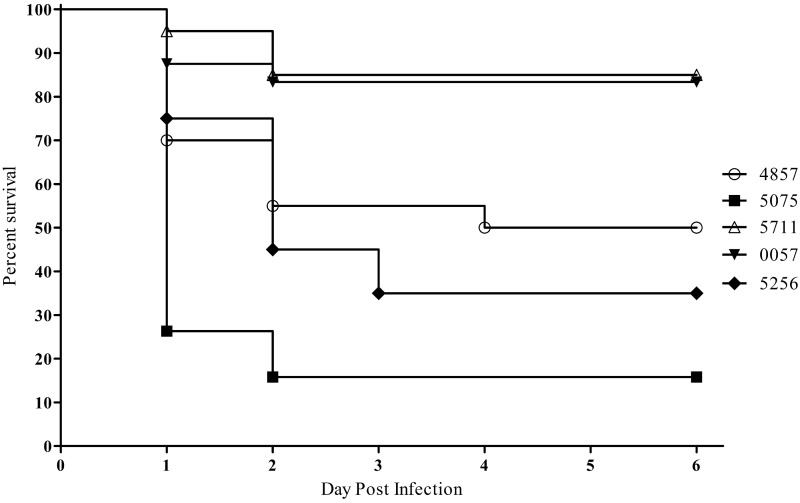
Strains were first tested in a Galleria mellonella infection model, as this model is well established to assess virulence and novel therapeutics for bacterial pathogens, including A. baumannii (27, 28). G. mellonella larvae were infected with an approximate dose of 1.0 × 105 CFU with each of the four sequenced A. baumannii isolates, AB4857, AB5075, AB5256, and AB5711, as well as control strain AB0057. Worms were observed for 6 days, and death was recorded. Within 24 h postinfection, approximately 25% of AB5075-infected worms remained, while the other four strains had survival rates of 70% or higher (Fig. 2). By the end of the 6-day study, AB5075-infected worms had a survival rate of 16%; strains AB4857 and AB5256 were considered moderately pathogenic in this model, with survival rates of 50% and 35%, respectively. The least lethal strains were AB5711 and AB0057, with survival rates of 85% and 83%, respectively. Phosphate-buffered saline (PBS)-injected control worms displayed 100% survival through the course of the study. Based on these data, it was hypothesized that AB5075 was more virulent than the other four strains tested. Using the Mantel-Cox test with Bonferroni correction for multiple comparisons, Kaplan-Meier curves were compared, and AB5075 was found statistically to be more lethal than AB4857, AB5711, and AB0057 (all P values < 0.0125). While AB5256 had a higher survival rate in this study than AB5075, the difference was not significant after the Bonferroni correction.
FIG 2 .
Survival of Galleria mellonella larvae infected with A. baumannii. Kaplan-Meier survival curves of G. mellonella infected with 1.0 × 105 CFU of selected strains of A. baumannii are shown. Curves were compared via the Mantel-Cox test with the Bonferroni correction for multiple comparisons. AB5075 showed significantly increased mortality compared to AB4857, AB5711, and AB0057 (P < 0.0125).
Separately, to compare the lethality of AB5075 to that of more commonly utilized A. baumannii model strains, the 50% lethal doses (LD50) of AB5075, ATCC 17978, and ATCC 19606T were determined in G. mellonella. The LD50 of AB5075 was 1.0 × 104 CFU. In contrast, the LD50 of ATCC 17978 and ATCC 19606T were 5.0 × 105 and 1.0 × 106, respectively.
Virulence assessed in the mouse pulmonary model.
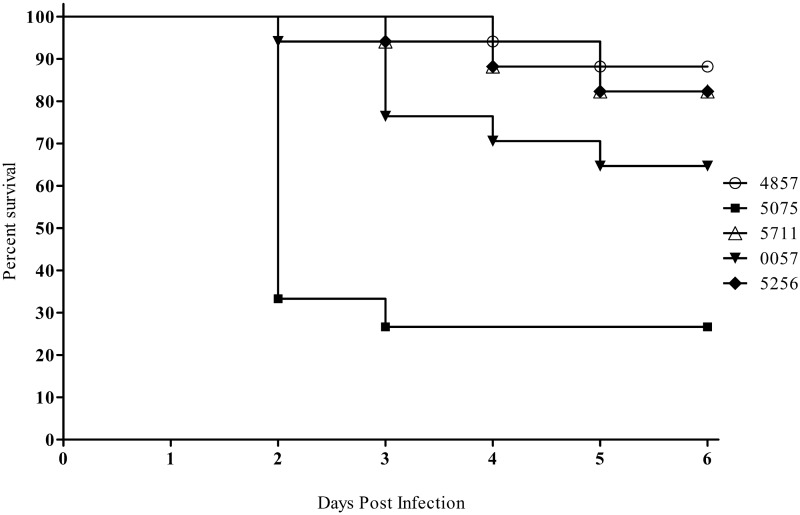
A. baumannii strains were examined in a murine pulmonary model of infection, because this model is commonly used to assess bacterial virulence and drug efficacy, and survival can be assessed rapidly after an inoculum is delivered (14, 29). The animals were immunocompromised with two doses of cyclophosphamide before inoculation, a treatment that allows A. baumannii to establish an infection (14). Mice were inoculated on day 0 with one of the five representative A. baumannii strains at a dose of 5.0 × 106 CFU and monitored for 6 days. Consistently, mice infected with AB5075 had a mortality rate of 70% within 48 to 72 h and a 6-day survival rate of 25% (Fig. 3). The other four strains tested, AB0057, AB5711, AB5256, and AB4857, were less lethal than AB5075 in this model, with 6-day survival rates of 65, 80, 80, and 85%, respectively. The log-rank (Mantel-Cox) test with Bonferroni correction determined that AB5075 time-to-death results were statistically significant compared to the other clinical isolates (P < 0.0125). The clinical scores of each infected animal also correlated with the survival plots; AB5075-infected mice displayed more severe illness than mice infected with the other strains. In a separate experiment, mice inoculated with 5.0 × 106 cells of ATCC 17978 or ATCC 19606T resulted in minimal clinical scores and no animal death (data not shown).
FIG 3 .
Assessment of A. baumannii virulence with the mouse pulmonary model. Kaplan-Meier survival curves of mice infected with 5.0 × 106 CFU of selected A. baumannii strains. Curves were compared via the Mantel-Cox test with the Bonferroni correction for multiple comparisons. AB5075 showed significantly increased mortality compared to AB4857, AB5711, AB0057, and AB5256 (P < 0.0125).
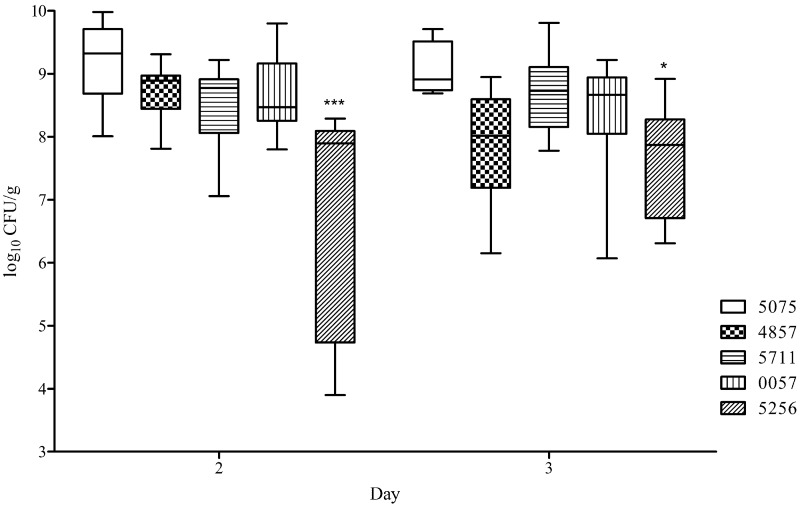
Lung bacterial burden in these infections was assessed on days 2 and 3, at the height of illness. The lung CFU/g values for the five infecting strains were compared using a Kruskal-Wallis test followed by a Dunn’s multiple comparison test. On day 2, the lungs of AB5256-infected mice had significantly less A. baumannii than AB4857-, AB0057-, and AB5075-infected lungs (P ≤ 0.05) but not AB5711-infected lungs. On day 3, AB5256 displayed a significant decrease in CFU/g compared only to AB5075 (Fig. 4). The four other strains assessed in the model had no significant difference in lung bacterial burden, with the median log CFU/g ranging from 8.5 to 9.3 on day 2 and 8.0 to 8.9 on day 3.
FIG 4 .
Bacterial levels in lung tissue in the mouse pulmonary model. Box-and-whisker plots of CFU burdens in lungs are shown for days 2 and 3 postinfection. Boxes show median and interquartile ranges, while whiskers represent the 95% confidence interval CI. Strains were compared via the Kruskal-Wallis test followed by Dunn’s multiple comparisons posttests. * and ***, P < 0.05 and 0.001, respectively.
Development of genetic tools in an A. baumannii model strain.
Our success in establishing AB5075 infections in multiple animal models suggests that the isolate would be an attractive model strain for studying A. baumannii virulence. However, in addition to an ideal model strain being highly virulent in animal models, the ability to genetically manipulate the isolate is vital for the study of A. baumannii pathogenicity. Because antibiotic resistance determinants are central to many types of genetic manipulations, such as transposon mutagenesis, the antibiotic sensitivity profile of AB5075 was examined in detail. It was observed that AB5075 is susceptible to tetracycline, doxycycline, and related antibiotics (see Table S1 in the supplemental material) and to high levels of erythromycin and hygromycin.
With these known susceptibilities in mind, a method previously developed in A. baumannii (30) was adapted for creating AB5075 isogenic mutants by utilizing the hph gene, encoding hygromycin resistance, from pMQ300 (31) and the EZ Tn5 (Epicentre Biotechnologies, Madison, WI) to develop a Tn5-based mutagenesis system. This system was used to generate a library of ~6,700 transposon mutants. DNA sequencing of the library was performed as previously described (32), yielding 2,548 unique transposon insertions and 68.5% coverage of the genome.
As a further means of modifying the genome, the same hygromycin cassette was inserted into the pUC18T-mini-Tn7T-Zeo vector, and this vector was introduced via conjugation into AB5075. Tn7 insertion into the chromosome was selected for by growth on 250 µg/ml hygromycin and confirmed by PCR across the attTn7 site on the 3′ end of the glmS gene in the AB5075 chromosome. As proof of concept for the use of Tn7 for gene insertion in the chromosome, the lux operon was inserted into the attTn7 site. This resulted in bioluminescence of this strain, and subsequent subculturing of AB5075::Tn7-lux over 7 days without antibiotic selection did not affect the bioluminescent signal (see Fig. S1A in the supplemental material), suggesting that the Tn7 insertion in the chromosome is stable. Additionally, when this strain was cultured in LB broth, there was no growth defect compared to the wild-type isolate AB5075 (see Fig. S1B in the supplemental material). These methods provide us with a means of interrupting and inserting genes on the chromosome, both of which are essential in studying bacterial pathogenesis.
Evaluation of rifampin as proof of concept.
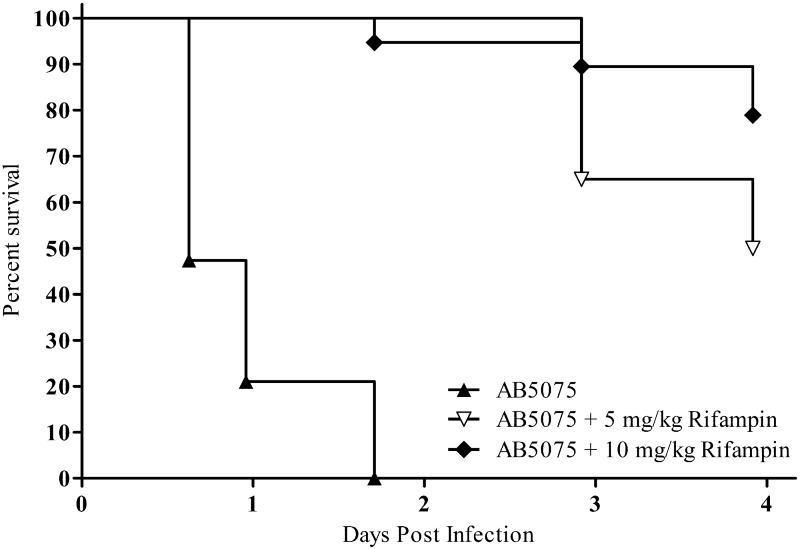
As a proof of concept for the use of AB5075 as a model strain for assessing novel antimicrobials, rifampin treatment against AB5075 was tested in the G. mellonella and murine pulmonary models. In G. mellonella, 30 min after worms were inoculated with 6.0 × 105 CFU of AB5075, they received a treatment injection of 5 or 10 mg/kg rifampin. By 41 h postinfection, all worms infected with AB5075 and receiving PBS treatment had died (Fig. 5). Conversely, worms treated with 5 or 10 mg/kg rifampin had survival rates of 94 and 100%, respectively, at this time point. At the end of the 4-day study, survival rates were 50 and 78%, respectively. The Mantel-Cox test determined that the time to death for AB5075-infected worms was statistically reduced compared to that for rifampin-treated worms (P < 0.001).
FIG 5 .
Rifampin as proof of concept in the G. mellonella model. Kaplan-Meier survival curves of G. mellonella infected with 6.0 × 105 CFU of AB5075 are shown. Worms received a single treatment, 30 min postinfection, of DMSO, 5 mg/kg rifampin, or 10 mg/kg rifampin. Curves were compared via the Mantel-Cox test. The control-treated worms showed significantly increased mortality compared to rifampin-treated worms (P < 0.001).
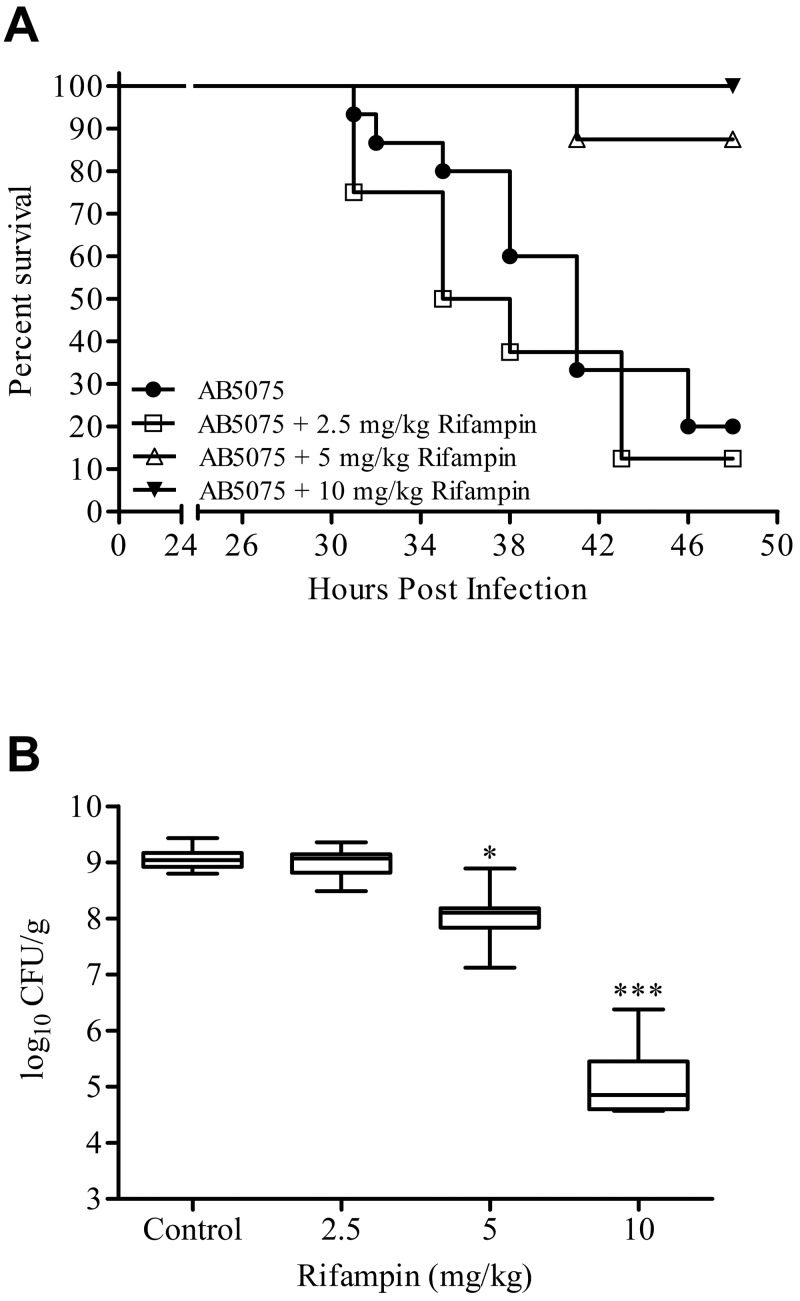
To further assess AB5075 as a model strain, the mouse lung model was used to examine the efficacy of rifampin. Mice infected intranasally with 5.0 × 106 CFU of AB5075 were treated once daily intraperitoneally (IP) with 2.5, 5, or 10 mg/kg rifampin, and survival was monitored for 48 h. At time of death, or euthanasia at 48 h postinfection, lungs were collected to determine bacterial load. As shown in Fig. 6A, at 48 h after inoculation, only 12.5% of mice infected but not treated with rifampin survived, whereas infected mice receiving 5 and 10 mg/kg rifampin had survival rates of 87.5 and 100%, respectively. These differences in survival were found by the Mantel-Cox test to be statistically significant (P = 0.0035 and 0.0008, respectively) compared with mice infected but not treated with rifampin. Mice treated with 2.5 mg/kg rifampin had a 48-h survival rate of 20%, which was not statistically different from that of the control group.
FIG 6 .
Rifampin as proof of concept in the mouse lung model. (A) Kaplan-Meier survival curves of mice infected with 5.0 × 106 CFU of selected strains of A. baumannii are shown. Mice were treated once daily IP with 0, 2.5, 5, or 10 mg/kg rifampin. Curves were compared via the Mantel-Cox test. Rifampin treatments of 5 and 10 mg/kg resulted in significantly increased survival compared to the untreated control (P = 0.0035 and 0.0008, respectively). (B) Box-and-whisker plots of CFU burdens within lungs are shown for day 2 postinfection. Boxes show median and interquartile ranges, while whiskers represent 95% CI. Treatments were compared via the Kruskal-Wallis test followed by Dunn’s multiple comparisons posttests. * and ***, P < 0.05 and 0.001, respectively.
The levels (CFU/g) of bacteria in lung tissue correlated with the survival curves. The median levels in the control and 2.5 mg/kg-treated groups were almost identical, at 9.04 and 9.07 log CFU/g, respectively. Figure 6B shows a decrease in the median log CFU/g for mice treated with 5 and 10 mg/kg rifampin, with values of 8.10 and 4.86, respectively. The Kruskal-Wallis test, followed by Dunn’s multiple comparison test, found a statistically significant difference in bacterial burden when the control group was compared to animals treated with 5 or 10 mg/kg rifampin (P < 0.05).
DISCUSSION
The goal of the work presented in this report was to identify a model strain of A. baumannii that represented current infection isolates, was highly virulent in multiple animal models, and was amenable to genetic manipulation. A strain fitting these characteristics would be clinically relevant and could be utilized in various models to study novel therapeutics, and the ability to create isogenic mutants would be critical in investigating genes required for virulence and the pathogenesis of severe infections in the human host. In our comprehensive studies of isolates representative of each major clonal complex group, we identified one such strain, AB5075.
Previous studies compared the virulence of A. baumannii strains in single infection models, including a murine pulmonary model (13, 14) and a G. mellonella model (33). Other work also compared the virulence of a single strain, ATCC 19606T, in multiple models in vivo (34); however, to our knowledge, this is the first study that used several infection models in parallel to compare multiple strains using representative isolates from each clonal group. Importantly, when we compared infections caused by other A. baumannii strains, including the widely used ATCC 19606T and ATCC 17978, we did not observe the same severe infection as was consistently observed with AB5075.
In the G. mellonella and mouse pneumonia models, AB5075 infection resulted in survival rates consistently below 25%, providing a wide spectrum between infected and uninfected animals to assess novel therapies or genetically mutated strains. Additionally, in the mouse pulmonary model, blood samples and lung histopathology from AB5075-infected mice were consistently positive for the presence of A. baumannii (data not shown) and, combined with the high lung bacterial burden observed, offer further means of evaluation in this model. Furthermore, while a recent publication claimed that A. baumannii could not cause osteomyelitis in a rat fracture model (35), our data showed that AB5075 could establish an infection in this model and that A. baumannii could still be cultured from the bone after 28 days postinfection (see the supplemental material). Recently, we also successfully developed a murine wound model of infection with AB5075, using a small inoculating dose (36). Therefore, the use of AB5075 in additional animal models further demonstrates the great utility of AB5075, as it can serve as a model strain for a variety of studies. The increased virulence of AB5075 in these models also results in larger differences between infected and uninfected animals compared to other tested strains, which allows for less ambiguous results, high statistical power, and thus a requirement for fewer animals to obtain publishable data.
In addition to the successful use of AB5075 in animal models, we were able to exploit the susceptibility of AB5075 to hygromycin to generate a Tn5 transposon insertion library and use a Tn7 transposon derivative to insert genes into the genome of this strain. In collaboration with the Manoil laboratory at the University of Washington, we were able to sequence the Tn5 transposon library using a convenient high-throughput sequencing method. Further, Manoil and his team illustrated the genetic utility of AB5075 by generating a Tn5 insertion library utilizing a tetracycline-based transposon system (C. Manoil, personal communication). It is expected that these sequenced transposon insertion libraries will be powerful tools for future investigations, as similar libraries have been utilized with success with other bacterial pathogens (37, 38). In the future, the Manoil laboratory will distribute wild-type and mutant derivatives of AB5075 (http://www.gs.washington.edu/labs/manoil/baumannii.htm).
The ability to interrupt or mutate specific genes via transposon insertion and complement mutations via Tn7-mediated chromosomal insertion provides a powerful toolkit to answer important questions, such as the nature of specific genes and gene products that are critical for virulence in the host. We believe that AB5075, and the genetic tools and animal models we have designed around this strain, will serve as a platform to readily and reproducibly test mutants in putative virulence factor genes and assess novel antimicrobials. Moreover, we encourage other research groups to use AB5075 as a model strain and to take advantage of the tools we are developing to test their own hypotheses about virulence determinants of A. baumannii.
MATERIALS AND METHODS
Bacterial strains, growth media, and clinical microbiology.
All work was carried out under biosafety level II or II+ conditions. All the A. baumannii strains used in this study can be found in Table 1. Routine growth and strain maintenance was carried out in Luria-Bertani Lennox (LB) broth and agar. Bacterial identification, antibiograms, and MIC were determined using the Vitek 2 (bioMérieux, France) and Phoenix (Becton, Dickinson and Co., Franklin Lakes, NJ) automated systems according to the manufacturer’s instructions. Rifampin was obtained from Sigma-Aldrich (St. Louis, MO), prepared in dimethyl sulfoxide (DMSO), and then further diluted in sterile saline.
Pulsed-field gel electrophoresis.
The pellet from an overnight broth culture was resuspended in 2 ml 10 mM Tris-HCl, 10 mM EDTA (pH 8.0) to a density equivalent to 0.5 McFarland. Suspensions were mixed with equal volumes of melted 1.6% SeaKem Gold agarose (Lonza, Walkersville, MD), dispensed into wells of a plug mold, and allowed to solidify. The plugs were incubated for 2 h in 20 mg/ml proteinase K and cell lysis buffer (50 mM Tris-HCl, 50 mM EDTA [pH 8.0], 1% N-lauroylsarcosine, sodium salt) at 54°C. Subsequently, the plugs were washed four times in TE buffer (10 mM Tris-HCl, 10 mM EDTA [pH 8.0]) and then incubated in a digestion buffer consisting of 50 U of ApaI restriction enzyme, NEBuffer 4, and bovine serum albumin (New England BioLabs, Ipswich, MA) at 25°C for at least 2 h. Electrophoresis was performed at a constant voltage of 200 V by the CHEF-DR II system (Bio-Rad Laboratories, Hercules, CA) with pulse times ramping from 7 to 20 s for 18.5 h. Gels were stained with ethidium bromide and photographed under UV light. PFGE clustering was determined by using the unweighted-pair group method with arithmetic averages (UPGMA) and Dice’s coefficient (BioNumerics version 6.0, created by Applied Maths NV).
Multiplex PCR assay.
Multiplex PCR methods were followed directly from the work of Turton et al. (24), with modified reaction volume and reagents. Briefly, A. baumannii templates were prepared from a single colony grown on Luria-Bertani (LB) agar and resuspended in Lyse-N-Go PCR reagent according to the manufacturer’s instructions (Thermo Scientific, Rockford, IL). PCRs were prepared in 20-µl volume using DreamTaq master mix (Thermo Scientific).
Genome sequencing and bioinformatic analysis.
Genome analysis was performed as previously described (17), with AB5075, AB5711, AB4857, and AB5256 included in the bioinformatic comparisons. Unique genes were determined using the BLAST score ratio analysis (26). BLAST score ratio (BSR) is an in silico approach to conduct comparative proteomic analyses based on proteins predicted to be encoded in a genome (26). BSR was used to compare the proteins encoded in the newly sequenced strains with three isolates from the University of Maryland (17) and eight previously sequenced reference isolates (SDF, AYE, ATCC 17978, ADP1, AB0057, ATCC 19606T, ACICU, and AB307). The BSRs were calculated as the ratio of raw BLASTP score for the query to the raw BLASTP score of the reference strain. BSR cutoffs of ≥0.8, <0.8 to >0.4, and <0.4 were used to determine whether a gene is conserved, divergent, or unique, respectively. A BSR value of 0.8 corresponds to ~85 to 90% identity over 90% of the length of a protein sequence, indicating a highly conserved sequence, while a BSR value of 0.4 corresponds to 30% identity over 30% of the length of a protein sequence, indicating a unique sequence (26).
Galleria mellonella infection model.
A. baumannii strains were grown overnight in an orbital shaker (37°C, 200 rpm), and overnight cultures were then diluted 100-fold into fresh medium and grown for 3 h. Cells were collected by centrifugation (5 min, 5,000 × g), washed once in phosphate-buffered saline solution (PBS), and resuspended in PBS to a final OD600 of 1.0. Further dilutions were done in PBS. The number of bacterial cells in the injected sample was enumerated by plating 10-fold serial dilutions on LB agar plates and counting CFU after overnight incubation.
G. mellonella larvae (Vanderhorst Wholesale, Saint Marys, OH) were used within 10 days of shipment from the vendor. Larvae were kept in the dark at 21°C before infection. Larvae weighing 200 to 300 mg were used in the LD50 and survival assays as described previously (28), with slight modifications. Briefly, 5 µl of the sample was injected into the last left proleg of the larvae using a 10-µl glass syringe (Hamilton, Reno, NV) fitted with a 30G needle (Novo Nordisk, Princeton, NJ). Each experiment included control groups of noninjected larvae or larvae injected with 5 µl sterile PBS. For rifampin experiments, approximately 30 min postinfection, worms were injected with 2 µl of rifampin in the second-to-last left proleg using a 10-µl glass syringe. Injected larvae were incubated at 37°C, assessing death at 24 h intervals over 6 days. Larvae were considered dead if they did not respond to physical stimuli. Experiments in which 10% or more of the larvae in either of the control groups died were omitted from the statistical analysis. Experiments were repeated three times using 10 or 20 larvae per experimental group.
The LD50 for A. baumannii strains AB5075, ATCC 19606T, and ATCC 17978 were determined by preparing a series of 2-fold dilutions of a PBS suspension of the bacterial strain, starting with a bacterial concentration that caused death of all the larvae within 24 h and going down to a concentration at which no deaths were recorded within this time frame. Twenty larvae were injected with 5 µl of the appropriate dilution, and larvae were determined to be alive or dead after 24 h. Two independent biological repeats of LD50 determination were performed. LD50 were determined using the Spearman-Karber method.
Murine pulmonary model.
The animal experimental procedures were approved by the Institutional Animal Care and Use Committee at the Walter Reed Army Institute of Research (IB02-10). All research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to principles stated in reference 39. Six-week-old female BALB/c mice (National Cancer Institute, Frederick, MD) were housed at 3 to 4 animals per cage and allowed access to food and water ad libitum throughout the experiment. The pulmonary infection model was adapted from reference 14. Briefly, to promote infection, mice were rendered neutropenic via intraperitoneal administration of 150 mg/kg and 100 mg/kg cyclophosphamide in sterile saline on day −4 and day −1 prior to infection (day 0), respectively.
A. baumannii isolates AB4857, AB5075, AB5711, AB0057, and AB5256 were grown overnight in LB broth with aeration at 37°C, subcultured to mid-exponential phase, washed, and resuspended in PBS with optical density at 600 nm (OD600) values corresponding to 2 × 108 CFU/ml. For infection, mice were anesthetized with oxygenated isoflurane immediately prior to intranasal inoculation with 25 µl of bacterial cultures, corresponding to 5 × 106 CFU. For rifampin experiments, mice were injected IP daily, starting at 4 h postinfection. Animal morbidity was scored twice daily for 6 days using a system evaluating mobility, coat condition, and conjunctivitis as previously described (14). As mice became exceedingly moribund based on clinical score, they were humanely euthanized according to protocol.
To assess CFU burden in the lungs, mice were humanely euthanized according to protocol on days 2 and 3 postinfection via an injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). To quantify the pulmonary CFU burden, lungs were homogenized in 1 ml PBS, and serial dilutions were plated using the Autoplate spiral plating system (Advanced Instruments, Norwood, MA) onto LB agar supplemented with 50 µg/ml carbenicillin. Bacterial load was reported as CFU per gram of lung tissue.
Transposon library generation.
Transposon mutants were constructed using the EZ-Tn5 transposon construction vector pMOD-5<R6Kγori/MCS> (Epicentre, Madison, WI). The transposable element was created by PCR amplifying hph, encoding hygromycin resistance, from vector pMQ300 (31) using the primers pMODHygForKPN (AAAAAAGGTACCggaaatgtgcgcggaacccc) and pMODHygRevPST (AAAAAACTGCAGttggtctgacaatcgatgcgaattgg). The amplicon was then cloned into the multicloning site (MCS) of pMOD-5<R6Kγori/MCS>. The transposome was constructed according to manufacturer’s instructions and introduced into cells via electroporation. The transformed cells were selected for on LB agar supplemented with 250 µg/ml hygromycin. Colonies were picked from plates and grown overnight in 96-well plates containing 100 µl of low-salt LB supplemented with 250 µg/ml hygromycin. After overnight incubation, 100 µl of 50% glycerol was added to each well, and plates were immediately moved to −80°C for storage. The Tn5 mutant library was subjected to high-throughput sequencing as previously described (32).
Construction of hygromycin mini-Tn7 vector and insertion onto the chromosome.
To make Tn7-based genetic tools usable in AB5075, the hph gene, coding for hygromycin resistance, was cloned into pUC18T-mini-Tn7T-Zeo (40). Briefly, hph was amplified from pMQ300 (31) using primers pMOD Hyg For (AAAGCATGCggaaatgtgcgcggaacccc) and pMOD Hyg Rev (AAAGCATGCttggtctgacaatcgatgcgaattgg) (lowercase letters represent the actual primer; capital letters are the restriction site for each primer and a poly-A overhang) and ligated into pUC18T-mini-Tn7T-Zeo, which was digested with NcoI and then blunted with the Klenow fragment of the DNA polymerase I (Fermentas). A derivative of the pUC18T-mini-Tn7T-hph vector containing the lux operon was constructed by amplifying the luxABCDE operon out of pUC18T-mini-Tn7T-Gm-lux (40) with the primers Lux For (TCAAGGTTCTGGACCAGTTG) and Lux Rev (AAAAAAAAGCTTGGTGTAGCGTCGTAAGCTAATA). The PCR product was digested with BamHI and HindIII and then cloned into the MCS of pUC18T-mini-Tn7T-hph.
The mini-Tn7 elements were transposed into the attTn7 site of AB5075 via the method of Kumar et al. (41). Conjugation mixtures were scraped from LB plates, resuspended in 1 ml of PBS, and plated on LB agar supplemented with 250 µg/ml of hygromycin and 25 µg/ml of chloramphenicol. Insertion into the attTn7 site was confirmed with the primers AB5075 attTn7 FWD (AACACAAGTGGAAGTGATTTCT) and AB5075 attTn7 REV (TGGCTTGCACCAATCATTTATAG), which flanked the attTn7 site.
Statistical analyses.
All statistical analyses were carried out using GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA). Survival curves were compared via Kaplan-Meier curve analysis with the Bonferroni correction for multiple comparisons. Recovered bacterial burdens were compared via either the Mann-Whitney U test or the Kruskal-Wallis test followed by Dunn’s multiple-comparison test. All results were considered significant at a P value of <0.05.
SUPPLEMENTAL MATERIAL
Tn7 insertion into the genome of AB5075. Wild-type AB5075 and five individual colonies of AB5075::Tn7-lux were subcultured on agar plates without antibiotic selection for 7 days. Pictured are the day 7 streaks after a 1-s exposure using an IVIS imaging system (PerkinElmer, Waltham, MA). Download
Tn7 insertion into the genome of AB5075. Growth curve of wild-type AB5075 and AB5075::Tn7-lux over 5 h. Download
Rat open fracture X-ray radiograph. A representative 4-week-postoperative radiograph of an AB5075-infected rat shows a defective K-wire implant (gap height closure) and displays typical signs of osteomyelitis with periosteal reaction (white arrow) and lytic bone changes (yellow arrows). Download
Antibiogram of isolates tested. Antibiogram results as determined by Vitek2 or Phoenix automated microbiology system are shown. Discrepancies between the machines are highlighted by an asterisk. In that case, the resistance was confirmed manually, and the incorrect sensitivity determination was still a high concentration at an MIC of 8 µg/ml. R (red), resistant; I (yellow), intermediate; S (green), susceptible.
Categorization of protein sequences based on BLAST score ratio comparison. BLAST score ratio (BSR) scores for A. baumannii clinical and reference isolates are shown. BSRs are calculated as the ratio of the raw BLASTP score for the query to the raw BLASTP score of the reference strain. A BSR of ≥0.8 indicates a conserved sequence, a BSR of >0.4 but <0.8 indicates a divergent sequence, and a BSR of <0.4 indicates a unique sequence.
Supplemental methods and results. Download
ACKNOWLEDGMENTS
We kindly acknowledge Colin Manoil for his collaboration in sequencing the Tn5 mutant library, his advice, and the critical reading of the manuscript. We thank Lenie Dijkshoorn for providing the clonal complex reference strains that were used as controls for the multiplex PCR assay. We are grateful to Robert Shanks for providing the gift of the pMQ300 plasmid. We thank COL Clint Murray and Mark Shirtliff for providing two additional isolates from Brooke Army Medical Center. We are grateful to Herbert Schweizer for providing the pUC18T-mini-Tn7T-Zeo plasmid, and Anthony Hay for providing the pUC18T-mini-Tn7T-Gm-lux plasmid.
The Zurawski laboratory and the research performed in this study were supported and funded via multiple grants from the Military Infectious Diseases Research Program (MIDRP) and the Defense Medical Research and Development Program (DMRDP).
The findings and opinions expressed herein belong to the authors and do not necessarily reflect the official views of the WRAIR, the U.S. Army, or the Department of Defense.
Footnotes
Citation Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5(3):e01076-14. doi:10.1128/mBio.01076-14.
REFERENCES
- 1. Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11:297–308. 10.1586/eri.13.12 [DOI] [PubMed] [Google Scholar]
- 2. Dizbay M, Tozlu DK, Cirak MY, Isik Y, Ozdemir K, Arman D. 2010. In vitro synergistic activity of tigecycline and colistin against XDR-Acinetobacter baumannii. J. Antibiot. 63:51–53. 10.1038/ja.2009.117 [DOI] [PubMed] [Google Scholar]
- 3. Murray CK, Hospenthal DR. 2008. Acinetobacter infection in the ICU. Crit. Care Clin. 24:237–248, vii. 10.1016/j.ccc.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 4. Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114–4123. 10.1128/AAC.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yun HC, Branstetter JG, Murray CK. 2008. Osteomyelitis in military personnel wounded in Iraq and Afghanistan. J. Trauma 64:S163–S168. 10.1097/01.ta.0000241143.71274.63 [DOI] [PubMed] [Google Scholar]
- 6. Tong MJ. 1972. Septic complications of war wounds. JAMA 219:1044–1047. 10.1001/jama.219.8.1044 [DOI] [PubMed] [Google Scholar]
- 7. Potron A, Munoz-Price LS, Nordmann P, Cleary T, Poirel L. 2011. Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob. Agents Chemother. 55:5946–5948. 10.1128/AAC.05124-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang T, Li D, Xie Y, Kang M, Chen Z, Chen H, Fan H, Wang L, Tao C. 2010. The microbiological characteristics of patients with crush syndrome after the Wenchuan earthquake. Scand. J. Infect. Dis. 42:479–483. 10.3109/00365541003671226 [DOI] [PubMed] [Google Scholar]
- 9. Keen EF, III, Murray CK, Robinson BJ, Hospenthal DR, Co EM, Aldous WK. 2010. Changes in the incidences of multidrug-resistant and extensively drug-resistant organisms isolated in a military medical center. Infect. Control Hosp. Epidemiol. 31:728–732. 10.1086/653617 [DOI] [PubMed] [Google Scholar]
- 10. Evans BA, Hamouda A, Amyes SG. 2013. The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 19:223–238. 10.2174/138161213804070285 [DOI] [PubMed] [Google Scholar]
- 11. Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, St Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, Waterman PE. 2013. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis. 208:1142–1151. 10.1093/infdis/jit293 [DOI] [PubMed] [Google Scholar]
- 12. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update. Infect. Dis. Soc. Am. Clin. Infect Dis. 48:1–12. 10.1086/591855 [DOI] [PubMed] [Google Scholar]
- 13. de Breij A, Eveillard M, Dijkshoorn L, van den Broek PJ, Nibbering PH, Joly-Guillou ML. 2012. Differences in Acinetobacter baumannii strains and host innate immune response determine morbidity and mortality in experimental pneumonia. PLoS One 7:e30673. 10.1371/journal.pone.0030673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eveillard M, Soltner C, Kempf M, Saint-André JP, Lemarié C, Randrianarivelo C, Seifert H, Wolff M, Joly-Guillou ML. 2010. The virulence variability of different Acinetobacter baumannii strains in experimental pneumonia. J. Infect. 60:154–161. 10.1016/j.jinf.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 15. Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614. 10.1101/gad.1510307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- 17. Sahl JW, Johnson JK, Harris AD, Phillippy AM, Hsiao WW, Thom KA, Rasko DA. 2011. Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genomics 12:291. 10.1186/1471-2164-12-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loehfelm TW, Luke NR, Campagnari AA. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 190:1036–1044. 10.1128/JB.01416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramirez MS, Don M, Merkier AK, Bistué AJ, Zorreguieta A, Centrón D, Tolmasky ME. 2010. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol. 48:1488–1490. 10.1128/JCM.01264-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russo TA, Beanan JM, Olson R, MacDonald U, Luke NR, Gill SR, Campagnari AA. 2008. Rat pneumonia and soft-tissue infection models for the study of Acinetobacter baumannii biology. Infect. Immun. 76:3577–3586. 10.1128/IAI.00269-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vila J, Pachón J. 2012. Therapeutic options for Acinetobacter baumannii infections: an update. Expert Opin. Pharmacother. 13:2319–2336. 10.1517/14656566.2012.729820 [DOI] [PubMed] [Google Scholar]
- 22. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46. 10.1111/j.1469-0691.2007.01786.x [DOI] [PubMed] [Google Scholar]
- 24. Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 25. Zurawski DV, Thompson MG, McQueary CN, Matalka MN, Sahl JW, Craft DW, Rasko DA. 2012. Genome sequences of four divergent multidrug-resistant Acinetobacter baumannii strains isolated from patients with sepsis or osteomyelitis. J. Bacteriol. 194:1619–1620. 10.1128/JB.06749-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasko DA, Myers GS, Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. 10.1186/1471-2105-6-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desbois AP, Coote PJ. 2012. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of new antimicrobial agents. Adv. Appl. Microbiol. 78:25–53. 10.1016/B978-0-12-394805-2.00002-6 [DOI] [PubMed] [Google Scholar]
- 28. Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53:2605–2609. 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manepalli S, Gandhi JA, Ekhar VV, Asplund MB, Coelho C, Martinez LR. 2013. Characterization of a cyclophosphamide-induced murine model of immunosuppression to study Acinetobacter baumannii pathogenesis. J. Med. Microbiol. 62:1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dorsey CW, Tomaras AP, Actis LA. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68:6353–6360. 10.1128/AEM.68.12.6353-6360.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalivoda EJ, Horzempa J, Stella NA, Sadaf A, Kowalski RP, Nau GJ, Shanks RM. 2011. New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol. Biotechnol. 48:7–14. 10.1007/s12033-010-9342-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. 2013. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio 4:e00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/journal.pone.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80:1015–1024. 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collinet-Adler S, Castro CA, Ledonio CG, Bechtold JE, Tsukayama DT. 2011. Acinetobacter baumannii is not associated with osteomyelitis in a rat model: a pilot study. Clin. Orthop. Relat. Res. 469:274–282. 10.1007/s11999-010-1488-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson MG, Black CC, Pavlicek RL, Honnold CL, Wise MC, Alamneh YA, Moon JK, Kessler JL, Si Y, Williams R, Yildirim S, Kirkup BC, Jr, Green RK, Hall ER, Palys TJ, Zurawski DV. 2014. Validation of a novel murine wound model of Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 58:1332–1342. 10.1128/AAC.01944-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchan BW, McLendon MK, Jones BD. 2008. Identification of differentially regulated Francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 74:2637–2645. 10.1128/AEM.02882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 40. Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- 41. Kumar A, Dalton C, Cortez-Cordova J, Schweizer HP. 2010. Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J. Microbiol. Methods 82:296–300. 10.1016/j.mimet.2010.07.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tn7 insertion into the genome of AB5075. Wild-type AB5075 and five individual colonies of AB5075::Tn7-lux were subcultured on agar plates without antibiotic selection for 7 days. Pictured are the day 7 streaks after a 1-s exposure using an IVIS imaging system (PerkinElmer, Waltham, MA). Download
Tn7 insertion into the genome of AB5075. Growth curve of wild-type AB5075 and AB5075::Tn7-lux over 5 h. Download
Rat open fracture X-ray radiograph. A representative 4-week-postoperative radiograph of an AB5075-infected rat shows a defective K-wire implant (gap height closure) and displays typical signs of osteomyelitis with periosteal reaction (white arrow) and lytic bone changes (yellow arrows). Download
Antibiogram of isolates tested. Antibiogram results as determined by Vitek2 or Phoenix automated microbiology system are shown. Discrepancies between the machines are highlighted by an asterisk. In that case, the resistance was confirmed manually, and the incorrect sensitivity determination was still a high concentration at an MIC of 8 µg/ml. R (red), resistant; I (yellow), intermediate; S (green), susceptible.
Categorization of protein sequences based on BLAST score ratio comparison. BLAST score ratio (BSR) scores for A. baumannii clinical and reference isolates are shown. BSRs are calculated as the ratio of the raw BLASTP score for the query to the raw BLASTP score of the reference strain. A BSR of ≥0.8 indicates a conserved sequence, a BSR of >0.4 but <0.8 indicates a divergent sequence, and a BSR of <0.4 indicates a unique sequence.
Supplemental methods and results. Download