ABSTRACT
Genetic engineering has contributed greatly to our understanding of Mycobacterium tuberculosis biology and has facilitated antimycobacterial and vaccine development. However, methods to generate M. tuberculosis deletion mutants remain labor-intensive and relatively inefficient. Here, methods are described that significantly enhance the efficiency (greater than 100-fold) of recovering deletion mutants by the expression of mycobacteriophage recombineering functions during the course of infection with specialized transducing phages delivering allelic exchange substrates. This system has been successfully applied to the CDC1551 strain of M. tuberculosis, as well as to a ΔrecD mutant generated in the CDC1551 parental strain. The latter studies were undertaken as there were precedents in both the Escherichia coli literature and mycobacterial literature for enhancement of homologous recombination in strains lacking RecD. In combination, these measures yielded a dramatic increase in the recovery of deletion mutants and are expected to facilitate construction of a comprehensive library of mutants with every nonessential gene of M. tuberculosis deleted. The findings also open up the potential for sophisticated genetic screens, such as synthetic lethal analyses, which have so far not been feasible for the slow-growing mycobacteria.
IMPORTANCE
Genetic manipulation of M. tuberculosis is hampered by laborious and relatively inefficient methods for generating deletion mutant strains. The combined use of phage-based transduction and recombineering methods greatly enhances the efficiency by which knockout strains can be generated. The additional elimination of recD further enhances this efficiency. The methods described herein will facilitate the construction of comprehensive gene knockout libraries and expedite the isolation of previously difficult to recover mutants, promoting antimicrobial and vaccine development.
Observation
Tuberculosis (TB) remains a tremendous global health problem, with recent estimates of almost 9 million new cases annually and 1.4 million deaths (1). In addition, multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains that resist the most effective chemotherapies have become increasingly common, and these strains are particularly lethal in the context of untreated HIV (2).
The development of techniques to genetically manipulate Mycobacterium tuberculosis, the causative agent of TB, and to generate defined mutants has revolutionized our ability to explore the biology and physiology of the bacterium and the pathogenesis of the disease. Despite the advances over the past 20 years, the genetic manipulation of M. tuberculosis has been limited by several factors, including the slow growth of the organism (doubling time of approximately 24 h), the need for biosafety level 3 (BSL3) containment, and most significantly, the relative inefficiency of existing techniques for targeted gene replacement compared with those used for “model” organisms, such as Escherichia coli and yeast (e.g., Saccharomyces cerevisiae), exacerbated by the relatively high frequency of illegitimate recombination that has been observed in slow-growing mycobacteria (3).
Following initial success with gene disruption in slow-growing mycobacteria by using linear DNA fragments, additional techniques were developed, including a “suicide vector” approach employing recombinant plasmids unable to replicate in mycobacteria and a two-step method using selectable and counterselectable markers located on replicating or nonreplicating plasmids (4). All of these approaches require electroporation to introduce the constructs into the mycobacterial cell. This can be problematic due to the relatively low efficiency with which DNA can be introduced into slow-growing mycobacteria by electroporation (5). This, together with the inefficiency of homologous recombination, results in significant challenges in recovery of mutants and also limits the technique to those strains that are most amenable to plasmid transformation. The development of methods for introducing the allelic exchange substrates (AESs) into recipient cells with in vitro generated specialized transducing phages represented a significant advance in the genetic manipulation of mycobacteria (6).
The specialized transducing phages consist of a cosmid cloning vector containing the AES—that is, upstream and downstream flanks of the targeted gene, separated by an antibiotic resistance cassette and in some instances a counterselectable marker—which is cloned into a conditionally replicating (temperature-sensitive) shuttle phasmid. The shuttle phasmid can replicate in E. coli as a plasmid and in mycobacteria as a phage and permits transfer of genetic material between the organisms (7). The transducing phage is amplified in Mycobacterium smegmatis at the permissive temperature of 30°C to generate a high-titer lysate, which is then used to transduce (infect) recipient mycobacteria at the nonpermissive temperature of 37°C. With appropriate adjustment of the multiplicity of infection (MOI), delivery of the AES by the phage to essentially every mycobacterial cell can be achieved (8). The inclusion of resolvase sites flanking the antibiotic resistance cassette allows the marker to be removed by transient expression of γδ-resolvase within the cell (6), permitting multiple rounds of gene deletion utilizing the same marker. This is important as the availability of only a few selection markers creates difficulties when knockouts of multiple genes are required to study the many redundant gene families in M. tuberculosis.
A recent advance in the genetic manipulation of mycobacteria and the creation of defined deletion mutants came with the successful application of recombineering (recombination-mediated genetic engineering) techniques to both rapidly growing and slowly growing mycobacteria (9). In E. coli, transient expression of phage-encoded recombination proteins, including the λ red system (Exo, Beta, and Gam proteins) and the Rac prophage RecE/RecT system have been exploited to enhance the efficiency of homologous recombination and the isolation of the desired mutants (10, 11). The exonuclease (Exo or RecE) degrades DNA processively from the 5′ ends of breaks in double-stranded DNA (dsDNA), while the single-stranded DNA annealing protein (Beta or RecT) binds to the 3′ single-stranded tail, preparing the DNA for strand invasion (11). Together these activities facilitate homologous exchange of dsDNA substrates. Through database searches of sequenced mycobacteriophage genomes, van Kessel and Hatfull identified genes 60 and 61 encoding putative recombinases in the genome of the Che9c phage. Although somewhat distant relatives of their Rac prophage counterparts, gp60 possesses a dsDNA-dependent exonuclease activity similar to RecE, while gp61 acts as a single-stranded DNA binding protein similar to RecT (9). Coexpression of the proteins in M. smegmatis under control of the inducible acetamidase promoter enhanced homologous recombination sufficiently to permit the efficient recovery of gene replacement mutants following electroporation of linearized DNA substrates, including PCR products with ~500 bp of homology upstream and downstream of the targeted gene (9). Beyond its use in rapidly growing mycobacteria, the recombineering system has been further applied to M. tuberculosis for deletion of groEL1 (9) and additional loci. Subsequently, expression of gp60 and gp61 under control of a nitrile-inducible promoter has been used by other groups for recombineering in M. smegmatis (12) and M. tuberculosis (13). However, as employed to date, these recombineering techniques are reliant upon introduction of recombination substrates into the bacterial cells by electroporation, which is subject to the limitations of transformability as discussed above.
Here we present methods that greatly enhance the efficiency by which targeted mutants can be generated in M. tuberculosis. The technique relies on specialized transduction—which, unlike electroporation, permits efficient phage-mediated delivery of the knockout construct to essentially the entire population of cells—but improves upon the method by taking advantage of the mycobacteriophage recombineering proteins. The pJV53 plasmid containing the gp60 and gp61 coding sequences under control of the inducible acetamidase promoter (9, 14) was introduced by electroporation into the M. tuberculosis CDC1551 strain, with transformants selected on medium containing kanamycin (see Table S1 in the supplemental material for a list of strains used in this study). The CDC1551 wild-type parental strain and the strain transformed with pJV53 [CDC1551(pJV53-1)], were grown to mid-log phase (optical density at 600 nm [OD600] of ~0.5) in 7H9 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.2% glycerol, 0.05% Tween 80 or tyloxapol, and 0.2% succinate; culture medium for the strain transformed with pJV53 also included kanamycin at 25 µg/ml. At this time, the cultures were divided into two aliquots: no additional supplement was added to one aliquot (“uninduced”), while the second aliquot was supplemented with 0.2% acetamide (“induced”). After overnight incubation, bacteria were pelleted by centrifugation, washed with MP buffer (50 mM Tris [pH 8], 150 mM NaCl, 10 mM MgCl2, 2 mM CaCl2), resuspended in MP buffer at a 10-fold concentration versus the initial culture volume (~3 × 109 bacteria per transduction), and incubated overnight at 37°C with ~0.5 ml high-titer phage lysate (109 to 1011 PFU/ml). The specialized transducing phages contained cosmid constructs targeting genes whose deletion had been previously demonstrated (15, 16) or would be predicted (17) to result in amino acid or vitamin auxotrophy: serA1 (Rv2996c [MT3074]), metA (Rv3341 [MT3444]), panC (Rv3602c [MT3707]), leuD (Rv2987c [MT3065]), and argB (Rv1654 [MT1692]), to induce auxotrophy for serine, methionine, pantothenate, leucine and arginine, respectively (see Table S2 in the supplemental material for sequences of primers used in cloning flanks for construction of AESs by using the p0004S vector [18]). Following the overnight incubation, bacteria were pelleted by centrifugation and resuspended in 7H9 medium supplemented with 0.05% Tween 80, and aliquots were plated in parallel onto 7H10 medium supplemented with hygromycin at 50 to 75 µg/ml alone or with additional supplements, including l-serine at 50 µg/ml, l-methionine at 50 µg/ml, d-pantothenate at 24 µg/ml, l-leucine at 50 µg/ml, and l-arginine at 50 µg/ml. (The precise combination of supplements was chosen depending upon the panel of phages used in the particular experiment.) In some experiments, the transductions designed to generate amino acid auxotrophs were also plated in parallel onto medium supplemented with 0.2% Casamino Acids, in addition to the plating with the individual amino acid supplements; because numbers of transductants were found to be similar for the two types of media, the results were pooled in calculating the numbers of colonies per transduction.
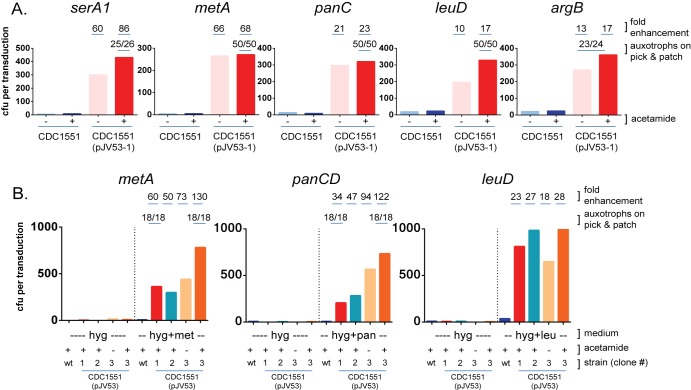
Results of representative experiments are shown in Fig. 1A. The strain harboring the pJV53 plasmid exhibited a dramatic increase in the number of transductants obtained, from ~10-fold up to ~86-fold more than the CDC1551 wild-type strain for the various loci. The loci that were targeted are distributed throughout the bacterial chromosome. Platings done in parallel to media with hygromycin but without other supplements yielded very few colonies—typically <10 per transduction—indicating that the colonies on the supplemented plates were very likely to be auxotrophs. This was verified by “pick and patch” replica plating of colonies from the original transduction plates (portions of the same colony streaked onto 7H10 plus hygromycin versus 7H10 plus hygromycin plus the single specific supplement required for the auxotroph in question), where the vast majority of the colonies were found to be auxotrophs (Fig. 1A), consistent with successful allelic replacement. Of note, the true enhancement of homologous recombination may be even greater than the 10- to 86-fold range depicted here, because the calculations of increased transduction frequency were determined by comparing numbers of colonies obtained on supplemented media, where a low background frequency of spontaneous hygromycin-resistant colonies can appear. However, “pick and patch” testing showed that the percentage of auxotrophs was in some cases greater for CDC1551(pJV53-1) than for the CDC1551 wild-type strain (see Fig. S1 in the supplemental material for an example with metA). However, this may depend as well on the locus under investigation, as panCD transductants were almost uniformly found to be auxotrophs for pantothenate for both the CDC1551 parental strain and CDC1551(pJV53-1) (data not shown).
FIG 1 .
The CDC1551 strain harboring the pJV53 recombineering plasmid exhibits an enhanced frequency of allelic replacement. (A) Mid-log-phase cultures of the CDC1551 wild-type strain or CDC1551 transformed with pJV53 [CDC1551(pJV53-1)] either were not treated [light blue bars for CDC1551, light red bars for CDC1551(pJV53-1)] or were subjected to overnight treatment with 0.2% acetamide [dark blue bars for CDC1551 and dark red bars for CDC1551(pJV53-1)]. Bacteria were then washed and incubated overnight at 37°C with specialized transducing phages to deliver allelic exchange substrates (AESs) for deletion of the serA1, metA, panC, leuD, or argB genes. Transductions were plated onto 7H10 medium containing 50 to 75 µg/ml hygromycin and the appropriate amino acid or vitamin supplement to allow recovery of auxotrophic deletion mutants. Colonies were counted after ~3 to 4 weeks of incubation. Fold enhancements in colony numbers versus the CDC1551 wild type untreated with acetamide are indicated. The auxotroph phenotypes of colonies appearing on the supplemented plates were further assessed by “pick and patch” testing, with a portion of each colony streaked onto 7H10 plus hygromycin or 7H10 plus hygromycin plus supplement. Shown are numbers of colonies growing only on supplemented media over the total numbers of colonies tested. In these studies, colonies for the argB mutants were slow to appear on solid media, requiring extended incubation; in a subsequent experiment, the l-arginine concentration of the supplemented media was increased to 200 µg/ml, and colonies then appeared in the typical 3 to 4 weeks. For serA1, the colonies obtained on Casamino Acid plates grew poorly on replica plate testing (even with the supplement), so the “pick and patch” results are for colonies obtained on transduction plates supplemented with l-serine. (B) Additional CDC1551 strains independently transformed with the pJV53 plasmid exhibit enhanced transduction frequencies similar to, or in some cases greater than, those of the original strain. Transductions were performed as in panel A above, with phages targeting the metA, panCD, and leuD loci and plated onto 7H10 medium plus hygromycin (hyg) and with or without l-methione (met), d-pantothenate (pan), and l-leucine (leu) supplementation. Shown are colony counts for the acetamide-treated CDC1551 wild type (dark blue bars), CDC1551(pJV53-1) (the original [first] transformed strain [dark red bars]), CDC1551(pJV53-2) (the second independent transformant [teal bars]), and CDC1551(pJV53-3) (the third independent transformant [dark orange bars]), as well as CDC1551(pJV53-3) untreated with acetamide (light orange bars). Fold enhancements in colony numbers versus the CDC1551 wild type are indicated, as are proportions of auxotrophs confirmed on “pick and patch” testing for the metA and panC-panD loci in CDC1551(pJV53-1) (original strain) and CDC1551(pJV53-3) (third transformant).
The findings indicate that expression of the mycobacteriophage Che9c recombination proteins in the CDC1551 strain of M. tuberculosis promotes a greatly improved efficiency of homologous recombination between the phage DNA and the host chromosome. This may be surprising, as recombineering has been demonstrated between linear substrates (such as PCR products) and circular replicons (such as plasmids or a bacterial chromosome) (19), and more recently, recombineering between two linear DNA substrates has been verified (20), but to our knowledge the process has not been demonstrated between two circular molecules. The mycobacteriophage DNA is expected to circularize upon cell entry via cos (cohesive end site) sequences, the complementary single-stranded cohesive ends, by analogy with the phage λ replication cycle (21). However, the mycobacteriophage used in these studies is a temperature-sensitive (ts) derivative of phage TM4, and although the mutations thought to be responsible for the ts phenotype have been defined (22), the functions of the corresponding gene products and the behavior of the ts constructs upon cell entry have not been characterized. In addition, it is also possible that expression of the Che9c recombinases within the bacterial cell may alter the TM4 phage life cycle. This is an area that requires further exploration and may ultimately shed light on mechanisms of both mycobacteriophage replication and mycobacterial recombination.
The numbers of transductants obtained for CDC1551(pJV53-1) were similar in the presence and the absence of acetamide (Fig. 1A), suggesting that, although the enhancement of homologous recombination requires pJV53, the inducer of the promoter (acetamide) is dispensable. There are several possible explanations for this observation. First, increased expression of gp60 and gp61 does not necessarily correlate with enhanced recombineering. In fact, in testing several constructs in M. smegmatis with electroporated linear substrates, it was noted that the number of colonies obtained was substantially increased for pJV53 compared with a similar plasmid construct that actually expressed higher levels of gp61 (9). This may be due in part to the toxicity of the recombinases, especially when expression is not tightly regulated (9). Regulation of the acetamidase promoter is complex and in M. smegmatis has been shown to be “leaky,” with a small amount of expression observed in both rich and minimal media in the absence of added inducer (14), while in M. tuberculosis, the situation may be further complicated by plasmid instability, as a vector containing the acetamidase promoter was found to be unstable and subject to deletions in the H37Rv strain (23). Because Parish et al. observed in M. smegmatis that the acetamidase promoter is controlled by a form of catabolite repression (14), we also tested transduction efficiency at the panCD (Rv3602c-Rv3601c [MT3707-MT3706.1]) locus in a more minimal medium, lacking the dextrose-containing OADC supplement and including 0.2% glycerol and 0.2% succinate as carbon sources (see Fig. S2 in the supplemental material). Transductions performed with this medium yielded a modest increase in colony numbers compared with the richer (glucose-containing) medium, suggesting a possible influence of catabolite repression on the system and some benefit for approaches that address this complication. In the minimal medium, as in rich medium, acetamide was largely dispensable for the enhanced efficiency of homologous recombination.
Because we observed enhanced specialized transduction in the CDC1551 strain harboring pJV53, even in the absence of acetamide, we sought to further demonstrate that the effect was due to the presence of the recombineering functions and not to a coincidental spontaneous mutation that may have occurred in the transformed strain. Therefore, we subsequently isolated two new independent pJV53 transformants of CDC1551 [CDC1551(pJV53-2) and CDC1551(pJV53-3)] and examined their transduction capacities in comparison with the original strain. We found, again using a panel of phages to generate auxotrophs (targeting loci metA, panCD, and leuD), that the new transformants yielded colony counts that were similar to or even greater than the original strain (Fig. 1B). One of the newly created strains [CDC1551(pJV53-3)] was tested in the presence and absence of inducer, and again there was only a relatively modest fold increase in the number of transductants obtained with addition of acetamide, similar to what was observed for the original CDC1551(pJV53-1) strain. The fact that large numbers of colonies were obtained only on supplemented media, and not on 7H10 plus hygromycin alone, indicated that they were likely to be true auxotrophs, which was verified by pick-and-patch testing for the metA and panCD loci (Fig. 1B). Our finding that the three CDC1551 strains harboring pJV53 obtained by independent transformations all share the phenotype of high transduction efficiency makes a coincidental spontaneous mutation an unlikely explanation.
Because it would be desirable to have a more tightly regulated expression system for the mycobacteriophage recombineering functions so as to avoid toxic effects within the cell (10), we conducted similar studies using a plasmid expressing the 60 and 61 genes under the control of a nitrile-inducible promoter (24), a system which has been exploited for recombineering-mediated gene replacement employing linear substrates in M. tuberculosis (13). M. tuberculosis CDC1551 was transformed with the pNIT::ET-sacB plasmid (generously provided by C. Sassetti) to generate strain CDC1551 (pNIT::ET-sacB), with transformants selected on kanamycin and screened by PCR for the presence of 60 and 61 sequences. A transduction assay in comparison with the parental strain for deletion of the leuD locus yielded an increased number of transductants, although the magnitude of the effects was less than for the CDC1551(pJV53-3) strain assayed in parallel (see Fig. S3 in the supplemental material). We once again observed that the effect appeared to be independent of the inducer (isovaleronitrile in this case). Although this remains to be experimentally verified, it is likely that, due to toxicity issues, increased expression levels of the recombineering functions may not lead to enhanced biological effects in M. tuberculosis, as we have observed a similar phenomenon in M. smegmatis (25). This alternative expression system using a different plasmid vector and promoter provides further reassurance that the enhanced recovery of transductants results from expression of the phage recombinases.
In the λ red recombineering system, Gam facilitates homologous recombination by interacting with the host RecBCD complex and inhibiting its various catalytic activities, including the exonuclease activity (26), preventing degradation of the incoming DNA. The RecBCD enzyme of E. coli possesses a complex set of activities and promotes recombination preferentially at short recognition sequences known as Chi sites (5′ GCTGGTGG 3′). Null mutations in recB and recC cause recombination-deficient phenotypes and sensitivity to DNA-damaging agents, while null mutations in recD yield cells which not only are recombination proficient but actually display enhanced recombination activity in the absence of Chi (27). It is thought that RecD functions as an inhibitor of recombination, perhaps due to interference by RecD with the loading of RecA (28), until the D subunit dissociates from the RecBCD enzyme upon encountering a Chi site (29).
Gam-related proteins do not appear to be encoded in mycobacteriophage genomes (9) and so are not available for testing in concert with gp60 and gp61; although not absolutely required for λ red-mediated recombination with linear double-stranded DNA substrates, Gam does improve the efficiency of this process (11). Therefore, we decided, as an alternative to including Gam in these studies, to examine the behavior of a recD null mutant of M. tuberculosis CDC1551 in our transduction assays. We targeted this component of the RecBCD complex due to the E. coli literature on the role of RecD as an inhibitor of homologous recombination. Specialized transduction was used to delete recD from the chromosome of the CDC1551 wild-type strain and also from the CDC1551 strain harboring pJV53, with deletion confirmed by Southern blot analysis (see Fig. S4 in the supplemental material); for the CDC1551(pJV53-1) strain, selection with kanamycin was included throughout to maintain the plasmid. The hygromycin resistance–sacB cassette was then removed by introducing γδ-resolvase via phage transduction (18), followed by plating on medium containing 10% sucrose, and the unmarking was confirmed by replica plating to demonstrate hygromycin sensitivity and by PCR analysis across the deleted region (see Fig. S4). The CDC1551 ΔrecD strain exhibited an enhanced transduction efficiency on the order of ~5- to 10-fold greater than that of the wild-type strain (Fig. 2A) for several loci tested; again, auxotrophy was confirmed by “pick and patch” testing. The experiment was repeated with deletion of panCD and yielded similar results (not shown). Furthermore, deletion of recD from the CDC1551 strain also expressing the phage recombineering functions [CDC1551(pJV53-1)] yielded a remarkably enhanced efficiency of gene replacement in this strain (Fig. 2A). Colony numbers increased substantially, from typically 10 or fewer per transduction for the wild-type strain to ~700 to 1,000 for deletion of the serA1 and metA loci in the presence of acetamide, for an approximate 100- to 150-fold enhancement of transduction efficiency.
FIG 2 .
Enhanced frequencies of allelic replacement in CDC1551 are further improved with deletion of recD. (A) Cultures prepared as described in the legend to Fig. 1A were incubated with specialized transducing phages for deletion of serA1, metA, or panC. Strains included CDC1551 untreated (light blue bars) or treated with acetamide (dark blue bars), CDC1551 ΔrecD untreated (light green bars) or treated with acetamide (dark green bars), CDC1551(pJV53-1) untreated (light red bars) or treated with acetamide (dark red bars), and CDC1551 ΔrecD(pJV53-1) untreated (light purple bars) or treated with acetamide (dark purple bars). The fold enhancement in terms of colonies recovered on supplemented plates and the numbers of auxotrophs verified on “pick and patch” testing are as described in the legend to Fig. 1A. In this experiment “pick and patch” analysis is unavailable for serA1 due to the poor growth of the serA1 transductants upon restreaking, even on supplemented media. (B) CDC1551(pJV53) demonstrates potential for high-throughput genetic screens. Transductions for deletion of the panCD locus were carried out using ~10% of typical amounts of mycobacterial cells (100 µl of 10×-concentrated log-phase cells) and of high-titer phage lysate (100 µl) in a total volume of 200 μl, washed after overnight incubation at 37°C, and plated onto agar in 24-well plates. Abundant growth on 7H10 supplemented with pantothenate is readily apparent for the CDC1551(pJV53-3) strain, while no colonies are evident on unsupplemented 7H10.
In interpreting these results, it should be noted that Chi sites have not been identified in mycobacteria, and although mycobacteria do encode homologues of RecBCD, deletion of recBCD from M. smegmatis did not result in increased sensitivity to UV damage (30), in contrast to the findings for E. coli. The situation for mycobacteria is complex, with three distinct DNA double-strand break repair pathways identified, and it appears that RecBCD is involved in the single-strand annealing (SSA) pathway and not the homologous recombination pathway, which instead requires the AdnAB helicase-nuclease (31). However, Gupta et al. noted that while the SSA pathway of double-strand break repair was abolished for an M. smegmatis ΔrecBCD mutant, the homologous recombination activity was actually significantly enhanced versus the wild type, indicating that mycobacterial RecBCD actually suppresses homologous recombination (31) via mechanisms yet to be defined. Therefore, this could provide an alternative explanation for the enhanced gene replacement that we observe in the CDC1551 recD deletion mutant, in both the absence and presence of recombineering functions.
Given the large numbers of deletion mutants we were able to obtain in M. tuberculosis using strains expressing the Che9C recombinases, we performed small-scale transductions using reduced amounts of mycobacteria and specialized transducing phage, in volumes that could be accommodated in a 96-well-plate high-throughput format. We plated the transductions onto agar deposited into wells of a 24-well plate to illustrate the capacity for screening for synthetic lethal or, as in this case, conditional essential genes (Fig. 2B). Using the CDC1551(pJV53-3) strain and a phage to target deletion of the panCD locus, colonies were recovered in abundance on 7H10 medium supplemented with hygromycin and pantothenate but not on 7H10 medium supplemented with hygromycin alone. Similar studies with panels of phages should provide valuable information as to gene essentiality under different growth conditions.
Implications.
Gene disruption is a basic tool for the geneticist and allows gene function to be elucidated. Inactivation of genes in a targeted fashion has been challenging for the slow-growing mycobacteria, including M. tuberculosis. We demonstrate here methods that greatly enhance the efficiency by which deletion mutants of M. tuberculosis can be generated. These methods will facilitate the construction of comprehensive mutant libraries. For example, in an effort to extend the potential of mycobacterial genetic analysis, a consortium of laboratories at the Albert Einstein College of Medicine (W. R. Jacobs, Jr.), Johns Hopkins University (W. Bishai), and University of Pittsburgh (G. Hatfull) in collaboration with the Genomic Institute of the Novartis Research Foundation (GNF) (A. Orth) seeks to generate a comprehensive set of specialized transducing phages that can be used to create bar-coded single-gene deletions in multiple strains of M. tuberculosis, such as the virulent H37Rv laboratory and CDC1551 clinical strains, as well as corresponding auxotrophic strains approved for use at biosafety level 2. The aim is to make these valuable resources openly available to the TB research community. Although the established methods for generating individual deletions in slowly growing mycobacteria are often successful, more facile methods amenable to high-throughput approaches would greatly facilitate the process of generating the complete bar-coded mutant collections. Such mutant libraries will make possible analyses that have been difficult or impossible to perform in a truly comprehensive fashion for M. tuberculosis. These include fitness profiling, or defining growth defects under specific environmental conditions (32), including growth in various in vivo models. The bar-coded library will offer a significant advantage over transposon site hybridization (TraSH) screens (33), where candidate genes can be identified, but the transposon insertions responsible for given phenotype are not recovered as bacteria, requiring the experimenter to separately construct and test specific mutants in the implicated region. The library of specialized transducing phages also provides a framework for undertaking systematic analyses of genetic interactions (34), such as synthetic lethal analysis to identify combinations of mutations in two or more genes that lead to loss of viability, often by affecting a common function or pathway. It should be possible to create an (unmarked) deletion mutant in the gene of interest, in the CDC1551(pJV53) strain, and then perform transductions with either the full complement of specialized transducing phages or a desired subset. Due to the greatly enhanced efficiency of gene replacement in this context, any transductions which yield colonies only with the CDC1551(pJV53) parental strain, but not with the strain with the gene of interest deleted, would be candidates for synthetic lethal interactions. Ultimately, examining the phenotypes of double mutants on a genome-wide scale should help to elucidate the components of essential biological pathways and their interactions within the cell and facilitate the development of novel antimycobacterial agents.
SUPPLEMENTAL MATERIAL
Appearance of transduction plates and of “pick and patch” testing results for metA. (A) Transductions of mid-log-phase bacteria, either uninduced or induced by overnight treatment with 0.2% acetamide, were carried out as described in the legend to Fig. 1A, using specialized transducing phage for deletion of metA. Shown are colonies appearing on 7H10 plates supplemented with 50 µg/ml hygromycin or additionally with 50 µg/ml l-methionine for the CDC1551 parental strain and the strain transformed with pJV53. Plates were photographed after ~5 weeks of incubation at 37°C. Colonies appearing on methionine-supplemented plates were patched onto 7H10-hygromycin or 7H10–hygromycin–l-methionine plates to determine the frequency with which auxotrophs were obtained. Plates were photographed ~3 weeks after streaking. Download
Efficiency of deleting panCD in rich versus minimal media, to examine the issue of catabolite repression. Transductions as described in the legend to Fig. 1A were carried out after growth of bacteria in 7H9 medium with 0.2% glycerol, 10% OADC supplement (oleic acid-albumin-dextrose-catalase), 0.2% succinate, and 0.05% Tween 80 (“rich”) or in a more minimal medium lacking OADC supplement and consisting of 7H9 base medium supplemented with 0.2% glycerol, 0.2% bovine serum albumin (BSA), 0.08% NaCl, 0.2% succinate, and 0.05% Tween 80 (“minimal”). Thus, this minimal medium did not contain any dextrose as a carbon source. Cultures of the strain transformed with pJV53 were also supplemented with 25 µg/ml kanamycin. Again, cultures were either uninduced or induced by overnight treatment with 0.2% acetamide, prior to incubation with a specialized transducing phage for the combined deletion of panC and panD. Fold enhancements in colony numbers versus the CDC1551 wild type in rich medium without acetamide are indicated. Download
CDC1551 harboring a plasmid expressing the phage recombineering proteins under control of the nitrile-inducible promoter exhibits an enhanced frequency of allelic replacement. CDC1551 transformed with the pNIT::ET-sacB plasmid was grown to the mid-log phase and then exposed to either 10 µM isovaleronitrile (induced) or an equivalent volume of dimethyl sulfoxide (DMSO) (uninduced vehicle control). Following overnight incubation, cells were washed and used in transductions with phage targeting the leuD locus. Comparison strains included the CDC1551 wild type (also exposed to isovaleronitrile) and acetamide-induced CDC1551 (pJV53-3). After overnight incubation with phage, equal numbers of cells were plated onto 7H10 plus 50 µg/ml hygromycin (top) and 7H10 plus 50 µg/ml hygromycin plus 100 µg/ml l-leucine (bottom). Download
Southern blot verifying deletion of recD from CDC1551 wild type and CDC1551 (pJV53) and PCR analysis to verify unmarking of the strains. (A) Southern blot analysis of CDC1551 ΔrecD clones. Specialized transduction was used to delete recD from the chromosome of CDC1551 wild type and CDC1551 (pJV53-1), with transductants plated onto 7H10 plus 50 µg/ml hygromycin. In addition, a portion of the CDC1551 (pJV53-1) transductant was plated onto 7H10 plus 50 µg/ml hygromycin plus 25 µg/ml kanamycin to maintain the plasmid. Genomic DNA was prepared from the following ΔrecD transductants: CDC1551 (lanes 1 to 4), CDC1551 (pJV53-1) plated on kanamycin (lanes 5 to 7), CDC1551 (pJV53-1) uninduced (lanes 8 to 11), and CDC1551 (pJV53-1) acetamide induced (lanes 12 to 15), as well as from the CDC1551 wild-type parental strain (lane 16). Genomic DNAs were digested with MluI; after separation by agarose gel electrophoresis, DNA was transferred to a charged nylon membrane (Hybond N+) using the alkaline transfer modification of the Southern blot protocol. Blots were probed with downstream sequences immediately flanking the recD gene (see Table S3 in the supplemental material). Probes were labeled with [α-32P]dCTP using the Amersham Rediprime II DNA labeling system. A schematic indicating expected sizes of hybridizing bands is shown above the blot. A smaller nonspecific band (smaller than the 5.9-kb band expected for the knockout) is present in all lanes, but the 5.9-kb band for the recD knockout and the 1.7-kb band for the wild type are clearly seen. All clones analyzed were mutants. The circled clones in lanes 1 and 5 were selected for unmarking and use in further studies. (B) PCR analysis of unmarked ΔrecD strains. The hygromycin resistance-sacB cassette was removed from the ΔrecD strains by introducing γδ-resolvase via phage transduction, followed by plating on medium containing 10% sucrose, and the unmarking was confirmed by replica plating to demonstrate hygromycin sensitivity. Genomic DNA was prepared from several hygromycin-sensitive clones and used as a template for PCR across the deleted region. All unmarked clones—five for CDC1551 ΔrecD (lanes 1 to 5) and five for CDC1551 ΔrecD (pJV53-1) (lanes 6 to 10)—gave a band of the expected size; the product was confirmed by sequencing. The CDC1551 ΔrecD::res-sacB-HygR-res parental strain yielded a larger product (lane 11). Approximate expected PCR product sizes are indicated in the diagram above the gel photograph. Download
M. tuberculosis strains used in this study.
Primers used in this study for generating allelic exchange substrates for gene deletion (Nucleotides in boldface indicate genomic sequences; the remainder represent restriction sites and tags.)
Primers used in this study (i) to generate probe for Southern blot confirmation of the recD deletion and (ii) to confirm CDC1551 ΔrecD unmarking by PCR.
ACKNOWLEDGMENTS
This work was supported by NIH/NIAID grant AI26170 (W.R.J.), NIH/NIAID ARRA award AI92760 (W.R.J. and G.F.H.), and by the Albert Einstein College of Medicine Center for AIDS Research (grant AI0-51519). We would also like to acknowledge generous support from the Bill and Melinda Gates Foundation (grant OPP1033104).
We extend sincere thanks to Anthony Orth and Myleen Medina (Genomics Institute of the Novartis Research Foundation) for sharing constructs from the GNF library collection prior to their publication. We are also grateful to Christopher Sassetti (University of Massachusetts Medical School) for providing the pNIT::ET plasmid expressing the mycobacteriophage recombineering functions under control of the nitrile-inducible promoter. We would also like to acknowledge the expert technical assistance of Annie Z. Dai in preparing high-titer phage lysates for use in this study. We thank Christopher Kerantzas for assistance with the genetic characterization of strains used in this study.
Footnotes
Citation Tufariello JM, Malek AA, Vilchèze C, Cole LE, Ratner HK, González PA, Jain P, Hatfull GF, Larsen MH, Jacobs WR, Jr. 2014. Enhanced specialized transduction using recombineering in Mycobacterium tuberculosis. mBio 5(3):e01179-14. doi:10.1128/mBio.01179-14.
REFERENCES
- 1. World Health Organization 2012. Global tuberculosis report. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580. 10.1016/S0140-6736(06)69573-1 [DOI] [PubMed] [Google Scholar]
- 3. Kalpana GV, Bloom BR, Jacobs WR., Jr. 1991. Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 88:5433–5437. 10.1073/pnas.88.12.5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinds J, Mahenthiralingam E, Kempsell KE, Duncan K, Stokes RW, Parish T, Stoker NG. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519–527. 10.1099/13500872-145-3-519 [DOI] [PubMed] [Google Scholar]
- 5. Parish T, Stoker NG. 1998. Electroporation of mycobacteria. Methods Mol. Biol. 101:129–144 [DOI] [PubMed] [Google Scholar]
- 6. Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 http://mic.sgmjournals.org/content/148/10/3007.long [DOI] [PubMed] [Google Scholar]
- 7. Jacobs WR, Jr, Tuckman M, Bloom BR. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532–535. 10.1038/327532a0 [DOI] [PubMed] [Google Scholar]
- 8. Jain P, Hartman TE, Eisenberg N, Odonnell MR, Kriakov J, Govender K, Makume M, Thaler DS, Hatfull GF, Sturm AW, Larsen MH, Moodley P, Jacobs WR., Jr 2012. φ2GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J. Clin. Microbiol. 50:1362–1369. 10.1128/JCM.06192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147–152. 10.1038/nmeth996 [DOI] [PubMed] [Google Scholar]
- 10. Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. 2007. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 421:171–199. 10.1016/S0076-6879(06)21015-2 [DOI] [PubMed] [Google Scholar]
- 11. Court DL, Sawitzke JA, Thomason LC. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36:361–388. 10.1146/annurev.genet.36.061102.093104 [DOI] [PubMed] [Google Scholar]
- 12. Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. 2012. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 8: e1002511. 10.1371/journal.ppat.1002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19:218–227. 10.1016/j.chembiol.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267–2276. 10.1099/00221287-143-7-2267 [DOI] [PubMed] [Google Scholar]
- 15. Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL, Jacobs WR., Jr 2002. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat. Med. 8:1171–1174. 10.1038/nm765 [DOI] [PubMed] [Google Scholar]
- 16. Hondalus MK, Bardarov S, Russell R, Chan J, Jacobs WR, Jr, Bloom BR. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect. Immun. 68:2888–2898. 10.1128/IAI.68.5.2888-2898.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 18. Jain P, Hsu T, Arai M, Biermann K, Thaler DS, Nguyen A, Gonzalez P, Tufariello JM, Kriakov J, Chen B, Larsen MH, Jacobs WR., Jr Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Buchholz F, Muyrers JP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123–128. 10.1038/2417 [DOI] [PubMed] [Google Scholar]
- 20. Narayanan K, Sim EU, Ravin NV, Lee CW. 2009. Recombination between linear double-stranded DNA substrates in vivo. Anal. Biochem. 387:139–141. 10.1016/j.ab.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skalka AM. 1977. DNA replication—bacteriophage lambda. Curr. Top. Microbiol. Immunol. 78:201–237 [PubMed] [Google Scholar]
- 22. Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, DeJong RJ, Elgin SC, Guilfoile FR, Forsyth MH, Harris AD, Harvey SE, Hughes LE, Hynes PM, Jackson AS, Jalal MD, MacMurray EA, Manley CM, McDonough MJ, Mosier JL, Osterbann LJ, Rabinowitz HS, Rhyan CN, Russell DA, Saha MS, Shaffer CD, Simon SE, Sims EF, Tovar IG, Weisser EG, Wertz JT, Weston-Hafer KA, Williamson KE, Zhang B, Cresawn SG, Jain P, Piuri M, Jacobs WR, Jr, Hendrix RW, Hatfull GF. 2011. Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4. PLoS One 6:e26750. 10.1371/journal.pone.0026750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown AC, Parish T. 2006. Instability of the acetamide-inducible expression vector pJAM2 in Mycobacterium tuberculosis. Plasmid 55:81–86. 10.1016/j.plasmid.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 24. Pandey AK, Raman S, Proff R, Joshi S, Kang CM, Rubin EJ, Husson RN, Sassetti CM. 2009. Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb.) 89:12–16. 10.1016/j.tube.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 6:851–857. 10.1038/nrmicro2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakaki Y, Karu AE, Linn S, Echols H. 1973. Purification and properties of the gamma-protein specified by bacteriophage lambda: an inhibitor of the host RecBC recombination enzyme. Proc. Natl. Acad. Sci. U. S. A. 70:2215–2219. 10.1073/pnas.70.8.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaudhury AM, Smith GR. 1984. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 81:7850–7854. 10.1073/pnas.81.24.7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Churchill JJ, Anderson DG, Kowalczykowski SC. 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev. 13:901–911. 10.1101/gad.13.7.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amundsen SK, Taylor AF, Smith GR. 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. U. S. A. 97:7399–7404. 10.1073/pnas.130192397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. 2007. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 189:5237–5246. 10.1128/JB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. 2011. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 79:316–330. 10.1111/j.1365-2958.2010.07463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 33. Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 98:12712–12717. 10.1073/pnas.231275498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baryshnikova A, Costanzo M, Dixon S, Vizeacoumar FJ, Myers CL, Andrews B, Boone C. 2010. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 470:145–179. 10.1016/S0076-6879(10)70007-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appearance of transduction plates and of “pick and patch” testing results for metA. (A) Transductions of mid-log-phase bacteria, either uninduced or induced by overnight treatment with 0.2% acetamide, were carried out as described in the legend to Fig. 1A, using specialized transducing phage for deletion of metA. Shown are colonies appearing on 7H10 plates supplemented with 50 µg/ml hygromycin or additionally with 50 µg/ml l-methionine for the CDC1551 parental strain and the strain transformed with pJV53. Plates were photographed after ~5 weeks of incubation at 37°C. Colonies appearing on methionine-supplemented plates were patched onto 7H10-hygromycin or 7H10–hygromycin–l-methionine plates to determine the frequency with which auxotrophs were obtained. Plates were photographed ~3 weeks after streaking. Download
Efficiency of deleting panCD in rich versus minimal media, to examine the issue of catabolite repression. Transductions as described in the legend to Fig. 1A were carried out after growth of bacteria in 7H9 medium with 0.2% glycerol, 10% OADC supplement (oleic acid-albumin-dextrose-catalase), 0.2% succinate, and 0.05% Tween 80 (“rich”) or in a more minimal medium lacking OADC supplement and consisting of 7H9 base medium supplemented with 0.2% glycerol, 0.2% bovine serum albumin (BSA), 0.08% NaCl, 0.2% succinate, and 0.05% Tween 80 (“minimal”). Thus, this minimal medium did not contain any dextrose as a carbon source. Cultures of the strain transformed with pJV53 were also supplemented with 25 µg/ml kanamycin. Again, cultures were either uninduced or induced by overnight treatment with 0.2% acetamide, prior to incubation with a specialized transducing phage for the combined deletion of panC and panD. Fold enhancements in colony numbers versus the CDC1551 wild type in rich medium without acetamide are indicated. Download
CDC1551 harboring a plasmid expressing the phage recombineering proteins under control of the nitrile-inducible promoter exhibits an enhanced frequency of allelic replacement. CDC1551 transformed with the pNIT::ET-sacB plasmid was grown to the mid-log phase and then exposed to either 10 µM isovaleronitrile (induced) or an equivalent volume of dimethyl sulfoxide (DMSO) (uninduced vehicle control). Following overnight incubation, cells were washed and used in transductions with phage targeting the leuD locus. Comparison strains included the CDC1551 wild type (also exposed to isovaleronitrile) and acetamide-induced CDC1551 (pJV53-3). After overnight incubation with phage, equal numbers of cells were plated onto 7H10 plus 50 µg/ml hygromycin (top) and 7H10 plus 50 µg/ml hygromycin plus 100 µg/ml l-leucine (bottom). Download
Southern blot verifying deletion of recD from CDC1551 wild type and CDC1551 (pJV53) and PCR analysis to verify unmarking of the strains. (A) Southern blot analysis of CDC1551 ΔrecD clones. Specialized transduction was used to delete recD from the chromosome of CDC1551 wild type and CDC1551 (pJV53-1), with transductants plated onto 7H10 plus 50 µg/ml hygromycin. In addition, a portion of the CDC1551 (pJV53-1) transductant was plated onto 7H10 plus 50 µg/ml hygromycin plus 25 µg/ml kanamycin to maintain the plasmid. Genomic DNA was prepared from the following ΔrecD transductants: CDC1551 (lanes 1 to 4), CDC1551 (pJV53-1) plated on kanamycin (lanes 5 to 7), CDC1551 (pJV53-1) uninduced (lanes 8 to 11), and CDC1551 (pJV53-1) acetamide induced (lanes 12 to 15), as well as from the CDC1551 wild-type parental strain (lane 16). Genomic DNAs were digested with MluI; after separation by agarose gel electrophoresis, DNA was transferred to a charged nylon membrane (Hybond N+) using the alkaline transfer modification of the Southern blot protocol. Blots were probed with downstream sequences immediately flanking the recD gene (see Table S3 in the supplemental material). Probes were labeled with [α-32P]dCTP using the Amersham Rediprime II DNA labeling system. A schematic indicating expected sizes of hybridizing bands is shown above the blot. A smaller nonspecific band (smaller than the 5.9-kb band expected for the knockout) is present in all lanes, but the 5.9-kb band for the recD knockout and the 1.7-kb band for the wild type are clearly seen. All clones analyzed were mutants. The circled clones in lanes 1 and 5 were selected for unmarking and use in further studies. (B) PCR analysis of unmarked ΔrecD strains. The hygromycin resistance-sacB cassette was removed from the ΔrecD strains by introducing γδ-resolvase via phage transduction, followed by plating on medium containing 10% sucrose, and the unmarking was confirmed by replica plating to demonstrate hygromycin sensitivity. Genomic DNA was prepared from several hygromycin-sensitive clones and used as a template for PCR across the deleted region. All unmarked clones—five for CDC1551 ΔrecD (lanes 1 to 5) and five for CDC1551 ΔrecD (pJV53-1) (lanes 6 to 10)—gave a band of the expected size; the product was confirmed by sequencing. The CDC1551 ΔrecD::res-sacB-HygR-res parental strain yielded a larger product (lane 11). Approximate expected PCR product sizes are indicated in the diagram above the gel photograph. Download
M. tuberculosis strains used in this study.
Primers used in this study for generating allelic exchange substrates for gene deletion (Nucleotides in boldface indicate genomic sequences; the remainder represent restriction sites and tags.)
Primers used in this study (i) to generate probe for Southern blot confirmation of the recD deletion and (ii) to confirm CDC1551 ΔrecD unmarking by PCR.