Abstract
Most eukaryotes develop close interactions with microorganisms that are essential for their performance and survival. Thus, eukaryotes and prokaryotes in nature can be considered as meta-organisms or holobionts. Consequently, microorganisms that colonize different plant compartments contain the plant’s second genome. In this respect, many studies in the last decades have shown that plant-microbe interactions are not only crucial for better understanding plant growth and health, but also for sustainable crop production in a changing world. This mini-review acting as editorial presents retrospectives and future perspectives for plant microbiome studies as well as information gaps in this emerging research field. In addition, the contribution of this research topic to the solution of various issues is discussed.
Keywords: meta-organisms, plant microbiome, plant-microbe interaction, biocontrol, stress protection, plant growth promotion
INTRODUCTION AND RETROSPECT ON THE STUDY OF PLANT-ASSOCIATED MICROORGANISMS
Many studies on plant-associated microorganisms reflect the enormous interest in this topic and the full effect of ongoing research (Bulgarelli et al., 2013). Due to the importance of the soil habitat of plants, the majority of research focuses on the rhizosphere, even though microorganisms are also able to readily colonize most plant compartments. Several recent reviews addressed particular aspects of plant microbiome research. The current knowledge of rhizosphere inhabitants, their function, and their promising biotechnological potential was summarized by Hirsch and Mauchline (2012), Bakker et al. (2013), Mendes et al. (2013). Berendsen et al. (2012) reviewed more specifically the plant microbiome and plant health relationship, while Berg et al. (2005a) focused on the occurrence of potential human pathogenic bacteria in the rhizosphere. The important question about the factors contributing to selective enrichment of microorganisms from the soil into the rhizosphere was addressed by Bais et al. (2006), Doornbos et al. (2012). It now appears that in addition to carbohydrates and even amino acids which act as general chemical determinants in the rhizosphere (Moe, 2013), secondary metabolites such as plant-specific flavonoids were identified as key drivers in the development of plant-specific microbial communities in the rhizosphere (Weston and Mathesius, 2013).
While the well-studied rhizosphere presents the soil-plant interface, the phyllosphere forms the air-plant interface. This microhabitat is also of special interest due to its large and exposed surface area and its connection to the air microbiome, especially air-borne pathogens (rev. in Lindow and Brandl, 2003; Vorholt, 2012; Rastogi et al., 2013). However, in addition to the well-studied rhizo- and phyllospheres, each plant can be divided into more microenvironments, e.g., the endorhiza (root), the anthosphere (flower), the spermosphere (seeds), and the carposphere (fruit). Moreover, we generally differentiate between the endosphere (inner tissues) and ectosphere (outer surfaces; Ryan et al., 2008). All these microenvironments provide specific biotic and abiotic conditions for microbial life, which also have a correspondingly specific function for the host. The potential of these findings and the use of plant growth-promoting bacteria and biocontrol agents for the development of sustainable forms of agricultural management were discussed by Leveau (2007), Köberl et al. (2012), Berg et al. (2013).
The first section of this editorial focuses on several historical milestones in plant microbiome research. Despite the enormous progress already made, many challenges still exist. We address some information gaps in the second section of this editorial, and conclude with an overview of the present contributions. The papers in this special issue focus mainly on the bacterial dimension of the plant-associated microbiome, and we will show how they complement and extend the current research and how they will spur further questions.
THE RHIZOSPHERE WAS DEFINED MORE THAN A CENTURY AGO
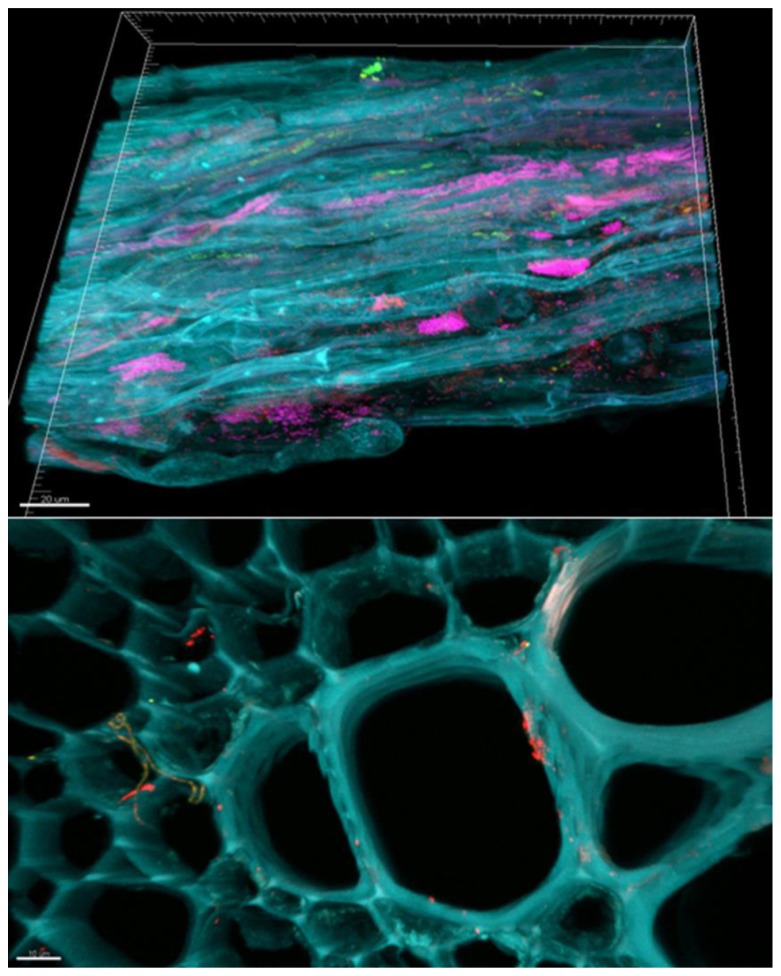
Hiltner (1904) defined the “rhizosphere” as root-surrounding soil influenced by root exudates (Hartmann et al., 2008). In addition, he was the first to suggest the importance of microbial root inhabitants for plant growth and health. The rhizosphere is of central importance not only for plant nutrition, health, and quality. Today we are aware of microorganism-driven carbon sequestration in this ecological niche, which has an important role in ecosystem functioning and nutrient cycling in terrestrial ecosystems. In contrast to the other microenvironment of plants, the rhizosphere is characterized by high microbial abundances (Berg et al., 2005b) and activities (Herron et al., 2013). Due to the densely colonized surface and surrounding soil (Figure 1), the rhizosphere was suggested as a protection shield against soil-borne pathogens (Weller et al., 2002).
FIGURE 1.
Bacterial micro-colonies in the rhizosphere and endosphere visualized by fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy(CLSM). The rhizosphere microbiome of lettuce is dominated by Betaproteobacteria (purple) forming dense colonies on the root surface. The endosphere is shown as section of the main root of a lettuce plant. While unspecifically labeled bacteria are in red, Alphaproteobacteria (orange-green) are to be seen as colonies as well as filamentous forms.
THE ENDOSPHERE IS A HABITAT FOR INTIMATE INTERACTIONS
Although endophytes were ignored or considered contaminants for a long time, many endophytic inhabitants of plants are now often recognized as symbionts with a unique and intimate interaction with the plant (Ryan et al., 2008; Reinhold-Hurek and Hurek, 2011; Mitter et al., 2013). In these and other more recent studies, evidence of the occurrence of endophytes was assessed by cultivation-independent analyses, and via fluorescence in situ hybridization-confocal laser scanning microscopy (FISH-CLSM; an example for endophytes in the lettuce endorhiza is shown in Figure 1). After the first definition by De Bary (1866) as “any organism occurring within plant tissues,” various researchers have defined endophytes in different ways, which were usually related to their own research context and perspective (Wilson, 1995; Hallmann et al., 1997; Bacon and White, 2000).
THE FUNCTIONS OF THE PLANT MICROBIOME ARE ESSENTIAL FOR THE HOST
Plant-associated microorganisms can help plants to suppress diseases, to stimulate growth, to occupy space that would otherwise be available to pathogens, to promote stress resistance, and influence crop yield and quality by nutrient mobilization and transport (Lugtenberg and Kamilova, 2009; Yang et al., 2009). Therefore, the plant microbiome is one of the key determinants of plant health and productivity. Additional essential roles of the plant microbiome for phenotypic and epigenetic plasticity as well as the evolution of plants were suggested by Partida-Martínez and Heil (2011).
SPECIFIC ENRICHMENT OF MICROORGANISMS IN PLANT-ASSOCIATED COMMUNITIES EXISTS
So far, research on the specificity of plant-associated microbiomes focused on the rhizosphere, while only few other compartments have been studied in this respect (Vorholt, 2012). Although plant specific microbiomes in the rhizosphere have already been postulated via cultivation-based approaches (Germida and Siciliano, 2001), molecular fingerprints provided the first clear evidence for plant-dependent microbial community compositions (Smalla et al., 2001). Differences in plant root exudates play an important role as both chemo-attractants as well as repellents (Badri and Vivanco, 2009), to which bacteria are especially responsive (Costa et al., 2006, 2007; DeAngelis et al., 2009). In addition, plant defense signaling plays a role in this process as well (Doornbos et al., 2012). Haichar et al. (2008) used a stable isotope probing (SIP) approach to show that plant host habitat and root exudates shape the soil bacterial community structure. Thus, the plant is clearly able to select microorganisms for rhizosphere colonization primarily from the large pool living in the surrounding soil. Lundberg et al. (2012), Bulgarelli et al. (2012) revealed that only a subset of the bacterial community in the soil is present around the plant roots of Arabidopsis thaliana through amplicon sequencing of 16S rRNA gene fragments. Furthermore, the use of catalyzed reporter deposition and in situ hybridization or FISH was used to confirm the co-localization and dynamics of dominant taxa determined by 454 pyrosequencing (Bulgarelli et al., 2012; Lundberg et al., 2012; Ofek et al., 2012). While the use of FISH and catalyzed reported deposition-fluorescence in situ hybridization (CARD-FISH) helped to unravel the spatial distribution of dominant indigenous bacterial communities, the use of marker and reporter genes was employed in several studies to localize inoculated potential biocontrol strains and to measure distributions of nutrients, metals, and organic exudates along the roots on a microscale (Sørensen et al., 2009).
However, the plant (species, cultivar, age, health, and developmental stage) is not the only factor that influences microbial communities in the rhizosphere: a multitude of abiotic factors modulate the structural and functional diversity of the rhizosphere microbiome, including soil properties, nutrient status, and climatic conditions (rev. in Berg and Smalla, 2009). Moreover, large-scale agricultural management such as manure application has a clear impact on the microbiome composition (Jechalke et al., 2014).
THE ORIGIN OF PLANT-ASSOCIATED BACTERIA IS DIFFERENT
Plants are in constant contact with diverse microorganisms originating either through soil, wind, and air, or water via the water cycle. After initial exposure, some of these microorganisms are able to colonize the plant and survive (Rastogi et al., 2012). In some cases, microorganisms can even be transferred vertically from the parent plants to their progeny. Endophytes present in plant seeds may subsequently colonize the roots and the rhizosphere (Johnston-Monje and Raizada, 2011; Links et al., 2014). In addition, generative organs such as anther pockets, producing pollen (Fürnkranz et al., 2012), and moss sporophytes (Bragina et al., 2012) share a microbiome containing beneficials with their host plant.
Pseudomonas AND Bacillus ARE MODEL PLANT-ASSOCIATED BACTERIA
Although we now know that plant-associated bacteria are phylogenetically diverse, Pseudomonas and Bacillus have been studied as models for beneficial plant-microbe interaction (Emmert and Handelsman, 1999; Weller et al., 2002; Raaijmakers et al., 2010) for a long time. Interestingly, the importance of both genera on plants has been corroborated in many metagenomic studies. While Pseudomonas is abundant under humid conditions (Mendes et al., 2012), Bacillus dominates plant microbiomes under arid conditions such as in Egypt where Pseudomonas cannot survive (Köberl et al., 2011). The more detailed information obtained for Pseudomonas–plant interactions now help in understanding the bigger picture of Pseudomonas genome–plant interaction in its entirety as shown in the excellent review by Loper et al. (2012).
Antibiotic production by plant-associated microorganisms, with the rhizosphere and endosphere as a “hot spot” for potential producers, is a further aspect of research, for which both model organisms again play an important role. Pseudomonas is known for its versatile antibiotic production, which has also been shown in situ in the rhizosphere (Bonsall et al., 1997). Yet, a lot has still to be learned about the diffusion and action of small molecule antibiotics. Antibiotics are not only acting in solutes, some bioactive compounds act as volatiles, both in antibiosis against pathogens as well as in communication with plants (Ryu et al., 2003). According to recent reports, antibiotics and lipopeptides of bacteria are regulators and support biofilm formation, signaling, motility, and acquisition of micronutrients at sub-inhibitory concentrations (Raaijmakers et al., 2010; Raaijmakers and Mazzola, 2012). An interesting regulatory network was also detected for redox-active antibiotics such as phenazine, which is also involved in the reduction of Fe3+(Raaijmakers and Mazzola, 2012). This high number of antibiotic producers associated with plants may have driven the evolutions of resistance genes as well (Allen et al., 2010).
Several studies, which focused primarily on Pseudomonas demonstrated bacterial intra- and interspecies communication in the plant-soil interface plant-microbe interaction via quorum sensing molecules such as N-acyl homoserine lactones (N-AHLs), or antibiotics at sub-inhibitory concentration (Steidle et al., 2001; DeAngelis et al., 2009; Hartmann and Schikora, 2012; Raaijmakers and Mazzola, 2012). Bacterial AHLs were demonstrated to change the plant transcriptome, modify root growth, and induce systemic resistance to phytopathogens (von Rad et al., 2008; Hartmann and Schikora, 2012; Raaijmakers and Mazzola, 2012); yet substantial differences were observed in the uptake, transport, and degradation of various AHLs for different plants (Götz et al., 2007).
HORIZONTAL GENE TRANSFER CONTRIBUTES TO PLASTICITY AND EVOLUTION OF PLANT-ASSOCIATED BACTERIA
Owing to the availability of various nutrients and surfaces, the plant-soil interface is also considered a hot spot for horizontal gene transfer processes via plasmids (Heuer and Smalla, 2012). The recent progress in microscopy tools has been extremely helpful in gaining further insight into the spatial distribution and dynamics of the plant-soil interface. Plant species-dependent differences were observed for the conjugation of a gfp-tagged IncP-1ε plasmid that did not express the gfp in its original host due to the presence of a lac-repressor (Mølbak et al., 2007). Through in situ visualization, these authors could demonstrate that both exudation patterns and root growth rates determined plasmid transfer in the pea and barley rhizospheres.
FUTURE PERSPECTIVES AND INFORMATION GAPS
Although the plant microbiome is recognized as an immense treasure trove of microbial diversity, numerous important crop species and their natural relatives have not yet been studied for their associated bacterial communities. With an approximate number of 500,000 plant species a lot of work lays ahead of plant microbiome research to explore new aspects about phylogenetic diversity of plant-associated microorganisms in the future. This might be particularly interesting with plants from extreme natural ecosystems or with unique life styles (carnivores, parasites, etc.).
Despite this enormous progress in the description of the plant microbiome, more fundamental and practical studies to address the processes leading to community assembly and function in and on plants are needed. Metagenomic analysis and comparison of plant-associated communities will lead to novel phylogenetic and functional insight. The first metagenomes, -proteomes, and -transcriptomes are currently published (Delmotte et al., 2009; Knief et al., 2012). An interesting example for a novel function is the detection of potential coexistence of microbial and plant photosynthesis on Tamarix leaves (Atamna-Ismaeel et al., 2012). Functional analysis will demonstrate whether the plants are able to benefit from the presence of certain microorganisms. In this context it should also be kept in mind that activation patterns and induction pathways can differ between ecotypes and strains.
Amplicon sequencing of 16S rRNA gene fragments provided valuable insight into the dominant colonizers, but too much emphasis on this locus may underdiagnose the potential biological variation. For example, biological functions provided from the mobilome (Eltlbany et al., 2012) do not correspond with 16S rRNA gene data. In addition, ribosomal gene amplicon quantities can depend on extraction methods, primer efficiency (Pinto and Raskin, 2012), and their copy-number variation (Kembel et al., 2012).
Although with the following articles in this special issue focus was given to the bacterial aspect of plant microbiomes we predict a future integration with fungal–bacterial interactions, specifically in the context of mycorrhiza (Bonfante and Anca, 2009; Song et al., 2010).
Plant microbiome discoveries could fuel advances in sustainable agriculture (Berg, 2009; Lugtenberg and Kamilova, 2009), such as the development of microbial inoculants as biofertilizers, biocontrol, or stress protection products (Berg, 2009; Berg et al., 2013).
In the future, the plant microbiome will have a greater importance for plant breeding and plant biotechnology. Until now, primarily plant pathogens were considered in these approaches. However, we suggest that the beneficial aspect of the entire microbiome should also be integrated as a biomarker.
A better understanding of the whole plant microbiome might be important to prevent outbreak of plant diseases or critical association of human pathogens with plants. We have learned that the human microbiome is much more involved in diseases than recently thought, and that pathogen outbreaks are associated with shifts in the entire community, including supporting pathogens (Blaser et al., 2013). While these processes are studied for human pathogens, much less is known about plant pathogens (Fürnkranz et al., 2012; Ottesen et al., 2013).
Furthermore, we envision the plant microbiome as an important source shaping other microbiomes. By the comparison of microbiome structures, a meaningful overlap of phylogenetic diversity can be recognized among microbiomes which are in some way linked to each other. This may also include the human habitat and plants. After we have received our first microbial inoculants by delivery and breast milk from our mother, our food becomes an important source not only of nutrients, but also of microorganisms (Blaser et al., 2013). Thus, digestive factors of plants and their microorganisms may modulate our own “second genome.” Observations of domestic microbiomes suggest that they are significantly influenced by their human inhabitants and by the surrounding vegetation (Oberauner et al., 2013). These connections, which we conceive as links in a complex network among microbiomes, are still little understood and need further attention.
WHAT IS THE CONTRIBUTION OF THIS RESEARCH TOPIC?
This special issue will close some of the information gaps in plant microbiome ecology. It includes studies about the microbial diversity of yet unknown plants. In medicinal plants, the production of bioactive plant metabolites leads to a highly pronounced specificity in the microbiome structure (rev. in Köberl et al., 2014). Interestingly a correlation between the bioactive substances (drimane sesquiterpenes) and the endophytic community of roots was shown for the medical tree Warburgia ugandensis (Drage et al., 2014). Although it is known that plant secondary metabolites play an important role as drivers for microbial community structure, these studies show for the first time the importance with medicinal plants. Vice versa – Schmidt et al. (2014) could show that Chamomile plants treated with selected Bacillus strains produced more bioactive substances than untreated controls, thus microbes might be able to induce production of secondary metabolites of interest.
To better understand the significance of the plant-associated microbiome in prevention of pathogen outbreaks several studies focused on the lettuce microbiome and connected aspects of plant– and human health (Erlacher et al., 2014; Schreiter et al., 2014). Erlacher et al. (2014) showed that pathogens as well as beneficals induce a shift in the structure of the microbial community. To our knowledge, this is the first study analyzing this background effect, which can be important for plant protection strategies. However, also soil type was identified as important driver of the lettuce-associated community as well as the corresponding biocontrol effect (Schreiter et al., 2014). In addition, also for lettuce plants the impact of plant secondary metabolites exudated by roots in different soil types was pointed out (Neumann et al., 2014).
Another contribution presents evidence that Escherichia coli and Salmonella enterica infections occur due to consumption of vegetables, sprouts, and occasionally fruits (van Overbeek et al., 2014). The authors described a new transmission route of pathogens via plants or products derived from plants, and defined this process as “phytonosis”.
The role of multitrophic interactions for plant diseases and the occurrence of the western corn rootworm were analyzed by Dematheis et al. (2014). In addition to biotic factors, the impact of abiotic factors on the plant microbiome was investigated. Elevated atmospheric O3 changed the community structure of biocontrol active actinobacteria in the rhizosphere of European beech (Haesler et al., 2014).
Two studies suggest members of the plant-associated Burkholderia cluster as model to study plant-microbe interactions. Oxalate acts as carbon source and as determinant in colonization processes in lupins and maize (Kost et al., 2014), while nitrogen-fixing Burkholderia populations are highly abundant in Sphagnum bogs (Bragina et al., 2014).
Two mini-reviews focus on the interplay of microbiomes as well as the importance of the plant microbiome for others. The connection between plant and our built environment microbiome is discussed by Berg et al. (2014), and another one highlighted similarities between the gut and root microbiome and suggested to transplant “healthy microbiomes” to avoid or therapy plant diseases (Gopal et al., 2013). A step forward to understand the plant-microbe networking was presented in the review by Hartmann et al. (2014). They come to the conclusion that functional interaction studies of holobiotic plant systems, including the plant host and its associated microbes, may result in a more profound understanding of the complicated social network of basic innate immune responses with specific effector molecules, if quorum sensing compounds of endophytic bacteria are integrated.
Overall, this issue presents new results about (i) the role of plant secondary metabolites for the microbiome and vice versa, (ii) health issues related to the consumption of raw-eaten plants, (iii) the interplay of microbiomes as well as within them and (iv) the impact of biotic and abiotic factors on the structure and function of plant-associated microbial communities.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Julian Quehenberger and Massimiliano Cardinale (Graz) for support with CLSM pictures. Margaret R. Starcher (Washington/Graz) and Ilse-Marie Jungkurth (Braunschweig) are gratefully acknowledged for language revision of the manuscript. This manuscript was supported by different grants from the Austrian Science Foundation FWF (I183-B16; I 882-B16) to Gabriele Berg. The cooperation of all authors was funded by a project in the Austrian Centre of Industrial Biotechnology, which has been supported by the Austrian BMWFJ, BMVIT, SFG, Standortagentur Tirol and ZIT through the Austrian FFG-COMET-Funding Program.
REFERENCES
- Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8 251–259 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Atamna-Ismaeel N., Finkel O. M., Glaser F., Sharon I., Schneider R., Post A. F., et al. (2012). Microbial rhodopsins on leaf surfaces of terrestrial plants. Environ. Microbiol. 14 140–146 10.1111/j.1462-2920.2011.02554.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C. W., White J. F. (2000). Microbial Endophytes. New York: Marcel Dekker Inc [Google Scholar]
- Badri D. V., Vivanco J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32 666–681 10.1111/j.1365-3040.2009.01926.x [DOI] [PubMed] [Google Scholar]
- Bais H. P., Weir T. L., Perry L. G., Gilroy S., Vivanco J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57 233–266 10.1146/annurev.arplant.57.032905.105159 [DOI] [PubMed] [Google Scholar]
- Bakker P. A., Berendsen R. L., Doornbos R. F., Wintermans P. C., Pieterse C. M. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 30:165. 10.3389/fpls.2013.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 478–486 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Berg G., Eberl L., Hartmann A. (2005a). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7 1673–1685 10.1111/j.1462-2920.2005.00891.x [DOI] [PubMed] [Google Scholar]
- Berg G., Krechel A., Ditz M., Sikora R. A., Ulrich A., Hallmann J. (2005b). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51 215–229 10.1016/j.femsec.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. J. Appl. Microbiol. Biotechnol. 84 11–18 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Berg G., Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68 1–13 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- Berg G., Zachow C., Müller H., Phillips J., Tilcher R. (2013). Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3 648–656 10.3390/agronomy3040648 [DOI] [Google Scholar]
- Berg G., Mahnert A., Moissl-Eichinger C. (2014). Beneficial effects of plant-associated microbes on indoor microbiomes and human health? Front. Microbiol. 5:15. 10.3389/fmicb.2014.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M., Bork P., Fraser C., Knight R., Wang J. (2013). The microbiome explored: recent insights and future challenges. Nat. Rev. Microbiol. 11 213–217 10.1038/nrmicro2973 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Anca I. A. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Ann. Rev. Microbiol. 63 363–383 10.1146/annurev.micro.091208.073504 [DOI] [PubMed] [Google Scholar]
- Bonsall R. F., Weller D. M., Thomashow L. S. (1997). Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A., Berg C., Cardinale M., Shcherbakov A., Chebotar V., Berg G. (2012). Sphagnum mosses harbor highly specific bacterial diversity during their whole lifecycle. ISME J. 6 802–813 10.1038/ismej.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A., Cardinale M., Berg C., Berg G. (2014). Vertical transmission explains the specific Burkholderia pattern in Sphagnum mosses at multi-geographic scale. Front. Microbiol. 4:394. 10.3389/fmicb.2013.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Rott M., Schlaeppi K., Ver Loren van Themaat E., Ahmadinejad N., Assenza F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488 91–95 10.1038/nature11336 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Costa R., Götz M., Mrotzek N., Lottmann J., Berg G., Smalla K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56 236–249 10.1111/j.1574-6941.2005.00026.x [DOI] [PubMed] [Google Scholar]
- Costa R., Gomes N. C. M., Krögerrecklenfort E., Opelt K., Berg G., Smalla K. (2007). Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ. Microbiol. 9 2260–2273 10.1111/j.1462-2920.2007.01340.x [DOI] [PubMed] [Google Scholar]
- DeAngelis K. M., Brodie E. L., DeSantis T. Z., Andersen G. L., Lindow S. E., Firestone M. K. (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J. 3 168–178 10.1038/ismej.2008.103 [DOI] [PubMed] [Google Scholar]
- Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106 16428–16433 10.1073/pnas.0905240106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dematheis F., Kurtz B., Vidal S., Smalla K. (2014). Multitrophic interactions among Western Corn Rootworm, Glomus intraradices and microbial communities in the rhizosphere and endorhiza of maize. Front. Microbiol. 4:357. 10.3389/fmicb.2013.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos R. F., Van Loon L. C, Bakker P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron. Sustain. Dev. 32 227–243 10.1007/s13593-011-0028-y [DOI] [Google Scholar]
- Drage S., Mitter B., Muchugi A., Jamnadass R. M., Sessitsch A., Hadacek F. (2014). Antimicrobial drimane sesquiterpenes and their effect on endophyte communities in the medical tree Warburgia ugandensis. Front. Microbiol. 5:13. 10.3389/fmicb.2014.00013 [DOI] [Google Scholar]
- Eltlbany N., Prokscha Z. Z., Castañeda-Ojeda P., Krögerrecklenfort E., Heuer H., Wohanka W., et al. (2012). A new bacterial disease on Mandevilla sanderi caused by Pseudomonas savastanoi – lessons learned for bacterial diversity studies. Appl. Environ. Microbiol. 78 8492–8497 10.1128/AEM.02049-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert E. A., Handelsman J. (1999). Biocontrol of plant disease: a (Gram+) positive perspective. FEMS Microbiol. Lett. 171 1–9 10.1111/j.1574-6968.1999.tb13405.x [DOI] [PubMed] [Google Scholar]
- Erlacher A., Cardinale C., Grosch R., Grube M., Berg G. (2014). The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Front. Microbiol. 5:175. 10.3389/fmicb.2014.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürnkranz M., Lukesch B., Müller H., Huss H., Grube M., Berg G. (2012). Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb. Ecol. 63 418–428 10.1007/s00248-011-9942-4 [DOI] [PubMed] [Google Scholar]
- Germida J., Siciliano S. (2001). Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils 33 410–415 10.1007/s003740100343 [DOI] [Google Scholar]
- Götz C., Fekete A., Gebefuegi I., Forczek S. T., Fuksová K., Li X., et al. (2007). Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal. Bioanal. Chem. 389 1447–1457 10.1007/s00216-007-1579-2 [DOI] [PubMed] [Google Scholar]
- Gopal M., Gupta A., Thomas G. V. (2013). Bespoke microbiome therapy to manage plant diseases. Front. Microbiol. 4:355. 10.3389/fmicb.2013.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler F., Hagn A., Engel M., Schloter M. (2014). Impact of elevated atmospheric O3 on the actinobacterial community structure and function in the rhizosphere of European beech (Fagus sylvatica L.). Front. Microbiol. 5:36. 10.3389/fmicb.2014.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haichar F. Z., Marol C., Berge O., Rangel-Castro J. I., Prosser J. I., Balesdent J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2 1221–1230 10.1038/ismej.2008.80 [DOI] [PubMed] [Google Scholar]
- Hallmann J., Quadt-Hallmann A., Mahaffee W. F., Kloepper J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43 895–914 10.1139/m97-131 [DOI] [Google Scholar]
- Hartmann A., Rothballer M., Schmid M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312 7–14 10.1007/s11104-007-9514-z [DOI] [Google Scholar]
- Hartmann A., Schikora A. (2012). Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactose with eucaryotes. J. Chem. Ecol. 38 704–713 10.1007/s10886-012-0141-7 [DOI] [PubMed] [Google Scholar]
- Hartmann A., Rothballer M., Hense B., Schroeder P. (2014). Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front. Microbiol. 5:131. 10.3389/fpls.2014.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron P. M., Gage D. J., Arango Pinedo C., Haider Z. K., Cardon Z. G. (2013). Better to light a candle than curse the darkness: illuminating spatial localization and temporal dynamics of rapid microbial growth in the rhizosphere. Front. Plant Sci. 4:323. 10.3389/fpls.2013.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H., Smalla K. (2012). Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36 1083–1104 10.1111/j.1574-6976.2012.00337.x [DOI] [PubMed] [Google Scholar]
- Hirsch P. R., Mauchline T. H. (2012). Who’s who in the plant root microbiome? Nat. Biotechnol. 30 961–962 10.1038/nbt.2387 [DOI] [PubMed] [Google Scholar]
- Jechalke S., Focks A., Rosendahl I., Groeneweg J., Siemens J., Heuer H., et al. (2014). Structural and functional response of the soil bacterial community to application of manure from difloxacin-treated pigs. FEMS Microbiol. Ecol. 87 78–88 10.1111/1574-6941.12191 [DOI] [PubMed] [Google Scholar]
- Johnston-Monje D., Raizada M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6:e20396. 10.1371/journal.pone.0020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel S. W., Wu M., Eisen J. A., Green J. L. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput. Biol. 8:e1002743. 10.1371/journal.pcbi.1002743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6 1378–1390 10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberl M., Ramadan E. M., Roßmann B., Staver C., Fürnkranz M., Lukesch B., et al. (2012). “Using ecological knowledge and molecular tools to develop effective and safe biocontrol strategies,” in Pesticides in the Modern World–Pests Control and Pesticides Exposure and Toxicity Assessment / Book 5. Available at: http://www.intechopen.com/about-intech.html [Google Scholar]
- Köberl M., Müller H., Ramadan E. M., Berg G. (2011). Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 6:e24452. 10.1371/journal.pone.0024452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberl M., Schmidt R., Ramadan E. M., Bauer R., Berg G. (2014). The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front. Microbiol. 4:400. 10.3389/fmicb.2013.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost T., Stopnisek N., Agnoli K., Eberl L., Weisskopf L. (2014). Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front. Microbiol. 4:421. 10.3389/fmicb.2013.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau J. H. J. (2007). The magic and menace of metagenomics: prospects for the study of plant growth-promoting rhizobacteria. Eur. J. Plant Pathol. 119 279–300 10.1007/s10658-007-9186-9 [DOI] [Google Scholar]
- Lindow S. E., Brandl M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69 1875–1883 10.1128/AEM.69.4.1875-1883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Links M. G., Demeke T., Gräfenhan T., Hill J. E., Hemmingsen S. M., Dumonceaux T. J. (2014). Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 202 542–553 10.1111/nph.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E., Hassan K. A., Mavrodi D. V., Davis E. W., II, Lim C. K., Shaffer B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth promoting rhizobacteria. Annu. Rev. Microbiol. 63 541–556 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 7409 86–90 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R., Kruijt M., De Bruijn I., Dekkers E., Van der Voort M., Schneider J. H. M., et al. (2012). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332 1097–1100 10.1126/science.1203980 [DOI] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37 634–663 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Mitter B., Petric A., Shin M. W., Chain P. S., Hauberg-Lotte L., Reinhold-Hurek B., et al. (2013). Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 4:120. 10.3389/fpls.2013.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe L. A. (2013). Amino acids in the rhizosphere: from plants to microbes. Am. J. Bot. 100 1692–1705 10.3732/ajb.1300033 [DOI] [PubMed] [Google Scholar]
- Mølbak L., Molin S., Kroer N. (2007). Root growth and exudates production define the frequency of horizontal plasmid transfer in the rhizosphere. FEMS Microbiol. Ecol. 59 167–176 10.1111/j.1574-6941.2006.00229.x [DOI] [PubMed] [Google Scholar]
- Neumann G., Bott S., Ohler M., Mock H. P., Lippmann R., Grosch R., et al. (2014). Root exudation and root development of lettuce (Lactuca sativa L. cv. Tizian) as affected by different soils. Front. Microbiol. 5:2. 10.3389/fmicb.2014.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberauner L., Zachow C., Lackner S., Högenauer C., Smolle K. H., Berg G. (2013). The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Sci. Rep. 3 1413 10.1038/srep01413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek M., Hadar Y., Minz D. (2012). Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 7:e40117. 10.1371/journal.pone.0040117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen A. R., González Peña A., White J. R., Pettengill J. B., Li C., Allard S., et al. (2013). Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13:114. 10.1186/1471-2180-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martínez L. P., Heil M. (2011). The microbe-free plant: fact or artifact? Front. Plant Sci. 2:100. 10.3389/fpls.2011.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. J., Raskin L. (2012). PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 7:e43093. 10.1371/journal.pone.0043093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J. M., de Bruijn I., Nybroe O., Ongena M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. 34 1037–1062 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., Mazzola M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50 403–424 10.1146/annurev-phyto-081211-172908 [DOI] [PubMed] [Google Scholar]
- Rastogi G., Sbodio A., Tech J. J., Suslow T. V., Coaker G. L., Leveau J. H. (2012). Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 6 1812–1822 10.1038/ismej.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi G., Coaker G. L., Leveau J. H. (2013). New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 348 1–10 10.1111/1574-6968.12225 [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Hurek T. (2011). Living inside plants: bacterial endophytes. Curr. Opin. Plant Biol. 14 435–443 10.1016/j.pbi.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Ryan R. P., Germaine K., Franks A., Ryan D. J., Dowling D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278 1–9 10.1111/j.1574-6968.2007.00918.x [DOI] [PubMed] [Google Scholar]
- Ryu C. M., Farag M. A., Hu C. H., Reddy M. S., Wei H. X., Paré P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100 4927–4932 10.1073/pnas.0730845100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Köberl M., Mostafa A., Ramadan E. M., Monschein M., Jensen K. B., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5:64. 10.3389/fmicb.2014.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiter S., Ding G., Heuer H., Neumann G., Sandmann M., Grosch R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. 10.3389/fmicb.2014.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla K., Wieland G., Buchner A., Zock A., Parzy J., Kaiser S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67 4742–4751 10.1128/AEM.67.10.4742-4751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J., Hauberg Nicolaisen M., Ron E., Simonet P. (2009). Molecular tools in rhizosphere microbiology – from single-cell to whole-community analysis. Plant Soil 321 483–512 10.1007/s11104-009-9946-8 [DOI] [Google Scholar]
- Song Y. Y., Zeng R. S., Xu J. F., Shen X., Yihdego W. G. (2010). Interplant communication of tomato plants through underground common mycorrhizal networks. PLoS ONE 5:e13324. 10.1371/journal.pone.0013324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidle A., Sigl K., Schuhegger R., Ihring A., Schmid M., Gantner S., et al. (2001). Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67 5761–5770 10.1128/AEM.67.12.5761-5770.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek L., van Doorn J., Wichers J., van Amerongen A., van Roermund H., Willemsen P. (2014). The arable plant ecosystem as battleground for emergence of human pathogens. Front. Microbiol. 5:104. 10.3389/fmicb.2014.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rad U., Klein I., Dobrev P. I., Kottova J., Zazimalova E., Fekete A., et al. (2008). Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229 73–85 10.1007/s00425-008-0811-4 [DOI] [PubMed] [Google Scholar]
- Vorholt J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10 828–840 10.1038/nrmicro2910 [DOI] [PubMed] [Google Scholar]
- Weller D. M., Raaijmakers J. M., Gardener B. B., Thomashow L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40 309–348 10.1146/annurev.phyto.40.030402.110010 [DOI] [PubMed] [Google Scholar]
- Weston L. A., Mathesius U. (2013). Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 39 283–297 10.1007/s10886-013-0248-5 [DOI] [PubMed] [Google Scholar]
- Wilson D. (1995). Endophyte: the evolution of a term, and clarification of its use and definition. Oikos 73 274–276 10.2307/3545919 [DOI] [Google Scholar]
- Yang J., Kloepper J. W., Ryu C. M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14 1–4 10.1016/j.tplants.2008.10.004 [DOI] [PubMed] [Google Scholar]