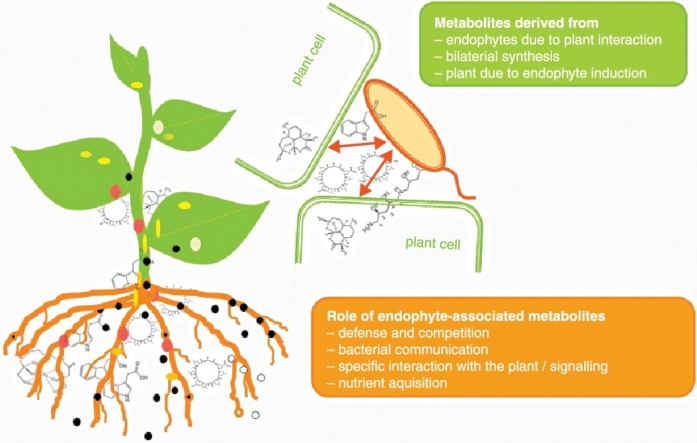
Graphical abstract
Highlights
-
•
Endophytes are a source of a plethora of biologically active substances.
-
•
Endophyte-associated metabolites may be needed for the interaction with the plant.
-
•
Some metabolites are produced jointly by plant and endophytes.
-
•
Endophytes may stimulate or alter metabolite production by the plant.
-
•
Metabolite functions include signalling and communication, nutrient acquisition and defense.
Abstract
The bacterial endophytic microbiome promotes plant growth and health and beneficial effects are in many cases mediated and characterized by metabolic interactions. Recent advances have been made in regard to metabolite production by plant microsymbionts showing that they may produce a range of different types of metabolites. These substances play a role in defense and competition, but may also be needed for specific interaction and communication with the plant host. Furthermore, few examples of bilateral metabolite production are known and endophytes may modulate plant metabolite synthesis as well. We have just started to understand such metabolic interactions between plants and endophytes, however, further research is needed to more efficiently make use of beneficial plant-microbe interactions and to reduce pathogen infestation as well as to reveal novel bioactive substances of commercial interest.
Current Opinion in Biotechnology 2014, 27:30–37
This review comes from a themed issue on Environmental biotechnology
Edited by Hauke Harms and Howard Junca
For a complete overview see the Issue and the Editorial
Available online 22nd October 2013
0958-1669/$ – see front matter, © 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
Introduction
Living organisms are the source for a vast diversity (>1 million) of different metabolites. The majority of these metabolites have been discovered in plants, but microorganisms are a particular rich source of more than 20 000 biologically active compounds, influencing the performance and survival of other organisms [1]. Of these active compounds the majority are derived from bacteria, mostly from the well investigated genus Streptomyces [1,2], which represents the microbial genus most thoroughly investigated for secondary metabolite production. Secondary metabolites are without known specific function in the organisms’ primary metabolism, but their high diversity reflects the biological role particularly in the interactions between organisms in their environment and shows their importance as signals and toxins.
In spite of the decreasing rate of discovery of active metabolites (e.g. antibiotics) in the last decades [3], the genomic revolution of the recent past clearly revealed that our knowledge on the structures and occurrence of metabolites of bacteria is far from saturated. Genome analysis of even well-known bacteria has revealed genes potentially involved in the production of yet unknown metabolites [4,5] and it is assumed that the metabolites identified so far encompass only a small fraction of the existing metabolic repertoire [2]. Another reason why it seems unlikely that the metabolic potential of bacteria is exhaustively known is the fact that so far only a small proportion of bacteria has been cultivated. In soils alone, different studies using DNA:DNA hybridization, Sanger sequencing of clone libraries and next generation sequencing suggest that only a very small percentage of bacteria has been cultivated so far [6]. Moreover, albeit Actinobacteria, most prominently the genus Streptomyces, proved to be an extremely rich source of secondary metabolites [7,8•], the potential of more ‘exotic’ and ‘rare’ actinobacterial taxa is less established [9,10,11] and similar considerations might also hold true for the large fraction of other far less well characterized bacterial taxa. Finally, certain niches, among others the bacteria living in association with plants and in particular inside plants (endophytes), are less well investigated for their metabolic potential than cultivable soil bacteria. Endophytes are also of special interest for their high number of microbial niches and environments they may inhabit and provide therefore a high potential as a less exploited resource. In the current review we understand endophytes as non-phytopathogenic organisms, which colonize plant tissues at least part of their lifetime [12]. Nevertheless, to discuss the potential and function of metabolites we briefly also take into account plant pathogenic microorganisms, which may be very closely related to non-pathogenic species.
Endophytes as a source of secondary metabolites
Considerable amount of information exists on the metabolic potential of endophytic fungi and exciting possibilities for exploiting endophytic fungi for the production of a plethora of known and novel biologically active secondary metabolites (reviewed by [13,14]). Bacteria can also thrive as endophytes in various plants and plant parts, but are less investigated for their metabolic potential. Various studies have shown that endophytic bacteria may, following rhizosphere soil colonization, be detected inside the endorhiza, in stems, leaves as well as inside plant reproductive organs of different host plants [15,16,17]. Endophytes have to be adapted to the specific plant environment, which they colonize and therefore, the metabolic potential of endophytes is likely to differ from their soil dwelling counterparts. As the resource-rich environment of the rhizosphere is extremely competitive, and bacteria need to survive in a competitor-rich and predator-rich environment, the rhizosphere microflora is likely to produce a rich arsenal of antibiotic and anti-nematodal compounds. In contrast, obligate endophytic bacteria face a lot less competition reflected in a less metabolite-rich arsenal [18], but they may produce other specific metabolites supporting the or needed for the interaction with the host. However, many endophytic bacteria are facultative plant colonizers and have to compete well in the rhizosphere before entering the plant [16] and might be therefore equipped with a rich arsenal of metabolites involved in defense as well as in interaction with the plant. In this context it has to be stated that the term ‘antibiotic’ as ‘defense weapon’ to other microbes may reflect a rather anthropocentric point of view and that the real function of these compounds in nature is not only the antibiotic function, but the compounds may also play a role in intraspecies and interspecies signalling processes [6,19,20,21].
Many bacteria with the capacity of colonizing plants utilize the nutrient niche of root surfaces in the rhizosphere and most of them might even actively switch from root surface to endophytic lifestyles [15,16]. These bacteria comprise several well characterized species of Bacillus and Pseudomonas and a number of metabolites, particularly lipopeptides synthesized by non-ribosomal peptide synthesases, have been described to be important for rhizosphere bacteria for antibiosis and for inducing plant defense mechanisms. The structures and functions of Bacillus and Pseudomonas lipopeptides have been recently thoroughly reviewed (e.g. [6,22,23]). Nevertheless the rich repertoire of metabolites found in endophytic Actinobacteria [8•] suggests that a large fundus of secondary metabolites produced by endophytic bacteria remains to be discovered. This is underlined by recently described multicyclic indolosesquiterpenes (Figure 1) found in the endophytic Streptomyces sp. HKI0595 of the mangrove tree Kandelia candel [24], antitrypanosomal alkaloids spoxazomicins A-C (Figure 1) produced by the endophytic actinomycete Streptosporangium oxazolinicum K07-0450T found in orchids [25,26] with structural similarities to siderophores from Pseudomonas aeruginosa and a series of NRPS (non-ribosomal peptide synthases) and PKS (polyketide synthases) gene clusters with uncharacterized metabolites were found to be produced by endophytes of Chinese medicinal plants [27,28]. The rich metabolic repertoire of endophytic bacteria is also shown in more than 100 actinobacterial isolates found as endophytes in Australian trees [29] and in more than 300 diverse actinobacterial strains found in the medicinal plant Maytenus austroyunnanensis [8•]. Furthermore, cultivation-independent analysis of bacterial endophytes of Chinese medicinal herbs based on the analysis NRPS and PKS gene fragments suggested the production of so far unknown metabolites [28]. Overall, only a tiny fraction of plant-associated Actinobacteria has been described so far representing a promising source of novel secondary metabolites.
Figure 1.
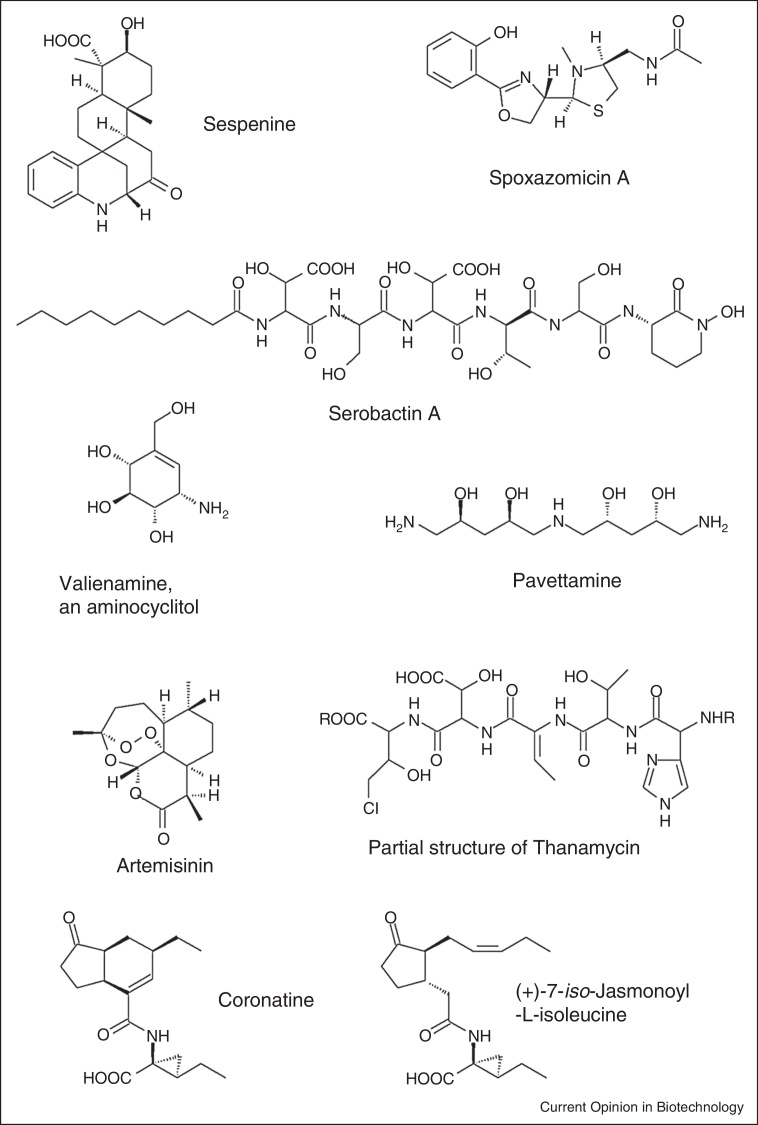
Metabolites of plant associated bacteria. Sespenine is derived from indolosesquiterpenes found in an endophyte (Streptomyces sp.) of mangrove trees. Spoxazomicins from an orchid endophyte (Streptosporangium oxazolinicum) with structural similarities to pyochelin, a siderophore from Pseudomonas aeruginosa, serobactin A, a siderophore from the grass endophyte Herbaspirillum seropedicae. Valienamine, as illustration of aminocyclitols, which might be produced by the endophytic C. Burkholderia kirkii. Pavettamine is the active toxic principle of South African Rubiaceae, where endophytic Burkholderia spp. are crucial for the biosynthesis in planta. The partial structure of thanamycine has been elucidated without isolation from bacterial colonies. Coronatine as an example of a plant hormone acting agent from the plant pathogenic Pseudomonas syringae and the structure of the actual plant hormone (+)-7-iso-jasmonoyl-l-isoleucine.
Function of metabolites in plant-bacteria interactions
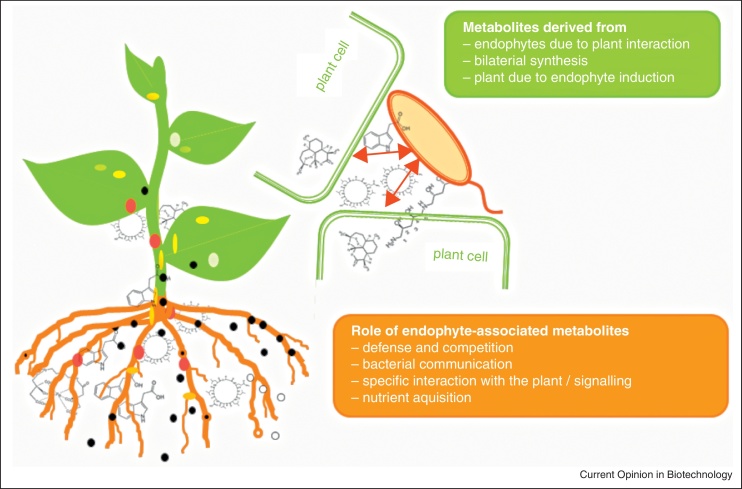
Many bacteria closely interacting with plants produce secondary metabolites as agents needed for nutrient uptake (for a schematic overview see Figure 2), in particular siderophores involved in iron acquisition (reviewed by [30]). Recently, in the diazotrophic endophyte Herbaspirillum seropedicae colonizing many grass crops, the structures of the amphiphilic lipopeptides serobactin A, B and C produced by NRPS (Figure 1) acting as siderophores have been described [31]. Moreover, metabolites acting as agents in biofilm formation and as toxins, virulence factors [6] or interfering with hormone signalling in plants [32,33] have been reported. The latter functions may be also important for plant pathogens. Generally, the boundary between pathogens and endophytes or phytohormones and toxins are not always clear-cut and especially hormone production is a widely spread characteristic of phytopathogens and plant growth-promoting bacteria (for a review see [32]). Plants produce several classes of phytohormones including auxins, cytokinins, brassinosteroids, gibberellins, abscisic acid, ethylene, jasmonates and strigolactones playing roles in development and stress responses. Cross talk and fine tuning of the different phytohormone pathways is essential for plant development, stress and defense responses ([34]; reviewed by [35]) and associated bacteria can interfere with plant signalling. In beneficial bacterial endophyte — plant interactions the production and modulation of auxins and ethylene play an essential role in plant development [32,36], but also stress (e.g. drought) tolerance has been reported to be influenced by endophyte-derived hormones. For example, abscisic acid and gibberellins produced by the endophyte Azospirillum lipoferum have been shown to be involved in alleviating drought stress symptoms in maize [37]. Interestingly, plant-associated bacteria do not only produce genuine plant hormones but also compounds mimicking the effect of the natural plant hormones as structural analogues (Figure 1). This is the case for coronatine produced by several plant pathogenic Pseudomonas species mimicking the active natural (+)-7-iso-jasmonoyl-l-isoleucine [38]. Coronatine acts as very active jasmonate finally showing phytotoxicity [33] and plays a role in suppressing stomatal closure and defense responses [39]. It will be interesting to see if plant hormone mimicry encoded by NRPS and PKS gene clusters with so far unknown function is a common feature in plant-associated bacteria.
Figure 2.
Schematic overview showing the different types of plant-endophyte interactions leading to the synthesis of metabolites, which are in many cases not produced by the macro- or microsymbiont alone or in different quantities. Furthermore, the different known functions of endophyte-associated metabolites are presented.
Adaptation to endophytic lifestyle and the potential to produce secondary metabolites
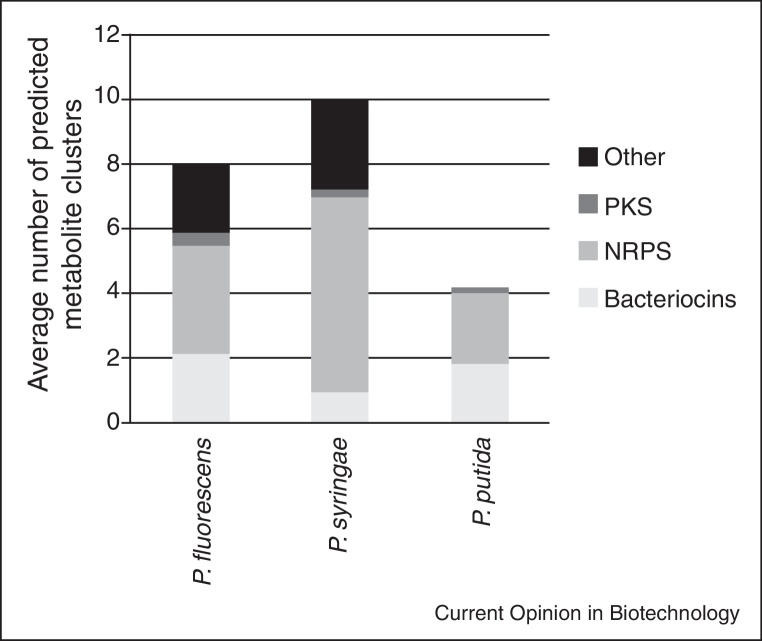
When comparing the amounts of predicted secondary metabolites of all completely sequenced Pseudomonas strains by antiSmash [40••], a prediction software for secondary metabolite production, it can be seen that pseudomonads associated with eukaryotes as plant pathogens (P. syringae) or as endophytes or epiphytes (P. fluorescens) host a higher number of gene clusters encoding for secondary metabolites, in particular NRPS and other metabolites (predicted quorum sensing signals, not further characterized metabolites) than free living P. putida strains (Figure 3). In the latter strains the number of bacteriocins potentially involved in competition with closely related species is higher [41]. It might be that the plant-associated lifestyle requires adaptation to several niches, in which different metabolites are required. On the other hand, specialized endophytes such as obligate endophytes or endophytes colonizing only specific niches may produce a lower number of potential secondary metabolites. Metabolites furthermore act as signals for interaction (communication) with the plant and host-specific signal exchange may occur as reported for plant–fungal interactions [42].
Figure 3.
Average numbers of metabolite gene clusters predicted by antiSmash 2.0 [40••]. The numbers are the mean of 6 Pseudomonas fluorescens (plant-associated) strains, 5 P. syringae (plant pathogens) and 9 P. putida strains (no association with plants) and contain all fully sequenced and published genomes in the given category.
Although genome reduction is a general mechanism of (intracellular) pathogens [43], and also highly adapted symbiotic and obligate endophytic bacteria like Candidatus Burkholderia kirkii show clear reduction of their genome compared to free living relatives [44•], this does not necessarily lead to a complete loss of the potential to produce secondary metabolites. Quite on the contrary, it has been speculated that C. Burkholderia kirkii produces metabolites to protect its host plant Psychotria kirkii (Rubiaceae) against pathogens or herbivores. The genome of C. Burkholderia kirkii contains several biosynthetic genes responsible for secondary metabolite production and especially two clusters for the biosynthesis and transport of sugar analogues of the C7N family of aminocyclitols (Figure 1). Several members of the C7N aminocyclitols display antibiotic, antifungal or insecticidal activity [45]. Several Psychotria species harbor Burkholderia spp. within specialized leaf nodules. It remains to be seen how far these endophytes contribute to the metabolic potential of Psychotria. Such specialized endophytes may play a role in plant defense by producing toxins active against herbivores as it is well known for endophytic fungi, especially for the genera Epichloe and Neotyphodium (see e.g. [13,46]). Also, bacteria living in association with marine eukaryotes are made responsible for the production of various toxins involved in the defense mechanism of the eukaryotic host, which include dinoflagellates and tunnicates [47]. Interestingly, the saprotrophic fungus Rhizopus microsporus produces rhizoxin and is responsible for rice seedling blight, but the actual producer of the toxin is the bacterium Burkholderia endofungorum [48–50]. It is remarkable that related Burkholderia spp. live in close association with Psychotria plants and it remains to be elucidated if plant toxin production might be in more cases related to bacterial endophytes. Other Rubiaceae plants, namely Fadogia, Pavetta and Vangueria, which can all cause a disease (called ‘gousiekte’) in ruminants feeding on these plants host different Burkholderia spp. suspected to play a role in production of the toxin causing the disease, the polyamine pavettamine (Figure 1) [51,52•].
Endophytes as contributors to plant metabolite production
So far we discussed the direct metabolic potential of endophytic bacteria. However, two other indirect ways exist, how endophytic bacteria play a role for the metabolic potential of plants (for a schematic overview see Figure 2). First, bacterial endophytes may strongly influence the performance, growth and stress tolerance of plants [16,53,54]. In this respect, it is remarkable that an endophytic actinobacterium, Pseudonocardia sp. strain YIM 63111, is able to enhance the production of the antimalarial compound artemisinin (Figure 1) in its host plant Artemisia annua [55•]. The induction of secondary metabolite production by endophytes might be a much more widespread phenomenon in aromatic and medicinal plants. Second, some metabolites are not only produced by a single organism, but might be produced by a plant in combination with associated bacteria. This has been discussed for the flavour of strawberries, where furanoids are responsible for the typical fragrant [56] and where it has been shown that plant-associated methylobacteria influence the quality and quantity of the flavour [57]. Also, for the biosynthesis of the polyamine pavettamine (Figure 1) of South African Rubiaceae, it has been discussed that the production might be because of bilateral biosynthesis as only nodulating plants produce the toxin. Furthermore, nodulating plants void of pavettamine production have been found and plant cell cultures without bacteria do not produce pavettamine but more common polyamines [58•].
Detection of metabolites in plant association
The majority of metabolites from endophytic bacteria have been characterized after isolating bacteria and growing them in vitro. Novel developments in the in situ analysis of metabolites might provide new opportunities to detect and to describe also metabolites specifically produced during the interaction with living plants. The overall concentration of compounds produced by plant-associated bacteria in roots and the rhizosphere is usually low (usually < 10 μg/g), making the direct structure elucidation challenging. Direct analysis of metabolites in situ has been achieved for antibiotic lipopeptides from several Bacillus subtilis and for pyrrolnitrin, 2,4-diacetylphloroglucinol and phenazine-1-carboxylic acid from Pseudomonas fluorescens strains [6]. Local concentrations might be still higher and biosensor-based approaches might be important tools to detect various metabolites in situ such as for Pseudomonas fluorescens CHA0 lipopeptides [59], but the detection of unknown compounds remains challenging, and for a quantitative approach mainly LC–MS based methods have been successful [6]. Apart from difficulties in detecting unknown compounds also the composition of already described metabolites may vary significantly in vitro and in planta. For example, the comparison of metabolic profiles produced in growth medium and in planta showed clear differentiation of lipopeptides produced by Bacillus amyloliquefaciens S499 with iturin and fengycin underrepresented in the root samples, while surfactins were stronger accumulated in roots. Combined electrospray and imaging mass spectrometry-based approaches were used to determine the detailed pattern of surfactins, iturins and fengycins [60]. Novel metabolites only produced in specific niches or low concentrations within the plant are not easily found, albeit genomic analysis can point to potential genes and help in the prediction of those metabolites. A breakthrough has been achieved here with the description of thanamycin (Figure 1) [61••]. After applying PhyloChip-based analysis secondary metabolites synthetized by a NRPS of Pseudomonas sp. strain SH-C52 were identified to be involved in suppressing sugar beet diseases caused by Rhizoctonia solani [62•]. On the basis of this discovery Watrous et al. [61••] could establish the partial structure of such a metabolite, thanamycin, with nanospray desorption electrospray ionization mass spectrometry (nanoDESI MS) combined with MS data alignment and molecular networking. This technology allowed the direct analysis and partial structure elucidation of a chlorinated lipopeptide thanamycin on Petri dishes without any sample preparation. NanoDESI MS or related technologies might in future even allow the detection of novel metabolites directly in environmental or root samples [63]. For example, MALDI-FTICR MS imaging has shown the production of fusaricidin lipopetides of Paenibacillus polymyxa in interaction with Fusarium oxysporum on plate [64]. These non-invasive methods have also the additional advantage to allow time-course metabolic analysis and can represent effective tools for the analysis of intermediate steps including less stable compounds of biosynthetic pathways [63–66].
Conclusions
Evidence is increasing that endophytic bacteria have a high potential in producing a wide range of so far undescribed metabolites. Partly, the concentration and circumstances under which these metabolites are produced are not well understood but the genomic revolution together with the steady development of analytic techniques will certainly accelerate the discovery of such cryptic compounds and the future will show how many novel chemical structures and compounds are encoded in endophytic bacterial genomes. Moreover, other known (plant) metabolites might turn out to be partly or fully derived from endophytic or associated bacterial metabolism. It remains to be seen how widespread this phenomenon might be or if it is restricted to genera of the Rubiaceae and certain fragrants. An additional challenge in future research is the detection and characterization of metabolites formed in niches on and in plants or under specific circumstances under natural conditions only, as indicated by the only partial realization of the metabolic potential of bacteria grown in vitro.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
This work was partly supported by the grant P24569-B25 provided by the Austrian Science Fund (FWF).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Demain A.L., Sanchez S. Microbial drug discovery: 80 years of progress. J Antibiot. 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao V., Davies J. Actinobacteria: the good, the bad, and the ugly. Anton Leeuwenhoek. 2010;98:143–150. doi: 10.1007/s10482-010-9440-6. 2010. [DOI] [PubMed] [Google Scholar]
- 3.Baltz R. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J Ind Microbiol Biotechnol. 2006;33:507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 4.Walsh C.T., Fischbach M.A. Natural roducts Version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traxler M.F., Kolter R. A massively spectacular view of the chemical lives of microbes. Proc Natl Acad Sci U S A. 2012;109:10128–10129. doi: 10.1073/pnas.1207725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaijmakers J.M., Mazzola M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol. 2012;50:403–424. doi: 10.1146/annurev-phyto-081211-172908. [DOI] [PubMed] [Google Scholar]
- 7.Zin N.M., Sarmin N.I., Ghadin N., Basri D.F., Sidik N.M., Hess W.M., Strobel G.A. Bioactive endophytic streptomycetes from the Malay Pensinsula. FEMS Microbiol Lett. 2007;274:83–88. doi: 10.1111/j.1574-6968.2007.00819.x. [DOI] [PubMed] [Google Scholar]
- 8•.Qin S., Xing K., Jiang J.-H., Xu L.-H., Li W.-J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl Microbiol Biotechnol. 2011;89:457–473. doi: 10.1007/s00253-010-2923-6. [DOI] [PubMed] [Google Scholar]; Comprehensive overview of the structures of metabolites produced by endophytic actinobacteria.
- 9.Bascom-Slack C.A., Ma C., Moore E., Babbs E., Fenn K., Greene J.S., Hann B.D., Keehner J., Kelley-Swift E.G., Kembaiyan V. Multiple, novel biologically active endophytic actinomycetes isolated from Upper Amazonian rainforests. Microb Ecol. 2009;58:374–383. doi: 10.1007/s00248-009-9494-z. [DOI] [PubMed] [Google Scholar]
- 10.Kurtböke D.I. Biodiscovery from rare actinomycetes: an eco-taxonomical perspective. Appl Microbiol Biotechnol. 2012;93:1843–1852. doi: 10.1007/s00253-012-3898-2. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Zhao G.Z., Huang H.Y., Qin S., Zhu W.Y., Zhao L.X., Xu L.H., Zhang S., Li W.J., Strobel G. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Anton Leeuwenhoek. 2012;101:515–527. doi: 10.1007/s10482-011-9661-3. [DOI] [PubMed] [Google Scholar]
- 12.Wilson D. Endophyte — the evolution of a term, and clarification of its use and definition. Oikos. 1995;73:274–276. [Google Scholar]
- 13.Aly A., Debbab A., Proksch P. Fungal endophytes: unique plant inhabitants with great promises. Appl Microbiol Biotechnol. 2011;90:1829–1845. doi: 10.1007/s00253-011-3270-y. [DOI] [PubMed] [Google Scholar]
- 14.Mousa W.K., Raizada M.N. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front Microbiol. 2013;4:65. doi: 10.3389/fmicb.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenblueth M., Martinez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- 16.Compant S., Clement C., Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–678. [Google Scholar]
- 17.Reinhold-Hurek B., Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–443. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Sturz A.V., Christie B.R., Nowak J. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit Rev Plant Sci. 2000;19:1e30. [Google Scholar]
- 19.Davies J., Spiegelman G.B., Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Fajardo A., Martinez J.L. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Monier J.M., Demanèche S., Delmont T.O., Mathieu A., Vogel T.M., Simonet P. Metagenomic exploration of antibiotic resistance in soil. Curr Opin Microbiol. 2011;14:229–235. doi: 10.1016/j.mib.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Ongena M., Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Raaijmakers J.M., De Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 24.Ding L., Maier A., Fiebig H.-H., Lin W.-H., Hertweck C. A family of multicyclic indolosesquiterpenes from a bacterial endophyte. Org Biomol Chem. 2011;9:4029–4031. doi: 10.1039/c1ob05283g. [DOI] [PubMed] [Google Scholar]
- 25.Inahashi Y., Iwatsuki M., Ishiyama A., Namatame M., Nishihara T.A., Matsumoto A., Hirose T., Sunazuka T., Yamada H., Otoguro K. Spoxazomicins A-C, novel antitrypanosomal alkaloids produced by an endophytic actinomycete, Streptosporangium oxazolinicum K07-0450T. J Antibiot. 2011;64:303–307. doi: 10.1038/ja.2011.16. [DOI] [PubMed] [Google Scholar]
- 26.Inahashi Y., Matsumoto A., Omura S., Takahashi Y. Streptosporangium oxazolinicum sp. nov., a novel endophytic actinomycete producing new antitrypanosomal antibiotics, spoxazomicins. J Antibiot. 2011;64:297–302. doi: 10.1038/ja.2011.18. [DOI] [PubMed] [Google Scholar]
- 27.Miller K.I., Qing C., Sze D.M.Y., Neilan B.A. Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS ONE. 2012;7:e35953. doi: 10.1371/journal.pone.0035953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller K., Qing C., Sze D.-Y., Roufogalis B., Neilan B. Culturable endophytes of medicinal plants and the genetic basis for their bioactivity. Microb Ecol. 2012;64:431–449. doi: 10.1007/s00248-012-0044-8. [DOI] [PubMed] [Google Scholar]
- 29.Kaewkla O., Franco C.M.M. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb Ecol. 2013;65:384–393. doi: 10.1007/s00248-012-0113-z. [DOI] [PubMed] [Google Scholar]
- 30.Barry S.M., Challis G.L. Recent advances in siderophore biosynthesis. Curr Opin Chem Biol. 2009;13:205–215. doi: 10.1016/j.cbpa.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Rosconi F., Davyt D., Martinez V., Martinez M., Abin-Carriquiry J.A., Zane H., Butler A., de Souza E.M., Fabiano E. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ Microbiol. 2013;15:916–927. doi: 10.1111/1462-2920.12075. [DOI] [PubMed] [Google Scholar]
- 32.Glick B.R. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012 doi: 10.6064/2012/963401. (article ID 963401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez M.A., Bannenberg G., Castresana C. Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr Opin Plant Biol. 2008;11:420–427. doi: 10.1016/j.pbi.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Durbak A., Yao H., McSteen P. Hormone signaling in plant development. Curr Opin Plant Biol. 2012;15:92–96. doi: 10.1016/j.pbi.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Robert-Seilaniantz A., Grant M., Jones J.D.G. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Ann Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 36.Hardoim P.R., van Overbeek L.S., van Elsas J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Cohen A.C., Travaglia C.N., Bottini R., Piccoli P.N. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany. 2009;87:455–462. [Google Scholar]
- 38.Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (+)-7-iso-jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 39.Lee S., Ishiga Y., Clermont K., Mysore K.S. Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by non-host bacterial pathogens. PeerJ. 2013;1:e34. doi: 10.7717/peerj.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Blin K., Medema M.H., Kazempour D., Fischbach M.A., Breitling R., Takano E., Weber T. Antismash 2.0 — a versatile platform for genome mining of secondary metabolite producers. Nucl Acid Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]; Description of an intuitive and versatile tool for mining for gene clusters involved in the production of secondary metabolites and provides an overview on software tools for the analysis of biosynthetic gene clusters.
- 41.Cotter P.D., Ross R.P., Hill C. Bacteriocins — a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 42.Lahrmann U., Ding Y., Banhara A., Rath M., Hajirezaei M.R., Döhlemann S., von Wirén N., Parniske M., Zuccaro A. Host-related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci U S A. 2013;110:13965–13970. doi: 10.1073/pnas.1301653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran N.A. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 44•.Carlier A.L., Eberl L. The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol. 2012;14:2757–2769. doi: 10.1111/j.1462-2920.2012.02763.x. [DOI] [PubMed] [Google Scholar]; The genome analysis of the symbiontic and endophytic Candidatus Burkholderia kirkii reveals the potential of production of secondary metabolites.
- 45.Mahmud T. The C7N aminocyclitol family of natural products. Nat Prod Rep. 2003;20:137–166. doi: 10.1039/b205561a. [DOI] [PubMed] [Google Scholar]
- 46.Schardl C.L., Florea S., Pan J., Nagabhyru P., Bec S., Calie P.J. The epichloae: alkaloid diversity and roles in symbiosis with grasses. Curr Opin Plant Biol. 2013;16:480–488. doi: 10.1016/j.pbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 48.Partida M.L.P., Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 49.Lackner G., Partida M.L.P., Hertweck C. Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 2009;17:570–576. doi: 10.1016/j.tim.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Lackner G., Moebius N., Partida M.L.P., Boland S., Hertweck C. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bode M.L., Gates P.J., Gebretnsae S.Y., Vleggaar R. Structure elucidation and stereoselective total synthesis of pavettamine, the causal agent of gousiekte. Tetrahedron. 2010;66:2026–2036. [Google Scholar]
- 52•.Verstraete B., Van Elst D., Steyn H., Van Wyk B., Lemaire B., Smets E., Dessein S. Endophytic bacteria in toxic South African plants: identification, phylogeny and possible involvement in gousiekte. PLoS One. 2011;6:e19265. doi: 10.1371/journal.pone.0019265. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with [58•] interesting observations pointing to a combined contribution of Burkholderia spp. and various member of the Rubiaceae in the biosynthesis of the cardiotoxin pavettamine.
- 53.Compant S., Duffy B., Nowak J., Clement C., Ait Barka E. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitter B., Brader G., Afzal M., Compant S., Naveed M., Trognitz F., Sessitsch A. Advances in elucidating beneficial interactions between plants, soil, and bacteria. In: Sparks D.L., editor. Advances in Agronomy. Burlington Academic Press; 2013. pp. 381–445. 121. [Google Scholar]
- 55•.Li J., Zhao G.-Z., Varma A., Qin S., Xiong Z., Huang H.-Y., Zhu W.-Y., Zhao L.-X., Xu L.-H., Zhang S., Li W.-J. An endophytic Pseudonocardia species induces the production of artemisinin in Artemisia annua. PLoS ONE. 2012;7:e51410. doi: 10.1371/journal.pone.0051410. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that an endophytic Actinobacterium (Pseudonocardia sp.) effectively stimulates the transcription of biosynthesis genes and the production of artemisinin in plants, which might represent also a promising tool in the enhancement of the production of this antimalarial agent in plants.
- 56.Zabetakis I. Enhancement of flavour biosynthesis from strawberry (Fragaria × ananassa) callus cultures by Methylobacterium species. Plant Cell Tiss Org. 1997;50:179–218. [Google Scholar]
- 57.Verginer M., Siegmund B., Cardinale M., Müller H., Choi Y., Miguez C.B., Leitner E., Berg G. Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol Ecol. 2010;74:136–145. doi: 10.1111/j.1574-6941.2010.00942.x. [DOI] [PubMed] [Google Scholar]
- 58•.Van Elst D., Nuyens S., van Wyk B., Verstraete B., Dessein S., Prinsen E. Distribution of the cardiotoxin pavettamine in the coffee family (Rubiaceae) and its significance for gousiekte, a fatal poisoning of ruminants. Plant Physiol Biochem. 2013;67:15–19. doi: 10.1016/j.plaphy.2013.02.022. [DOI] [PubMed] [Google Scholar]; Together with [52•] interesting observations pointing to a combined contribution of Burkholderia spp. and various member of the Rubiaceae in the biosynthesis of the cardiotoxin pavettamine.
- 59.Rochat L., Pechy-Tarr M., Baehler E., Maurhofer M., Keel C. Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by a biocontrol pseudomonad on cereals with flow cytometry. Mol Plant-Microbe Interact. 2010;23:949–961. doi: 10.1094/MPMI-23-7-0949. [DOI] [PubMed] [Google Scholar]
- 60.Nihorimbere V., Cawoy H., Seyer A., Brunelle A., Thonart P., Ongena M. Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499. FEMS Microbiol Ecol. 2012;79:176–191. doi: 10.1111/j.1574-6941.2011.01208.x. [DOI] [PubMed] [Google Scholar]
- 61••.Watrous J., Roach P., Alexandrov T., Heath B.S., Yang J.Y., Kersten R.D., van der Voort M., Pogliano K., Gross H., Raaijmakers J.M. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper demonstrates the partial structure elucidation of the lipopeptide thanamycin from living bacterial colonies using nanospray desorption electrospray ionization mass spectrometry, sequence information and molecular networking representing novel opportunities for the discovery of metabolites produced in situ and in interaction with other organisms.
- 62•.Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J.H.M., Piceno Y.M., DeSantis T.Z., Andersen G.L., Bakker P.A.H.M., Raaijmakers J.M. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]; Using PhyloChip-based metagenomics of the rhizosphere microbiome and culture-dependent functional analyses the authors detected key bacterial taxa associated with disease suppression. Such -omics based approaches might be very useful also for identification of key members of endophytic communities.
- 63.Watrous J.D., Dorrestein P.C. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Debois D., Ongena M., Cawoy H., Pauw E. MALDI-FTICR MS imaging as a powerful tool to identify Paenibacillus antibiotics involved in the inhibition of plant pathogens. J Am Soc Mass Spectromet. 2013;24:1202–1213. doi: 10.1007/s13361-013-0620-2. [DOI] [PubMed] [Google Scholar]
- 65.Vater J., Wilde C., Kell H. In situ detection of the intermediates in the biosynthesis of surfactin, a lipoheptapeptide from Bacillus subtilis OKB 105, by whole-cell cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in combination with mutant analysis. Rapid Commun Mass Spectromet. 2009;23:1493–1498. doi: 10.1002/rcm.4031. [DOI] [PubMed] [Google Scholar]
- 66.Hoefler B.C., Gorzelnik K.V., Yang J.Y., Hendricks N., Dorrestein P.C., Straight P.D. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc Natl Acad Sci U S A. 2012;109:13082–13087. doi: 10.1073/pnas.1205586109. [DOI] [PMC free article] [PubMed] [Google Scholar]