Summary
Tissue regeneration is an orchestrated progression of cells from an immature state to a mature one, conventionally represented as distinctive cell subsets. A continuum of transitional cell states exists between these discrete stages. We combine the depth of single-cell mass cytometry and an algorithm developed to leverage this continuum by aligning single cells of a given lineage onto a unified trajectory that accurately predicts the developmental path de novo. Applied to human B cell lymphopoiesis, the algorithm (termed Wanderlust) constructed trajectories spanning from hematopoietic stem cells through to naïve B cells. This trajectory revealed nascent fractions of B cell progenitors and aligned them with developmentally-cued regulatory signaling including IL-7/STAT5 and cellular events such as immunoglobulin rearrangement, highlighting checkpoints across which regulatory signals are rewired paralleling changes in cellular state. This study provides a comprehensive analysis of human B lymphopoiesis, laying a foundation to apply this approach to other tissues and “corrupted” developmental processes including cancer.
Introduction
Most complex organisms start as a single cell that matures through coordinated stages of development into a diverse set of transitional and terminal cell types, many of which have yet to be defined. There is a continuous relationship between maturing cell subsets: more potent cells divide and differentiate into more functionally restricted cells. The challenge is to devise approaches that analyze and order the cells of these complex tissues to reveal their developmental relationships, behavior, and the mechanisms that govern their differentiation.
One technique that has been used to effectively determine immune system hierarchies is fluorescence activated cell sorting (FACS) which traditionally relies on surface antigen expression for purifying cells of a given population (Hardy et al., 1984). One drawback of this technology is the limited number of simultaneously assayed markers (generally < 12) which confines experiments to isolating a narrow “slice” of the overall cellular pool, thereby restricting the ability to characterize transitional populations and the relationships between them. Emerging single-cell technologies (Bendall et al., 2011; Jaitin et al., 2014), however, can measure a large number of simultaneous features in individual cells with unprecedented resolution. Mass cytometry (Bendall et al., 2011) can now quantify more than 40 features simultaneously in thousands to millions of individual cells per experiment. Thus, an opportunity exists to assay nearly all cell types, from the earliest to the most mature within a given system, and by simultaneously measuring a sufficient number of identifying markers in a single sample, enable direct inference of a continuous developmental trajectory of primary cells in situ.
A case in point is the early development of human B lymphocytes. Early B cells originate from the hematopoietic stem cell, followed by a common lymphoid progenitor cell, pro-B cell, pre-B cell, and finally an immature B cell, which migrates out of the marrow (LeBien and Tedder, 2008). While these early developmental hallmarks have been described in the mouse (Rolink et al., 1999) the exact nature of cell types and timing of critical events such as IgH rearrangement and clonal expansion remain elusive in human B lymphopoiesis. As hematopoiesis is both continuous and asynchronous throughout life, the full spectrum of cell types exists in a single sample of bone marrow from a healthy individual. It was reasoned that by combining mass cytometry with computation, we could construct a putative B-lineage trajectory (an ordering of cells according to their most likely developmental chronology) representing in vivo development, from primary human bone marrow. Then, this trajectory could be used to characterize the order of key molecular and cellular events during development.
B cell centric, 44 parameter single cell mass cytometry data was collected from human bone marrow, simultaneously measuring multiple cellular features, including phenotypic proteins, transcription factors, regulatory enzymes, cell state indicators, and activation of regulatory signaling molecules. Sufficient cells were measured to encompass a complete spectrum of B cell lymphopoiesis that could be reassembled into a continuous progression from a single sample. Experimental design was tailored to maximize physiologic interpretability of the data by allowing for minimal ex vivo manipulation. The resulting high dimensional data was ordered using a graph-based trajectory detection algorithm, Wanderlust, that orders cells to a unified trajectory based on their maturity, thus predicting the developmental path de novo, which was subsequently validated.
Wanderlust generated remarkably consistent trajectories across multiple individuals that were largely congruent with prior knowledge. Using the trajectory, we determined the timing and order of key molecular and cellular events across development, including identifying previously unrecognized subsets of B cell progenitors that pinpoint the timing of DJ and V(D)J recombination of the immunoglobulin heavy chain (IgH). Surveying the dynamic changes in cellular expression across the Wanderlust trajectory, we identified ‘coordination points’, where re-wiring of the signaling network occurs concurrently with the rise and fall of multiple proteins. These coordination points and their characteristic signaling were further aligned with cell cycle status, apoptosis, and germline IgH locus rearrangement, together forming a deeply detailed map of human B lymphopoiesis. By exploiting the cellular heterogeneity of the human system while monitoring both single-cell identity and behavior, a holistic model ordered by developmental chronology was created.
Results
Aligning cells to a developmental trajectory
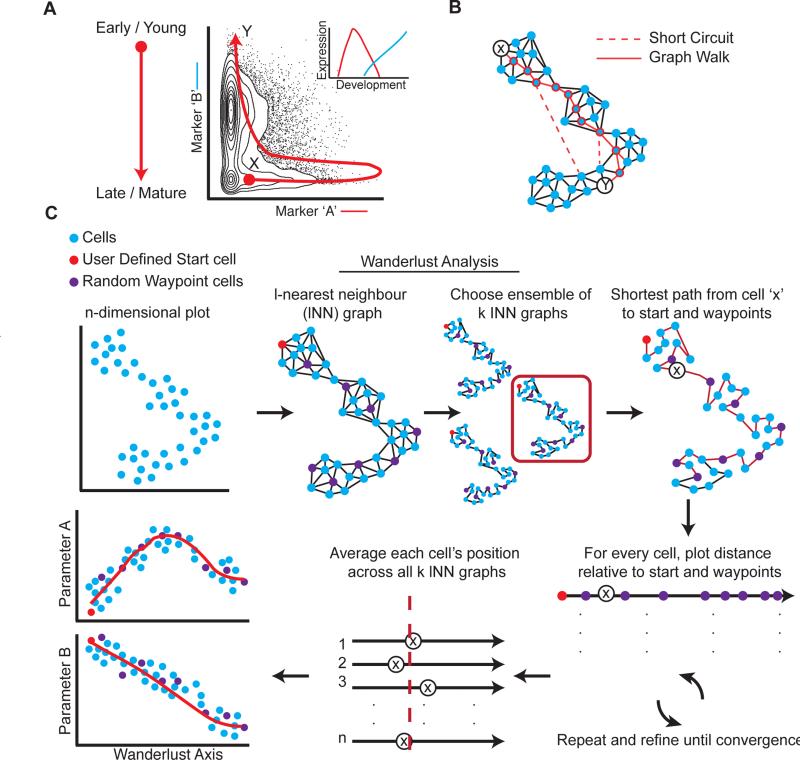
Primary human tissues are a rich source of cellular diversity as they contain both multi-potent progenitors and mature specialized cells. Previously, it has been shown that the transitional cooccurrence of an extended suite of phenotypic markers, measured simultaneously in individual cells, can be used to roughly order cells along a developmental hierarchy (Bendall et al., 2011; Qiu et al., 2011). However, previous approaches were limited, either by false assumptions of linearity (Figure 1A), or stochastic partitioning of cell populations into overly-coarse clusters, losing directionality and single cell resolution, and thus the ability to accurately order cellular relationships (see Supplementary methods). To address these limitations, we developed a robust algorithm that uses high dimensional single cell data to map individual cells onto a trajectory representing the chronological order of development in fine detail.
Figure 1. Developmental trajectory detection.
A) Non-linear relationships between developmentally related cells. Markers ‘A’ and ‘B’ represent sequentially expressed phenotypic epitopes on cells in a developing system (inset). The red line shows the expected developmental trajectory from the earliest (cell ‘X’) to the most mature cell type (cell ‘Y’). Developmentally, the distant cell types can be close in Euclidean space. B) Determining the shortest path through a graph of the data reflects temporal distance between cells (solid red line between early (cell ‘X’) and target (cell ‘Y’)) better than standard metrics (e.g. Euclidian norm or correlation). Short circuits (dashed red line) impede a naïve shortest path-based algorithm. C) Description of the Wanderlust algorithm. The input data is single cells in N-dimensional space (top left). Wanderlust transforms the data into an ensemble of graphs and selects random waypoints (purple). Each graph is independently analyzed (single graph, red box) where a user-defined starting cell (red) is used to calculate an orientation trajectory. The orientation trajectory is iteratively refined using the waypoint cells. The final trajectory is an average over all graphs. To examine trends, the trace of each marker can be plotted according to trajectory position. See also Figure S1 for evaluation of Wanderlust on simulated data.
Several assumptions are made regarding the data. First, the sample includes cells representative of the entire developmental process, including most transient and rare populations. Second, the developmental trajectory is non-branching: cells are placed along a one-dimensional path. Third, changes in protein expression are gradual during development. Ordering single cells onto a trajectory is based on continuous tracking of the progressive rise and fall of phenotypic markers during development. This trajectory provides a framework to infer the order and transition between additional key molecular and cellular events.
A fundamental challenge to constructing an accurate trajectory is that the relationships between markers cannot be assumed to be linear. Thus, determining the distance between two individual cells using standard metrics based on marker levels (e.g. Euclidian norm or correlation) results in poor measures of their chronological distance in development, except in the case of very similar cells. Figure 1A demonstrates the non-linearity that manifests from using only two markers; while cells X and Y are close based on Euclidian distance, they are quite distant in terms of developmental chronology. The complexity of such non-linear behavior only increases as more instances occur in high dimensions.
A graph-based representation of the data overcomes such problems and helps construct a distance metric that corresponds to developmental chronology (Figure 1B). In the graph, each cell is represented as a node connected to its neighbors—the cells most similar to it—by a series of edges. Conversion into this graph structure represents a new geometry for the data: distances between cells are defined as shortest paths on the graph (Figure 1B), composed of steps (edges) between neighbors, where each step traverses similar cells that are likely adjacent in their developmental chronology. Moreover, because the model is based on similarity between cells, rather than relationships between parameters, it can more naturally handle the non-linearity.
Wanderlust, a robust graph-based trajectory detection algorithm
We developed Wanderlust, a graph-based trajectory detection algorithm that receives multi-parameter single-cell events as input and maps them onto a one-dimensional developmental trajectory (Figure 1C). Cells are ordered along a trajectory that represents their most likely placement along a developmental continuum. A key challenge for any such algorithm is that most data is rife with noise from biological and technical sources. Wanderlust determines a cell's position based on steps between neighboring cells, but noise accumulates with each step, so longer paths (a series of steps) are less reliable than shorter paths. To construct a more accurate trajectory, Wanderlust incorporates random waypoint cells, each of which helps refine estimations for the positions of nearby cells. An initial estimation of each cell's position, including the waypoint cells, is set to its distance from a pre-chosen ‘early cell’. Next, each cell's position is refined using its distance to nearby waypoint cells. Since the refinement affects the positions of waypoint cells themselves, it is repeated iteratively until each cell's position converges (Figure 1C).
The most harmful effect of noise is “short circuits” (Figure 1B): i.e., spurious edges between developmentally distant cells which nevertheless have similar marker measurements. Even a single short-circuit in a graph of thousands of cells can impede construction of a correct trajectory, as all shortest paths will “cut through” this short-circuit. Wanderlust overcomes short circuits by building an ensemble of graphs (Figure 1C). The exact set of neighbors varies between each graph, so any randomly occurring short circuit appears in very few graphs in the ensemble. A trajectory is constructed separately for each graph in the ensemble and the final trajectory is found by taking the average over the positions from all graphs, thus averaging out the influence of short circuits in the final trajectory. Full details of the Wanderlust algorithm can be found in the Supplemental Methods section.
Wanderlust's performance was initially evaluated using synthetic data (see Supplementary Methods). Wanderlust faithfully recovered the correct trajectory (ρ=0.97), even under increasing magnitude of noise, including noise levels that exceed those typically found in biological data (Figure S1A). To emulate the short circuits that render biological data challenging, false edges were randomly added between distant points across the entire synthetic data set following graph construction. The algorithm successfully detected the solution trajectory, even in the increasing presence of short circuits (Figure S1B). In summary, Wanderlust robustly recovered the correct trajectory from synthetic data, despite increasing noise and short circuits, providing confidence in the trajectories it derives from real data.
Constructing a trajectory for B cell lymphopoiesis
B cell lymphopoiesis, a non-branching process occurring entirely within bone marrow, represented an ideal test case for Wanderlust. Mass cytometry was applied to a cohort of healthy primary human marrow aspirates (lineage negative bone marrow mononuclear cells (BMMC)) using a B cell centric marker panel (Table S1). Forty-four markers were simultaneously measured for each individual cell, including both phenotypic surface markers and internal functional proteins involved in signaling, cell cycle, apoptosis and genome rearrangement. Both surface and intracellular markers were chosen based on prior indications of their utility in defining developmental states in B cell maturation. Not all markers were used to construct the trajectory. Certain markers were used to validate stages in the trajectory or to discover new principles in the maturation process that had been previously obscured.
BMMC were enriched for B cells or their precursors, resulting in ~200,000 cells for analysis from each individual (Methods). Wanderlust was applied to each marrow independently, using a ‘starting position’ of hematopoietic progenitors (Lin-CD34+CD38-) and the expression of seventeen phenotypic markers (Table S1) to order all cells along a one-dimensional trajectory. To characterize marker trends over the course of this trajectory, we used a sliding window over slices of cells, as ordered by Wanderlust; the median marker level, for each marker, was computed over all cells in each window (Supplementary Methods).
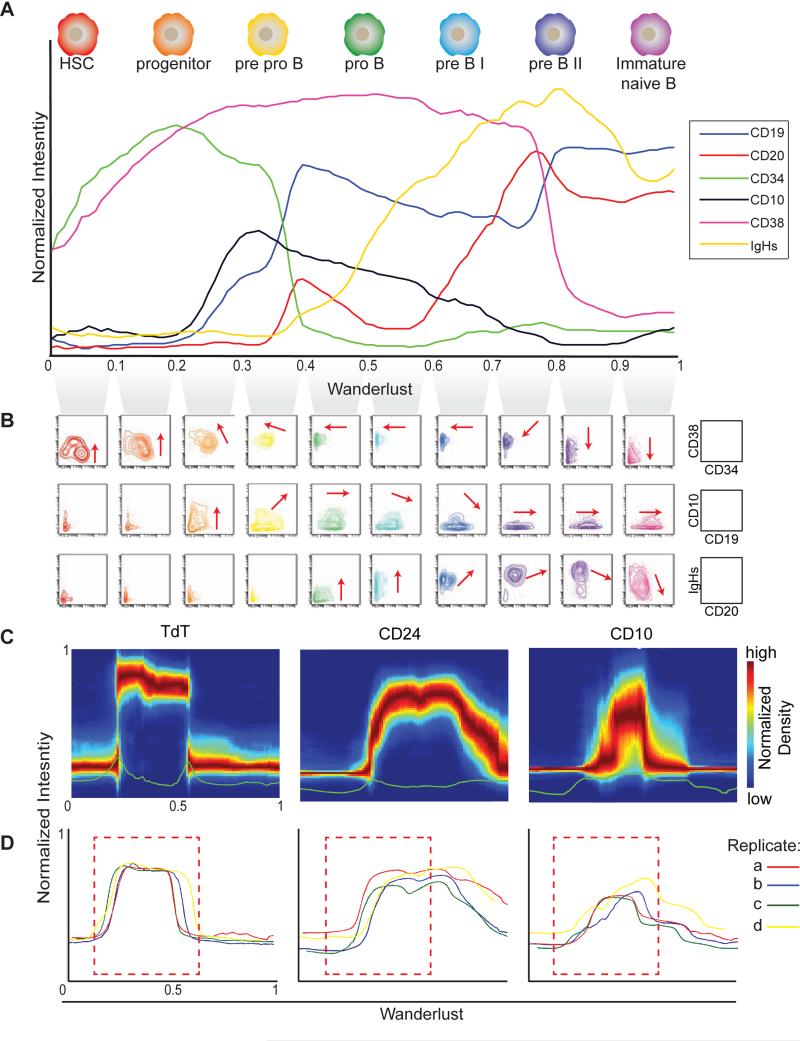
To evaluate the resulting trajectory, we examined the expression of canonical markers of B cell development as they align with expected cell populations (Figure 2A). The rise and fall of phenotypic markers along the resulting trajectory matched prior knowledge, starting with CD34, followed by CD38, CD10 (the earliest canonical Pro/Pre-B cell marker), CD19, CD20, and ending with immunoglobulin heavy chain IgH expression, indicative of immature B cells ready to leave the marrow. Developmental ordering was further cross-checked using biaxial plots. Examining ten percentile slices of cells, as ordered across the trajectory, demonstrated the expected progression (red arrows) of phenotypic markers CD34/38, CD10/19, and CD20/IgH, respectively (Figure 2B, biaxial plots). Together, these observations indicate that, from a starting point of hematopoietic stem cells, the primary phenotypic landmarks of B cell lymphopoiesis were correctly reconstructed and ordered. This was accomplished de novo and without ex vivo manipulation or synchronization in a single primary sample.
Figure 2. Wanderlust confirms known hallmarks of human B cell development and is consistent across healthy individuals.
A) The Wanderlust trajectory is fixed to an arbitrary scale where the most immature cells are at 0 and the most mature cells at 1. The traces (based on median marker levels within a sliding window) demonstrate the relative expression patterns of CD34, CD38, CD10, CD19, IgH (s)urface, and CD20 across development. The approximate position of progenitors and B cell fractions is indicated. B) Biaxial plots demonstrate the two-dimensional progression of cellular marker expression (red arrow) across the Wanderlust trajectory taken in segments of 0.1 C) Distribution of marker expression across the trajectory for CD24, TdT, and CD10. D) Marker traces across the trajectory for four different samples (denoted a to d) aligned using cross-correlation. Pearson's ρ > 0.9 between the trajectories of different samples. The red box demarcates the expression of CD24, which bisects the TdT expression prior to CD10 expression across all four healthy individuals. See also Figure S2 for traces on full marker panel and additional robustness analysis.
The Wanderlust trajectory is robust across parameter choice
The algorithm's sensitivity to the user-defined initial cell is a crucial feature. To evaluate this feature, initiating cells selected from evenly spaced points along the entire trajectory were each used to seed an independent trajectory determination by the algorithm. Wanderlust's iterative approach correctly detected the trajectory, even when using more mature starting points (Figure S1C). For example, using a starting point of 0.3 on the trajectory results in a correlation of ρ=0.98 with the original trajectory. With mature cells, the trajectory reverses, but remains congruent, with a correlation of ρ=-0.98 between the forward and reverse trajectory (Figure S1C).
To test the robustness of the algorithm to the selection of phenotypic features used to map the trajectory, Wanderlust was run removing one marker at a time, and the correlation with the original trajectory was assessed. Exclusion of any one individual marker had little effect on the overall trajectory as evidenced by the strong correlation (ρ > 0.97) with the original model, except HLA-DR (Figure S2A). Wanderlust was then run removing all of 6 key B cell markers (CD19, CD20, IgM-i, IgM-s, CD79b and CD10), and the correlation with the original trajectory remained (ρ > 0.95, Figure S2B). Thus even without canonical markers, Wanderlust was able to correctly order the events and phenotypic progression of B cell lymphopoiesis. Note that only three of the seven markers in this trace were actually used for the Wanderlust analysis (Figure S2C,D).
Additionally, we tested the algorithm's robustness by varying all free parameters. We compared trajectories generated independently over a wide range of input values to our original model, constructed using default parameters, and found a correlation of (ρ > 0.99) between trajectories (See supplementary materials). In conclusion, the Wanderlust algorithm constructs a remarkably consistent trajectory and is robust to variation in the parameters used for its construction.
Importantly, not only do the median marker levels follow expected trends over the trajectory, variation in marker expression within each window was remarkably low (Figure 2C,S2E). This tightness is especially apparent with TdT, which was not used as input to the algorithm. At any given point, the distribution of B cell centric epitopes was tight around the median, indicating the algorithm's ability to leverage multi-dimensional information to create a highly-organized trajectory of cellular development in silico. Thus, the quality of the trajectory is demonstrated by its robustness and marker tightness.
The trajectory is consistent across individuals
Having demonstrated Wanderlust's robustness when applied to a single healthy bone marrow sample, we investigated whether the trajectory of human B cell development is consistent across independent human samples. Altered proportions of cell subtypes due to outside factors (genetics, exposure to pathogens, etc.) could lead to scaling discrepancies between the output trajectories, yet we expected to see the general shape, order and co-expression of given markers maintained. To account for the scaling variations expected due to subpopulation frequency differences, we used cross-correlation (see methods) to compare the trajectories of four independent bone marrow samples (using the same experimental procedures). The four trajectories were completely overlaid (Figure 2D, S2D), demonstrating that Wanderlust consistently recapitulates the developmental trajectory across independent samples despite distinct genotypes and environmental backgrounds.
In addition, we observed qualitative agreement on the order of molecular events. Focusing on the less-characterized emerging B cell populations at the beginning of the trajectory, Wanderlust revealed that CD24 consistently bisected the wave of terminal deoxynucleotidyl transferase (TdT)—an enzyme that participates in rearrangement of the IgH locus (Figure 2D). Both CD24 and TdT were reliably expressed earlier than expected, before the rise of CD10, a canonical surface marker believed to be the earliest identifying surface marker of emerging human B cells committed to the lineage.
Ordering of emerging B cell precursors
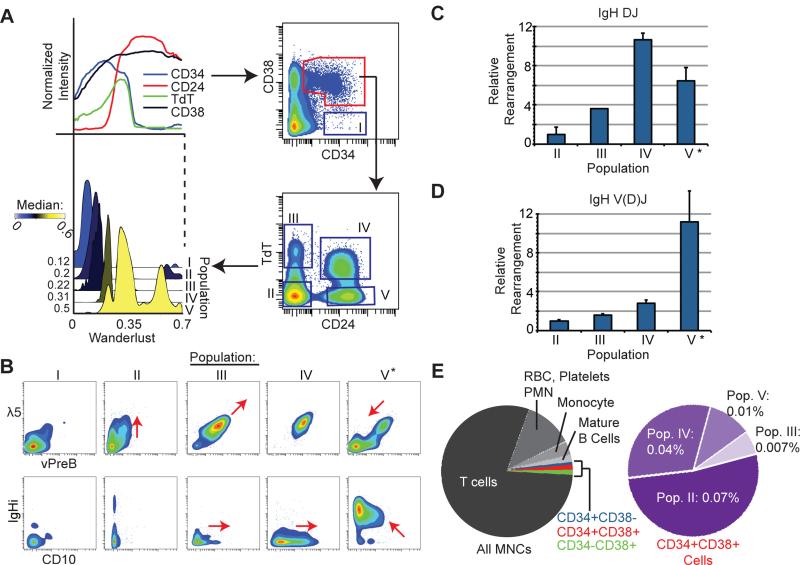
The Wanderlust trajectory guided the identification of distinct, early populations and determined their relative ordering across development. The expression of both TdT and CD24 increased prior to the expression of any canonical B cell surface markers (i.e. CD10 or CD19) in every bone marrow sample examined (Figure 2D). Given TdT's defining role in mammalian B cell emergence, we hypothesized that TdT, in combination with CD24 and other progenitor markers, could serve as a novel set of identifiers to dissect early populations of human B cells in the marrow. We used Wanderlust to guide the selection of a series of biaxial gates based on CD34, CD38, CD24 and TdT, revealing four distinct populations of cells (Figure 3A). According to Wanderlust, these were early cellular fractions sequentially occupying populations labeled II-V (Figure 3A, S3A). Additional phenotypic markers, including λ5 (CD179b), vPreB, CD10 and intra-cellular IgH protein (Figure 3B), were used to support the determined progression of these populations and their identity as definitive early B cells. Thus, the Wanderlust trajectory of these populations (II-V) is confirmed by protein co-expression patterns typical of B-lineage development.
Figure 3. Wanderlust uncovers rare B cell progenitors prior to the expression of CD10 or CD19.
A) Wanderlust trace showing the expression of CD24, CD34, CD38, and TdT (upper left panel). Density plots of four distinct TdT and CD24 populations defined within the CD34+CD38+ fraction (right). A histogram overlay of the Wanderlust values for cells contained in population I-V (dark blue boxes). The median Wanderlust values are indicated. B) Expression of the surrogate light chain of the pre-B cell receptor, λ5 and VpreB (top), and CD10 and IgH (i)ntracellular (bottom) across populations I though V. ‘*” denotes the uniform expression of IgHi in population V. C) Relative to population II, the amount of (C) IgH DJ and (D) IgH V(D)J rearrangement by qPCR of genomic DNA from prospectively isolated cells from populations II-V. Triplicate analysis of two biological replicates. Error bars are standard deviation. ‘*’ denotes uniform IgHi expression implying complete IgH V(D)J rearrangement. E) Pie charts summarizing the cellular contribution of human BMMNCs. The purple pie denotes CD34+CD38+ cells. Percentages are relative to all BMMNCs. See also Figure S3 for supporting data.
VH(D)JH Recombination confirms ordering of novel early human B cell populations
To independently confirm the developmental ordering of the hypothesized early B cell fractions, we used the rearrangement of the germline IgH locus, the molecular target of TdT, as a measure of developmental stage and B cell identity. A quantitative polymerase chain reaction (qPCR) assay was developed to quantify the relative proportions of DJH and VH(D)JH arranged cells and validated by assaying mixtures of cells containing known proportions of mature (fully rearranged) B cells (Figure S3B-C). FACS was then used to isolate populations II-V (Figure S3D) from BMMC preparations of two additional subjects. Genomic DNA was extracted from each fraction, and the relative IgH rearrangement status of each fraction was quantified using the qPCR assay.
As anticipated, relative to population II, there was a progressive rearrangement of the IgH locus towards population V. Most cells had detectable DJH rearrangement upon reaching population IV and VH(D)JH rearrangement upon reaching population V (Figure 3C and D). This was consistent with the observation that virtually all cells in fraction V displayed intracellular expression of IgH protein (Figure 3B‘*’). Establishing the progressive rearrangement of IgH in these populations confirmed that the Wanderlust trajectory not only facilitated the identification of the earliest human B lymphocytes, but also accurately ordered their developmental timing, all from the analysis of a single human marrow, without synchronization or manipulation.
This ability to identify and order cells was particularly notable given the sparsity of cells in these early fractions. Figure 3E highlights the rarity of these early B cell populations relative to total BMMCs. In particular, population III comprised only 0.007% of total BMMCs. The fact that population III occurs prior to CD19 expression (Figure 2A-B), in combination with inconsistent expression of CD10 (Figure 3B), suggests why these populations had not been described previously.
pSTAT5 response to IL-7 is confined to rare B cell precursors
Mass cytometry allows simultaneous measurement of surface markers, as well as internal functional proteins and their modifications, in the same cells. To functionally characterize early B cells and how they respond to stimuli, data was collected following multiple cellular signaling perturbations, including the cytokine IL-7 (Table S2). The activation of STAT5 by IL-7 via its phosphorylation site has a critical regulatory role in mouse lymphopoiesis (Corfe and Paige, 2012): disruption of this pathway results in arrest of B cell maturation at the pro-B cell stage (Malin et al., 2009). However, in human, the precise developmental timing of this pathway and its regulatory role remain unclear.
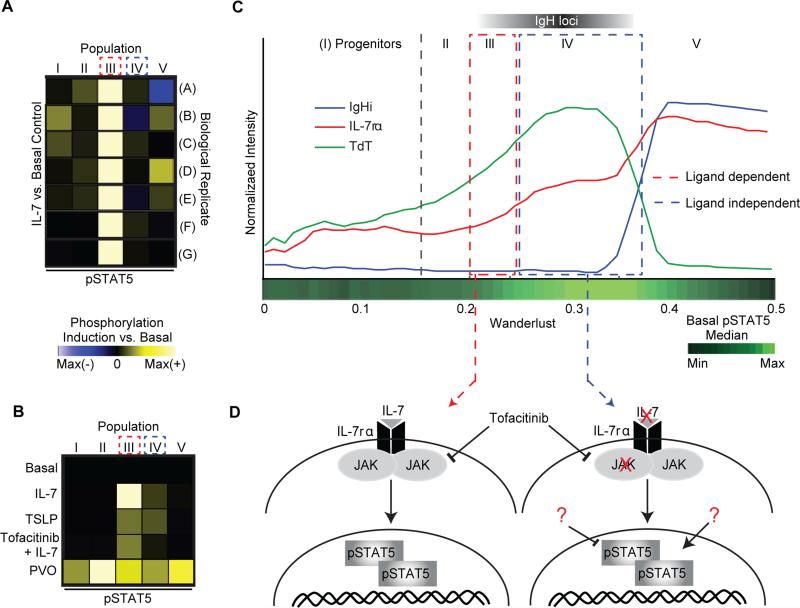
Investigation of signaling response to IL-7 across the four early B cell populations II-V revealed that cells within population III displayed an almost exclusive ~5 fold induction of STAT5 phosphorylation versus basal (Figure 4A) – a striking observation considering population III represents seven in 10,000 cells in the marrow (Figure 3E). Moreover, this pinpointed response was consistent across seven distinct marrows from independent human subjects. We note that pSTAT5 and other functional markers were not used to construct the Wanderlust trajectory and therefore this pattern of pSTAT5 induction was not enforced by the algorithm, but rather was revealed due to its precise phenotypic ordering of cells.
Figure 4. Regulatory signaling re-wires across development.
A) Compared to basal control, the IL-7 induction of pSTAT5 in populations I through V. Population III (CD34+CD38+TdT+) had consistently the largest induction across seven replicate BM. Induction is arcsinh difference versus basal, scaled individually. Maximum differences were 0.8, 2.2, 1.7, 1.3, 0.3, 0.8, and 1.8 for A to G, respectively. B) The effect of TSLP, tofacitinib (JAK1/3i), and pervanadate (PVO) on induction of pSTAT5 across populations I to V in the same individual versus the basal. The maximum difference is 1.3. C) Early Wanderlust trace (trajectory 0-0.5) showing expression of IL-7rα (CD127), TdT, and IgHi. The green heat bar indicates relative pSTAT5 phosphorylation in the basal sample. Relative positions of populations I to V and IgH V(D)J rearrangement are indicated. D) Network rewiring between population III and IV. The schematics show proposed regulatory networks of pSTAT5; population III activates in a ligand and JAK-dependent manner whereas population IV becomes ligand independent, but maintains a high pSTAT5 level in the absence of stimulation. See also Figure S4 and table S2 for additional perturbations and full distributions representing the IL-7 perturbation.
STAT5 network rewiring occurs during immunoglobulin rearrangement
Since the IL-7/STAT5 response was limited to a specific fraction, STAT5 regulation was further characterized relative to adjacent cell fractions across the developmental progression. We used the JAK inhibitor Tofacitinib, combined with IL-7 stimulation, to confirm a Janus kinase mediated mechanism of STAT5 control (Johnson et al., 2005). As expected, within population III, STAT5 activation was attenuated by treatment with Tofacitinib, indicating a JAK mediated mechanism (Figure 4B, S4A).
Populations III's induction of pSTAT5 coincides with the cells gaining expression of the IL-7 receptor (CD127), where all CD127 positive cells of population III strongly induce pSTAT5 in response to IL-7 (Figure S4B). IL-7 receptor levels continue to rise in populations IV and V (Figure 4C), yet ex vivo IL-7 stimulation no longer induces pSTAT5 in these later populations (Figure S4B). However, cells occupying population IV display a higher basal level of pSTAT5 (Figure 4C). To test if pSTAT5 levels are saturated in later populations, the pan tyrosine phosphatase inhibitor pervanadate (PVO) was tested. In the presence of PVO, the levels of pSTAT5 rose in all CD34+ progenitor B cell fractions, across biological replicates (Figure 4B, S4A). Additionally, cells in populations III and partially IV yielded a similar STAT5 phosphorylation pattern in response to thymic stromal lymphoprotein (TSLP) (Figure 4B & S4A), a ligand that shares the IL-7rα chain (CD127) and activates STAT5 (Kang and Der, 2004).
Together these observations illustrate a STAT5 network rewiring over the development of B cell precursors (Figure 4D). STAT5 phosphorylation is initially dependent upon an exogenous ligand (i.e. IL-7, TSLP or others) in a JAK mediated mechanism (population III). Then, despite continued expression of the IL-7 receptor, STAT5 phosphorylation becomes ligand independent (population IV-V), yet remains basally high relative to developmentally adjacent cells.
Previous studies in mouse have implicated the IL-7-dependent STAT5 induction in the initiation of genomic rearrangement (Malin et al., 2009). The peak expression of TdT (Figure 4C) indicated that cells in population IV are actively rearranging the IgH locus of the immunoglobulin gene (Figure 3C-D). Therefore, the switch in regulation of STAT5 activation overlaps with germline gene rearrangement, a cell state in which successful outcome requires careful monitoring by the cell. Thus, when cells were organized into a progression, we observed the coordinated rewiring of the regulatory signaling network in the rare, early B cell populations (Figure 4D).
Derivative analysis of the trajectory reveals coordination points in B cell development
The coordinated expression of phenotypic markers coupled with re-wiring of regulatory signaling suggested that these events coalesced around developmental checkpoints controlling the progression of B cell lymphopoiesis. Because this highly multiplexed dataset, combined with the developmental ordering revealed by Wanderlust, allows examination of the concurrent timing of protein expression across B cell development, we used derivative analysis to determine the rates by which given markers changed at each point along the trajectory. The derivative for each marker along the Wanderlust trajectory was approximated using a sliding window (Figure 5A).
Figure 5. Coordination of protein expression across B cell development.
A) The first derivative was calculated in windows across the trajectory for each marker. These values are expressed as a heat map with red indicating a positive slope (increasing expression) and blue indicating a negative slope (decreasing expression). B) Heat map summary of the first derivative analysis, rows are markers and columns the progression of the trajectory. Markers were hierarchically clustered using absolute values of the first derivative. ‘*’ indicates decline in kappa light chain expression coincident with increase in lambda light chain expression. Coordination points: C) at ~0.25 Wanderlust (red dashed line and box) rise in VpreB, λ5, TdT, CD10, and CD24 and fall in CD117 expression; D) 0.3-0.4 (blue dashed line and box) drop in CD34 expression and increases in CD19, CD20, IgHi, and Pax5; E) ~0.6 (purple dashed line and box) showing increases in CD20, kappa, and lambda light chain protein expression. F) ~0.8 (black dashed line and box) with drop in CD38 expression and increases in CD40, IgD, and CD22. See also Figure S5 for additional replicates.
The derivatives were examined to see if multiple phenotypic features changed in a coordinated fashion. Clustering the parameters based on the absolute value of their derivative across the trajectory uncovered several striking ‘coordination points’ where the changes in expression of multiple proteins coalesced across B cell development (Figure 5B). At least four major coordination points were identified across the trajectory (Figure 5B, dashed boxes), and were consistent across samples from independent human subjects (Figure S5). The first (Figure 5B and C, red) coincides with population III, the ligand-dependent pSTAT5 cells (Figure 4), representing cells at the early pro-B cell stage of development just prior to IgH locus rearrangement. The second (Figure 5B and D, blue) is consistent with cells that are passing through the pre-B cell stage and are preparing to rearrange the light chain locus of the immunoglobulin (Cobaleda and Sanchez-García, 2009).
Light chain rearrangement is crucial to the latter two coordination points. The first of these (Figure 5B and E, purple) coincides with kappa light chain protein expression, which mirrors the trajectory of CD20, signifying that the expression of CD20 occurs in concert with BCR light chain rearrangement and expression. Cells that do not successfully express kappa switch to lambda light chain, both consistent with the known biology and correctly ordered by Wanderlust (Figure 5B ‘*’ and E). The last coordination point (Figure 5B and F, black) cements the emerging cells as naïve, immature B cells preparing to enter peripheral lymphoid organs.
In summary, Wanderlust successfully organized a dynamically asynchronous cellular system, providing a holistic view of the coordination of a complex system, even for transient and rare cell types. Derivative analysis reveals a closely coordinated series of regulatory and cellular events, suggesting coordination points that might act as checkpoints between shifts in cellular states and fate determinations.
Coordination points reveal a checkpoint for B cell developmental progression
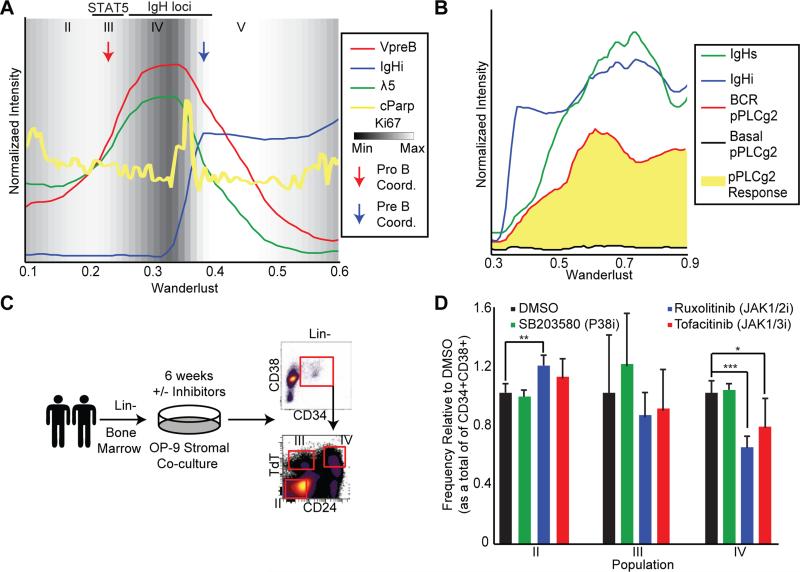
Using Wanderlust to overlay simultaneously measured indicators of cell proliferation (Ki67) and apoptosis (cleaved poly ADP ribose polymerase–cPARP) revealed a further level of functional coordination across nascent human B cell populations (Figure 6A). The first coordination point (Figure 6A, red arrow) marks a transition from a state of high to low proliferation, as assessed by decreasing Ki67 expression (Figure 6A, background shade). This drop in proliferation leads directly into population IV (Figure 6A), signifying the transition into pro-B cells, suggesting a checkpoint that has never been clearly demonstrated in the human.
Figure 6. Regulatory signaling influences cell fate decisions in developing B cells.
A) Wanderlust traces of VpreB, λ5, IgHi, and cleaved PARP across the early trajectory (0.1-0.6). The background is shaded for Ki67 (proliferative antigen) expression. Indicated are the relative positions of population II to V. The red and blue arrows indicate putative timing of pro- and pre-B cell coordination points, respectively. B) Wanderlust traces across the late trajectory (0.3-0.9) showing IgH (i)ntracellular and (s)urface expression with the basal and receptor cross-linked levels of pPLCϒ2, an intermediary of B cell receptor signaling. pPLCϒ2 induction in response to cross-link shown in yellow. C) Using two human BM samples, B cell differentiation co-cultures were performed, following lineage depletion, for 6 weeks on an OP-9 stromal layer in the presence or absence of inhibitors. After, cells were analyzed by flow cytometry for the frequencies of populations II through IV. D) Frequency of populations II through IV, as a proportion of CD34+CD38+ cells, after six weeks of culture in the indicated condition. Each treatment is normalized to the DMSO control for that population. Two biological replicates analyzed in quadruplicate cultures for each condition. Error bars are standard deviation. A two-tailed t-test was performed to determine statistical significance *p<0.05, **p<0.01, ***p<0.001. See also Figure S6 and Table S4 for supporting data.
As the cells pass through the second coordination point, which occurs after the IgH locus has been completely rearranged, Ki67 levels show that the cells re-enter a state of proliferation, expanding the pool of pre-B cells, which have productively formed an IgH (Figure 6A, blue arrow). Just preceding this pre-B cell expansion there is a discrete spike in cell death, indicated by a surge in single cells with higher cPARP (Figure 6A, yellow line), consistent with cells that could not form a productive IgH rearrangement and thus were unable to pass through this checkpoint.
In concert with expression of VpreB and λ5, the newly expressed IgH now composes a complete pre-B cell receptor (preBCR). Mapping cells following B cell receptor cross-linking onto Wanderlust demonstrates that precisely paralleling the surface expression of the IgH (IgHs), cells are able to induce massive phospholipase C (PLC) gamma 2 phosphorylation (Figure 6B, red) as compared to the basal state (Figure 6B, black). Thus, with pre-BCR on the surface of the cells, they have yet again re-wired their regulatory signaling and have become responsive to receptor cross-linking (Figure 6B).
Ex vivo differentiation assay confirms pro-B cell checkpoint
To determine the role these checkpoints play in a cell's developmental progression, the earliest pro-B cell checkpoint was interrogated using an ex vivo differentiation assay (Figure 6C, Tables 3,4). The re-wiring of STAT5 regulation across fractions II to IV (Figure 4) suggests that a blockade of STAT5 phosphorylation could alter the progression of cells through the pro-B checkpoint proposed here. To test this, Lin- human BMMCs from two donors were differentiated on OP-9 stromal cell feeders for six weeks (Sanz et al., 2010), after which the relative proportions of fractions II through IV were assessed.
Both JAK inhibitors used, Ruxolitinib (JAK1/2 inhibitor) and Tofacitinib (JAK1/3 inhibitor), restricted progression of cells from population II through to population IV (Figure 6D), significantly decreasing the frequency of cells in population IV, relative to a DMSO control. At the same time, there was a significant accumulation of cells in population II. The P38 inhibitor, did not have a significant influence on the allocation of cells across the three fractions, though it did promote significant, albeit compartment independent, cellular expansion (Figure S6B). Collectively, these ex vivo culture assays imply that STAT5 promotes the developmental progression of early B cell precursors, where the ligand dependent phosphorylation in population III likely represents a critical (pro-B cell) transition point for initiating IgH V(D)J rearrangement and progression to later stages of maturity.
Discussion
By leveraging the massively multiplexed, single-cell analysis of a complex primary sample, algorithmic ordering of cellular processes can detect the underlying temporal element in the system and be used for novel biological inquiries. The Wanderlust algorithm described here is resilient to noise, consistent between samples, and scalable to up to tens of millions of cells. It extracts a trajectory from a snapshot of the system rather than from time-series data and only requires an approximate starting point as prior information. The Wanderlust trajectory is continuous; in addition to mapping stable cell states, it also provides information about the transitions between states. The combination of these characteristics makes this an ideal approach for the exploration of any system undergoing a continuous developmental process.
As single cell measurements amass due to new technologies such as mass cytometry and single-cell RNA-seq, researchers are faced with the novel challenge of organizing this volume of data. The solution most commonly used is clustering, but by averaging cells into groups, this approach loses the richness of single cell resolution. Using the graph approach proposed here, rather than assigning single cells to a group of similar cells, each cell is mapped to a unique position in a graph structure that can be easily navigated. This structure affords many of the advantages of clustering while preserving much of the cell's individual information. This graph-based representation of single cell data can be adapted to a wide range of additional applications.
Wanderlust determined the developmental trajectory of human B cell emergence in bone marrow by simultaneously examining the features of this primary tissue from progenitor to maturity, requiring no cellular synchronization, purification, or manipulation. The trajectory is consistent with the traditional understanding of this process. Furthermore, Wanderlust provides a quantitative, high-resolution ordering of surface marker expression, signaling and recombination events, including markers whose timing and relevance were previously unappreciated. The determined trajectory unifies virtually all relevant cellular features and regulatory behaviors of early B cell development in the human with discrete cell subsets that can now be demarcated using conventional cytometric methods (Figure 3).
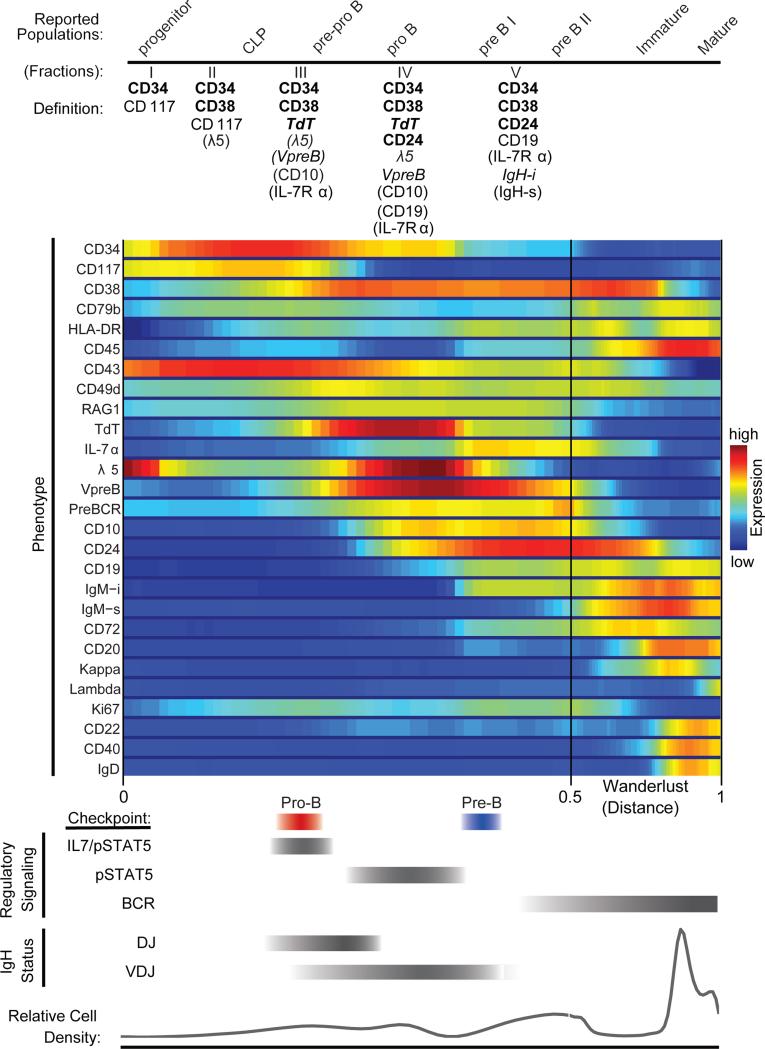
A unifying model of mammalian B cell development
Because it is difficult to obtain and experimentally manipulate human bone marrow, the understanding of mammalian B cell development comes mostly from murine systems. The Wanderlust trajectory identified a precise ordering of key events and explicitly pinpointed the developmental hallmarks of B cell development, previously assumed to exist based on the murine system, in the human. A more precise overview of human B cell development in the bone marrow is now made possible by aligning phenotype with regulatory signaling and key developmental events such as immunoglobulin rearrangements (Figure 7). Moreover, the progression identified by Wanderlust is maintained across all donor bone marrow specimens examined, such that the developmental timing of key coordination points ordered by the Wanderlust trajectory are consistent across independent marrows and analyses (Figure S7).
Figure 7. Summary of phenotypic marker expression, regulatory signaling, and IgH recombination status and proposed pro- and pre- B cell checkpoints aligned to reported populations across human B cell development as ordered by the Wanderlust trajectory.
Under ‘Definition’ (top) the expected marker localization and expression is listed for subsequent cytometric characterization. ‘Bold type’ markers are the minimum required to positively select cells by cytometric analysis. ‘Regular type’ markers are uniformly expressed on the surface. ‘Italic type’ markers are intracellularly expressed. Markers in parentheses ‘()’ are only partially expressed on that fraction. Red and blue bars indicate the pro- and pre- B cell coordination points, respectively. For regulatory signaling and IgH status (bottom), the grey bars indicate the developmental regions where cells are prone to the listed behaviors. See also Figure S7 for additional replicates.
In addition, the Wanderlust trajectory facilitated an in-depth examination of rare and transient early stages of B cell development: previously unrealized populations of B cell progenitors based on the combined expression of CD34, CD38, CD24 and TdT. Notably, the earliest of these B cell precursors expressed neither CD10 nor CD19, the earliest markers conventionally used in human B cell identification (Cobaleda and Sanchez-García, 2009). Previous experimental studies of early B cells, which relied on identification based on expression of CD10 or CD19, would have entirely excluded the earlier fraction of B cells highlighted here.
Moreover, the overlay of multiple markers onto a single trajectory offered a holistic picture of the coordination of a complex system, even for transient and rare cell types; in particular, identifying regulatory signal re-wiring of STAT5 regulation across populations III through V. Remarkably, IL-7 induced activation of STAT5 was a limited regulatory state, only active in a rare population of the cells. While population III comprises only 0.007% of the BMMCs, the results demonstrated that it serves as a checkpoint to ensure successful initiation of IgH rearrangement. Through this lens, coordination points appear as a hallmark of developmental progression.
A discrete versus continuous concept of cellular development
Much effort has been devoted to the taxonomic characterization of cellular populations across development in virtually all tissues, with new cell subsets constantly being described based on increasingly complex patterns of expression. Although it is easy to conceptualize this process as a series of discrete steps, in reality, it is continuous and characterized by transitional stages. In our approach, the trajectory captures expression as trends: markers rise and fall in patterns that correspond to the cell's behavior, capturing transitional behavior. Applying Wanderlust to high-dimensional single-cell data from a primary human tissue determined a developmental ordering (trajectory) of cells without any time-point experiments or genetic manipulations. Furthermore, as seen in Figure S1, the algorithm was able to begin from a late cell and map the trajectory from a known finale back to its beginning. This variation is relevant in the context of non-hematopoietic development, where the stem cells are not known but the mature cells are plentiful and easily identified, such as in mesenchymal development.
A foundation upon which to understand disease
Many human diseases can be considered corruptions of normal development. Indeed, pathologic examination of tissues often reports findings as the degree of divergence from normal tissue architecture. However, our understanding of the underpinnings of a disease is only as good as our understanding of the normal, healthy condition. Wanderlust provides the ability to infer regulatory events across the healthy developmental trajectory, so it is now possible to use the precise foundation of healthy tissue ordering to further understand corrupted developmental disease processes.
In the case of lymphopoiesis, as demonstrated here, the identification of coordination points across development coupled to critical regulatory signaling that influence cell fate decisions (including survival and proliferation), highlights specific developmental periods of risk for malignant transformation. Understanding the critical network configurations that surround these transitions may provide important insight into disease, especially when a developmental state may serve as an additional diagnostic or classification metric.
There are several possible extensions to the concepts presented here. Wanderlust assumes that the developmental process is composed of a series of consecutive stages, with no branching. Incorporation of a more sophisticated model that allows for branching will enable the analysis of more complex systems, such as the complete immune system. Given its flexibility and minimal experimental requirements, this study lays the foundation for applying these methods to other tissue types and corrupted developmental processes, such as cancer, in the future.
Materials and Methods
Mass Cytometry Analysis
Processing of primary human bone marrow and mass cytometry analysis including data pre-processing is as previously described (Bendall et al., 2011; Fienberg et al., 2012; Finck et al., 2013; Kotecha et al., 2010). Extended description of these methods can be found in the supplementary material.
Analysis of Primary Human B Cells
Lin- BMMCs were stained for CD34, CD38, TdT, and CD24 and populations II through V were collected on a FACS Aria (BD Bioscience). Unamplified genomic DNA from the sorted cell populations was assessed for the level of IgH (D)J and V(D)J rearrangement using a qPCR approach adapted from adapted from Van Dongen et. al. (van Dongen et al., 2003). OP-9 progenitor cell co-cultures for B cell specification were performed as previously described (Sanz et al., 2010). Extended description of these methods can be found in the supplementary material.
The Wanderlust algorithm
Input and initialization
The Wanderlust trajectory detection algorithm receives as input the high-dimensional sample data and a user-defined initial cell (for example, a stem cell), referred to here as the “early” cell. The output is a continuous trajectory score for each cell that provides the cell's temporal positioning across development; undifferentiated cells have low scores whereas mature cells have high scores. Wanderlust is composed of two steps: initialization and trajectory calculation, which is performed iteratively, see outline below. Please refer to the supplementary methods for a full and detailed description of the Wanderlust algorithm.
Wanderlust Outline Description
Wanderlust receives as input the single-cell measurements and a user-defined early cell. The algorithm begins with a two-step initialization step (Figure 1C, top left). First, a set of cells is randomly chosen as waypoints. Then, the data is transformed into a randomly generated ensemble of graphs. The algorithm proceeds by calculating a trajectory separately in each graph. For each cell (called a target), its position along the trajectory is first set to the shortest-path distance from the early cell. The target's position is refined according to the shortest-path distance from each waypoint using a weighted average. Waypoints closer to the target contribute more to it's location as they are less susceptible to the noise inherent in the shortest-path distance. However, the waypoints are themselves targets. Therefore, their position will change following this same refinement step. Since cell positions depend on waypoint positions, the shift in waypoints might change the newly calculated positions. Therefore, the refinement step is repeated with the new waypoint positions until the positions of all cells converge. Once the trajectory calculation step completes in all of the graphs, the output trajectory is set to the average over all graph trajectories.
Simulated data
We applied Wanderlust to a series of simulated datasets. Each dataset included the same curved, one-dimensional simulated trajectory that was embedded in 3 dimensions. The simulated trajectory was generated by starting at position (1, 1, 1) and randomly traversing the space for 10,000 steps. After each step the current position was added to the trajectory as a point. Seven additional dimensions of normally-distributed noise were added to each dataset. The magnitude of the noise dimensions (defined as the standard deviation divided by the range of the solution trajectory) varied between datasets. Additionally, some datasets included short circuits; the number of short circuits varied between datasets, and their distances were exponentially distributed with mean again varying between datasets. Full details are available in the supplementary methods.
Wanderlust analysis
Wanderlust was run on 21 phenotypic markers in each sample using the following parameters: nl = 20, dist = angular distance, p = 2, ng = 20, k = 5, l = 30. The early cell was chosen as the cell expressing the highest level of CD34. The output trajectory was normalized to the [0, 1] range by subtracting the 5th percentile and dividing by the value of the 95th percentile minus the 5th percentile. We defined the trace of each marker as the median marker intensity in overlapping windows across the trajectory. One hundred windows were uniformly distributed across the [0, 1] range. Each window included all cells whose trajectory score was within +/-0.08 of the center of the window.
Wanderlust cross-correlation between individuals
Given a marker, for each sample, we calculated the cross-correlation between the trace of the marker in the sample and its trace in an arbitrarily chosen sample (sample A). The trajectory was shifted such that the mean of all cross-correlations was maximized.
Wanderlust derivative analysis
Given the trace of a marker tm over the Wanderlust trajectory w, we calculated an approximation of the derivative of the trace at a given point p using:
Supplementary Material
Research Highlights.
-Wanderlust aligns single cells on a trajectory according to their developmental path
-Mass cytometry plus Wanderlust infers path from HSCs to naïve B cells
-Trajectory identifies precursor B-cell populations where rearrangement occurs
-Discovery of coordination points dictating cell fate decisions
Acknowledgements
We would like to thank Omer Angel, Antonio de-la-Hera, Astraea Jager, Ulf Klein, Smita Krishnaswamy, Jacob Levine, Eva Sanz, Peter Sims and Angelica Trejo for their intellectual and technical contributions. KLD is supported by a St. Baldrick's Foundation Scholar Award. SCB is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09) and the NIH K99GM104148-01. EDA is a Howard Hughes Medical Institute International Student Research Fellow. This work was supported by 0158 G KB065; 1R01CA130826; 5U54CA143907NIH; CIRM: DR1-01477; HEALTH.2010.1.2-1; HHSF223201210194C - FDA: BAA-12-00118; HHSN272200700038C; N01-HV-00242; NIH 41000411217; NIH 5-24927; P01 CA034233-22A1; PN2EY018228; RB2-01592; RFA CA 09-009; RFA CA 09-011; U19 AI057229; U54CA149145; W81XWH-12-1-0591 OCRP-TIA NWC; NIH S10 SIG S10RR027582-01 to GPN. This work was supported by NSF MCB-1149728, NIH DP2-OD002414-01, NIH U54CA121852-01A1 to DP. DP holds a Packard Fellowship for Science and Engineering. GPN has personal financial interest in and SCB is a paid consultant for the company DVS Sciences, the manufacturers that produced some of the reagents and instrumentation used in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
EDA, SCB, KLD, GPN, DP conceived the study. EDA, DJS, DP designed and developed Wanderlust. EDA, MDT, DP developed analysis tools and performed statistical analysis of Wanderlust. SCB, KLD, EFS developed reagents and performed all data acquisition experiments. TJC conceived of the derivative analysis. SCB, KLD designed and performed all functional validation experiments. SCB, KLD, EDA, DP performed the biological analysis and interpretation. SCB, KLD, EDA, GPN, and DP wrote the manuscript.
References
- Amir E-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013 doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir E-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Sanchez-García I. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. BioEssays. 2009;31:600–609. doi: 10.1002/bies.200800234. [DOI] [PubMed] [Google Scholar]
- Corfe SA, Paige CJ. The many roles of IL-7 in B cell development; Mediator of survival, proliferation and differentiation. Semin Immunol. 2012;24:198–208. doi: 10.1016/j.smim.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe'er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K, Parks DR, Herzenberg LA, Herzenberg LA. Murine B cell differentiation lineages. J Exp Med. 1984;159:1169–1188. doi: 10.1084/jem.159.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SE, Shah N, Panoskaltsis-Mortari A, LeBien TW. Murine and human IL-7 activate STAT5 and induce proliferation of normal human pro-B cells. J Immunol. 2005;175:7325–7331. doi: 10.4049/jimmunol.175.11.7325. [DOI] [PubMed] [Google Scholar]
- Kang J, Der SD. Cytokine functions in the formative stages of a lymphocyte's life. Current Opinion in Immunology. 2004;16:180–190. doi: 10.1016/j.coi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2009;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink AG, Boekel ten, E., Yamagami T, Ceredig R, Andersson J, Melchers F. B cell development in the mouse from early progenitors to mature B cells. Immunol. Lett. 1999;68:89–93. doi: 10.1016/s0165-2478(99)00035-8. [DOI] [PubMed] [Google Scholar]
- Sanz E, Munoz-A N, Monserrat J, Van-Den-Rym A, Escoll P, Ranz I, Alvarez-Mon M, de-la-Hera A. Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways. Proc Natl Acad Sci USA. 2010;107:5925–5930. doi: 10.1073/pnas.0907942107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, García-Sanz R, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.