Abstract

Eukaryotic translation initiation factor 2B (eIF2B), the guanine nucleotide exchange factor for the G-protein eIF2, is one of the main targets for the regulation of protein synthesis. The eIF2B activity is inhibited in response to a wide range of stress factors and diseases, including viral infections, hypoxia, nutrient starvation, and heme deficiency, collectively known as the integrated stress response. eIF2B has five subunits (α–ε). The α, β, and δ subunits are homologous to each other and form the eIF2B regulatory subcomplex, which is believed to be a trimer consisting of monomeric α, β, and δ subunits. Here we use a combination of biophysical methods, site-directed mutagenesis, and bioinformatics to show that the human eIF2Bα subunit is in fact a homodimer, at odds with the current trimeric model for the eIF2Bα/β/δ regulatory complex. eIF2Bα dimerizes using the same interface that is found in the homodimeric archaeal eIF2Bα/β/δ homolog aIF2B and related metabolic enzymes. We also present evidence that the eIF2Bβ/δ binding interface is similar to that in the eIF2Bα2 homodimer. Mutations at the predicted eIF2Bβ/δ dimer interface cause genetic neurological disorders in humans. We propose that the eIF2B regulatory subcomplex is an α2β2δ2 hexamer, composed of one α2 homodimer and two βδ heterodimers. Our results offer novel insights into the architecture of eIF2B and its interactions with the G-protein eIF2.
Eukaryotic translation initiation factor 2 (eIF2) is a G-protein that in its GTP-bound form binds to the initiator methionyl-tRNA (Met-tRNAi), yielding a ternary complex (TC), which is then recruited to the translation initiation complex. Upon start codon recognition, eIF2 hydrolyzes GTP and is released from the initiation complex (reviewed in refs (1−4)).
The eIF2 TC is regenerated after every cycle of translation initiation through a process catalyzed by the guanine nucleotide exchange factor (GEF) eIF2B, which is one of the main targets for the regulation of translation. Phosphorylation of serine 51 in the α subunit of eIF2 converts eIF2 from a substrate into a competitive inhibitor of the GEF eIF2B. eIF2α phosphorylation is mediated by a group of related kinases: the dsRNA-activated protein kinase (PKR), PKR-like ER kinase (PERK), heme-regulated inhibitor (HRI), and general control nonderepressible 2 (GCN2). eIF2α is phosphorylated in response to a number of different stress factors, such as viral infection, unfolded protein response, hypoxic stress, heme deficiency, amino acid starvation, etc., collectively known as the integrated stress response (ISR). The result is inhibition of translation in the cell, which can range from modestly slowing to nearly completely shutting off protein synthesis and induction of apoptosis. At the same time, translation of the mRNA encoding transcription factor ATF4, which mediates the stress response, is turned on through a mechanism called translation reinitiation. Phosphorylation of eIF2α by PKR in response to viral infection causes translation shut-off and apoptosis and serves as a powerful defense mechanism in the cell. The ISR triggered by amino acid starvation or heme deficiency typically causes more modest translation inhibition (mediated by GCN2 or HRI activation, respectively) and serves to balance the supply and demand of metabolites. PERK is involved in controlling the accumulation of misfolded proteins as part of the unfolded protein response (UPR). Persistent PERK activation leads to cell death in prion diseases and other neurodegenerative disorders, and PERK inhibition was recently shown to have neuroprotective effects in mice.5 ISR induction via PERK in the hypoxic environment inside solid tumors is important for cancer cell survival (reviewed in refs (6−8)).
The yeast Saccharomyces cerevisiae has only one of the eIF2α kinases, GCN2, whose main role is in balancing amino acid supply and demand. Phosphorylation of eIF2α by GCN2, in response to amino acid starvation, not only decreases translation rates but also turns on translation of a number of proteins, including the transcription factor GCN4, which in turn stimulates expression of amino acid biosynthetic enzymes (reviewed in refs (9) and (10)).
eIF2B is composed of five different subunits (α–ε). eIF2Bγ and eIF2Bε form the catalytic subcomplex. eIF2Bα, -β, and -δ form the regulatory subcomplex.11,12 The α, β, and δ subunits of eIF2B are homologous to each other over the entire eIF2Bα sequence; eIF2Bδ has an N-terminal tail (NTT) not found in the other two subunits. All the eIF2B subunits except for eIF2Bα are essential in S. cerevisiae (reviewed in refs (2) and (10)). The essential functions of eIF2Bβ, -γ, and -ε are in nucleotide exchange, whereas eIF2Bδ is important for the recruitment of Met-tRNAi to eIF2-GTP.13 While eIF2Bα plays an accessory role in the GEF function, it is required for the regulation of eIF2B activity by phosphorylation of its substrate eIF2. The eIF2Bα subunit is not tightly associated with the rest of eIF2B, especially when eIF2B is not bound to its substrate eIF2, and is sometimes partially or completely lost during purification (see, for example, refs (14−16)). α-Less eIF2B has been reported to have activity either lower than16 or similar to17 that of intact eIF2B and is not inhibited by phosphorylated eIF2 [eIF2(α-P)].11,17
Mutations that decrease the activity of eIF2B in S. cerevisiae lead to lower TC concentrations, mimicking the effect of eIF2α phosphorylation and derepressing the translation of GCN4 in the absence of amino acid starvation. Such a phenotype is called general amino acid control derepressed (Gcd–), and Gcd– mutations have been found in all five eIF2B subunits. Mutations that prevent derepression of GCN4 translation by amino acid starvation, called general amino acid control nonderepressible (Gcn–), have been found in all three subunits of the eIF2Bα/β/δ regulatory subcomplex. The Gcn– mutations in eIF2Bα, -β, and -δ prevent inhibition of eIF2B activity by eIF2(α-P) (reviewed in refs (9) and (10)).
Mutations in human eIF2B have been associated with a genetic neurodegenerative disorder known as childhood ataxia with CNS hypomyelination (CACH) or leukoencephalopathy with vanishing white matter (VWM). The CACH/VWM mutations seem to lead to decreased eIF2B levels or activity in the cell; however, the underlying molecular mechanisms are not understood (reviewed in refs (7), (18), and (19)).
Despite decades of research, the architecture of eIF2B remains unknown. It has typically been reported to have an apparent molecular mass between 250 and 350 kDa, based on size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) experiments (see, for example, refs (20−22)). Because the combined mass of its five subunits is ∼260 kDa, it has always been thought to be a heteropentamer composed of one each of the five subunits. More recently, mass spectrometry data have confirmed that the five subunits are present in equimolar amounts.23 Models for the architecture of the catalytic eIF2Bγε subcomplex have been proposed, based on distantly related enzymes.24,25 However, no viable model exists for the regulatory eIF2Bα/β/δ subcomplex, and it is not known how the individual subunits interact with each other (see also the Discussion).
Because eIF2B is believed to be a heteropentamer, the regulatory eIF2Bα/β/δ subcomplex is presumed to be a 1:1:1 trimer, and all three subunits (α, β, and δ) are presumed to be monomers. However, it is difficult to reconcile this notion with recent data about the structures of homologous proteins, all of which are dimers. More recently, when the crystal structure of human eIF2Bα was determined,26 the authors did not discuss the possibility of eIF2Bα being a dimer, likely because that would conflict with the notion that it is a monomer (because eIF2Bα/β/δ is believed to be a trimer). However, close inspection of the deposited structure reveals that the eIF2Bα molecules found in the asymmetric unit appear to form dimers.
Here we show that eIF2Bα is indeed a homodimer, like all its homologs with known structures, using the same dimerization interface. We also present evidence indicating that the interaction between eIF2Bβ and -δ likely occurs along the same interface, forming a heterodimer similar to the eIF2Bα2 homodimer. These results allow us to propose a model about the possible architecture of the regulatory eIF2Bα/β/δ subcomplex. They also provide insights into the molecular basis of a number of mutations in human eIF2B that cause leukoencephalopathy with vanishing white matter. While this article was being prepared, Proud and co-authors reported that eIF2Bα is a dimer and eIF2B as a whole is a decamer,27 at odds with a number of previous reports,20−22 but in line with the results presented here. While the interactions between eIF2Bβ and -δ were not addressed in this recent report,27 the data presented there are consistent with our conclusions.
Experimental Procedures
Vectors, Protein Expression, and Purification
Human eIF2Bα was cloned in a pET21a derivative vector with a short 11-residue C-terminal His tag. The eIF2BαI210E/V217E/A221D triple mutant (eIF2Bα-TM) and the eIF2Bα deletion mutant missing residues 256–266 (eIF2Bα-Δarm) were derived from the wild-type (WT) eIF2Bα plasmid using site-directed mutagenesis. Human eIF2Bβ was cloned in a pET21a derivative vector with an N-terminal protein G IgG-binding domain 1 (GB1) tag, followed by a His tag and a TEV cleavage site (GH-eIF2Bβ), or in a pRSFDuet-1 vector (Novagen) with a short 16-residue N-terminal His tag (H-eIF2Bβ). The eIF2Bβ1–143 construct (eIF2Bβ-NTD) was derived from the WT eIF2Bβ plasmid using site-directed mutagenesis.
WT eIF2Bα and its derivatives were expressed in Rosetta2(DE3) cells overnight (O/N) at 20 °C. eIF2Bβ and eIF2Bβ-NTD were expressed in Rosetta2(DE3)pLysS cells. eIF2Bβ was grown for 3 h at 37 °C; eIF2Bβ-NTD was grown O/N at 20 °C. 15N-labeled eIF2Bβ-NTD was expressed in minimal medium using a standard protocol, as described previously,28 under the same conditions that were used for the expression of the unlabeled proteins. Proteins were purified using His tag affinity chromatography on TALON CellThru resin (Clontech), followed by SEC on a Superdex 75 column (GE Healthcare) in 10 mM sodium phosphate buffer (pH 7.0), 150 mM NaCl, 2 mM DTT, 1 mM EDTA, and 0.1 mM AEBSF. The short C-terminal His tag of WT and mutant eIF2Bα (34.9 kDa) and the short N-terminal His tag of H-eIF2Bβ (40.7 kDa) are not cleavable. The GB1/His tag of GH-eIF2Bβ and GH-eIF2Bβ-NTD was cleaved with TEV protease, yielding untagged eIF2Bβ (39.0 kDa) and eIF2Bβ-NTD (16.2 kDa). The tag was removed using His tag affinity chromatography on TALON resin or GB1 affinity chromatography on IgG resin (GE Healthcare).
Size Exclusion Chromatography (SEC)
Analytical SEC of WT and mutant eIF2Bα (∼35 kDa) and of untagged eIF2Bβ (∼39 kDa) was performed on a Superdex 75 10/300 GL column (GE Healthcare). Apparent molecular weights (MWapp) were estimated using a set of protein standards: bovine serum albumin (BSA, 67 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa). Proteins showing concentration-dependent increases in MWapp were run at a series of concentrations, and their MWapp was plotted as a function of concentration. Apparent KD values for dimerization were calculated as described previously.29 In cases in which the protein was mostly or entirely dimeric at the lowest concentration tested, only an upper limit of the KD could be obtained.
Small Angle X-ray Scattering (SAXS)
SAXS data of eIF2Bα (∼35 kDa) were measured on beamline X9 at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY). No reference protein was used, and therefore, no absolute MW was determined. SAXS data at three concentrations (57, 143, and 287 μM) were measured for eIF2Bα. The low-angle region of the SAXS profile at low concentrations was merged with the high-angle region of the high-concentration profile to compensate for interparticle repulsion. The SAXS data were processed using the ATSAS software suite.30 The radius of gyration (Rg) was measured using PRIMUS, and the maximum dimension (Dmax) was determined from the pair distance distribution function P(r) calculated using GNOM. The SAXS profile of eIF2Bα was used for fitting dimer and tetramer assemblies from the eIF2Bα crystal structure [Protein Data Bank (PDB) entry 3ecs] using FoXS.31
Size Exclusion Chromatography–Multiangle Laser Light Scattering (SEC–MALLS) Determination of Native Protein Molecular Weights
SEC–MALLS of eIF2Bα (∼35 kDa) and H-eIF2Bβ (∼41 kDa) was performed at the W. M. Keck Biotechnology Resource Facility at Yale University (New Haven, CT). The samples were run on a Superdex 200 HR10/300 GL SEC column (GE Healthcare), connected inline to a DAWN-EOS LS detector and an RI detector (Wyatt Technology). Absolute molecular weights (MWs) were calculated using ASTRA (Wyatt Technology). Proteins showing concentration-dependent self-association were run at a series of concentrations, and their MW was plotted as a function of concentration. The starting sample concentration was used for plotting, instead of the estimated concentration at the apex, because it was impossible to determine the degree to which the complexes were able to reequilibrate at the gradually decreasing concentrations during the SEC run.
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR spectra were recorded at 298 K on a Bruker 500 MHz instrument. Samples for NMR measurements were in buffer containing 10 mM sodium phosphate buffer (pH 7.0), 150 mM NaCl, 2 mM DTT, 1 mM EDTA, 0.1 mM AEBSF, and 10% D2O. NMR chemical shift perturbation assays were performed as previously described.321H–15N heteronuclear single-quantum coherence (HSQC) spectra of 15N-labeled eIF2Bβ-NTD were recorded in the presence and absence of excess eIF2Bα and compared.
Bioinformatics, Modeling, and Structure Analysis
To obtain multiple-sequence alignments for eIF2Bα, -β, and -δ families, as well as among the three families, we performed sequence homology searches with PSI-BLAST.33,34 A representative set of complete sequences with <90% pairwise sequence identity was obtained with the help of HHfilter from the HHsuite.35−37 Multiple-sequence alignments were obtained using CLUSTAL W38 and T-COFFEE.39 Sequence alignments among eIF2Bα, -β, and -δ were also obtained with HHpred from the HHsuite.35−37 All alignment approaches yielded essentially the same results for the C-terminal Rossmann fold domain (CTD) of eIF2Bα, -β, and -δ. The sequence alignments for the N-terminal helical domain (NTD) obtained from CLUSTAL W and T-COFFEE were different from each other and from the HHpred and PSI-BLAST results and showed poor agreement with the eIF2Bα structure (insertions and/or deletions in secondary structure elements, buried uncompensated charges), as well as with the multiple-sequence alignments within the eIF2Bα, -β, and -δ families. HHpred and PSI-BLAST yielded comparable sequence alignments for the NTD, which were in good agreement with the eIF2Bα crystal structure and with the multiple-sequence alignments within the eIF2Bα, -β, and -δ families. The resulting eIF2Bα/β and eIF2Bα/δ pairwise sequence alignments were used to generate homology models for the structures of eIF2Bβ and -δ with the alignment mode of SWISS-MODEL.40 eIF2Bβ and -δ segments that had no counterparts in the eIF2Bα sequence or corresponded to segments not visible in the eIF2Bα structure were modeled de novo (colored black in Figure 3A). Their predicted conformation was thus considered unreliable. Sequence alignments were colored with ESPript,41 using the BLOSUM62 homology scoring matrix.
Figure 3.
Models for the structure of eIF2Bβ and -δ. (A) Model for the proposed dimeric structure of eIF2Bβ (yellow) and eIF2Bδ (red), based on the structure of eIF2Bα2 and the sequence alignment shown in Figure S4 of the Supporting Information. The first eight residues of eIF2Bβ and the first 200 residues of eIF2Bδ were not modeled. Segments that have no counterpart in eIF2Bα or are not visible in the eIF2Bα crystal structure were modeled de novo and are colored black, because their real conformation is unknown. (B) The putative eIF2Bβ and -δ dimerization surfaces are highly conserved among eIF2Bβ and eIF2Bδ homologs. The eIF2Bβδ dimer model is shown in surface representation. Amino acids are colored by sequence conservation from white (<30% conservation) to yellow (65% conservation) to green (100% conservation). To show the dimerization surfaces, eIF2Bδ was omitted from the middle left panel and eIF2Bβ from the middle right panel. The dimerization interface is marked with a light blue line. The conserved surface unique to eIF2Bδ is circled in the right panel. (C) The putative eIF2Bβ and -δ dimerization surfaces are highly hydrophobic. The display is as in panel B, except that the surface is colored by hydrophobicity and charge. Backbones are colored dark gray, hydrophobic side chains yellow, positively charged side chains blue, negatively charged side chains red, and the remaining side chains light gray.
Structure analysis was conducted in MOLMOL.42 Structure alignments between the CTDs of eIF2Bα, aIF2B, MTNA, and RBPI structures were created in MOLMOL using structure alignments from the DALI server.43 One CTD from each dimer was used for the alignment, while the rest of the dimer was not used in the alignment and was aligned indirectly. This approach allowed for an unbiased analysis of the similarity between two dimers, which could have been overemphasized by global alignment over the entire dimers. In all cases, the alignment between the indirectly aligned second pair of CTDs (from the second subunits in the dimer) was almost as good as that between the directly aligned CTDs, illustrating the remarkably high degree of conservation of the dimer interface among eIF2Bα, aIF2B, MTNA, and RBPI structures. The angle between the NTD and the CTD varies slightly among the different families, as has been previously observed.26 The buried surface area was calculated using PISA.44 The eIF2Bα structure and the eIF2Bβ and -δ models were colored by conservation with the help of ProtSkin,45 based on multiple-sequence alignments of ∼50 to ∼90 sequences with <90% pairwise sequence identity. The coloring scheme was from white (<30% conservation) to yellow (65% conservation) to green (100% conservation), similar to that used by the Burley lab (see, e.g., ref (46)). The BLOSUM62 matrix was used to calculate sequence homology scores. Figures were generated in MOLMOL.42
Molecular Docking
Model Building
Models of eIF2Bβ and -δ used for molecular docking were based on chain C (the most complete chain) in the X-ray crystal structure of eIF2Bα (PDB entry 3ecs). We used Modeller version 9v8 and modeled only residues at positions that were present in the X-ray structure of eIF2Bα. This was done for two reasons. First, the conservative approach improves the reliability of the model. Second, because all the systems have the same number of residues (i.e., differ only by side chains), the calculated relative affinity scores are comparable across the different pairs. No further energy minimization was performed for the model structures prior to docking.
Docking
Models of subunits were docked using the ClusPro protein docking server, which consists of systematic global sampling of all mutual orientations of the two proteins on a rotational/translational grid.47 The conformations are evaluated using an energy-based scoring function that includes attractive and repulsive contributions to the van der Waals interaction energy, an electrostatic energy term, and a pairwise interaction potential representing desolvation effects. The N (≥1000) lowest-energy structures are clustered using the pairwise rmsd as the distance measure and a 9 Å clustering radius.48 The biophysical meaning of clustering is isolating highly populated low-energy basins of the energy landscape.49 The largest clusters indicate the most likely models, as shown previously using approximate partition functions.50
Stability Analysis Using Focused Sampling
As stable structures are expected to be located at energy minima surrounded by a funnel-shaped energy distribution,51 the stability of a model can be investigated by focused sampling of structures in its neighborhood, thereby determining the local energy landscape. The resampling is based on the same algorithm as global docking but uses a denser rotational grid, and the translational space is limited to the region defined by the cluster of interest identified in the docking stage. We have previously shown that focused sampling provides detailed information about the energy landscape and improves relative affinity estimation.52
Results
The Human eIF2Bα Crystal Structure Indicates That It Is a Dimer
Most archaeons with an eIF2Bα/β/δ homolog have a single protein, archaeal translation initiation factor 2B (aIF2B), which is a homodimer. Throughout this paper, the “a” in aIF2B is in bold, to aid in the distinction between aIF2B and eIF2B. aIF2B dimerization is mediated by the C-terminal domains (CTDs) of the two subunits (Figure 1A and Figure S1B of the Supporting Information). A long loop (“arm” region) protrudes from each subunit and packs against the other subunit in the dimer, effectively extending its β-sheet.53 More distant relatives, such as ribose-1,5-bisphosphate isomerases (RBPIs)54 and methylthioribose-1-phosphate isomerases (MTNAs),55,56 are also dimeric and utilize the same dimer interface as aIF2B, including the arm region (Figure S1C,D of the Supporting Information). All these dimeric structures cast doubt on the widely held view that the regulatory eIF2Bα/β/δ subcomplex is a trimer, which requires the eIF2Bα, -β, and -δ subunits to be monomers.
Figure 1.
eIF2Bα dimerizes along the same interface as all its homologs with known structures. (A) Crystal structure of the archaeal eIF2Bα/β/δ homolog, aIF2B (PDB entry 1vb5), which is a homodimer.53 The two aIF2B subunits are colored red and yellow. The arm regions, which interact with the other subunit in the dimer, are labeled with arrows. (B) Crystal structure of eIF2Bα (PDB entry 3ecs)26 showing a dimer with a large buried surface. One subunit is colored cyan and the other blue. The arm regions, which interact with the other subunit in the dimer, are labeled with arrows. (C) eIF2Bα dimerizes using the same interface as aIF2B. Structure alignment of aIF2B and eIF2Bα, with the same orientation and coloring as in panels A and B, respectively. The CTDs of the eIF2Bα and aIF2B subunits were aligned [Cα rmsd of 1.38 Å (excluding the arm region)]. The interdomain orientations differ somewhat between eIF2Bα and aIF2B, as noted previously,26 and the NTDs of the proteins were not used in the alignment. Structure alignments were done in MOLMOL.42 (D) The eIF2Bα dimerization surface is highly conserved among eIF2Bα homologs. eIF2Bα is shown in surface representation. Amino acids are colored by sequence conservation from white (<30% conservation) to yellow (65% conservation) to green (100% conservation). The dimerization interface is marked with a light blue line. One subunit is omitted in the middle panel, to show the dimerization surface. (E) The eIF2Bα dimerization surface is highly hydrophobic. The display is as in panel D, except that the surface is colored by hydrophobicity and charge. Backbones are colored dark gray, hydrophobic side chains yellow, positively charged side chains blue, negatively charged side chains red, and the remaining side chains light gray. The three residues at the dimerization surface, whose mutation (designated eIF2Bα-TM) abolishes dimerization (see Figure 2), are labeled.
The crystal structure of human eIF2Bα was published recently.26 The authors reported that eight molecules were found in the asymmetric unit but did not discuss the subject further. Close inspection of the eIF2Bα structure shows that the eight molecules in the asymmetric unit form four identical dimers (shown in different colors in Figure S1E of the Supporting Information). Dimerization is along the same interface as in all homologs with known structures (Figure 1B,C and Figure S1 of the Supporting Information). The arm region (marked with arrows in Figure 1B) is also part of the dimerization interface, again as in all homologous structures. The dimer interface is extensive, with >1500 Å2 of buried surface area. The dimerization surface observed in the crystal structure is among the best-conserved surfaces in the protein, with significant hydrophobicity, as expected for a protein–protein interaction surface (Figure 1D,E). Hydrophobic amino acid side chains are typically found in the protein core and at protein interaction surfaces, because exposing them is unfavorable and burying them contributes to the binding energy. Hydrophobic side chains also contribute to the specificity of interaction, because the binding energy is proportional to the sixth power of the distance and helps “lock” the complex into place, compared to electrostatic interactions, which are proportional to the first power of the distance. Compared to the highly conserved putative eIF2α-P binding surface,57 the eIF2Bα dimerization surface shows a comparable degree of sequence conservation and even greater hydrophobicity, consistent with both surfaces being important for protein–protein interactions and the dimerization surface being constitutively buried. These observations led us to hypothesize that eIF2Bα is a homodimer, utilizing the same dimerization surface as all its homologs with a known structure.
eIF2Bα Is a Dimer at Physiological Concentrations
To test our hypothesis that eIF2Bα is a dimer, we used SEC. We found that eIF2Bα (∼35 kDa) migrates with an apparent molecular weight (MWapp) of ∼60 kDa, roughly as expected for a dimer (Figure 2A). The MWapp of eIF2Bα did not change as its concentration was varied between 0.6 and 3 μM (Figure 2B), indicating that there is no significant fraction of monomer within this concentration range. While these results do not allow us to estimate the KD of dimerization, the absence of detectable amounts of monomer (below a few percent) provides an upper limit of ∼1 nM for the KD: if there is no detectable monomer at 0.6 μM, the protein concentration must be more than 2 orders of magnitude higher than the KD of dimerization. Therefore, eIF2Bα is a dimer at physiological concentrations, estimated to be in the low micromolar range.58 The MWapp of eIF2Bα gradually increased as its concentration was varied between 3 and 230 μM (Figure 2B), indicating that the dimer was in equilibrium with higher-order species at concentrations above 3 μM. To corroborate the SEC results, we used SAXS at concentrations of 57, 143, and 287 μM to characterize the oligomeric state of eIF2Bα (Figure S2 of the Supporting Information). The radius of gyration (Rg) and the maximum dimension (Dmax) of the eIF2Bα particle at 57 μM could be determined from SAXS data to be ∼44 and ∼150 Å, respectively (Figure S2 of the Supporting Information). Comparison of these results with the values calculated from the crystal structure for a monomer (Rg = 21 Å; Dmax = 78 Å) and dimer (Rg = 31 Å; Dmax = 132 Å) showed that the average complex size in the eIF2Bα sample at 57 μM was greater than a dimer. Because there are eight molecules in the asymmetric unit in the eIF2Bα crystal, forming four identical dimers, there is more than one possible tetrameric arrangement (and it is also possible that the interdimer orientations could be different from those observed in the crystal). The calculated values of Rg and Dmax for two of the possible tetrameric arrangements found in the crystal are 37 and 133 Å (both values smaller than experimentally determined) and 41 and 163 Å (Dmax larger than experimentally determined), respectively. For reference, the values of Rg and Dmax calculated for the octameric arrangement in the crystal were 45 and 167 Å, respectively. We were unable to fit the SAXS data to a dimer, a tetramer, or a mixture thereof with a χ2 of better than 6 by using FoXS.31 Nevertheless, the calculated Rg and Dmax show that at the high protein concentrations used for SAXS, eIF2Bα is clearly not a monomer and is larger than a dimer, in agreement with the SEC results.
Figure 2.

Size exclusion chromatography (SEC) of eIF2Bα and -β. (A) SEC traces of WT eIF2Bα (blue) and eIF2Bα-TM (green) at a concentration of 15 μM. The theoretical MWs for WT eIF2Bα and eIF2Bα-TM monomers are both ∼35 kDa. The apparent molecular weights (MWapp) from SEC are 60 kDa for WT eIF2Bα (dimer) and 33 kDa for eIF2Bα-TM (monomer). The positions of the markers used to calculate MWapp are shown with vertical dashed lines. (B) Calculated MWapp as a function of protein concentration. WT eIF2Bα (data from two independent sets of experiments are colored blue and light blue) is dimeric up to 3 μM, but its MWapp starts to gradually increase at higher concentrations, indicative of the formation of larger complexes. eIF2Bα-Δarm (red) is in equilibrium between the monomer and dimer, and possibly higher-order complexes, in the concentration range tested; eIF2Bα-TM (green) is clearly monomeric. (C) SEC trace of untagged eIF2Bβ at a concentration of 15 μM. The theoretical MW for an eIF2Bβ monomer is 39 kDa. The apparent molecular weight (MWapp) from SEC is 41 kDa (monomer). The positions of the markers used to calculate MWapp are shown with vertical dashed lines.
To determine unambiguously the oligomeric state of eIF2Bα in solution, we used SEC–MALLS, which allows the determination of absolute molecular weights and can also be performed at physiological protein concentrations. The SEC–MALLS experiment was performed at three different eIF2Bα concentrations: at a physiological concentration58 of 1 μM [within the concentration range in which eIF2Bα mobility in SEC is independent of concentration (see Figure 2B)] as well as at 10 and 100 μM [within the concentration range in which there is a concentration-dependent increase in MWapp as determined via SEC (see Figure 2B)]. When eIF2Bα was loaded at a concentration of 1 μM, the MW determined by SEC–MALLS was 67 kDa, as expected for a dimer (Table 1). At starting concentrations of 10 and 100 μM, the MWs determined at the apex of the peak by SEC–MALLS were 83 and 195 kDa, respectively [at these concentrations, the MW decreased toward the tail side of the peak, likely because of the lower eIF2Bα concentrations leading to partial complex dissociation (see Figure S3 of the Supporting Information)]. Therefore, the average complex size in the eIF2Bα samples was larger than a dimer at 10 μM and corresponded to a hexamer at 100 μM. Comparison of the SEC–MALLS results with those from the SEC and SAXS experiments shows that SEC and SAXS results qualitatively agree with SEC–MALLS results, with SEC results (both stand-alone and the SEC portion of the SEC–MALLS) tending to underestimate the size of the eIF2Bα oligomers. These results also explain our inability to fit the SAXS data to a structure model, because at the concentrations used in SAXS (57–287 μM), eIF2Bα likely exists as a mixture of dimers, tetramers, hexamers, and possibly also octamers. In conclusion, these results show that eIF2Bα is a dimer at physiological concentrations and oligomerizes at higher concentrations.
Table 1. Native Molecular Weights of eIF2Bα and eIF2Bβ Determined by SEC–MALLS.
| protein | starting concentrationa (μM) | theoretical MWb (monomer) (kDa) | MW at the apexc (kDa) |
|---|---|---|---|
| eIF2Bα | 1 | 34.9 | 66.7 (dimer) |
| eIF2Bα | 10 | 34.9 | 83.0 (>dimer) |
| eIF2Bα | 100 | 34.9 | 195 (∼hexamer) |
| H-eIF2Bβ | 1 | 40.7 | 39.5 (monomer) |
| H-eIF2Bβ | 10 | 40.7 | 39.7 (monomer) |
Concentration at which the samples were loaded onto the SEC column. Sample dilution at the end of the run was estimated to be between 2- and 20-fold.
Calculated on the basis of the protein sequence.
The SEC peak of eIF2Bα at 10 and 100 μM was polydisperse, with the MW determined by MALLS gradually decreasing along the tail of the peak, likely because of the lower protein concentrations in the tail affecting the average oligomer size (see also Figure S3 of the Supporting Information).
eIF2Bα Dimerizes through the Same Interface as Its Homologs
As described above, the dimer interface observed in the crystal structure of eIF2Bα, as well as all of its homologs with known structures, is composed of a large interface between the CTDs of two subunits and a long loop (or arm), which protrudes across the dimer interface and packs against the other subunit in the dimer (Figure 1). To confirm that eIF2Bα dimerizes through the interface observed in the crystal structure, we used site-directed mutagenesis. We replaced three hydrophobic residues at the predicted dimerization surface of the CTD with negatively charged residues (I210E, V217E, and A221D) to generate an eIF2BαI210E/V217E/A221D triple mutant (eIF2Bα-TM). Using SEC, we found that eIF2Bα-TM migrated with an MWapp of ∼33 kDa, as expected for a monomer (Figure 2A). To assess the contribution of the arm to eIF2Bα dimerization, we generated an eIF2Bα deletion mutant missing residues 256–266 (eIF2Bα-Δarm). SEC shows that at 1.25 μM, eIF2Bα-Δarm migrates with an MWapp of ∼50 kDa (Figure 2B), a value intermediate between that of a monomer (∼33 kDa) and that of a dimer (∼67 kDa), indicating that at this concentration eIF2Bα-Δarm is in equilibrium between monomers and dimers. As the eIF2Bα-Δarm concentration is increased to 12.5 and 125 μM, its MWapp gradually increases to ∼67 kDa (Figure 2B). These results show that both the main dimer interface and the arm play important roles in eIF2Bα dimerization.
eIF2Bβ Is a Monomer
Having determined that eIF2Bα is a dimer, we explored the possibility that the homologous eIF2Bβ and eIF2Bδ subunits could also dimerize through their corresponding surfaces. We generated homology models for the structures of eIF2Bβ and eIF2Bδ (Figure 3), using the eIF2Bα structure as a template and the sequence alignment shown in Figure 4. The ∼200-residue eIF2Bδ N-terminal tail is not homologous to eIF2Bα and -β and was not modeled. As noted previously,26,57 the level of homology among eIF2Bα, -β, and -δ was greater for the Rossmann fold C-terminal domain (CTD) than for the helical N-terminal domain (NTD) (Figure 4). Analysis of the putative dimerization surfaces in the CTDs of the eIF2Bβ and -δ homology models shows that they are among the best-conserved surfaces in the proteins (Figure 3B). They exhibit significant hydrophobicity (Figure 3C), similar to the corresponding surface of eIF2Bα (Figure 1DE), as expected for a protein–protein interaction surface (see also the sequence alignment in Figure 4). The putative dimerization surface of eIF2Bδ is particularly hydrophobic (Figure 3C). Both eIF2Bβ and -δ have the arm region (Figure 3A), which in eIF2Bα and the more distant homologs forms part of the dimer interface. The conservation and charge and hydrophobicity patterns of the rest of eIF2Bβ and -δ are also similar to those of eIF2Bα (Figure 1D,E), including the highly conserved putative eIF2α-P binding surfaces,57 where a number of Gcn– mutations in yeast eIF2Bα, -β, and -δ have been found26,57 (Figure 3B,C, left panels, and Figure 1D,E; see also Figure 6). A notable exception is the surface in eIF2Bδ visible in the right panels of panels B and C of Figure 3, which shows a greater degree of conservation than its counterparts in eIF2Bα (Figure 1D,E, right panels) or eIF2Bβ (Figure 3B,C, right panels).
Figure 4.
Sequence alignment of human eIF2Bα, -β, and -δ. Identical positions are shown as white letters on a red background; conserved positions are shown as red letters. The secondary structure and residue numbering above the alignment are for eIF2Bα. Amino acids located at the dimer interface are marked with a black asterisk below the alignment. The position of V183 in eIF2Bα is marked with a red asterisk. V183 is buried just under the dimerization surface and surrounded by residues that are part of the dimer interface (see also Figure 6A). The V183F mutation causes CACH/VWM19,59 and was recently shown to affect eIF2Bα dimerization.27 The sequence alignment was obtained with HHpred from the HHsuite.35−37 This figure was generated with ESPript,41 using the BLOSUM62 homology scoring matrix.
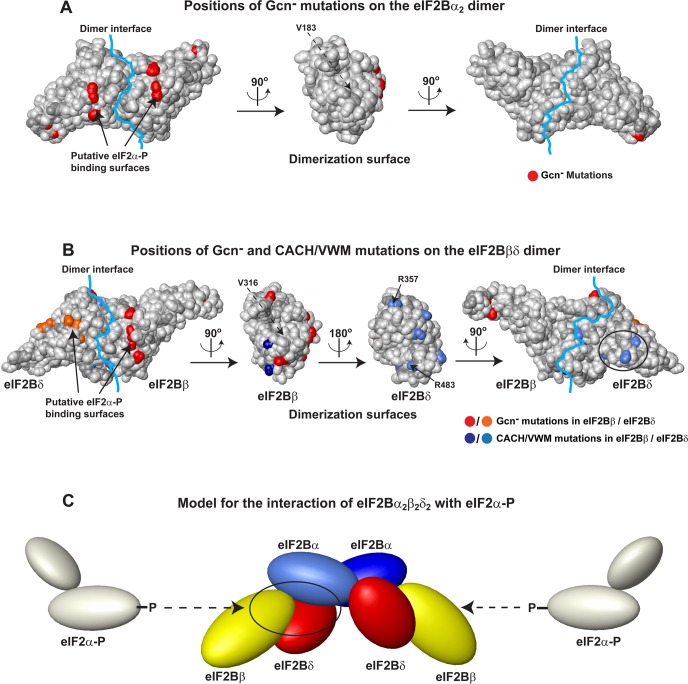
Figure 6.
Model for the interaction of eIF2Bα2β2δ2 with eIF2α-P. (A) Positions of Gcn– mutations (red) on the surface of the eIF2Bα2 dimer. The orientations are the same as in panels D and E of Figure 1. The dimerization interface is marked with a light blue line. In the central panel, only one eIF2Bα subunit is shown, to show the dimerization surface. Only mutations of surface-exposed residues that do not involve a glycine or a proline are shown, because these are least likely to affect the protein structure or stability. The approximate position of V183 (invisible because it is buried under the surface) is labeled in the central panel with a dashed arrow. (B) Positions of Gcn– and CACH/VWM mutations on the surface of the eIF2Bβδ dimer, in the same orientation as the eIF2Bα2 dimer in panel A. The eIF2Bβδ dimer is in the same orientation as in Figure 3. The dimerization interface is marked with a light blue line. The two central panels show the dimerization surfaces of eIF2Bβ (left) and eIF2Bδ (right). Sites of Gcn– mutations are colored red (eIF2Bβ) and orange (eIF2Bδ), and CACH/VWM mutations are colored navy (eIF2Bβ) and blue (eIF2Bδ). The clusters of CACH/VWM mutations at the eIF2Bβ and eIF2Bδ dimerization surfaces are visible in the two central panels. The cluster of CACH/VWM mutations in eIF2Bδ-NTD can be seen in the right panel (circled). Residues discussed in the text are labeled. The position of V316 (buried under the surface) is labeled in the left central panel with a dashed arrow. (C) Model for the interaction of eIF2Bα2β2δ2 with eIF2α-P. One eIF2Bα subunit is colored light blue, the other dark blue, eIF2Bβ yellow, and eIF2Bδ red. eIF2α-P (light gray) binds in a pocket (circled on the left) formed between one eIF2Bα subunit and one eIF2Bβδ dimer. The proteins are represented by solids, drawn approximately to scale with their respective sizes. Unphosphorylated eIF2α should bind to an overlapping surface on eIF2Bα2β2δ2.13
Using SEC, we found that eIF2Bβ (∼39 kDa) migrates with an MWapp of ∼41 kDa, as expected for a monomer (Figure 2C). This result was later confirmed using SEC–MALLS, which yielded an MW of ∼40 kDa, as expected for a monomer, at both 1 and 10 μM (Table 1). Unfortunately, we were unable to express eIF2Bδ in soluble form in Escherichia coli and thus could not determine whether it is monomeric or study its interaction with eIF2Bβ.
Molecular Docking Indicates an eIF2Bβδ Heterodimer along the Same Interface as eIF2Bα2
Unlike eIF2Bα, free eIF2Bβ is a monomer (Figure 2C), despite a high degree of conservation in its putative dimerization surface. The interaction between eIF2Bβ and -δ is important for the stability of eIF2Bδ in vivo in S. cerevisiae, because eIF2Bβ depletion causes codepletion of eIF2Bδ.13 Therefore, we considered the possibility that eIF2Bβ and -δ form a heterodimer using the same extensive dimerization surfaces as eIF2Bα2. Because we were unable to produce soluble eIF2Bδ, we could not test this hypothesis directly.
Because eIF2Bα, -β, and -δ are homologous, we built models of β and δ subunits based on the crystal structure of eIF2Bα (PDB entry 3ecs). Residues were modeled only at positions present in the eIF2Bα structure. We used molecular docking with the ClusPro server48 to evaluate the potential for homo- or heterodimerization of all combinations of eIF2Bα, -β, and -δ. For the eIF2Bα2 homodimer, the highest-scoring model differs by only a 3.5 Å interface root-mean-square deviation (rmsd) from the dimer interface seen in the crystal structure (Figure 5A), thus reproducing the experimentally confirmed eIF2Bα2 homodimer. The calculated interaction energy values show a funnel-shaped distribution in the vicinity of this best model (Figure 5B), indicative of a stable structure. The interface in the highest-scoring model of the eIF2Bβδ heterodimer was the same as seen in the eIF2Bα2 homodimer, with a funnel-like local energy landscape (Figure 5C). This supports the hypothesis that eIF2Bβ and -δ interact with each other along an interface corresponding to the homodimerization interface in eIF2Bα2 (Figure 1B), forming a stable heterodimer similar to the eIF2Bα2 homodimer. In contrast, docking generated relatively few models with the putative interface for eIF2Bδ2, eIF2Bαβ, and eIF2Bαδ, placing the highest-scoring models at least 25 Å interface rmsd away in each case, confirming that these combinations do not form dimers. The remaining combination, eIF2Bβ2, was found to be an interesting case: although the best-scoring model included the putative dimer interface, the energy values did not exhibit any funnel-like behavior (Figure 5D). This indicates that interactions between two eIF2Bβ proteins do not result in the formation of a stable homodimer, in good agreement with the experimental data.
Figure 5.
Molecular docking indicates an eIF2Bβδ heterodimer utilizing the same interface as the eIF2Bα2 homodimer. (A) Highest-scoring model of the eIF2Bα2 dimer superimposed on the eIF2Bα2 dimer observed in the crystal structure of eIF2Bα (PDB entry 3ecs).26 The left subunits from each dimer (model and crystal structure) are aligned (colored gray). The second subunit in the modeled dimer is colored blue; the second subunit in the dimer found in the crystal structure is colored yellow. The interface rmsd between the two structures is 3.5 Å. None of the high-scoring models showed any similarity to any other crystal contacts observed in the eIF2Bα structure26 (PDB entry 3ecs). (B) Local energy landscape of the eIF2Bα2 models generated by docking, plotted with a model having the observed eIF2Bα2 crystal interface placed at the origin. The plot shows that such models are surrounded by a well-defined energy funnel. (C) Local energy landscape of the eIF2Bβδ models generated by docking, plotted with a model having the eIF2Bα2 crystal interface placed at the origin. The plot shows that, along the energy funnel, the models converge to ones with the interface seen in the eIF2Bα2 structure. (D) Local energy landscape of the eIF2Bβ2 models generated by docking, again plotted with a model having the eIF2Bα2 crystal interface placed at the origin. Unlike for the dimers shown in panels B and C, no funnel-shaped energy distribution is observed in the vicinity of such models, indicating that no stable eIF2Bβ2 homodimer is formed.
All eIF2Bα/β/δ homologs with known structures are dimers utilizing the same dimer interface as we show here for eIF2Bα2 (Figures 1 and 2 and Figure S1 of the Supporting Information). Therefore, this dimerization interface has been conserved through evolution of the eIF2B homolog and distantly related metabolic enzymes. Because the corresponding surfaces in eIF2Bβ and -δ are also well-conserved (Figure 3B), it is logical to expect that they are not exceptions to the rule and are also involved in dimerization (either homo- or heterodimerization). The molecular docking results presented here are in full agreement with our experimental data and indicate that, while eIF2Bα is a homodimer, eIF2Bβ and -δ form a heterodimer.
Discussion
Implications for the Architecture of eIF2B
Since it was first discovered, eIF2B has been presumed to be a heteropentamer with equimolar amounts of its α, β, γ, δ, and ε subunits. Accordingly, the regulatory eIF2Bα/β/δ subcomplex has always been presumed to be a 1:1:1 heterotrimer. However, the architecture of eIF2B and its regulatory subcomplex has remained a mystery, despite decades of research.
The results presented here show that eIF2Bα is a homodimer and eIF2Bβ is a monomer. Our data also indicate that that eIF2Bβ and -δ form a heterodimer along the same interface as the eIF2Bα2 dimer. What is then the architecture of eIF2B and the regulatory eIF2Bα/β/δ subcomplex in particular? There are two alternative models that are consistent with eIF2Bα being a homodimer and eIF2Bβ a monomer, as well as with the utilization of the conserved hydrophobic C-terminal surfaces of eIF2Bβ and -δ for dimerization. One possible model is that the regulatory subcomplex is an eIF2Bα2βδ tetramer, made up of an eIF2Bα2 dimer and an eIF2Bβδ dimer, which would imply that eIF2B as a whole is a hexamer (α2βδγε). This model fits well with the majority of previously published SEC and AUC data about the size of eIF2B, which has been reported to have an MWapp of 250–350 kDa.20−22 However, it contradicts a recent mass spectrometry study, which showed that all eIF2B subunits are present in equimolar amounts.23 Alternatively, the eIF2B regulatory subcomplex could be an α2β2δ2 hexamer built up of one α2 homodimer and two βδ heterodimers, which would imply that eIF2B is a decamer. Such a model is at odds with most SEC and AUC data20−22 but is consistent with the recent mass spectrometry report about the stoichiometry of eIF2B subunits.23 The results presented here alone cannot distinguish between these two models. However, Proud and co-authors recently reported that eIF2B is a decamer.27 Therefore, in view of their results and the earlier mass spectrometry report,23 we favor the latter model: a hexameric eIF2B regulatory subcomplex, composed of an eIF2Bα2 homodimer and two eIF2Bβδ heterodimers. These authors also found that eIF2Bα is a dimer, in agreement with the results presented here. While they did not study the interactions of eIF2Bβ and eIF2Bδ, their observation that eIF2Bα stabilizes the interaction between two eIF2Bβγδε tetramers27 is consistent with the existence of two eIF2Bβδ heterodimers, rather than an eIF2Bβ2 and an eIF2Bδ2 homodimer in eIF2B.
Alternative Models for the Architecture of eIF2Bα/β/δ
The only attempt to date for a structure model for the regulatory eIF2Bα/β/δ subcomplex was by Kakuta and co-authors, based on crystal packing contacts in the structure of the archaeal eIF2Bα/β/δ homolog aIF2B from Pyrococcus horikoshii (which is a dimer in solution).53 As discussed above, the structure shows dimerization along the same interface as all other homologs.26,54−56 Therefore, when looking for possible implications for the structure of the presumed trimeric eIF2Bα/β/δ complex, the authors had to ignore the known dimerization interface (>1500 Å2 of buried surface area, light blue line in Figure S4A,B of the Supporting Information). Instead, they focused on hexameric-like crystal packing contacts among three dimers (red line in Figure S4B of the Supporting Information) to propose that the eIF2Bα/β/δ complex forms a symmetrical trimer, leaving the conserved hydrophobic dimerization surfaces unoccupied. Any trimeric model of the eIF2Bα/β/δ regulatory subcomplex in general is incompatible with the results presented here, because an even number of subunits is needed to occupy all dimerization surfaces. An eIF2Bα/β/δ trimer would leave at least one of the extensive conserved and hydrophobic dimerization surfaces in eIF2Bα, -β, and -δ unoccupied, while the model of Kakuta and co-authors specifically would leave all three dimerization surfaces unoccupied. In view of the data presented here and in the recent report by Proud and co-authors,27 the Kakuta model could be considered in the context of a hexameric eIF2Bα2β2δ2 regulatory subcomplex: as a possible interface between three dimers, instead of three monomers, especially because it was in fact based on crystal packing contacts among three dimers (Figure S4B of the Supporting Information).53 However, although the coordinates for the proposed trimeric arrangement were not deposited, it is clear from Figure S4B of the Supporting Information that the buried surface area is small and insignificant. It is thus highly unlikely that such a small interface could produce a stable complex. In support of this conclusion, we found using an NMR chemical shift perturbation assay that eIF2Bα does not bind to eIF2Bβ-NTD, because adding unlabeled eIF2Bα did not affect the NMR spectra of 15N-labeled eIF2Bβ-NTD (Figure S4C of the Supporting Information). Therefore, this possibility is not supported by experimental data.
Implications for eIF2B Function and Interactions with Its Substrate, eIF2
Most Gcn– mutations in eIF2Bα, -β, and -δ map to the same conserved surface in the CTD26,57 (Figure 6A,B, left panels) and have been proposed to be the binding sites for phosphorylated eIF2α (eIF2α-P).57 How these putative eIF2α-P binding surfaces on the eIF2Bα, -β, and -δ subunits come together to simultaneously interact with eIF2α has remained an open question. Remarkably, in the eIF2Bα2 dimer, the C-terminal surfaces from each subunit are adjacent to each other across the dimer interface (Figure 6A, left panel). Likewise, the two C-terminal surfaces of eIF2Bβ and -δ end up adjacent to each other in the model of the proposed eIF2Bβδ dimer (Figure 6B, left panel). The important implication of this observation is that bringing these surfaces closer together in eIF2Bα2 and eIF2Bβδ makes it much easier for eIF2α-P to contact them simultaneously, as indicated by the mutational studies. Binding between an eIF2Bα2 dimer and two eIF2Bβδ dimers can form two binding sites by bringing the putative eIF2α contact surfaces close together (Figure 6C). It should be noted that because the mutual orientation of eIF2Bα2 and eIF2Bβδ is not known, their arrangement in Figure 6C is purely hypothetical and not based on experimental evidence. We did not observe a stable interaction between eIF2Bα and eIF2Bβ using SEC and affinity pull-down experiments (data not shown). Therefore, in the model, eIF2Bα contacts mainly eIF2Bδ. eIF2Bβ likely also binds to eIF2Bα, but with an affinity too low to observe by these assays in the absence of eIF2Bδ. Although in the model shown in Figure 6C the two eIF2Bβδ dimers do not contact each other, they could do so, depending on the actual architecture of the eIF2B regulatory subcomplex.
Gcn– mutations map to several additional surfaces in the regulatory eIF2Bα2βδ subcomplex. However, it is likely that not all of these regions contact eIF2 directly: some of the mutations could have indirect effects. We did not include in our analysis the vast number of Gcd– mutations, because a Gcd– phenotype could arise not only from an impaired mode of binding to unphosphorylated eIF2 but also from lower eIF2B activity or stability. Accordingly, most Gcd– mutations in eIF2Bβ and eIF2Bα affect buried residues (not shown in Figure 6) and are thus likely to affect the structure or stability of the protein, consistent with a loss-of-function mutation lowering the amount or activity of eIF2B.
CACH/VWM Mutations Map to the Proposed eIF2Bβ/δ Dimer Interface
The results presented here help explain the molecular basis of CACH/VWM mutations found in the human eIF2Bα/β/δ regulatory subcomplex. All known CACH/VWM mutations reported to date are recessive: either homozygous or heterozygous with another mutation or complete deletion of the gene. Therefore, it is clear that they lower eIF2B activity. Because complete disruption or inactivation of the eIF2B complex would be lethal, the mutant eIF2B complexes must retain at least a certain level of activity. For those mutations that affect a residue buried in the hydrophobic core of a folded domain, one can speculate that their effect on eIF2B is mediated by destabilization of protein structure. It is thus more interesting to consider mutations in surface-exposed residues. Only three CACH/VWM mutations have been reported in eIF2Bα,19,59 two of them affecting a buried residue and the third affecting a proline. One of these CACH/VWM mutations, V183F,19,59 was recently reported to disrupt eIF2Bα dimerization in vitro.27 This mutation affects a residue buried under the eIF2Bα dimerization surface (its approximate location is marked with a dashed arrow in the central panel of Figure 6A).
As shown in Figure 6B (center panels), a number of CACH/VWM mutations in eIF2Bβ and -δ map to the predicted dimerization surfaces of these two subunits. Therefore, they most likely weaken the interaction between eIF2Bβ and -δ. Two of these mutations, eIF2Bδ-R357W and eIF2Bδ-R483W, both causing a severe form of the disease, were reported to destabilize the association of eIF2Bδ with the rest of eIF2B.60 Another mutation, eIF2Bβ-V316D (not visible in Figure 6B), affects a residue buried just under the predicted eIF2Bδ-binding surface of eIF2Bβ and was reported to weaken interactions with the rest of eIF2B.61 The corresponding mutation in yeast was found to destabilize association with other subunits, especially eIF2Bδ, and to have a slow-growth phenotype that was partially rescued by eIF2Bδ overexpression.62 Therefore, not only does our model for eIF2Bβδ dimerization help explain the phenotypes of many CACH/VWM mutations, but these phenotypes in turn support the model.
A small group of CACH/VWM mutations map to residues in eIF2Bδ-NTD, which appear to cluster on one surface of the domain, away from the proposed eIF2α interface (circled in Figure 6B, right panel). These mutations could interfere either with binding to other eIF2B subunits or with the role of eIF2Bδ in promoting eIF2 ternary complex formation. Consistent with this hypothesis, the surface of eIF2Bδ where these mutations map shows a high degree of sequence conservation and hydrophobicity (circled in Figure 3B,C, right panels), unlike the corresponding surfaces of eIF2Bβ (Figure 3B,C, right panels) and eIF2Bα (Figure 1D,E, right panels). The sequence conservation in this surface and the clustering of CACH/VWM mutations there have not been previously identified. Thus, our results indicate that surface-exposed CACH/VWM mutations in the regulatory eIF2Bα/β/δ subcomplex affect eIF2B complex assembly or stability, with the majority of them mapping to the predicted eIF2Bβδ interaction interface.
In summary, we show here that eIF2Bα is a dimer at physiological concentrations while eIF2Bβ is a monomer. We also present evidence that eIF2Bβ and -δ likely use the same evolutionarily conserved dimer interface as eIF2Bα2 to form a heterodimer similar to the eIF2Bα2 homodimer. These findings indicate that the eIF2B regulatory subcomplex is most likely an α2β2δ2 hexamer composed of an eIF2Bα2 homodimer and two eIF2Bβδ heterodimers. While this conclusion contradicts a number of previous reports about the size of eIF2B (see, for example, refs (20−22)), it is consistent with most of the remaining experimental data about eIF2B and its homologs.23,26,27,53−56 The resulting model for the architecture of the eIF2Bα/β/δ regulatory subcomplex shows how all the surfaces in the regulatory complex predicted to play a direct role in eIF2α binding could indeed contact eIF2α simultaneously and also helps explain the molecular basis for a number of CACH/VWM mutations in eIF2Bβ and -δ. These mutations map at or near the proposed eIF2Bβ/δ dimer interface and are thus likely to affect eIF2B complex formation and/or stability. A key question that remains unanswered is how the regulatory eIF2Bα2β2δ2 subcomplex discriminates between the substrate eIF2 and the inhibitor eIF2(α-P). More work is needed to elucidate the architecture of eIF2B and its interactions with phosphorylated and unphosphorylated eIF2 as well as to understand the detailed molecular mechanisms of the action and regulation of eIF2B.
Acknowledgments
We thank Boriana Marintcheva, Katherine Edmonds, and Nabanita Nag for helpful discussions. We thank Ewa Folta-Stogniew (Director of Biophysics Resource of Keck Laboratory, Yale School of Medicine) for running the SEC–MALLS experiments and for thorough analysis of the results.
Glossary
Abbreviations
- eIF
eukaryotic translation initiation factor
- Met-tRNAi
initiator methionyl-tRNA
- TC
ternary complex
- GEF
guanine nucleotide exchange factor
- PKR
dsRNA-activated protein kinase
- PERK
PKR-like ER kinase
- HRI
heme-regulated inhibitor
- GCN2
general control nonderepressible 2
- ISR
integrated stress response
- NTT
N-terminal tail
- CACH
childhood ataxia with CNS hypomyelination
- VWM
leukoencephalopathy with vanishing white matter
- SEC
size exclusion chromatography
- SEC–MALLS
size exclusion chromatography–multiangle laser light scattering
- AUC
analytical ultracentrifugation
- eIF2Bα-TM
eIF2BαI210E/V217E/A221D triple mutant
- eIF2Bα-DM
eIF2BαV217E/A221D double mutant
- eIF2Bα-Δarm
eIF2BαΔ256–266 deletion mutant
- NTD
N-terminal domain
- CTD
C-terminal domain
- MWapp
apparent molecular weight
- HSQC
heteronuclear single-quantum coherence
- aIF2B
archaeal translation initiation factor 2B
- RBPI
ribose-1,5-bisphosphate isomerase
- MTNA
methylthioribose-1-phosphate isomerase
- rmsd
root-mean-square deviation
- eIF2α-P
phosphorylated eIF2α
- eIF2(α-P)
eIF2 phosphorylated on its α-subunit.
Supporting Information Available
Supplemental Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by National Institutes of Health (NIH) Grants R01 GM095720 to A.M., GM93147 to D.K., and GM61687 to S.V. and National Science Foundation Grant DBI1047082 to S.V. and D.K. The SEC−MALLS instrumentation was supported by NIH Grant 1S10RR023748-01 to the Biotechnology Resource Facility at Yale University.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jackson R. J.; Hellen C. U.; Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A.; Wagner G. (2004) Translation initiation: Structures, mechanisms and evolution. Q. Rev. Biophys. 37, 197–284. [DOI] [PubMed] [Google Scholar]
- Pestova T. V., Lorsch J. R., and Hellen C. U. T. (2007) The Mechanism of Translation Initiation in Eukaryotes. In Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., Eds.) pp 87–128, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Sonenberg N.; Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J. A.; Halliday M.; Molloy C.; Radford H.; Verity N.; Axten J. M.; Ortori C. A.; Willis A. E.; Fischer P. M.; Barrett D. A.; Mallucci G. R. (2013) Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 5, 206ra138. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Dar A. C., and Sicheri F. (2007) The eIF2α kinases. In Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., Eds.) pp 319–344, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Ron D., and Harding H. P. (2007) eIF2α phosphorylation in cellular stress responses and disease. In Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., Eds.) pp 345–368, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Wek R. C.; Jiang H. Y.; Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Dever T. E., and Asano K. (2007) Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., Eds.) pp 225–268, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Pavitt G. D.; Yang W.; Hinnebusch A. G. (1997) Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17, 1298–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Hinnebusch A. G. (1996) Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol. Cell. Biol. 16, 6603–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K.; Qiu H.; Dong J.; Zhang F.; Barthlme D.; Hinnebusch A. G. (2010) The β/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol. Cell. Biol. 30, 5218–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A.; Safer B. (1983) Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J. Biol. Chem. 258, 3402–3408. [PubMed] [Google Scholar]
- Oldfield S.; Proud C. G. (1992) Purification, phosphorylation and control of the guanine-nucleotide-exchange factor from rabbit reticulocyte lysates. Eur. J. Biochem. 208, 73–81. [DOI] [PubMed] [Google Scholar]
- Williams D. D.; Price N. T.; Loughlin A. J.; Proud C. G. (2001) Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J. Biol. Chem. 276, 24697–24703. [DOI] [PubMed] [Google Scholar]
- Kimball S. R.; Fabian J. R.; Pavitt G. D.; Hinnebusch A. G.; Jefferson L. S. (1998) Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α. Role of the α- and δ-subunits of eiF2b. J. Biol. Chem. 273, 12841–12845. [DOI] [PubMed] [Google Scholar]
- Bugiani M.; Boor I.; Powers J. M.; Scheper G. C.; van der Knaap M. S. (2010) Leukoencephalopathy with vanishing white matter: A review. J. Neuropathol. Exp. Neurol. 69, 987–996. [DOI] [PubMed] [Google Scholar]
- Fogli A.; Boespflug-Tanguy O. (2006) The large spectrum of eIF2B-related diseases. Biochem. Soc. Trans. 34, 22–29. [DOI] [PubMed] [Google Scholar]
- de Haro C.; Ochoa S. (1979) Further studies on the mode of action of the heme-controlled translational inhibitor. Proc. Natl. Acad. Sci. U.S.A. 76, 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranu R. S.; London I. M. (1979) Regulation of protein synthesis in rabbit reticulocyte lysates: Additional initiation factor required for formation of ternary complex (eIF-2·GTP·Met-tRNAf) and demonstration of inhibitory effect of heme-regulated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 76, 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekierka J.; Mitsui K. I.; Ochoa S. (1981) Mode of action of the heme-controlled translational inhibitor: Relationship of eukaryotic initiation factor 2-stimulating protein to translation restoring factor. Proc. Natl. Acad. Sci. U.S.A. 78, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito K.; Ota K.; Fujita T.; Ito T. (2007) A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J. Proteome Res. 6, 792–800. [DOI] [PubMed] [Google Scholar]
- Reid P. J.; Mohammad-Qureshi S. S.; Pavitt G. D. (2012) Identification of intersubunit domain interactions within eukaryotic initiation factor (eIF) 2B, the nucleotide exchange factor for translation initiation. J. Biol. Chem. 287, 8275–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Wortham N. C.; Liu R.; Proud C. G. (2012) Identification of residues that underpin interactions within the eukaryotic initiation factor (eIF2) 2B complex. J. Biol. Chem. 287, 8263–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama T. B.; Ito T.; Imataka H.; Yokoyama S. (2009) Crystal structure of the α subunit of human translation initiation factor 2B. J. Mol. Biol. 392, 937–951. [DOI] [PubMed] [Google Scholar]
- Wortham N. C.; Martinez M.; Gordiyenko Y.; Robinson C. V.; Proud C. G. (2014) Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J. 28, 2225–2237. [DOI] [PubMed] [Google Scholar]
- Marintchev A.; Edmonds K. A.; Marintcheva B.; Hendrickson E.; Oberer M.; Suzuki C.; Herdy B.; Sonenberg N.; Wagner G. (2009) Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B.; Maciejewski M. W.; Marintchev A.; Mullen G. P. (2001) Solution structure of the catalytic domain of γδ resolvase. Implications for the mechanism of catalysis. J. Mol. Biol. 310, 1089–1107. [DOI] [PubMed] [Google Scholar]
- Petoukhov M. V.; Svergun D. I. (2007) Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 17, 562–571. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D.; Hammel M.; Sali A. (2010) FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 38, W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A.; Frueh D.; Wagner G. (2007) NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 430, 283–331. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schaffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.; Zaretskaya I.; Raytselis Y.; Merezhuk Y.; McGinnis S.; Madden T. L. (2008) NCBI BLAST: A better web interface. Nucleic Acids Res. 36, W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960. [DOI] [PubMed] [Google Scholar]
- Soding J.; Biegert A.; Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M.; Biegert A.; Hauser A.; Soding J. (2011) HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175. [DOI] [PubMed] [Google Scholar]
- Thompson J. D.; Higgins D. G.; Gibson T. J. (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C.; Higgins D. G.; Heringa J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Arnold K.; Bordoli L.; Kopp J.; Schwede T. (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Gouet P.; Courcelle E. (2002) ENDscript: A workflow to display sequence and structure information. Bioinformatics 18, 767–768. [DOI] [PubMed] [Google Scholar]
- Koradi R.; Billeter M.; Wuthrich K. (1996) MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]; 29 - 32.
- Holm L.; Rosenstrom P. (2010) Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Deprez C.; Lloubes R.; Gavioli M.; Marion D.; Guerlesquin F.; Blanchard L. (2005) Solution structure of the E. coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 346, 1047–1057. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A.; Cao C.; Dever T. E.; Burley S. K. (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: Conformational changes on GDP and GTP binding. Cell 103, 781–792. [DOI] [PubMed] [Google Scholar]
- Kozakov D.; Brenke R.; Comeau S. R.; Vajda S. (2006) PIPER: An FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406. [DOI] [PubMed] [Google Scholar]
- Kozakov D.; Hall D. R.; Beglov D.; Brenke R.; Comeau S. R.; Shen Y.; Li K.; Zheng J.; Vakili P.; Paschalidis I.; Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D.; Clodfelter K. H.; Vajda S.; Camacho C. J. (2005) Optimal clustering for detecting near-native conformations in protein docking. Biophys. J. 89, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S.; Hall D. R.; Kozakov D. (2013) Sampling and scoring: A marriage made in heaven. Proteins 81, 1874–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D.; Schueler-Furman O.; Vajda S. (2008) Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins 72, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden M. S.; Cote S. M.; Sayeg M.; Zerbe B. S.; Villar E. A.; Beglov D.; Sazinsky S. L.; Georgiadis R. M.; Vajda S.; Kozakov D.; Whitty A. (2013) Comprehensive experimental and computational analysis of binding energy hot spots at the NF-κB essential modulator/IKKβ protein-protein interface. J. Am. Chem. Soc. 135, 6242–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y.; Tahara M.; Maetani S.; Yao M.; Tanaka I.; Kimura M. (2004) Crystal structure of the regulatory subunit of archaeal initiation factor 2B (aIF2B) from hyperthermophilic archaeon Pyrococcus horikoshii OT3: A proposed structure of the regulatory subcomplex of eukaryotic IF2B. Biochem. Biophys. Res. Commun. 319, 725–732. [DOI] [PubMed] [Google Scholar]
- Nakamura A.; Fujihashi M.; Aono R.; Sato T.; Nishiba Y.; Yoshida S.; Yano A.; Atomi H.; Imanaka T.; Miki K. (2012) Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1. J. Biol. Chem. 287, 20784–20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumann M.; Djafarzadeh S.; Oberholzer A. E.; Bigler P.; Altmann M.; Trachsel H.; Baumann U. (2004) Crystal structure of yeast Ypr118w, a methylthioribose-1-phosphate isomerase related to regulatory eIF2B subunits. J. Biol. Chem. 279, 37087–37094. [DOI] [PubMed] [Google Scholar]
- Tamura H.; Saito Y.; Ashida H.; Inoue T.; Kai Y.; Yokota A.; Matsumura H. (2008) Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism. Protein Sci. 17, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K.; Santangelo T. J.; Rothenburg S.; Neculai D.; Dey M.; Sicheri F.; Dever T. E.; Reeve J. N.; Hinnebusch A. G. (2009) Archaeal aIF2B interacts with eukaryotic translation initiation factors eIF2α and eIF2Bα: Implications for aIF2B function and eIF2B regulation. J. Mol. Biol. 392, 701–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T.; McCarthy J. E. (2002) Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 46, 531–544. [DOI] [PubMed] [Google Scholar]
- Pronk J. C.; van Kollenburg B.; Scheper G. C.; van der Knaap M. S. (2006) Vanishing white matter disease: A review with focus on its genetics. Mental Retardation and Developmental Disabilities Research Reviews 12, 123–128. [DOI] [PubMed] [Google Scholar]
- Liu A. R.; van der Lei H. D.; Wang X.; Wortham N. C.; Tang H.; van Berkel C. G.; Mufunde T. A.; Huang W.; van der Knaap M. S.; Scheper G. C.; Proud C. G. (2011) Severity of Vanishing White Matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum. Mutat. 32, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Li W.; Wang X.; Van Der Knaap M. S.; Proud C. G. (2004) Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 24, 3295–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P.; Mohammad S. S.; Pavitt G. D. (2004) Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 24, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.