Graphical abstract
Highlights
-
•
Flagellar and non-flagellar T3SS are built assembling homologous protein machineries.
-
•
Unified nomenclature for non-flagellar T3SS.
-
•
New model of the T3SS needle is consistent with the flagellar filament, both in terms of helical parameters and orientation.
-
•
Structural and functional implication of the new architecture of the T3SS export apparatus and ATPase complex.
Abstract
To fulfill complex biological tasks, such as locomotion and protein translocation, bacteria assemble macromolecular nanomachines. One such nanodevice, the type III secretion system (T3SS), has evolved to provide a means of transporting proteins from the bacterial cytoplasm across the periplasmic and extracellular spaces. T3SS can be broadly classified into two highly homologous families: the flagellar T3SS which drive cell motility, and the non-flagellar T3SS (NF-T3SS) that inject effector proteins into eukaryotic host cells, a trait frequently associated with virulence. Although the structures and symmetries of ancillary components of the T3SS have diversified to match requirements of different species adapted to different niches, recent genetic, molecular and structural studies demonstrate that these systems are built by arranging homologous modular protein assemblies.
Current Opinion in Structural Biology 2014, 25:111–117
This review comes from a themed issue on Macromolecular Machines
Edited by Karl-Peter Hopfner and Tom Smith
For a complete overview see the Issue and the Editorial
Available online 1st April 2014
0959-440X/$ – see front matter, © 2014 The Authors. Published by Elsevier Ltd. All rights reserved.
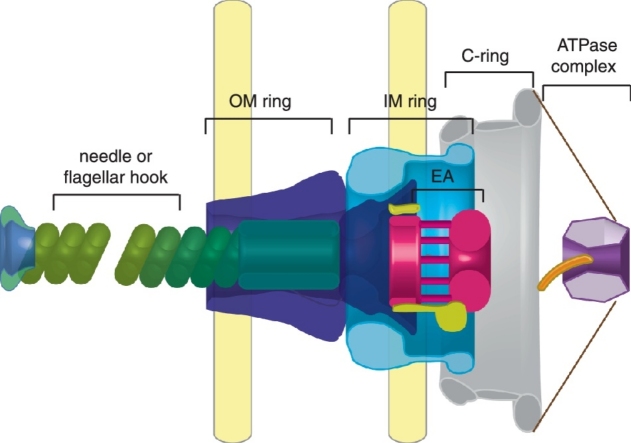
The core of both flagellar-and NF-T3SSs shows an evolutionary related architecture [1] consisting of a multi-ring basal structure embedded in both inner and outer bacterial membranes, with its proximal end connected to an export apparatus (EA) and to an ATPase complex in the cytosol. The T3SS is welded at its distal end to a needle to translocate virulence proteins directly into the host or a flagellar hook onto which polymerizes an extracellular filament dedicated to locomotion [2,3] (Figure 1a). In this review we focus on the recent breakthroughs about the supramolecular structure of NF-T3SS key subassemblies, highlighting analogies to the equivalent flagellar components, when appropriate. For clarity, we will refer to the proteins of the NF-T3SS according to their secretion and cellular translocation (Sct) abbreviation (Table 1), a unified naming system previously proposed [4].
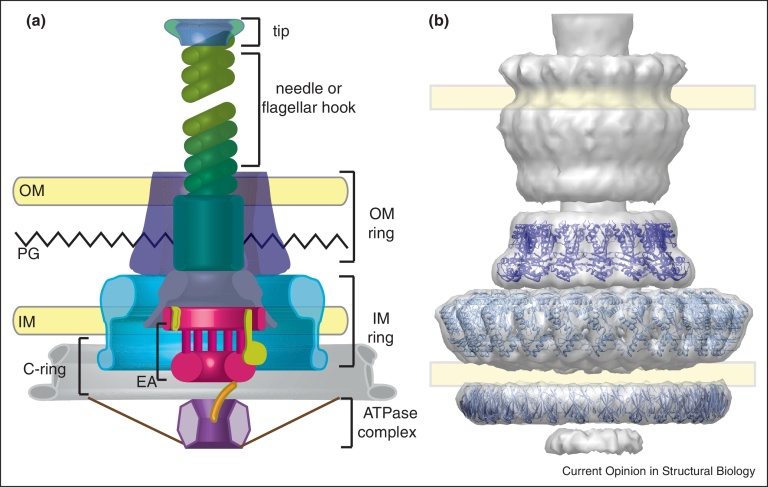
Figure 1.
General architecture of the T3SS. (a) The cartoon schematizes the major subassemblies of the T3SS, showing their relative localization with respect to the outer membrane (OM), the peptidoglycan layer (PG) and the inner membrane (IM). (b) Surface view of the T3SS (C3) from Salmonella typhimurium (EMDB-1875; [22••]) with fitted atomic models [31] of SctC (PDB-3j1v), SctD-N (PDB-3j1w), SctD-C (PDB-3j1x).
Table 1.
Summary of names of homologous proteins in different type three systems including the unified, Sct naming system
| Functional name | Sct name | Yersinia | Shigella | Salmonella SPI-1 | Flagellar homologue |
|---|---|---|---|---|---|
| Needle filament protein | SctF | YscF | MxiH | PrgI | – |
| Inner rod protein | SctI | YscI | MxiI | PrgJ | – |
| OM secretin ring | SctC | YscC | MxiD | InvG | – |
| IM outer ring | SctD | YscD | MxiG | PrgH | – |
| IM inner ring | SctJ | YscJ | MxiJ | PrgK | FliF |
| Minor export apparatus protein | SctR | YscR | Spa24 | SpaP | FliP |
| Minor export apparatus protein | SctS | YscS | Spa9 | SpaQ | FliQ |
| Minor export apparatus protein | SctT | YscT | Spa29 | SpaR | FliR |
| Export apparatus switch protein | SctU | YscU | Spa40 | SpaS | FlhB |
| Major export apparatus protein | SctV | YscV | MxiA | InvA | FlhA |
| Accessory cytosolic protein | SctK | YscK | MxiK | OrgA | FliG (?) |
| C-ring protein | SctQ | YscQ | Spa33 | SpaO | FliM + FliN |
| Stator (ATPase regulator) | SctL | YscL | MxiN | OrgB | FliH |
| ATPase | SctN | YscN | Spa47 | InvC | FliI |
| Stalk | SctO | YscO | Spa13 | InvI | FliJ |
| Needle length regulator | SctP | YscP | Spa32 | InvJ | FliK |
One area in which major strides have been taken in the last two years is in the study of the needles of the NF-T3SS, the major extracellular component formed by the helical assembly of multiple copies of a single protein (SctF) in an analogous manner to the assembly of the flagellar hook and filament [5]. However, the small size of SctF (∼9 kDa), combined with the natural propensity toward polymerization, has made high resolution structural studies challenging. Initial studies on the Shigella flexneri needle by negative stain EM demonstrated that it shared similar helical parameters (∼5.5 subunits/turn; 4.6 Å axial rise/subunit) to the flagellar hook/filament [6]. Subsequently, X-ray crystallography and NMR [7–13] showed that SctF consisted of a conserved two helix coiled-coil fold, similar to the filament building D0 domains of the flagellar components. However, fitting of an X-ray crystallographic structure of the monomer into a 16 Å EM map produced a model for the needle in which the N-terminal helix lined the channel [7], at odds with the orientation of the D0 helices of the flagellar filament model built in a 4–5 Å cryo-EM map. Further confusion was introduced with the publication of two independent studies of the Salmonella typhimurium needle [14]. The first, using cryo-EM analyses, suggested very different helical parameters (∼6.3 subunits/turn) to the S. flexneri needle and S. typhimurium flagellum. The second, using a combination of techniques, suggested that a double point mutant of S. typhimurium SctF underwent a structural rearrangement upon polymerization, whereby the C-terminal 25% of the protein went from α-helical to β-strand, again at odds with the polymerization of the flagellum [9].
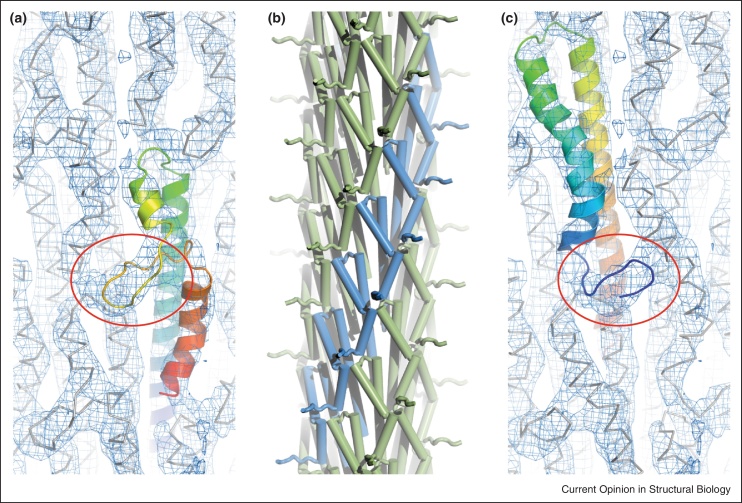
Several elegant high resolution studies have now gone a long way toward resolving these various inconsistencies. Early in 2012 a 7.7 Å cryo-EM map of the S. flexneri needle confirmed the flagellum-like parameters and demonstrated that 90% of the protein was α-helical [15•]. Interestingly a novel, non-helical protrusion was observed and based on this the authors proposed an alternative model for the S. flexneri needle in which residues 51-65 of SctF were remodeled from the α-helix seen in the high resolution structures to a β-hairpin structure (Figure 2a). Shortly after this, a new needle architecture was proposed for the S. typhimurium needle [16••]. Using a combination of solid state NMR and Rosetta modeling the authors produced a needle in which the C-terminal helix of SctF lines the needle channel, i.e., opposite to the previous models (Figure 2b). This orientation was validated by immuno-EM and was subsequently confirmed for the S. flexneri needle [17•]. Gratifyingly, the new model also fits well to the S. flexneri cryo-EM map, with the non-helical protrusion likely to be explained by the extended N-terminus (Figure 2c). These studies therefore return the NF-T3SS needle to being consistent with the flagellar filament, both in terms of helical parameters and orientation, with the highly conserved C-terminal helix lining the channel and forming the majority of the inter-subunit connections.
Figure 2.
Alternate models for the helical needle assembly. (a) The high resolution EM map (EMDB-5352) and C-terminal out subunit fitting for the Shigella flexneri needle (PDB-3j0r; [15•]). (b) A cartoon representation of the Salmonella enterica needle with N-terminal out subunit built using Rosetta by Loquet et al. (PDB-2lpz; [16••]). (c) Trivial remodeling of the flexible N-terminus of the Loquet et al. [16••] subunit position demonstrates a good fit of this independently derived needle model to the Fuji et al. EM map. The circled density in (a) and (c) highlights the alternate interpretations of this region by either an unwound piece of the C-terminal helix [15•] or the flexible N-terminus (this work).
As well as demonstrating the power of combining multiple high resolution structural techniques, this new needle model has implications for the structures of the periplasmic rod and distal tip assemblies. Structural information regarding the rod component SctI is scant, with NMR studies showing that the monomeric protein is mostly unfolded in solution with a 15 residue stretch of α-helix in the C-terminal half [18•]. However, this combined with the homology to the needle monomer in the C-terminal helix suggests a model for the rod being assembled in much the same way, with the more variable N-terminal half decorating the outside. At the other end of the needle, despite a plethora of high resolution structures of tip protein monomers, confusion reigns over the structure of the tip assembly. A number of models have been proposed based on low resolution EM projections/maps and crystal structures [10,19,20], but they suffer from a lack of consistency even at the most basic level as to whether they display the helical symmetry of the needle or pure rotational symmetry. This is clearly another area where higher resolution information is required.
More recently, evidence from both high and low resolution structural techniques has demonstrated an overall threefold symmetry for the NF-T3SS, with an either 12-fold [21] or 15-fold [22••] outer membrane (OM) secretin ring through a 24-fold inner membrane (IM) ring [22••] to the 9-fold of the major component of the EA [23••] which is functionally coupled to the hexameric ATPase [24,25,26••]. Despite three-dimensional reconstructions from negative stain and cryo-EM, together with top views of selective disassembled basal body rings of isolated S. typhimurium and Yersinia enterocolitica T3SS, the symmetry of the OM ring remains ambiguous with genuine inter-species differences proposed as one resolution of the conflicts between different symmetric arrangements of SctC [21,22••,27]. However, the IM ring, which was previously debated as being 12-fold, 20-fold or 24-fold symmetric [28–30] has been revealed to be two concentric rings, each made of 24 subunits (the integral bitopic SctD outside and the lipidated SctJ inside) (Figure 1b [22••]). Crosslinking and interface disruptive mutations also support some of the proposed symmetries and ring interactions [22••,31,32]. All three of the proteins that dominate the periplasmic regions are modular and contain one or more copies of two ‘ring forming motifs’ (α-β-β-α-β or β-α-β-β-α), through which the backbone of each ring is built [31,33]. More recently, the in situ tomographic reconstruction of the Y. enterocolitica T3SS, and the crystal structure of the equivalent SctD periplasmic domains [34•], seem to suggest that both the OM and IM components are capable of stretching vertically along the axis of the basal body. However the functional implications of such a mechanism are yet to be resolved. Understanding the molecular details of the symmetry mismatch in these assemblies and its effect on the formation and disruption of a web of non-covalent interactions among subunits is a crucial step to learn how the design of these nanomachines conjugates plasticity and integrity of the holostructure with its efficient assembly [35,36••].
Electron density maps also reveal that the SctD ring in the periplasmic region is less mobile than the ring formed by the SctD N-terminal cytosolic domain (SctD-N). The structure of this domain has been recently determined for several organisms [31,37,38•,39,40], revealing a forkhead-associated (FHA) fold with 4-stranded and 5-stranded β-sheets packing against each other in a globular structure that is conserved despite the low sequence identity amongst family members. However, all homologues lack the full-repertoire of highly conserved residues required for phospho-threonine binding, suggesting that any interactions with these FHA domains are occurring in a phosphorylation-independent manner. Although one study reported an interaction between S. flexneri SctD-N and phosphorylated peptides [39], other studies indeed indicate that these domains are unable to bind phosphorylated substrates and thus are likely to function as non-canonical FHA domains [31,37,38•,40].
Although a NF-T3SS C-ring has not yet been imaged, molecular evidence and sequence homology strongly suggest the existence of a structure analogous to the flagellar C-ring beneath the basal body [41]. This has been postulated to be involved in secretion substrate sorting through differential affinities for substrate–chaperone complexes [42]. As in the flagellar T3SS, the crucial role of the cytoplasmic IM ring could be to act as a scaffold in the formation of the putative NF-T3SS C-ring. Indeed, pull-down assays have indicated an interaction between S. flexneri SctD-N and the C-ring component SctQ [39,41], whilst deletion of either S. typhimurium SctD-N or SctQ results in the formation of basal bodies lacking the needle appendage [31,41], indicating that these proteins are acting at a similar point in T3SS assembly. The structural flexibility of the adjoining cytoplasmic SctD-N ring [22••] and observed dynamic nature of the flagellar C-ring [43] could explain why the NF-T3SS C-ring has thus far remained elusive in EM reconstructions.
SctQ, the essential component of this sorting platform, shows sequence homology to the two flagellar C-ring components: FliM, which is of an equivalent size, and FliN, which encompasses the C-terminal third of the protein sequence. Recently, it has been discovered that in the pathogenic NF-T3SS background an internal initiation site within the gene enables this class of proteins to be produced as two alternative translational forms [44•,45•], a full length and a shorter C-terminal variant. Although a chaperone role for the shorter variant has been proposed [45•], these proteins could also be directly equivalent to FliM and FliN respectively as integral structural components of the C-ring, suggesting that the molecular arrangement of the putative NF-C-ring could be more similar to the flagellar C-ring than previously anticipated. Indeed, structures of the C-terminal portion of NF-T3SS SctQ [44•,46] show an intertwined dimeric assembly highly similar to that already observed for FliN [47], where each protomer is made of 5 β-strands, with the first and the second strand undergoing domain swapping within the dimer. This homodimer binds to monomeric full-length SctQ to form a 1:2 complex that could be acting as the building block of the putative NF-C-ring, in contrast to the proposed 1:4 stoichiometry of the flagellar FliM-FliN building block [44•]. Furthermore, in the current model of the flagellar T3SS, the C-ring is connected to the basal body by FliG [3] and thus it is tempting to speculate that the proposed interaction between the SctD-N ring and C-ring is indirect. Given the requirement for the SctK family in C-ring localization [48], and its ability to interact with SctQ, assignment of a FliG-like role to these homologues could be hypothesized [49].
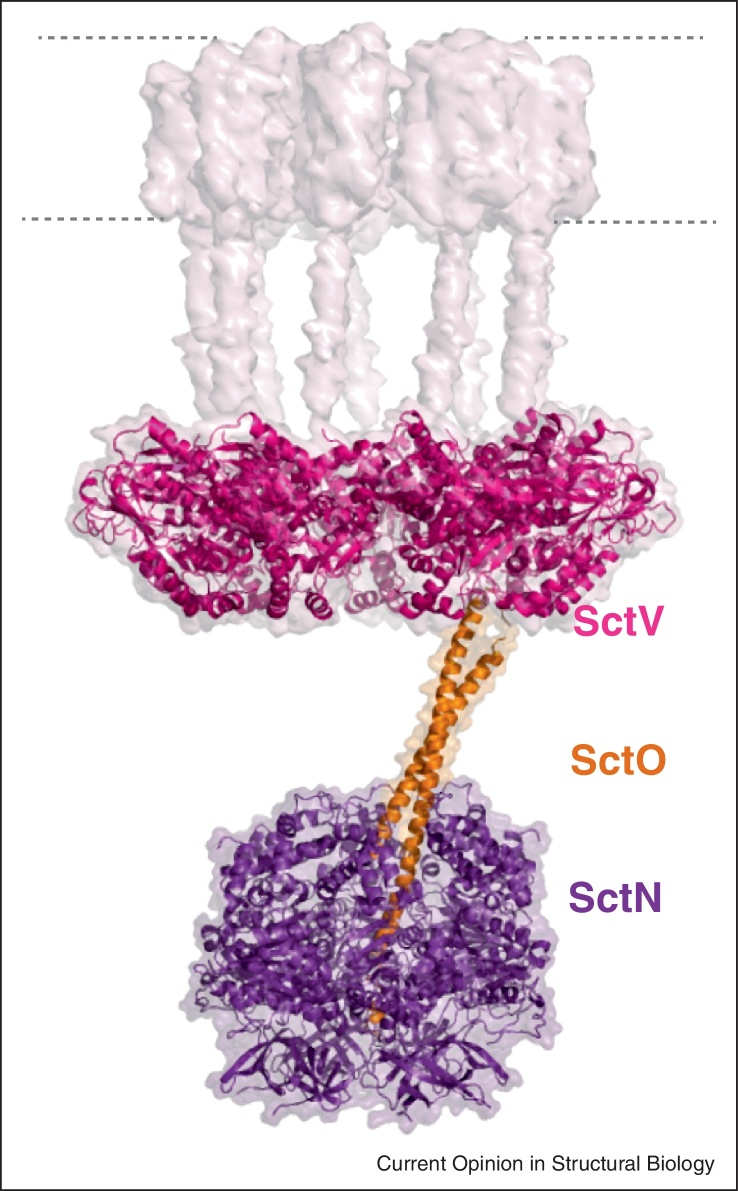
Finally, the combination of crystal structures with electron cryotomography (ECT) has significantly advanced our understanding of the architecture of the T3SS EA and ATPase complex [23••]. The EA is made up of 5 integral IM proteins, with two of them (SctV and SctU) having globular C-terminal domains (SctV-C and SctU-C) protruding into the EA cytosol. It has been proposed to act as a secretion gate where substrates are gathered and uncoupled from their cognate chaperone to be routed out to the secretion path. Structurally the cytoplasmic domain of SctU is well characterized across species [50] and it is proposed to be mainly involved in export substrate selection in conjunction with SctP/FliK, a supposed molecular ruler for which structural information is only just becoming available [51]. Insights into the rest of the EA have advanced in the last few years. The structure of SctV-C is highly conserved in flagellar and non-flagellar homologues [52–55] and recently the crystal structure of the S. flexneri SctV-C demonstrated a homo-nonameric ring assembly [23••]. Crucially, not only did ring assembly mutations disrupt secretion, but variants containing mutations on the inner surface of the ring considerably affected protein export, suggesting that the route through the SctV ring is the path toward secretion. Fitting this structure into a torus of density in ECT maps of flagellar motors [56••] has begun the process of defining the geometry of the export machinery, placing the SctV-C ring midway between the IM and the ATPase complex (Figure 3). Interestingly, the presence of the SctV-C ring is independent of the ATPase and vice-versa. The ATPase complex is made of three soluble components, SctN, SctO and SctL, and structures of SctN/FliI [24,57] and SctO/FliJ [26••,58] reveal homology with the α/β and γ subunits of the F0F1-ATP synthase, respectively. Although previously characterized as a general chaperone, it has now been shown that SctO/FliJ has a coiled-coil structure that inserts into the central cavity of the ATPase hexamer, in a manner directly analogous to the ATP synthase, and that it is capable of rotation within a chimeric V1V0 A3B3J assembly [25]. In addition, the observed correlation between FliH length and ATPase/C-ring distance in flagellar motors [23••] combined with fluorescence data suggests that the ATPase complex is held underneath the SctV-C ring by SctL/FliH, which at its opposite end interacts with the hydrophobic patch at the dyad axis of the FliN dimer of the putative C-ring [59]. However, despite the recent progress in decoding the biophysical properties of its major components, the molecular architecture of the T3SS EA and C-ring clearly require further structural characterization in order to address mechanistic issues, such as the role of the protonmotive force [60], and localization of the crucial, low copy number components.
Figure 3.
Geometry of the export machinery. The export machinery as built by Abrusci et al. [23••]. The SctV nonameric cage is represented as surface in light pink with fitted atomic model of the Shigella flexneri SctV-C (PDB-4a5p), in magenta. ATPase and its stalk are modeled using the atomic model form the Escherichia coli SctN (PDB-2obm) and the flagellar homologue of SctO form Salmonella typhimurium (PDB-3ajw) and colored in purple and orange, respectively.
In summary, cumulative work over the last few years has highlighted the architectural similarities between the flagellar-T3SS and NF-T3SS. Intriguingly, especially in light of the ATPase complex structures, a recent study has suggested a proton-motive force dependent rotation of the NF-T3SS needle filament, thereby drawing further parallels to the flagellum [61••] and the latest work proposing a general mechanism for flagellar export [62••] may therefore also provide insight into the NF-T3SS. Future research concerning the molecular mechanisms of assembly and secretion by these nanomachines will undoubtedly provide further insight into the extent of the diversification of T3SSs.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
PA and MAM are funded by grant 083599/Z/07/Z from the Wellcome Trust; SJ is funded by grant G0900888 from the UK Medical Research Council, both granted to SML.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1.Abby S.S., Rocha E.P. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erhardt M., Namba K., Hughes K.T. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hueck C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blocker A.J., Deane J.E., Veenendaal A.K., Roversi P., Hodgkinson J.L., Johnson S., Lea S.M. What's the point of the type III secretion system needle? Proc Natl Acad Sci U S A. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordes F.S., Komoriya K., Larquet E., Yang S., Egelman E.H., Blocker A., Lea S.M. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 7.Deane J.E., Roversi P., Cordes F.S., Johnson S., Kenjale R., Daniell S., Booy F., Picking W.D., Picking W.L., Blocker A.J. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc Natl Acad Sci U S A. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Wang Y., Picking W.L., Picking W.D., De Guzman R.N. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Poyraz O., Schmidt H., Seidel K., Delissen F., Ader C., Tenenboim H., Goosmann C., Laube B., Thunemann A.F., Zychlinsky A. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol. 2010;17:788–792. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- 10.Lunelli M., Hurwitz R., Lambers J., Kolbe M. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Ouellette A.N., Egan C.W., Rathinavelan T., Im W., De Guzman R.N. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun P., Tropea J.E., Austin B.P., Cherry S., Waugh D.S. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinaud M., Ple S., Job V., Contreras-Martel C., Simorre J.P., Attree I., Dessen A. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci U S A. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galkin V.E., Schmied W.H., Schraidt O., Marlovits T.C., Egelman E.H. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J Mol Biol. 2010;396:1392–1397. doi: 10.1016/j.jmb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Fujii T., Cheung M., Blanco A., Kato T., Blocker A.J., Namba K. Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms. Proc Natl Acad Sci U S A. 2012;109:4461–4466. doi: 10.1073/pnas.1116126109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports the highest resolution EM reconstruction of a NF-T3SS needle.
- 16••.Loquet A., Sgourakis N.G., Gupta R., Giller K., Riedel D., Goosmann C., Griesinger C., Kolbe M., Baker D., Becker S. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uses in silico modelling to generate a model for the NF-T3SS needle topologically consistent with the flagellar models.
- 17•.Demers J.P., Sgourakis N.G., Gupta R., Loquet A., Giller K., Riedel D., Laube B., Kolbe M., Baker D., Becker S. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog. 2013;9:e1003245. doi: 10.1371/journal.ppat.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extends the model from [16••] to other species.
- 18•.Zhong D., Lefebre M., Kaur K., McDowell M.A., Gdowski C., Jo S., Wang Y., Benedict S.H., Lea S.M., Galan J.E. The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J Biol Chem. 2012;287:25303–25311. doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structural studies of rod component and first evidence of inherent disorder in T3SS components.
- 19.Mueller C.A., Broz P., Cornelis G.R. The type III secretion system tip complex and translocon. Mol Microbiol. 2008;68:1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- 20.Epler C.R., Dickenson N.E., Bullitt E., Picking W.L. Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol. 2012;420:29–39. doi: 10.1016/j.jmb.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowal J., Chami M., Ringler P., Muller S.A., Kudryashev M., Castano-Diez D., Amstutz M., Cornelis G.R., Stahlberg H., Engel A. Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure. 2013;21:2152–2161. doi: 10.1016/j.str.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 22••.Schraidt O., Marlovits T.C. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]; Highest resolution imaging of the entire NF-T3SS assembly.
- 23••.Abrusci P., Vergara-Irigaray M., Johnson S., Beeby M.D., Hendrixson D.R., Roversi P., Friede M.E., Deane J.E., Jensen G.J., Tang C.M. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence for the assembly and location of the SctV homononameric ring.
- 24.Zarivach R., Vuckovic M., Deng W., Finlay B.B., Strynadka N.C. Structural analysis of a prototypical ATPase from the type III secretion system. Nat Struct Mol Biol. 2007;14:131–137. doi: 10.1038/nsmb1196. [DOI] [PubMed] [Google Scholar]
- 25.Kishikawa J., Ibuki T., Nakamura S., Nakanishi A., Minamino T., Miyata T., Namba K., Konno H., Ueno H., Imada K. Common evolutionary origin for the rotor domain of rotary ATPases and flagellar protein export apparatus. PLoS ONE. 2013;8:e64695. doi: 10.1371/journal.pone.0064695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Ibuki T., Imada K., Minamino T., Kato T., Miyata T., Namba K. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol. 2011;18:277–282. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]; Identification of the gamma subunit of the T3SS ATPase.
- 27.Bayan N., Guilvout I., Pugsley A.P. Secretins take shape. Mol Microbiol. 2006;60:1–4. doi: 10.1111/j.1365-2958.2006.05084.x. [DOI] [PubMed] [Google Scholar]
- 28.Hodgkinson J.L., Horsley A., Stabat D., Simon M., Johnson S., da Fonseca P.C., Morris E.P., Wall J.S., Lea S.M., Blocker A.J. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16:477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlovits T.C., Kubori T., Sukhan A., Thomas D.R., Galan J.E., Unger V.M. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yip C.K., Kimbrough T.G., Felise H.B., Vuckovic M., Thomas N.A., Pfuetzner R.A., Frey E.A., Finlay B.B., Miller S.I., Strynadka N.C. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- 31.Bergeron J.R., Worrall L.J., Sgourakis N.G., DiMaio F., Pfuetzner R.A., Felise H.B., Vuckovic M., Yu A.C., Miller S.I., Baker D. A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog. 2013;9:e1003307. doi: 10.1371/journal.ppat.1003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross J.A., Plano G.V. A C-terminal region of Yersinia pestis YscD binds the outer membrane secretin YscC. J Bacteriol. 2011;193:2276–2289. doi: 10.1128/JB.01137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spreter T., Yip C.K., Sanowar S., Andre I., Kimbrough T.G., Vuckovic M., Pfuetzner R.A., Deng W., Yu A.C., Finlay B.B. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol. 2009;16:468–476. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Kudryashev M., Stenta M., Schmelz S., Amstutz M., Wiesand U., Castano-Diez D., Degiacomi M.T., Munnich S., Bleck C.K., Kowal J. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2013;2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]; First tomograms of the NF-T3SS.
- 35.Loquet A., Habenstein B., Lange A. Structural investigations of molecular machines by solid-state NMR. Acc Chem Res. 2013;46:2070–2079. doi: 10.1021/ar300320p. [DOI] [PubMed] [Google Scholar]
- 36••.Deeds E.J., Bachman J.A., Fontana W. Optimizing ring assembly reveals the strength of weak interactions. Proc Natl Acad Sci U S A. 2012;109:2348–2353. doi: 10.1073/pnas.1113095109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mathematical modelling demonstrates that weak interactions are key to correct assembly of highly symmetric homooligomers.
- 37.Lountos G.T., Tropea J.E., Waugh D.S. Structure of the cytoplasmic domain of Yersinia pestis YscD, an essential component of the type III secretion system. Acta Crystallogr D Biol Crystallogr. 2012;68:201–209. doi: 10.1107/S0907444911054308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.McDowell M.A., Johnson S., Deane J.E., Cheung M., Roehrich A.D., Blocker A.J., McDonnell J.M., Lea S.M. Structural and functional studies on the N-terminal domain of the Shigella type III secretion protein MxiG. J Biol Chem. 2011;286:30606–30614. doi: 10.1074/jbc.M111.243865. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structure of the cytoplasmic domain of the major component of the inner membrane ring.
- 39.Barison N., Lambers J., Hurwitz R., Kolbe M. Interaction of MxiG with the cytosolic complex of the type III secretion system controls Shigella virulence. FASEB J. 2012;26:1717–1726. doi: 10.1096/fj.11-197160. [DOI] [PubMed] [Google Scholar]
- 40.Gamez A., Mukerjea R., Alayyoubi M., Ghassemian M., Ghosh P. Structure and interactions of the cytoplasmic domain of the Yersinia type III secretion protein YscD. J Bacteriol. 2012;194:5949–5958. doi: 10.1128/JB.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morita-Ishihara T., Ogawa M., Sagara H., Yoshida M., Katayama E., Sasakawa C. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281:599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 42.Lara-Tejero M., Kato J., Wagner S., Liu X., Galan J.E. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown M.T., Delalez N.J., Armitage J.P. Protein dynamics and mechanisms controlling the rotational behaviour of the bacterial flagellar motor. Curr Opin Microbiol. 2011;14:734–740. doi: 10.1016/j.mib.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 44•.Bzymek K.P., Hamaoka B.Y., Ghosh P. Two translation products of Yersinia YscQ assemble to form a complex essential to type III secretion. Biochemistry. 2012;51:1669–1677. doi: 10.1021/bi201792p. [DOI] [PMC free article] [PubMed] [Google Scholar]; With [45•], this study demonstrates that the NF-T3SS use alternate translation initiation to generate proteins homologous to FliM and FliN. This study also suggests a novel stoichiometry for the association of these homologues within the NF-T3SS.
- 45•.Yu X.J., Liu M., Matthews S., Holden D.W. Tandem translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J Biol Chem. 2011;286:36098–36107. doi: 10.1074/jbc.M111.278663. [DOI] [PMC free article] [PubMed] [Google Scholar]; With [44•], this study demonstrates that the NF-T3SS use alternate translation initiation to generate proteins homologous to FliM and FliN.
- 46.Fadouloglou V.E., Tampakaki A.P., Glykos N.M., Bastaki M.N., Hadden J.M., Phillips S.E., Panopoulos N.J., Kokkinidis M. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc Natl Acad Sci U S A. 2004;101:70–75. doi: 10.1073/pnas.0304579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown P.N., Mathews M.A., Joss L.A., Hill C.P., Blair D.F. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., Cornelis G.R. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson S., Blocker A. Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol Lett. 2008;286:274–278. doi: 10.1111/j.1574-6968.2008.01284.x. [DOI] [PubMed] [Google Scholar]
- 50.Deane J.E., Abrusci P., Johnson S., Lea S.M. Timing is everything: the regulation of type III secretion. Cell Mol Life Sci. 2010;67:1065–1075. doi: 10.1007/s00018-009-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizuno S., Amida H., Kobayashi N., Aizawa S., Tate S. The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus. J Mol Biol. 2011;409:558–573. doi: 10.1016/j.jmb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Worrall L.J., Vuckovic M., Strynadka N.C. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 2010;19:1091–1096. doi: 10.1002/pro.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saijo-Hamano Y., Imada K., Minamino T., Kihara M., Shimada M., Kitao A., Namba K. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol Microbiol. 2010;76:260–268. doi: 10.1111/j.1365-2958.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 54.Moore S.A., Jia Y. Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J Biol Chem. 2010;285:21060–21069. doi: 10.1074/jbc.M110.119412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bange G., Kummerer N., Engel C., Bozkurt G., Wild K., Sinning I. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci U S A. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Chen S., Beeby M., Murphy G.E., Leadbetter J.R., Hendrixson D.R., Briegel A., Li Z., Shi J., Tocheva E.I., Muller A. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]; Survey using tomography to highlight conserved and varying structures making up the flagellar T3SS.
- 57.Imada K., Minamino T., Tahara A., Namba K. Structural similarity between the flagellar T3SS ATPase FliI and F1-ATPase subunits. Proc Natl Acad Sci U S A. 2007;104:485–490. doi: 10.1073/pnas.0608090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenzini E., Singer A., Singh B., Lam R., Skarina T., Chirgadze N.Y., Savchenko A., Gupta R.S. Structure and protein–protein interaction studies on Chlamydia trachomatis protein CT670 (YscO homolog) J Bacteriol. 2010;192:2746–2756. doi: 10.1128/JB.01479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul K., Harmon J.G., Blair D.F. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J Bacteriol. 2006;188:5240–5248. doi: 10.1128/JB.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minamino T., Morimoto Y.V., Hara N., Namba K. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun. 2011;2:475. doi: 10.1038/ncomms1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Ohgita T., Hayashi N., Hama S., Tsuchiya H., Gotoh N., Kogure K. A novel effector secretion mechanism based on proton-motive force-dependent T3SS apparatus rotation. FASEB J. 2013;27:2862–2872. doi: 10.1096/fj.13-229054. [DOI] [PubMed] [Google Scholar]; This paper proposes that rotation of the NF-T3SS needle is required for secretion.
- 62••.Evans L.D.B., Poulter S., Terentjev E.M., Hughes C., Fraser G.M. A chain mechanism for flagellum growth. Nature. 2013;504:287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]; Proposes that a chaining mechanism underlies secretion through T3SS.