Abstract
Reovirus attachment to cells is mediated by the binding of viral attachment protein σ1 to junctional adhesion molecule 1 (JAM1). The crystal structure of the extracellular region of human JAM1 (hJAM1) reveals two concatenated Ig-type domains with a pronounced bend at the domain interface. Two hJAM1 molecules form a dimer that is stabilized by extensive ionic and hydrophobic contacts between the N-terminal domains. This dimeric arrangement is similar to that observed previously in the murine homolog of JAM1, indicating physiologic relevance. However, differences in the dimeric structures of hJAM1 and murine JAM1 suggest that the interface is dynamic, perhaps as a result of its ionic nature. We demonstrate that hJAM1, but not the related proteins hJAM2 and hJAM3, serves as a reovirus receptor, which provides insight into sites in hJAM1 that likely interact with σ1. In addition, we present evidence that the previously reported structural homology between σ1 and the adenovirus attachment protein, fiber, also extends to their respective receptors, which form similar dimeric structures. Because both receptors are located at regions of cell–cell contact, this similarity suggests that reovirus and adenovirus use conserved mechanisms of entry and pathways of infection.
Attachment to specific host molecules is the initial step in viral infection and plays a key role in target-cell selection in the infected host. In the case of mammalian reoviruses, this is accomplished by interactions between the viral attachment protein, σ1, and junctional adhesion molecule 1 (JAM1) (1). Reoviruses are nonenveloped, icosahedral viruses that infect most mammalian species, including humans. The capacity of reovirus strains to engage distinct types of receptors in the host strongly influences tissue tropism and disease after reovirus infection (2). JAM1 is a member of the Ig superfamily with two extracellular Ig-like domains, a single transmembrane region, and a short cytosolic tail. The crystal structure of the extracellular region of murine JAM1 (mJAM1) revealed a dimer stabilized by interactions involving the membrane-distal Ig-like domain (3). The protein is expressed in a variety of tissues and cell types, including circulating platelets and lymphocytes, and it has been postulated to function as a regulator of endothelial and epithelial tight junction formation (4–6). The human homolog of JAM1 (hJAM1) serves as a ligand for the integrin αLβ2 (LFA-1) (7), an interaction that likely plays an important role in host inflammatory responses by mediating transmigration of leukocytes. Two hJAM1 homologs, hJAM2 and hJAM3, have been recently identified (8–13). Additional evidence that JAM family members are involved in the inflammatory response is provided by the observations that hJAM2 interacts with α4β1 integrins (14), and hJAM3 interacts with αMβ2 (MAC-1) integrins (15).
Reovirus attachment protein σ1 is a long, fibrous molecule with head-and-tail morphology and several defined regions of flexibility within its tail (16, 17). The σ1 tail inserts into the 12 vertices of the icosahedral virion, whereas the JAM-binding σ1 head extends away from the particle surface (18, 19). Laboratory-adapted and field-isolate strains of all three reovirus serotypes bind JAM1 (ref. 1; J.A.C. and T.S.D., unpublished observations). Some reoviruses also use additional carbohydrate-based coreceptors for cell attachment (20, 21). For type 3 reoviruses, this coreceptor is α-linked sialic acid (22), and its binding site has been mapped to a region close to the midpoint of the σ1 tail (20, 23). The finding that reoviruses bind to different receptors by using distinct domains within the σ1 protein has led to the suggestion that reoviruses use a multiple-step adhesion-strengthening mechanism to engage the cell surface (21). In this scenario, reovirus binding to carbohydrate facilitates viral attachment through low-affinity adhesion. This interaction places the virus on the cell surface where access to the higher affinity, but lower abundance, JAM1 protein is thermodynamically favored.
The recently determined crystal structure of a JAM1-binding fragment of σ1 revealed numerous structural and functional similarities to the adenovirus attachment protein, fiber, suggesting an evolutionary link in the receptor-binding strategies of reoviruses and adenoviruses (17). Most adenovirus serotypes initiate infection by binding to the coxsackievirus and adenovirus receptor (CAR) (24). The crystal structure of the adenovirus fiber knob in complex with CAR is known (25). Although no structural information is currently available for a reovirus-receptor complex, analysis of the crystallized σ1 fragment revealed a region that is likely involved in the interaction with JAM1 (17). This putative JAM1-binding site forms a recessed groove at the lower edge of the σ1 head that contains many of the residues conserved in prototype strains of the three reovirus serotypes. The location of this site suggests that each σ1 monomer can independently interact with a JAM1 molecule.
To enhance an understanding of how reovirus mediates cell tropism and initiates organ-specific disease, we determined the crystal structure of the hJAM1 ectodomain. Analysis of this structure allows us to identify regions of the receptor that are most likely involved in the binding of σ1. Moreover, comparison of the structures of hJAM1 and mJAM1 reveals differences in the dimeric arrangements of the molecules despite absolute conservation of residues at the interface, suggesting that the JAM1 dimer is dynamic and can undergo rearrangement and perhaps dissociation. Finally, we show that the structure of the JAM1 dimer closely resembles that of the CAR dimer, mirroring the close resemblance of the reovirus and adenovirus attachment proteins and suggesting that the similarities extend beyond conservation of structure toward conserved strategies of attachment and entry.
Methods
Protein Expression, Purification, and Crystallization.
A cDNA corresponding to the extracellular region of hJAM1 (residues 27–233) was cloned into the pGEX-4T-3 expression vector (Amersham Pharmacia), which encodes N-terminal GST followed by a thrombin cleavage site. GST–hJAM1 fusion protein was expressed in Escherichia coli and purified by affinity chromatography using glutathione beads. hJAM1 was released from the beads by thrombin cleavage and further purified by anion-exchange chromatography. The cleaved protein contains three additional non-native amino acids (Gly-24, Ser-25, and Met-26) at the N terminus. A final gel filtration step resulted in a homogenous peak that corresponded to a dimer of 48 kDa. Higher-order oligomers were not observed. Crystals were obtained by using 8 mg/ml protein and 16% PEG 6K, 18% isopropanol, 0.1 M sodium citrate as precipitant. The final pH of the mixture was 6.0.
Structure Determination.
The crystals belong to space group C2 (a = 116.8 Å, b = 61.8 Å, c = 82.9 Å, β = 120.01°) and contain two molecules in their asymmetric unit. Before data collection, crystals were cryoprotected with 15% glycerol and then flash-frozen in liquid nitrogen. Diffraction data were collected at NSLS beamline X25 and processed with hkl (26). The structure was determined by molecular replacement using the structure of mJAM1 (3). Rotation and translation searches were performed separately with the N- and C-terminal domains of mJAM1 in amore (27), which yielded two clear solutions for each domain. The free R factor (28) for the combined solutions was 45.1% (8–3.5 Å) after rigid body refinement. Alternating rounds of model building in o (29) and refinement in x-plor (30) produced a model with good refinement statistics (Table 1). Bulk solvent correction and noncrystallographic symmetry constraints were used throughout the refinement. The final model contains residues 25–233 of both chains and 124 water molecules. procheck (27) analysis shows no residues in disallowed regions in the Ramachandran plot.
Table 1.
Data collection and refinement statistics
| Data set | Native |
|---|---|
| Diffraction data* | |
| Resolution range, Å | 30–2.9 |
| Completeness, % | 85.1 (51.0) |
| Total reflections | 27,850 |
| Unique reflections | 9,736 |
| Rmerge,† % | 11.4 (13.5) |
| I/σ | 8.8 (3.7) |
| Refinement statistics | |
| Rcryst, %; work set‡ | 22.0 (no I/σI cutoff) |
| Rcryst, %; free set‡ | 30.5 (no I/σI cutoff) |
| rms deviation bond lengths, Å | 0.02 |
| rms deviation bond angles, ° | 2.7 |
| Number of waters | 124 |
Data sets were collected at 100 K and a wavelength of 1.1 Å. Values in parentheses refer to the outermost resolution shell.
Rmerge = ∑hkl|I − 〈I〉|/∑hklI, where I is the intensity of a reflection hkl, and 〈I〉 is the average over symmetry-related observations of hkl.
Rcryst = ∑hkl|Fobs − Fcalc|/∑hklFobs, where Fobs and Fcalc are observed and calculated structure factors, respectively. Free set (28) contains 10% of the data.
Infectivity Assay.
Chinese hamster ovary (CHO) cells (60–80% confluence) were transfected with 0.4 μg of plasmid encoding hJAM1, hJAM2, hJAM3, or pEGFP-N1 (transfection control) by using Lipofectamine Plus (GIBCO/BRL). Surface expression was confirmed by flow cytometry using antibodies specific for hJAM1 (31), hJAM2 (M.A.L. and B.A.I., unpublished observations), or hJAM3 (32). Transfected cells were infected with reovirus type 1 Lang (T1L) at a multiplicity of infection of 1 fluorescent focus unit per cell in a total volume of 150 μl. Adsorptions were terminated after incubation at room temperature for 1 h by washing with PBS. Infected cells were identified 18 h after adsorption by indirect immunofluorescence using rabbit antireovirus sera (1).
Results and Discussion
Overall Structure.
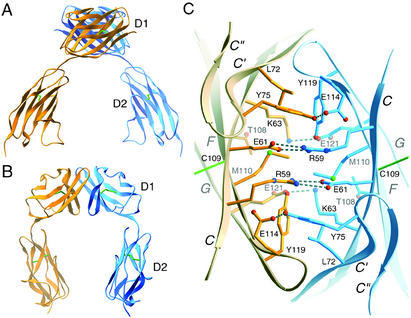
The polypeptide chain of the extracellular region of hJAM1 folds into two concatenated Ig-like domains, termed D1 and D2 (Fig. 1 A and B). The N-terminal D1 domain contains two antiparallel β-sheets (strands ABED and GFCC′C"), which classifies it as an Ig-like domain of the variable type (V-set). Although the fold of D2 is very similar to that of D1, this domain has a much shorter C′ strand, and it lacks the C′′ strand. These differences indicate that D2 is an intermediate type (I-set) Ig-like domain (33) and not a V-set Ig-like domain, as reported for mJAM1 D2 (3). Analysis with dali (34) shows that the prototypical I-set domain, telokin (33), is among the closest structural homologs of hJAM1 D2. Both D1 and D2 of hJAM1 have the classical disulfide bond between β-strands B and F. The hJAM1 structure exhibits a pronounced elbow angle of ≈125° between the two domains. As expected, the overall structure of each hJAM1 D1D2 monomer resembles that of mJAM1 D1D2 (3), although there are conformational differences between the two proteins in surface loops and at the interdomain interface. Our structure also allows us to trace a long loop between β-strands C′ and D (C′D loop) in D2 that was disordered in the model for mJAM1 D1D2.
Fig 1.
Structure of hJAM1 D1D2. (A and B) Ribbon drawings of the hJAM1 dimer, with one monomer shown in orange and the other in blue. Two orthogonal views are displayed. Disulfide bonds are shown in green. (C) View of the interface between two hJAM1 monomers. The interface is formed by residues on the GFCC′ faces of two D1 domains. The view is along a crystallographic dyad. Hydrogen bonds and salt bridges are represented by broken cylinders. Amino acids are labeled in single-letter code.
The asymmetric unit of the hJAM1 crystals contains two independent but virtually identical chains (termed A and B). Each chain assembles into a U-shaped homodimer with crystallographic 2-fold symmetry. The dimer interface features an extensive contact between the D1 domains and is reminiscent of an arm-wrestling grip, with the GFCC′ faces of the two N-terminal domains interlocking at an angle of ≈90° (Fig. 1 A and B). Two crystallographically independent dimers are observed. One is formed by chain A and its symmetry mate A, and the other is formed by chains B and B. Two observations suggest that the hJAM1 dimer is physiologically relevant: (i) the purified protein elutes at the expected molecular mass for a dimer (48 kDa) by size-exclusion chromatography and (ii) a similar dimeric structure was seen in the crystals of mJAM1 (3). With the exception of the dimeric interaction, the arrangements of the molecules in the hJAM1 and mJAM1 crystals are not related.
Structure of the Dimer.
Residues involved in hJAM1 homodimer formation are exclusively located on the concave GFCC′ face of D1. The dimer interface includes four buried salt bridges (Arg-59–Glu-61, Glu-61–Arg-59, Lys-63–Glu-121, and Glu-121–Lys-63) as well as several hydrophobic contacts (Leu-72–Tyr-119, Met-110–Met-110, and Tyr-119–Leu-72) (Fig. 1C). Hydrogen bonds exist between the Tyr-75 and Glu-114 side chains.
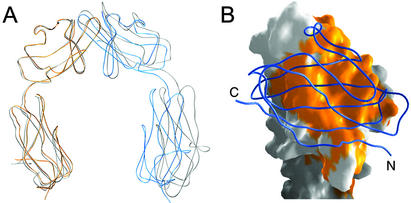
Although the GFCC′ face of D1 also mediates homodimer formation in the mJAM1 D1D2 structure (3), the relative arrangements of the D1 domains in the two dimers are noticeably different (Fig. 2A). The mJAM1 interdomain interface contains only two salt bridges and fewer additional contacts (3). The Lys-63–Glu-121 and Met-110–Met-110 interactions at the center of the hJAM1 interface are absent in mJAM1, as are the hydrogen bonds involving Tyr-75, Glu-114, and their symmetry-related counterparts. These differences result in a smaller interface in mJAM1 (Fig. 2A). Using the program surface (27) and a standard probe radius of 1.4 Å, we calculate buried surface areas of 1,380 and 1,200 Å2 for the hJAM1 and mJAM1 dimers, respectively.
Fig 2.
Comparison of dimeric arrangements in hJAM1 and mJAM1 (3). (A) Superposition of dimeric structures of hJAM1 D1D2 (colored as in Fig. 1) and mJAM1 D1D2 (gray). The superposition is based on D1 residues of one monomer only (the orange monomer of hJAM1), which yielded a rms deviation of 0.55 Å for 2 × 90 Cα atoms. The matrix derived from this superposition was applied to the entire dimer. (B) Conservation of residues at the mJAM1 and hJAM1 D1 domain dimer interfaces. One hJAM1 monomer is shown in surface representation, and the other is shown as a blue ribbon. Residues that are strictly conserved in mJAM1 are shown in orange and cover the entire dimer interface.
The observed differences in the hJAM1 and mJAM1 dimers are noteworthy given the near absolute conservation of residues at the dimer interfaces (Fig. 2B). The GFCC′ faces of hJAM1 and mJAM1 (residues 58–75 and 105–122 of hJAM1) can be superimposed onto each other with an rms deviation of 0.4 Å for the 36 Cα atoms. There are only six substitutions among these 36 aa, and none of the substituted residues engage in dimer contacts (distances >4 Å). Thus, differences in the arrangement of the hJAM1 and mJAM1 dimer interfaces cannot be explained by altered contacts mediated by substituted residues. Instead, these differences are likely caused by crystal packing forces involving other regions of the molecules, and they indicate that small movements of one monomer with respect to the other can occur.
The dynamic nature of the JAM1 dimer interface may result in part from its dependence on ionic interactions, which is unusual for protein–protein interfaces. Low pH and moderately high ionic strength lead to dissociation of the mJAM1 dimer (35), indicating that ionic interactions represent the principal means of association. We note that both structures were obtained by using conditions of low ionic strength and at pH values at which acidic side chains are expected to be deprotonated. Thus, salt bridges are expected to exist in both cases. However, the free energy contribution of salt bridges in an aqueous environment is small (36). The JAM1 dimer interface, which is stabilized primarily by salt bridges and contains several solvent molecules, is therefore especially likely to undergo rearrangement or dissociation.
Implications for the Structures of hJAM2 and hJAM3.
The conservation of key features of the dimer interface in hJAM1 and mJAM1 strongly suggests that this interface is responsible for the dimeric structure of the protein in solution, as suggested (3). Thus, it offers a framework for predicting the oligomeric state of other JAM family members. Sequence alignments show that most of the residues mediating dimer formation are conserved in the two additional hJAM family members, hJAM2 and hJAM3, which has led to the suggestion that these two molecules form similar dimeric structures (3). Analysis of conserved residues at the dimer interface, however, shows that several key residues are not conserved in hJAM2 or hJAM3. Met-110, which is located at the center of the hJAM1 interface (Fig. 1C) and conserved in mJAM1, is replaced with a glutamic acid in both hJAM2 and hJAM3. The nearby residue Thr-108 (Fig. 1C), which also is conserved in hJAM1 and mJAM1, is replaced with an arginine in hJAM2 and hJAM3. The presence of two additional charged residues is almost certain to impart different characteristics to the structure and stability of hJAM2 and hJAM3 dimers. hJAM2 and hJAM3 have been shown to engage in heterophilic interactions, with hJAM2 serving as a counterreceptor for hJAM3 (12, 13). The precise distribution of residues at the respective GFCC′ faces and secondary interactions may favor heterodimeric contacts rather than homodimeric interactions between these proteins. It is also possible that formation of an hJAM2–hJAM3 dimer uses molecular regions other than those described, although the D1 domain of hJAM2 is clearly involved in facilitating this arrangement (14).
The hJAM1 residues involved in salt bridge formation (Arg-59, Glu-61, Lys-63, Glu-121) and hydrophobic contacts (Leu-72, Tyr-119) are highly conserved not only in the JAM1 sequences of other mammals but also in those of nonmammalian vertebrates such as zebrafish and the African clawed frog (data not shown). This observation suggests that the dimeric structure of JAM1 is present throughout vertebrates. We note that some nonmammalian sequences have features that render them more similar to JAM2 or JAM3 (e.g., substitution of Met-110 with Glu), suggesting that versions of all three JAM variants exist in vertebrates.
Implications for Homophilic Interaction at Tight Junctions.
Analysis of the crystal-packing arrangement of the mJAM1 molecules has led to a model of JAM1 interactions at tight junctions (3). In this model, a JAM1 dimer located at the surface of one cell engages a dimer on the opposing cell via contacts in D1, producing an extensive network of contacts between dimers. Although our crystals contain two crystallographically nonequivalent copies of hJAM1, we do not observe a similar contact involving either of these molecules in our crystals, and thus our structural analysis does not provide supporting evidence for the model presented by Kostrewa et al. (3). In the hJAM1 crystals, two alternative contacts between hJAM1 dimers that could form the basis for interactions in tight junctions exist. However, neither of these contacts is observed in the mJAM1 crystal lattice. One interpretation of the available crystallographic data is that contacts between JAM1 dimers in tight junctions involve low-affinity interactions that depend on the presence of additional proteins, and these interactions cannot be easily reproduced by using conditions that promote crystallization. Another interpretation is that the JAM1 dimer itself, which is observed in the crystals of hJAM1 and mJAM1, represents a physiologic contact present in tight junctions. In this interpretation, JAM1 monomers would engage JAM1 monomers on apposing cells to help mediate homophilic cell–cell interactions. The dimensions for such a model suggest that it deserves consideration, because it would lead to a separation of cells of ≈85 Å, similar to the observed distances at tight junctions of ≈100 Å. CAR is also thought to mediate homophilic interactions between cells (37–39), and the homodimeric structure of CAR has been interpreted to depict an interaction between CAR monomers from apposing cells (40).
Similarities to CAR.
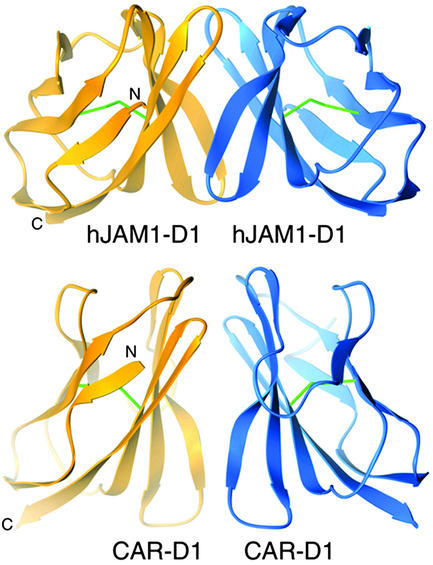
The attachment proteins of adenovirus and reovirus share structural and functional properties, which has led to speculation that they have a common ancestor (17). Remarkably, the receptors for both viruses, CAR and JAM1, respectively, also share key structural properties. CAR forms a homodimer (40) that is structurally similar to the hJAM1 homodimer and also is formed by interactions between the GFCC′ β-sheets of the N-terminal D1 domains (Fig. 3). Although the contacts between CAR monomers are somewhat more hydrophobic and do not directly involve salt bridges (40), several charged residues are present at the CAR–CAR interface (Glu-56, Asp-68, Lys-121). It is possible that these side chains interact via water molecules. Moreover, the relative arrangement of the two CAR monomers is highly similar to that observed for the two hJAM1 monomers (Fig. 3). Both contacts involve concave GFCC′ β-sheets that face each other at an angle of ≈90° and bury an almost identical amount of solvent [1,300 Å2 for the CAR–CAR dimer (40) and 1,380 Å2 for hJAM1–hJAM1].
Fig 3.
Dimeric structures of virus receptors hJAM1 and CAR. The D1 domains of hJAM1 (Upper) and CAR (Lower) engage in a conserved mode of dimerization based on interactions between the concave GFCC′ β-sheets. Disulfide bonds are in green.
Homodimeric structures have been observed in a number of other Ig superfamily proteins (41–45), but only one of these proteins, CD2, forms dimers via the GFCC′ β-sheet of D1 (45, 46). However, it is noteworthy that several Ig superfamily receptors engage viral ligands via the GFCC′ β-sheet. The HIV glycoprotein gp120 binds to residues on the GFCC′ face of its receptor CD4 (47), and the same region of CD4 also interacts with its natural ligand MHC class II (48). Complexes of rhinoviruses and coxsackievirus A21 with their receptor intercellular adhesion molecule-1 (ICAM-1) also show that the interactions involve residues at the top of ICAM-1 D1, a region that includes part of the GFCC′ face (49, 50). Moreover, the adenovirus fiber knob engages the same area of the GFCC′ face that mediates formation of the CAR–CAR dimer (25, 40). The structure of the complex between fiber knob and CAR shows a trimeric knob decorated with three copies of monomeric CAR (25, 40).
Interaction of hJAM1 with the Reovirus Attachment Protein σ1.
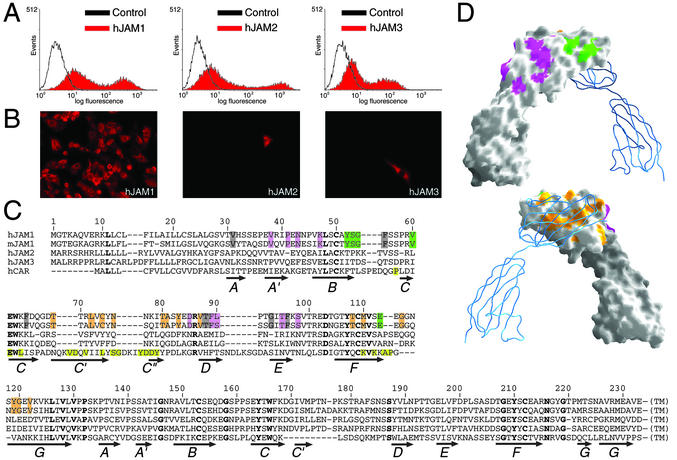
The crystal structure of the reovirus attachment protein σ1 revealed a putative binding site for JAM1 at the lower edge of the σ1 head domain (17). To investigate whether other JAM family members serve as reovirus receptors, we transiently transfected CHO cells with hJAM1, hJAM2, or hJAM3 and assayed transfected cells for the capacity to support reovirus infection. Expression of JAM proteins at the cell surface was confirmed by flow cytometry (Fig. 4A). In contrast to cells transfected with hJAM1, cells transfected with hJAM2 or hJAM3 were not infected by reovirus (Fig. 4B). Our findings clearly demonstrate that reovirus recognizes structural features that are present in hJAM1 but not in hJAM2 or hJAM3.
Fig 4.
Interaction of reovirus with hJAM1. (A) Transiently transfected CHO cells display surface expression of JAM family members. CHO cells were transiently transfected with hJAM1, hJAM2, or hJAM3 and screened for surface expression of JAM proteins by flow cytometry. (B) Transfection of CHO cells with hJAM1, but not hJAM2 or hJAM3, rescues infection by reovirus strain T1L. Shown are infected cells as detected by indirect immunofluorescence using rabbit antireovirus sera. (C) Sequence alignment, with residues conserved in hJAM1 and mJAM1, but not in hJAM2 or hJAM3, highlighted in orange, magenta, and green. Arrows indicate β-strands. The sequence of CAR was aligned with the JAM sequences using clustalw (www.ebi.ac.uk/clustalw). CAR residues contacting the adenovirus fiber knob (distance <4 Å) in the complex (25) are highlighted in yellow. Residues conserved in all sequences are shown in bold. (D) The hJAM1 D1D2 dimer viewed from opposite angles, with one monomer shown in surface representation and the other shown as a blue ribbon. The side chains of conserved residues from C were mapped onto the hJAM1 D1D2 surface by using the same color code. Residues colored in orange, green, or magenta cluster in three different surface areas. Residues shaded gray in C are buried and not visible in this representation.
To define structural features unique to JAM1 and potentially involved in contacting σ1, we identified conserved sequences in hJAM1 and mJAM1 that are not conserved in the other two JAM family members (Fig. 4C). Both hJAM1 and mJAM1 serve as reovirus receptors (1), whereas hJAM2 and hJAM3 do not. Because the D1 domain of hJAM1 is necessary and sufficient for interaction with σ1 (J.C.F. and T.S.D., unpublished observations), our analysis was restricted to that domain. Residues unique to hJAM1 D1 and mJAM1 D1, and therefore likely to participate in σ1 binding, cluster in three main areas (Fig. 4D): a region at the dimer interface (shown in orange) and two surface-exposed regions at the “back” of the molecule, opposite the dimer interface (shown in green and magenta, respectively). All three areas are candidates for interaction with σ1.
Interestingly, the “top” of the hJAM1 dimer does not contain conserved residues, and therefore we hypothesize that σ1 either engages the side of the hJAM1 dimer (via the magenta or green surfaces) or the JAM1 dimer interface (via the orange surface). The green surface near the top of D1 comprises the BC loop and the beginning of strand C. This surface is most complementary in shape and size to the proposed JAM1-binding region of σ1 (17) and, thus, it is a good candidate for σ1–JAM1 interactions. The equivalent regions in JAM2 and JAM3 likely have a different structure because of two-residue insertions (Fig. 4C), perhaps explaining the inability of these proteins to serve as reovirus receptors. The magenta-colored regions at the “back” of the protein contain three solvent-exposed side chains, Lys-47, Thr-88, and Thr-95, which also could participate in the interaction with σ1.
The interdimer interface of JAM1 (the orange surface in Fig. 4D) is not accessible to σ1 in the context of a JAM1–JAM1 dimer. However, σ1 might engage this surface in monomeric forms of JAM1. Such a mechanism of binding is identical to the strategy used by the adenovirus fiber knob to bind the monomeric form of CAR (25). Indeed, comparison of the sequences of JAM1 and CAR shows that JAM1 residues highlighted in orange cluster in the same region as the CAR residues known to bind fiber (Fig. 4C). Although we have no evidence to suggest that σ1 binds to JAM1 in a similar manner, the similarities exhibited by σ1 and fiber, and between JAM1 and CAR, indicate that such an interaction might occur. The affinity of the σ1 head domain for hJAM1 is in the nanomolar range (6 × 10−8 M) (1). Although the affinity of JAM1 for itself is not known, studies using recombinant mJAM1 demonstrated that a significant portion of this protein is monomeric under physiologic conditions (35). This finding suggests that JAM1–JAM1 interactions are weak. Therefore, it seems plausible that σ1 binds to monomeric JAM1, perhaps by engaging residues that form the JAM1–JAM1 interface.
Conclusions
The crystal structure of the hJAM1 ectodomain provides insights into how JAM1 functions in tight junction formation and viral attachment. A key feature of JAM1 is its capacity to form dimers via an extensive interface in its N-terminal domain. This interface is distinguished from traditional protein–protein interfaces by its highly polar character and its capacity to accommodate substantial rearrangements. The latter is evidenced by the different dimeric structures of hJAM1 and mJAM1 despite absolute conservation of residues at the interface. We think it possible that the dynamic nature of the interface plays a role in mediating and perhaps facilitating transitions between monomeric and dimeric forms of JAM1. Moreover, the dynamic nature of the interface likely distinguishes JAM1 from the other JAM family members, JAM2 and JAM3, both of which contain substitutions that are predicted to alter the stability of the interface.
Previous work from our laboratories has shown that the adenovirus and reovirus attachment proteins share many structural and functional features (17). Here we show that the similarities also extend to their receptors. The crystal structure of hJAM1 features a dimeric arrangement that closely resembles that seen in the adenovirus receptor, CAR. Parallels in the structures of these molecules are especially intriguing in light of the recent observation that CAR, like JAM1, is a component of cell–cell junctions (37–39). Thus, both viral and cellular determinants of adenovirus and reovirus binding exhibit striking structural similarities, which suggest conserved strategies of attachment among these viruses.
Acknowledgments
We thank members of our laboratories for review of the manuscript. This research was supported by Public Health Service Awards T32 CA09385 (to J.A.C. and J.C.F.), R01 AI38296 (to T.S.D.), R01 AI45716 (to T.S.), and R01 GM67853 (to T.S.D. and T.S.), the Vanderbilt University Research Council (to J.C.F.), the Swiss National Science Foundation (to P.S.), the Vanderbilt Medical Scholar's Program (to M.J.W.), the Howard Hughes Medical Institute (to T.R.P.), the Milton Foundation (to A.E.P.), and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service Awards CA68485 for the Vanderbilt Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
Abbreviations
CAR, coxsackievirus and adenovirus receptor
JAM, junctional adhesion molecule
mJAM, murine JAM
hJAM, human JAM
CHO, Chinese hamster ovary
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Barton E. S., Forrest, J. C., Connolly, J. L., Chappell, J. D., Liu, Y., Schnell, F. J., Nusrat, A., Parkos, C. A. & Dermody, T. S. (2001) Cell 104, 441-451. [DOI] [PubMed] [Google Scholar]
- 2.Tyler K. L. (2001) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), pp. 1729–1745.
- 3.Kostrewa D., Brockhaus, M., D'Arcy, A., Dale, G. E., Nelboeck, P., Schmid, G., Mueller, F., Bazzoni, G., Dejana, E., Bartfai, T., et al. (2001) EMBO J. 20, 4391-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin-Padura I., Lostaglio, S., Schneemann, M., Williams, L., Romano, M., Fruscella, P., Panzeri, , Stoppacciaro, A., Ruco, L., Villa, A., et al. (1998) J. Cell Biol. 142, 117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malergue F., Galland, F., Martin, F., Mansuelle, P., Aurrand-Lions, M. & Naquet, P. (1998) Mol. Immunol. 35, 1111-1119. [DOI] [PubMed] [Google Scholar]
- 6.Williams L. A., Martin-Padura, I., Dejana, E., Hogg, N. & Simmons, D. L. (1999) Mol. Immunol. 36, 1175-1188. [DOI] [PubMed] [Google Scholar]
- 7.Ostermann G., Weber, K. S., Zernecke, A., Schroder, A. & Weber, C. (2002) Nat. Immunol. 3, 151-158. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham S. A., Arrate, M. P., Rodriguez, J. M., Bjercke, R. J., Vanderslice, P., Morris, A. P. & Brock, T. A. (2000) J. Biol. Chem. 275, 34750-34756. [DOI] [PubMed] [Google Scholar]
- 9.Naik U. P., Naik, M. U., Eckfeld, K., Martin-DeLeon, P. & Spychala, J. (2001) J. Cell Sci. 114, 539-547. [DOI] [PubMed] [Google Scholar]
- 10.Aurrand-Lions M., Duncan, L., Ballestrem, C. & Imhof, B. A. (2001) J. Biol. Chem. 276, 2733-2741. [DOI] [PubMed] [Google Scholar]
- 11.Aurrand-Lions M., Johnson-Leger, C., Wong, C., Du Pasquier, L. & Imhof, B. A. (2001) Blood 98, 3699-3707. [DOI] [PubMed] [Google Scholar]
- 12.Arrate M. P., Rodriguez, J. M., Tran, T. M., Brock, T. A. & Cunningham, S. A. (2001) J. Biol. Chem. 276, 45826-45832. [DOI] [PubMed] [Google Scholar]
- 13.Liang T. W., Chiu, H. H., Gurney, A., Sidle, A., Tumas, D. B., Schow, P., Foster, J., Klassen, T., Dennis, K., DeMarco, R. A., et al. (2002) J. Immunol. 168, 1618-1626. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham S. A., Rodriguez, J. M., Arrate, M. P., Tran, T. M. & Brock, T. A. (2002) J. Biol. Chem. 277, 27589-27592. [DOI] [PubMed] [Google Scholar]
- 15.Santoso S., Sachs, U. J., Kroll, H., Linder, M., Ruf, A., Preissner, K. T. & Chavakis, T. (2002) J. Exp. Med. 196, 679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser R. D., Furlong, D. B., Trus, B. L., Nibert, M. L., Fields, B. N. & Steven, A. C. (1990) J. Virol. 64, 2990-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappell J. D., Prota, A. E., Dermody, T. S. & Stehle, T. (2002) EMBO J. 21, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furlong D. B., Nibert, M. L. & Fields, B. N. (1988) J. Virol. 62, 246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dryden K. A., Wang, G., Yeager, M., Nibert, M. L., Coombs, K. M., Furlong, D. B., Fields, B. N. & Baker, T. S. (1993) J. Cell Biol. 122, 1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chappell J. D., Duong, J. L., Wright, B. W. & Dermody, T. S. (2000) J. Virol. 74, 8472-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barton E. S., Connolly, J. L., Forrest, J. C., Chappell, J. D. & Dermody, T. S. (2001) J. Biol. Chem. 276, 2200-2211. [DOI] [PubMed] [Google Scholar]
- 22.Paul R. W., Choi, A. H. & Lee, P. W. (1989) Virology 172, 382-385. [DOI] [PubMed] [Google Scholar]
- 23.Chappell J. D., Gunn, V. L., Wetzel, J. D., Baer, G. S. & Dermody, T. S. (1997) J. Virol. 71, 1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergelson J. M., Cunningham, J. A., Droguett, G., Kurt-Jones, E. A., Krithivas, A., Hong, J. S., Horwitz, M. S., Crowell, R. L. & Finberg, R. W. (1997) Science 275, 1320-1323. [DOI] [PubMed] [Google Scholar]
- 25.Bewley M. C., Springer, K., Zhang, Y. B., Freimuth, P. & Flanagan, J. M. (1999) Science 286, 1579-1583. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computing Project No. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 28.Brünger A. T. (1992) Nature 355, 472-475. [DOI] [PubMed] [Google Scholar]
- 29.Jones T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Kuriyan, J. & Karplus, M. (1987) Science 235, 458-460. [DOI] [PubMed] [Google Scholar]
- 31.Liu J. H., Nusrat, A., Schnell, F. J., Reaves, T. A., Walsh, S., Pochet, M. & Parkos, C. A. (2000) J. Cell Sci. 113, 2363-2374. [DOI] [PubMed] [Google Scholar]
- 32.Palmeri D., van Zante, A., Huang, C. C., Hemmerich, S. & Rosen, S. D. (2000) J. Biol. Chem. 275, 19139-19145. [DOI] [PubMed] [Google Scholar]
- 33.Harpaz Y. & Chothia, C. (1994) J. Mol. Biol. 238, 528-539. [DOI] [PubMed] [Google Scholar]
- 34.Holm L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 35.Bazzoni G., Martinez-Estrada, O. M., Mueller, F., Nelboeck, P., Schmid, G., Bartfai, T., Dejana, E. & Brockhaus, M. (2000) J. Biol. Chem. 275, 30970-30976. [DOI] [PubMed] [Google Scholar]
- 36.Schulz G. E. & Schirmer, R. H., (1976) Principles of Protein Structure (Springer, New York).
- 37.Honda T., Saitoh, H., Masuko, M., Katagiri-Abe, T., Tominaga, K., Kozakai, I., Kobayashi, K., Kumanishi, T., Watanabe, Y. G., Odani, S. & Kuwano, R. (2000) Brain Res. Mol. Brain Res. 77, 19-28. [DOI] [PubMed] [Google Scholar]
- 38.Cohen C. J., Shieh, J. T., Pickles, R. J., Okegawa, T., Hsieh, J. T. & Bergelson, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters R., Freimuth, P., Moninger, T., Ganske, I., Zabner, J. & Welsh, M. (2002) Cell 110, 789-799. [DOI] [PubMed] [Google Scholar]
- 40.van Raaij M. J., Chouin, E., van der Zandt, H., Bergelson, J. M. & Cusack, S. (2000) Structure (London) 8, 1147-1155. [DOI] [PubMed] [Google Scholar]
- 41.Ostrov D. A., Shi, W., Schwartz, J. C., Almo, S. C. & Nathenson, S. G. (2000) Science 290, 816-819. [DOI] [PubMed] [Google Scholar]
- 42.Kasper C., Rasmussen, H., Kastrup, J. S., Ikemizu, S., Jones, E. Y., Berezin, V., Bock, E. & Larsen, I. K. (2000) Nat. Struct. Biol. 7, 389-393. [DOI] [PubMed] [Google Scholar]
- 43.Casasnovas J. M., Stehle, T., Liu, J., Wang, J. & Springer, T. (1998) Proc. Natl. Acad. Sci. USA 95, 4134-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H., Kwong, P. D. & Hendrickson, W. A. (1997) Nature 387, 527-530. [DOI] [PubMed] [Google Scholar]
- 45.Jones E. Y., Davis, S. J., Williams, A. F., Harlos, K. & Stuart, D. I. (1992) Nature 360, 232-239. [DOI] [PubMed] [Google Scholar]
- 46.Bodian D. L., Jones, E. Y., Harlos, K., Stuart, D. I. & Davis, J. P. (1994) Structure (London) 2, 755-766. [DOI] [PubMed] [Google Scholar]
- 47.Kwong P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1998) Nature 393, 648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J. H., Meijers, R., Xiong, Y., Liu, J. H., Sakihama, T., Zhang, R., Joachimiak, A. & Reinherz, E. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10799-10804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bella J., Kolatkar, P. R., Marlor, C. W., Greve, J. M. & Rossmann, M. G. (1998) Proc. Natl. Acad. Sci. USA 95, 4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao C., Bator, C. M., Bowman, V. D., Rieder, E., He, Y., Hebert, B., Bella, J., Baker, T. S., Wimmer, E., Kuhn, R. J. & Rossmann, M. G. (2001) J. Virol. 75, 2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]