SUMMARY
Biofilm cells are less susceptible to antimicrobials than their planktonic counterparts. While this phenomenon is multifactorial, the ability of the biofilm matrix to reduce antibiotic penetration into the biofilm is thought to be of limited importance, as previous studies suggest that antibiotics move fairly rapidly through biofilms. In this study, we monitored the transport of two clinically relevant antibiotics, tobramycin and ciprofloxacin, into non-mucoid P. aeruginosa biofilms. To our surprise, we showed that the positively charged antibiotic tobramycin is sequestered to the biofilm periphery, while the neutral antibiotic ciprofloxacin readily penetrated. We provide evidence that tobramycin in the biofilm periphery both stimulated a localized stress response and killed bacteria in these regions, but not in the underlying biofilm. Although it is unclear which matrix component binds tobramycin, its penetration was increased by the addition of cations in a dose-dependent manner, which led to increased biofilm death. These data suggest that ionic interactions of tobramycin with the biofilm matrix limit its penetration. We propose that tobramycin sequestration at the biofilm periphery is an important mechanism in protecting metabolically active cells that lie just below the zone of sequestration.
INTRODUCTION
Bacterial biofilms cause many persistent chronic infections. Biofilm growth of the human pathogen Pseudomonas aeruginosa is involved in the infection of burn wounds, urinary tracts, corneas, and ears, as well as the lungs of cystic fibrosis patients. Because biofilm bacteria are much less susceptible to antimicrobial agents than their planktonic counterparts, these infections are difficult, if not impossible, to eradicate (Costerton et al., 1999). Although biofilm growth is associated with the genetic diversification of bacteria, many investigators have reported that biofilm antimicrobial tolerance is not linked to heritable genetic changes. For instance, surviving bacteria isolated from antimicrobial exposed biofilms can be used to grow new biofilms with susceptibility that is identical to that of the original inoculating strain (Harrison et al., 2005). Moreover, the antibiotic susceptibility of biofilm bacteria returns upon dispersion of the biofilm cells back to the planktonic state (Stewart, 2002). Therefore, the physical and physiological attributes of the biofilm growth environment protect resident cells from the lethal action of antimicrobials (Costerton et al., 1999).
Several mechanisms have been proposed to account for the increased antimicrobial tolerance of biofilms. Since antibiotics are generally more effective against actively dividing cells (Brown et al., 1988), slow growing subpopulations are thought to be a major factor contributing to the reduced antimicrobial susceptibility of biofilms. Oxygen and nutrient limitation play a key role in this process (Brown et al., 1988; Xu et al., 1998; Anderl et al., 2003; Walters et al., 2003; Borriello et al., 2004; Werner et al., 2004; Pamp et al., 2008; Kim et al., 2009; Williamson et al., 2012). Furthermore, not only does nutrient limitation slow growth, but it also activates a starvation response that mediates antibiotic tolerance (Nguyen et al., 2011). Additional mechanisms could involve the formation of “persister” cells, which arise at high frequency in biofilm populations (Brooun et al., 2000; Spoering and Lewis, 2001; Keren et al., 2004; Allison et al., 2011). The reduced antimicrobial susceptibility of P. aeruginosa biofilms has also been linked to genes encoding efflux pumps (Gillis et al., 2005; Zhang and Mah, 2008), a transcriptional regulator (brlR) (Liao and Sauer, 2012), a two-component system (amgRS) (Lee et al., 2009), a phosphodiesterase involved in the degradation of the intracellular signaling molecule cyclic-di-GMP (arr) (Hoffman et al., 2005), extracellular matrix polysaccharides (pel, psl, and alg operons) (Hodges and Gordon, 1991; Colvin et al., 2011) and periplasmic glucans (ndvB) (Mah et al., 2003). NdvB is also important for the expression of ethanol oxidation genes, which are also implicated in the ability of the biofilm to tolerate antibiotics (Beaudoin et al., 2012). The regulation and biochemical function of all these systems in biofilm antibiotic tolerance, however, are not completely understood.
Limited antimicrobial penetration into biofilms, due to interactions with the matrix, was originally thought to be an important mechanism in antibiotic tolerance (Suci et al., 1994; Vrany et al., 1997). Over the years, however, only a few special cases of this mechanism have been identified (Stewart, 2003). The penetration of multiple antibiotics through biofilms of different species has been measured using a variety of methodologies (Hoyle et al., 1992; Dunne et al., 1993; Yasuda et al., 1993; Kumon et al., 1994; Suci et al., 1994; Shigeta et al., 1997; Vrany et al., 1997; Anderl et al., 2000; Stone et al., 2002; Zheng and Stewart, 2002; Walters et al., 2003; Jefferson et al., 2005; Singh et al., 2010; Ortiz-Perez et al., 2011). In general, antibiotics are found to readily penetrate biofilms at time scales that suggest against limited penetration contributing to antimicrobial tolerance. One notable exception involves mucoid P. aeruginosa and aminoglycoside antibiotics. Positively charged aminoglycosides, such as tobramycin, have been shown to interact with the negatively charged exopolysaccharide alginate (Gordon et al., 1988; Nichols et al., 1988; Hatch and Schiller, 1998). Although tobramycin penetration is hindered (Walters et al., 2003), it is generally not considered to play an important role in the increased tolerance of biofilms due to the expected eventual penetration of the antibiotic over long exposure times (Stewart, 2003). The slow penetration of antibiotics into biofilms, however, has been proposed to afford resident cells the time to physiologically adapt to the antibiotic and adopt a more antimicrobial tolerant state before killing concentrations of the antibiotic can be achieved (Jefferson et al., 2005; Szomolay et al., 2005; Anderson and O'Toole, 2008).
The purpose of this work was to examine antibiotic penetration of two clinically relevant antibiotics, the aminoglycoside tobramycin and the fluoroquinolone ciprofloxacin, through non-mucoid P. aeruginosa biofilms. The penetration of these antibiotics has been studied in clinically relevant, alginate over-expressing mucoid variants of P. aeruginosa (Nichols et al., 1989; Shigeta et al., 1997; Walters et al., 2003). While alginate impedes tobramycin penetration into mucoid biofilms, ciprofloxacin penetration is relatively rapid (Gordon et al., 1988; Nichols et al., 1988; Hatch and Schiller, 1998; Walters et al., 2003). Antibiotic penetration through non-mucoid biofilms, however, has not been systematically measured. We directly observed and tracked the penetration of fluorescently labeled tobramycin and ciprofloxacin through a PAO1 biofilm using time-lapse confocal microscopy. While we originally predicted that non-mucoid biofilms, which produce little to no alginate, would not impede tobramycin penetration, our data show that the non-mucoid biofilm sequestered tobramycin to the periphery via ionic interactions. Furthermore, our results suggest that limited penetration is a significant factor in the aminoglycoside tolerance of non-mucoid P. aeruginosa biofilms.
RESULTS
Non-mucoid P. aeruginosa biofilms limit the penetration of tobramycin, but not ciprofloxacin
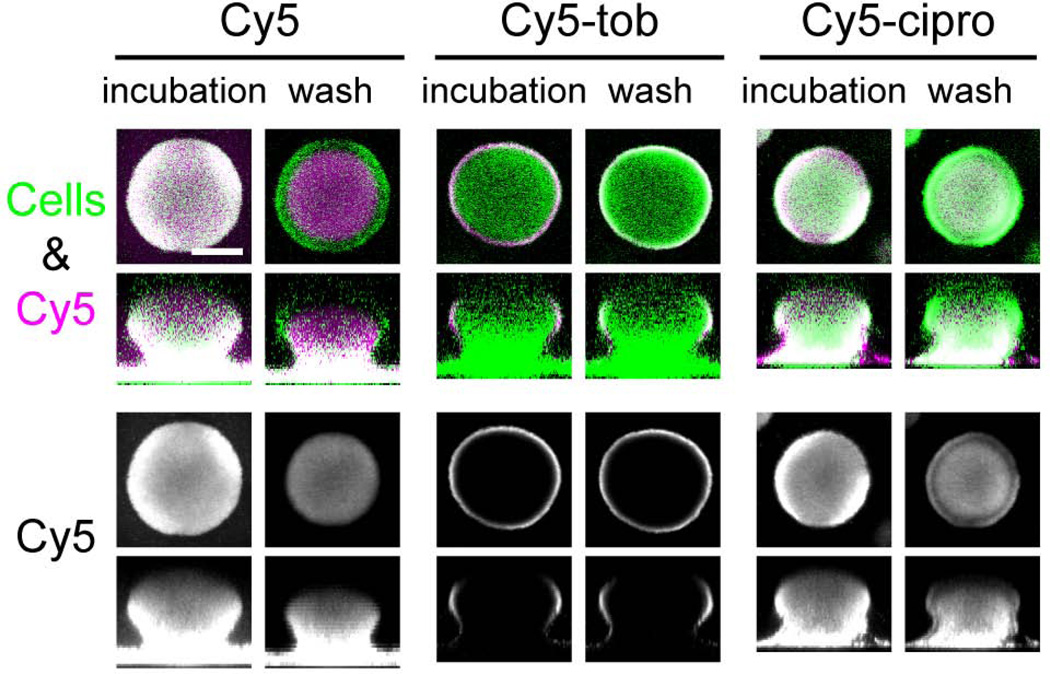
To study the penetration kinetics of these antibiotics, we used a Cy5-conjugated form of both drugs (Cy5-tobramycin and Cy5-ciprofloxacin). We cultivated PAO1 biofilms that exhibited significant structure, with microcolonies several microns thick. We acquired images every 2.5 minutes during a 30-minute static exposure to the Cy5-conjugated antibiotic followed by a subsequent 30-minute wash. Representative images after the static incubation and after the wash are shown in Fig. 1A. Both Cy5 conjugated antibiotics, as well as Cy5 alone, accumulated in the biofilm during the static incubation, since Cy5 fluorescence in the biofilm was higher than that of the bulk solution. While Cy5 alone and Cy5-ciprofloxacin quickly penetrated through the biofilm, Cy5-tobramycin remained at the periphery of the biofilm, producing a shell-like pattern of localization (Fig. 1 and 2). On average, Cy5-tobramycin penetrated only 4.57 ± 0.54 µm (S.D.; N = 63). Although the Cy5 conjugation affected the antibacterial activity of tobramycin (the minimum inhibitory concentration (MIC) of Cy5-tobramycin was 128 µg/ml, while the MIC of unlabeled tobramycin was 0.5 µg/ml), the reduced transport was not due to the Cy5 conjugation, since Cy5 alone readily penetrated the biofilm (Fig. 1 and 2B). In addition, although we conducted these experiments with equivalent molar amounts of Cy5, Cy5-tobramycin, and Cy5-ciprofloxacin, much more fluorescent label accumulated in the biofilms treated with Cy5 and Cy5-ciprofloxacin than Cy5-tobramycin (Fig. 2B), indicating that the reduced penetration of Cy5-tobramycin is not due to a limitation in the amount of labeled antibiotic in the bulk fluid. Furthermore, while the Cy5 fluorescence in the bulk fluid for the Cy5-tobramycin exposed biofilm dropped by ∼70% over the 30 minute static incubation, the remaining fluorescence (∼30%) was well above background (Fig. S1), further suggesting that depletion of Cy5-tobramycin in the bulk liquid did not influence the observed tobramycin penetration. Finally, we found that static antibiotic exposure and exposure to Cy5-tobramycin under flow produced similar results (data not shown).
Figure 1. Biofilms limit the penetration of tobramycin into the biomass.
Biofilms of a GFP-expressing PAO1 strain were treated statically with 21.4 µM Cy5 (left), Cy5-tobramycin (center), or Cy5-ciprofloxacin (right) for 30 min. Confocal laser scanning micrographs were acquired every 2.5 min during the static incubation and a subsequent 30-min wash. Representative images after the static incubation and after the wash are shown. In the top row, the biomass is pseudo-colored green, and Cy5, purple. The bottom row shows the Cy5 channel alone in grayscale. Bar, 50 µm.
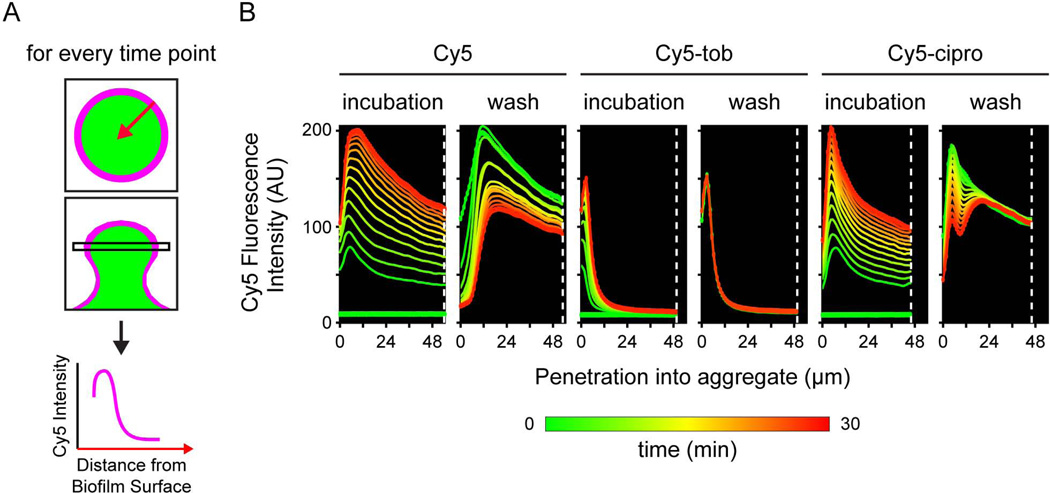
Figure 2. Kinetics of antibiotic penetration.
A. Schematic of experiment. For every time point, the micrograph containing the widest part of the biofilm was used. The average Cy5 fluorescence intensity of pixels at the same distance from the edge of the biofilm was quantified for each time point. A graph of the Cy5 fluorescence intensity versus the distance from the exterior of the biofilm was then created with the origin of the x-axis representing the exterior of the biofilm. B. Graphs of the penetration kinetics of the representative biofilm shown in Figure 1. On the left is Cy5; center, Cy5-tobramycin; and right, Cy5-ciprofloxacin. The dashed white line represents the center of the biofilm. Each colored line on the graph represents one time point and is color-coded to show increasing time with green as the starting time point and red as the ending time point.
During the 30-minute post-treatment wash, the signal intensity of both unconjugated Cy5 and Cy5-ciprofloxacin within the biofilm decreased progressively over time, suggesting that these molecules were washed out rapidly and that both interact weakly with the biofilm. By contrast, neither the signal intensity nor localization of Cy5-tobramycin changed during the flush period (Fig. 2B). In fact, the Cy5-tobramycin signal remained even after 24 hours of resumed media flow, without significant redistribution within the biofilm or loss to the bulk fluid (data not shown), indicating that the tobramycin was immobilized in the biofilm. Together, these data show that the biofilm limits the penetration of tobramycin, but not ciprofloxacin. To gain insight on how the biofilm limits antibiotic penetration, we focused our further studies on tobramycin.
Known determinants of biofilm antibiotic tolerance cannot account for the limited tobramycin penetration
We hypothesized that a component of the biofilm matrix may be responsible for the tobramycin sequestration. We thus investigated the role of the three matrix exopolysaccharides (EPS) that P. aeruginosa is known to produce: alginate, Pel and Psl (Ryder et al., 2007). Similar tobramycin penetration was observed in biofilms of strains unable to produce any one of the three EPS components (Fig. S2A). Furthermore, overproduction of either Pel or Psl did not influence the penetration of Cy5-tobramycin (Fig. S2B and C).
Several genetic elements that affect P. aeruginosa biofilm susceptibility to tobramycin have been identified. We investigated whether these previously identified genetic determinants of biofilm antibiotic susceptibility might be responsible for the limited penetration. Biofilms of strains containing mutations in amgRS (Lee et al., 2009), arr (Hoffman et al., 2005), brlR (Liao and Sauer, 2012) and ndvB (Mah et al., 2003) have been reported to be more susceptible to tobramycin than wild-type biofilms. However, none of these genes, when deleted, resulted in biofilms with increased tobramycin penetration (Fig. S2A).
A hypothesis that arises from these findings is that restricted penetration may involve a general chemical interaction with diverse functional groups present in the biofilm matrix. At neutral pH, tobramycin is highly positively charged. We, therefore, hypothesized that tobramycin may be binding to a negatively charged component in the biofilm matrix, such as LPS. Thus, we constructed mutants of the lipid A (ΔphoQ) and the O-antigen (ΔwbpL) components, both which impact the overall charge of LPS. We hypothesized that since a phoQ deletion mutant has a more negatively charged LPS than that of wild type (Guo et al., 1997; Ernst et al., 1999), the mutant biofilm may further decrease tobramycin penetration. While phoQ mutants are less susceptible to aminoglycosides in planktonic culture than wild type cells (Macfarlane et al., 2000), a ΔphoQ biofilm displayed a tobramycin penetration phenotype similar to wild type (Fig. S2A). Similarly, the ΔwbpL mutant, which lacks the A- and B-band (Rocchetta et al., 1998) and should also have a more negatively charged LPS relative to that of wild type (Makin and Beveridge, 1996), also limited tobramycin penetration similar to wild type (Fig. S2A).
Ability of the biofilm to limit penetration can be overcome with high concentrations of tobramycin
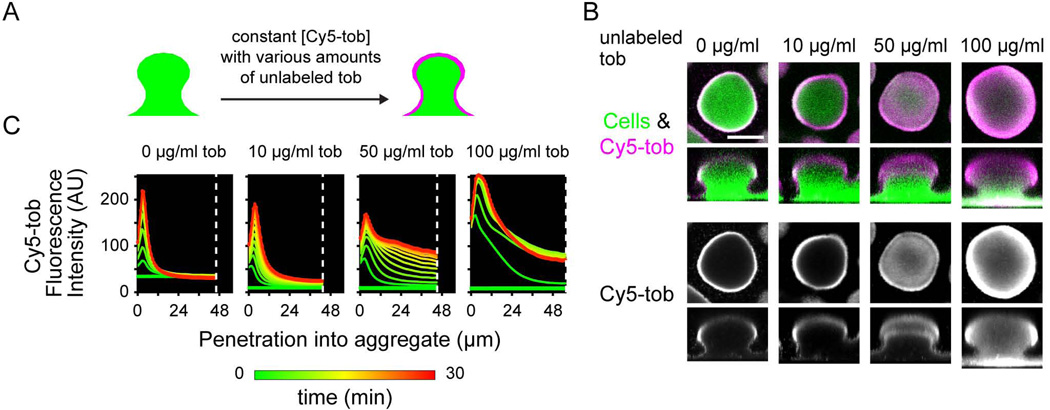
Our results suggest that tobramycin penetration is hindered by an interaction with the biofilm matrix. To test if the tobramycin binding sites could be saturated, we exposed biofilms to Cy5-tobramycin in conjunction with various concentrations of unlabeled tobramycin. A constant concentration of Cy5-tobramycin was used in these experiments to ensure that the fluorescence intensity of the Cy5 could be compared across micrographs. While an extra 10 µg/ml unlabeled tobramycin had little effect on tobramycin penetration, addition of 50 or 100 µg/ml unlabeled tobramycin dramatically increased penetration (Fig. 3). This difference was readily observed in the penetration plots as an increase in fluorescence intensity at the center of the biofilm over time (Fig. 3C, at white dashed line). Cy5-tobramycin partially penetrated a biofilm concurrently treated with 50 µg/ml unlabeled tobramycin, and fully penetrated in the presence of 100 µg/ml tobramycin (Fig. 3B). This difference in Cy5-tobramycin penetration between biofilms treated with the 50 µg/ml versus 100 µg/ml unlabeled tobramycin suggests that the ability to prevent the penetration of tobramycin can be titrated by saturating the tobramycin binding sites in the biofilm matrix.
Figure 3. Ability of the biofilm to limit penetration can be overcome with high concentrations of tobramycin.
A. Schematic of experiment. Biofilms of a GFP-expressing PAO1 strain were treated statically with 20 µg/ml Cy5-tobramycin and 0, 10, 50, or 100 µg/ml tobramycin concurrently. Confocal laser scanning micrographs were acquired every 2.5 min during a static 30-min incubation. B. Representative micrographs, as described as in Figure 1, of biofilms co-treated with Cy5-tobramycin and the indicated unlabeled tobramycin concentration. Bar, 50 µm. C. Graphs, as described in Figure 2, representing the Cy5-tobramycin penetration kinetics.
This change in tobramycin penetration is unlikely due to the killing of biofilm cells by the high concentrations of antibiotic, since the effect on penetration was immediate (Fig. 3C). The increased Cy5-tobramycin penetration into the biofilm concurrently treated with 50 µg/ml tobramycin was observed within the first 2.5 minutes of static incubation. A similar timescale of penetration was noted for the biofilm co-treated with 100 µg/ml tobramycin. While more Cy5-tobramycin accumulated in the biofilm over the 30-minute static incubation for both samples, the rapid change in localization suggests that the effect is physicochemical, instead of biological. Furthermore, the 30-minute exposure to 10 – 100 µg/ml tobramycin did not result in substantial cell death (see Fig. S6). In addition, Cy5-tobramycin penetration was not affected by concurrent treatment with 100 µg/ml ciprofloxacin (Fig. S3). Since both ciprofloxacin and tobramycin are known to kill cells in the periphery of the biofilm (Walters et al., 2003; Williamson et al., 2012), cell death cannot explain the difference in Cy5-tobramycin penetration patterns in the presence of high concentrations of unlabeled tobramycin versus ciprofloxacin.
Metal cations facilitate the penetration of tobramycin into biofilms
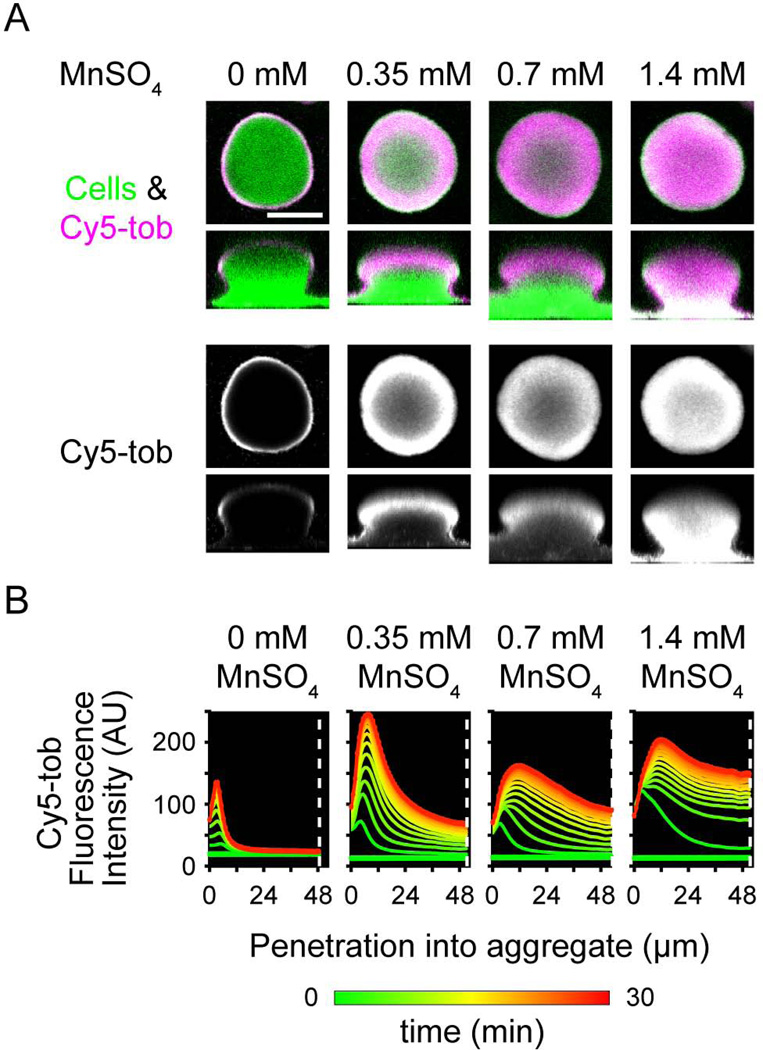
We hypothesized that tobramycin was interacting with the biofilm on the basis of its charge. Therefore, we tested whether other cations could compete with tobramycin for binding sites in the biofilm and thus promote tobramycin penetration. We chose the essential divalent cation Mn2+ for these studies, as it has low toxicity in many bacteria, including P. aeruginosa (Harrison et al., 2004). While little effect was observed with 0.35 and 0.7 mM MnSO4, concurrent treatment of Cy5-tobramycin with 1.4 mM MnSO4 allowed Cy5-tobramycin to fully penetrate the biofilm (Fig. 4). Furthermore, the fluorescence intensity in the center of the biofilm increased with increasing amounts of MnSO4 (Fig. 4B, at white dashed line). We observed a similar effect with the divalent essential cation Mg2+ (Fig. S4). As the MICs of MnSO4 and MgSO4 for P. aeruginosa (8 mM and >256 mM, respectively) are much higher than the concentrations used here, the change in tobramycin penetration is unlikely a result of metal toxicity. Furthermore, the timescale of Cy5-tobramycin penetration for these metal-treated biofilms are similar to biofilms concurrently treated with 100 µg/ml tobramycin (Fig. 4C), suggesting that death of the biomass did not play a role in the change in Cy5-tobramycin transport. Together, our results show that the penetration of tobramycin into the biofilm is inhibited via ionic interactions and suggest that tobramycin interacts with a negatively charged component of the biofilm matrix.
Figure 4. Metal cations facilitate the penetration of tobramycin into biofilms.
Biofilms of a GFP-expressing PAO1 strain were treated statically with 20 µg/ml Cy5-tobramycin and 0, 0.35, 0.7, or 1.4 mM manganese sulfate. Confocal laser scanning micrographs were acquired every 2.5 min during a static 30-min incubation. Representative images are displayed as in Figure 1. Bar, 50 µm. B. Graphs, as described in Figure 2, representing the Cy5-tobramycin penetration kinetics.
Response of biofilm cells to tobramycin is localized to the region of penetration
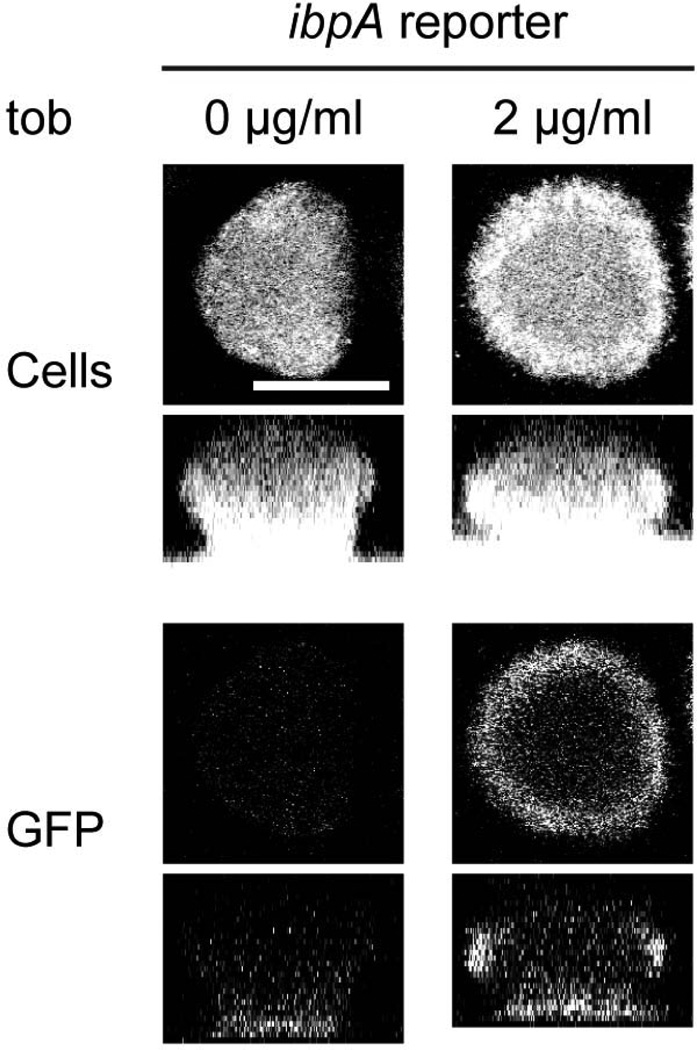
We next sought to determine if biofilm cells perceive and respond to the presence of tobramycin. Therefore, we constructed a tobramycin responsive reporter strain, in which a chromosomally integrated gfp is driven by the promoter of ibpA, a gene encoding a heat shock protein whose expression is induced in cells exposed to tobramycin (Kindrachuk et al., 2011). To confirm that our reporter strain responds to tobramycin, planktonic cells were treated with increasing concentrations of tobramycin and assayed for fluorescence over several hours. In comparison to a control strain that contains a promoterless gfp, the ibpA reporter strain showed GFP fluorescence that peaked 2 hours after tobramycin treatment, with increasing concentrations of tobramycin producing increased amounts of GFP fluorescence (Fig. S5). These results confirm that this reporter strain does respond to the presence of tobramycin.
In a biofilm produced by the ibpA reporter strain, cells in the periphery expressed GFP upon exposure to tobramycin (Fig. 5). While the exact mechanism leading to tobramycin-induced activation of ibpA is unknown, this cellular stress response in the biofilm is likely due to intracellular tobramycin, as Cy5-tobramycin can penetrate into both planktonic and biofilm cells (data not shown). Furthermore, the region of this stress response corresponded to the region where we observed Cy5-tobramycin sequestration in the biofilm. In comparison, as expected, an untreated control biofilm of the reporter strain and a tobramycin-treated biofilm harboring a promoter-less gfp control strain did exhibit fluorescence (Fig. 5 and data not shown). Using a control strain that has gfp under the control of an arabinose-inducible promoter, we found that most cells in the biofilm are capable of producing fluorescent GFP (data not shown). Since oxygen levels as low as 0.1 ppm have been shown to be sufficient for GFP fluorescence (Hansen et al., 2001), oxygen limitation is likely not influencing the pattern of GFP expression that is observed in Fig. 5. These results suggest two important points. First, cells perceiving and responding to tobramycin are found in regions where tobramycin is sequestered. Second, cells within the interior of the biofilm are shielded from tobramycin.
Figure 5. Cells at the periphery of the biofilm respond to tobramycin.
Biofilms of PAO1 Tn7::PibpAgfp (ibpA reporter) were treated with 0 or 2 µg/ml tobramycin sulfate for 9 h and then stained with Syto62 to visualize the biomass. Top row, biomass alone; bottom row, GFP fluorescence alone. Bar, 50 µm.
Tobramycin sequestration at the biofilm periphery is a protective mechanism for cells within the biofilm interior
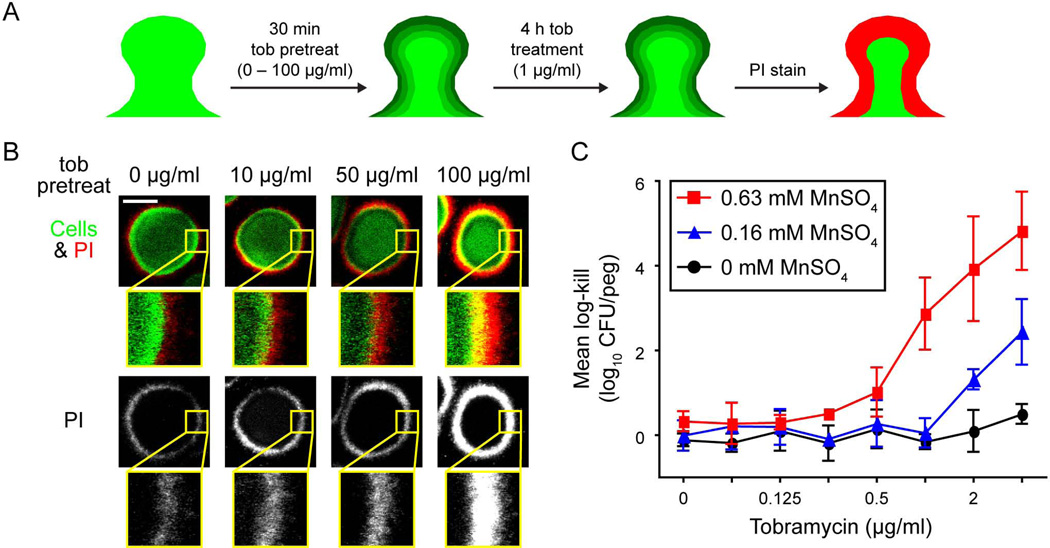
We predicted that limiting tobramycin penetration could confer a measure of protection to cells in the biofilm interior. To test this, we pretreated biofilms for a short period of time with different levels of tobramycin to saturate tobramycin binding sites in the biofilm to different degrees. To do this, biofilms were subjected to a 0, 10, 50, or 100 µg/ml tobramycin exposure for 30 minutes, which was shown to influence tobramycin penetration to different degrees (Fig. 3). This short pretreatment had a minimal effect on cell viability, with killing observed only at a low level for the highest concentration of tobramycin (Fig. S6).
Following pretreatment, the biofilms were exposed to a continuous 4-hour treatment of 1 µg/ml tobramycin, after which we directly visualized the distribution of dead cells by viability staining (Fig. 6A). In agreement with the literature (Walters et al., 2003; Bjarnsholt et al., 2005), treatment of biofilms with 1 µg/ml tobramycin killed cells only in the periphery of the biofilm (Fig. 6B). The 4-hour treatment with 1 µg/ml tobramycin in the absence of a pretreatment killed only a thin layer of cells at the biofilm periphery. Slightly more death was observed following pretreatment with 10 µg/ml tobramycin. In contrast, pretreatment with 50 or 100 µg/ml tobramycin produced killing drastically deeper into the interior, supporting our initial prediction that tobramycin sequestration protects biofilm-resident cells.
Figure 6. Limiting penetration of tobramycin protects biofilm cells.
A. Schematic of experiment. Biofilms of PAO1 were treated with 0, 10, 50, or 100 µg/ml tobramycin sulfate for 30 min followed by continuous treatment of 1 µg/ml tobramycin sulfate for 4 h. Biofilms were then stained with propidium iodide (PI) before imaging. Green shading, penetration of tobramycin due to pretreatment; red, area of cell death from 4 h tobramycin treatment. B. Representative images are shown. In the top row, the biomass is pseudo-colored green and PI, red. The bottom row is PI alone in grayscale. Underneath each cross-sectional image is a 4.25x magnified region (inset yellow box). Bar, 50 µm. C. Synergistic killing of tobramycin with MnSO4 using the MBEC assay. Biofilms were grown on polystyrene pegs and challenged with various combinations of tobramycin and MnSO4. Viable cell counts were determined by spot dilution plating. Mean log-kill was determined by subtracting the final from the initial log10-transformed cell counts. For untreated control samples, the log10-transformed mean viable cell count was 5.4 ± 0.2 CFU per peg at the end of the experiment. For clarity, only three concentrations of MnSO4 are shown. Black circles, tobramycin with 0 mM MnSO4; blue triangles, tobramycin with 0.16 mM MnSO4; and red squares, tobramycin with 0.63 mM MnSO4. Error bars represent standard deviation of results from three independent trials.
The addition of metal salts, such as MgSO4 and MnSO4, also facilitated tobramycin penetration (Fig. 4 and S4). Therefore, we postulated that combining metal treatment with tobramycin would enhance tobramycin killing by enhancing its penetration. To test this, we grew biofilms using the Calgary Biofilm Device and assayed for viability after exposure to various concentrations of MnSO4 and tobramycin. Treating biofilms with sub-lethal quantities of MnSO4 resulted in a ≥8-fold reduction in the minimum bactericidal concentration (MBC) of tobramycin, and moreover, caused massive synergistic killing that was up to 2,100-fold greater than the antibiotic alone (Fig. 6C and S7). In comparison, much less synergistic killing was observed in planktonic cells treated with MnSO4 and tobramycin (Fig. S7). Our results suggest this synergistic killing in biofilms is due to manganese increasing the penetration of tobramycin, and further supports our conclusion that increased penetration of tobramycin increases the efficacy of the antibiotic. Collectively, these data show that limiting tobramycin penetration can serve as a protective mechanism.
DISCUSSION
The high antimicrobial tolerance of biofilm cells is a major barrier to the eradication of chronic infections. While heightened tolerance is clearly due to multiple mechanisms (Stewart, 2002), here we showed that non-mucoid P. aeruginosa biofilms protect resident cells by limiting the penetration of tobramycin via ionic interactions with the biofilm. Our results clearly demonstrate that tobramycin becomes sequestered near the surface and does not fully penetrate the biofilm, while ciprofloxacin transport is uninhibited (Fig. 1 and 2). While we have yet to identify what is interacting with tobramycin, our results suggest that the positively charged antibiotic ionically interacts with negatively charged matrix components, as addition of excess cations substantially increased tobramycin penetration into the biofilm (Fig. 3 and 4). Increasing penetration produced greater killing (Fig. 6), indicating that the limited penetration of tobramycin can protect biofilm cells.
We demonstrated that tobramycin, but not ciprofloxacin, had limited penetration into a non-mucoid biofilm. This was surprising, since alginate, produced mainly by mucoid strains, was previously thought to be the primary antibiotic-binding biofilm matrix component produced by P. aeruginosa (Gordon et al., 1988; Nichols et al., 1988; Hodges and Gordon, 1991; Hatch and Schiller, 1998). Thus, the matrix was thought to afford protection against antibiotics primarily in mucoid strains (Hodges and Gordon, 1991; Hatch and Schiller, 1998). Our results suggest that the protection against tobramycin afforded by the matrix does not depend specifically on the production of alginate (Fig. S2A). Furthermore, our observations provide one possible explanation for the discrepancy in killing between ciprofloxacin and tobramycin towards P. aeruginosa biofilms observed by Preston and colleagues (Preston et al., 1996), who found that the concentration of ciprofloxacin required to eliminate biofilms is ten times the MIC, while for tobramycin, 75 to 100 times the MIC is required. Although it is difficult to directly compare the amount of biofilm killing by the two antibiotics due to differences in mechanism and efficacy, our results suggest that the increased efficacy of ciprofloxacin over tobramycin in eliminating biofilms is in part due to the difference in their ability to penetrate into the biofilm. Ionic interactions of tobramycin, but not ciprofloxacin, with the biofilm matrix appear to dictate the ability of these antibiotics to penetrate the biomass and may affect the efficacy of these antibiotics in killing biofilm cells.
The ability of the biofilm to limit tobramycin penetration is most likely correlated with the structure and maturity of the biofilm. We used fully matured biofilms consisting of large cell aggregates for our studies. Results from Landry and colleagues suggest that the difference in tobramycin tolerance observed in flat, homogenous biofilms versus biofilms characterized by large cellular aggregates is due to differences in antibiotic penetration (Landry et al., 2006). Furthermore, the stage of biofilm development may be a key variable. The availability and quantity of matrix components for tobramycin sequestration may differ depending upon the stage of biofilm development, and several factors could potentially impact tobramycin sequestration.
Two negatively charged components in the biofilm matrix that may interact with tobramycin are LPS or extracellular DNA (eDNA). Our results with ΔphoQ and ΔwbpL suggest that LPS is not involved (Fig. S2A). We did not test mutants that have altered eDNA production, such as pqsA, as these mutations are highly pleiotrophic and produce biofilms that are grossly different in structure than that of wild type strains (Allesen-Holm et al., 2006). Interestingly, Chiang and colleagues have recently published results showing that biofilms that have been treated with exogenously added DNA are less sensitive to tobramycin (Chiang et al., 2013). Their results suggest that the added DNA directly protected the biofilm from tobramycin. It is unclear, however, if eDNA in the biofilm affords similar protection.
Since our results suggest that tobramycin binds to a component of the biofilm matrix and we could not demonstrate an interaction with LPS (Fig. S2A) or eDNA, we systematically tested biofilms formed by mutants of known EPS matrix components. Tobramycin penetration was similar in all of these biofilms (Fig. S2). These results are not entirely surprising. While alginate does interact with tobramycin (Gordon et al., 1988; Nichols et al., 1988), it is not an integral component of the non-mucoid biofilm (Wozniak et al., 2003). Psl is a neutral polysaccharide (Byrd et al., 2009) and our results suggest that tobramycin interacts with negatively charged components of the biofilm matrix. PAO1 does not produce Pel (Colvin et al., 2012), so it is not surprising that little difference was observed with the ΔpelF mutant. Further, while over-expression of Pel in the PAO1 background increases the tolerance of biofilm cells to tobramycin (Colvin et al., 2011), we found no effect of Pel over-expression on tobramycin penetration (Fig. S2B and C), suggesting that the increased tolerance that Pel imparts is not due to reducing tobramycin penetration.
We propose the following mechanism, which is consistent with our results: sequestration of tobramycin at the periphery of the biofilm protects metabolically active cells that lie just underneath the zone of sequestration (indirectly supported by Fig. 6). In addition to the direct effect of protecting the underlying active cells, limiting the antibiotic penetration may also have an indirect secondary effect on increasing tolerance. As previously proposed (Jefferson et al., 2005; Szomolay et al., 2005; Anderson and O'Toole, 2008), limiting the penetration of antibiotics may also allow cells in the interior of the biofilm to sense sub-lethal concentrations of the antibiotic for a sufficient period of time to adapt to a more tolerant state. Furthermore, sub-lethal concentrations of tobramycin have been shown to up-regulate expression of defensive mechanisms (Karlowsky et al., 1997; Bagge et al., 2004; Hoffman et al., 2005; Kindrachuk et al., 2011), and adaptive tolerance has been documented in patients with chronic P. aeruginosa infections being treated with tobramycin (Barclay et al., 1996). The results depicted in Fig. 4 indicate that cells in the biofilm are responding to the presence of tobramycin, supporting the notion that a subpopulation of cells may be able to adapt physiologically in the presence of tobramycin. While this mechanism might be of limited importance at very high concentrations of tobramycin, the tobramycin levels that can be achieved in CF airways (16 to 204 µg/ml tobramycin in the fluid lining of the lung epithelia in patients treated with inhaled tobramycin (Rosenfeld et al., 2001)) suggest that this mechanism could be of importance in clinical settings. Our results also have potential implications for therapy. Addition of cationic adjuvants to tobramycin treatment of biofilm-based infections may reduce bacterial burden not only by directly increasing the efficacy of the antibiotic against the biofilm, but also by decreasing the time for an adaptive response.
EXPERIMENTAL PROCEDURES
Bacterial strains, growth conditions, and antibiotics
Bacterial strains, plasmids, and primers used in this study are listed in Table 1, S1 and S2. Bacteria were propagated at 37°C in Luria-Bertani (LB) medium (Difco). PAO1 Tn7::PrrnBP1gfpAGA was created with pBK-mini-Tn7-rrnBP1-gfpAGA using the mini-Tn7 system as previously described (Lambertsen et al., 2004; Choi and Schweizer, 2006). Cy5-conjugated tobramycin and ciprofloxacin, containing one Cy5 molecule conjugated to an amino group of one molecule of antibiotic, were custom made by Bio-Synthesis. Tobramycin sulfate and ciprofloxacin was obtained from Research Products International and Hospira, respectively. Antibiotics were used at the following concentrations: for E. coli, 50 mg/L kanamycin, 10 mg/L gentamicin; for P. aeruginosa, 30 mg/L gentamicin for chromosomally integrated strains and 100 mg/L gentamicin for plasmid-borne strains.
Table 1.
Bacterial strains used in this study.
| STRAINS | Relevant characteristics | Source |
|---|---|---|
| Escherichia coli | ||
| ccdB Survival™ 2 T1R | F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG fhuA::IS2, Smr | Invitrogen |
| NEB5α | cloning strain; fhuA2Δ(argF-lacZ)U169 phoA glnV44 ϕ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England BioLabs |
| S17.1 (λpir) | conjugation donor; F– RP4-2-Tc::Mu aphA::Tn7 recA λpir lysogen, Smr, Tcr | Lab Archive (Simon et al., 1983) |
| Pseudomonas aeruginosa | ||
| PAO1 | Wild-type | Lab Archive (Jacobs et al., 2003) |
| BTPA92 | Wild-type PAO1 tagged with the transcriptional reporter fusion PrrnBP1–gfp[AGA] integrated at attTn7; Gmr | This study |
| BTPA146 | Wild-type PAO1 tagged with the transcriptional reporter fusion PibpA–gfp integrated at attTn7; Gmr | This study |
| BTPA147 | Wild-type PAO1 tagged with promoterless gfp integrated at attTn7; Gmr | This study |
| PA14 | Wild-type; can not produce Psl (Friedman and Kolter, 2004) | Lab Archive (Rahme et al., 1995) |
| BTPA156 | PAO1 with a markerless in-frame deletion of algD; can not produce alginate (Wozniak et al., 2003) | This study |
| PAO1 ΔamgRS | PAO1 with a markerless in-frame deletion of amgRS; mutant is more sensitive to tobramycin (Lee et al., 2009) | (Lee et al., 2009) |
| BTPA158 | PAO1 with a markerless in-frame deletion of arr; biofilm of mutant is more sensitive to tobramycin (Hoffman et al., 2005) | This study |
| JJH782 | PAO1 with a markerless in-frame deletion of brlR; biofilm of mutant is more sensitive to tobramycin (Liao and Sauer, 2012) | This study |
| BTPA155 | PAO1 with a markerless in-frame deletion of ndvB; biofilm of mutant is more sensitive to tobramycin (Mah et al., 2003) | This study |
| JJH485 | PAO1 with a markerless in-frame deletion of pelF; can not produce Pel (Vasseur et al., 2005) | This study |
| JJH688 | PAO1 with a markerless in-frame deletion of phoQ; more negatively charged LPS (Ernst et al., 1999) | This study |
| JJH784 | PAO1 with a markerless in-frame deletion of PA0615 – PA0629 (R2 pyocin biosynthetic genes) | This study |
| JJH827 | PAO1 with a markerless in-frame deletion of PA0615 – PA0629 and wbpL; lacks O-antigen component of LPS (Rocchetta et al., 1998) | This study |
| PAO1 PBADpel | PAO1 with a chromosomal replacement of the native pel promoter with the araC-PBAD promoter | (Colvin et al., 2011) |
| BTPA48 | PAO1 with a chromosomal replacement of the native psl promoter with the araC-PBAD promoter | Zhao et al., in press |
Flow cell biofilm preparation
Continuous flow cell biofilm reactors were prepared and assembled as previously described (Christensen et al., 1999). Log phase cultures, grown in Tryptic Soy Broth (TSB; Difco), were diluted in 1% TSB to a final OD600 of 0.01 for PAO1-based strains and 0.05 for PA14. Flow cell chambers were then inoculated with these diluted cultures and incubated inverted for 1 h before initiation of flow. Biofilms, which were continuously supplied with fresh 1% TSB at 10 ml/h, were grown for four days at room temperature. A Zeiss LSM 510 confocal laser scanning microscope was used to image the biofilms and Volocity software (Improvision) was used for compiling image series as well as for quantification.
For the reporter biofilm experiment, four-day-old biofilms were treated with 0 or 2 µg/ml tobramycin for 9 h. The biomass was then stained with 2.5 µM Syto62 (Molecular Probes) to visualize the entire biomass before imaging.
Transport of Cy5-conjugated antibiotics
Four-day-old biofilms were treated with 21.4 µM of Cy5, Cy5-tobramycin (20 µg/ml), or Cy5-ciprofloxacin (17 µg/ml). Images of the same field of view were acquired every 2.5 min during a 30-min static incubation and a subsequent 30-min flush.
Time-lapse images were used to analyze the penetration kinetics. For each time point, the widest z-slice of the biofilm aggregate was chosen and then processed using the image processing software BioSPA (Biofilm Spatial Pattern Analysis, to be released). The edge of each biofilm was defined using the GFP signal from the cells. The average intensity of the Cy5 signal for pixels equidistant from the edge of the biofilm was quantified for each time point. A graph of the fluorescence intensity versus the distance from the exterior of the biofilm was created (Fig. 2A). Each line on the graph represents one time point and is color-coded to show increasing time with green as the starting time point and red as the ending time point.
To quantify the depth of penetration, the z-slice containing the widest part of the biofilm aggregate was again used. The ring of penetration was defined by pixels that were one standard deviation brighter than the average pixel for that field of view. The width of the ring was quantified using Volocity (Improvision).
For the concurrent cation treatment experiments, biofilms were treated with 20 µg/ml Cy5-tobramycin and the cation at the same time. Tobramycin was used at 0, 10, 50, or 100 µg/ml; manganese sulfate (SAFC) at 0.35 mM, 0.7 mM, and 1.4 mM; and magnesium sulfate (Fisher) at 1.4 mM.
MIC determination
The MIC of tobramycin, Cy5-tobramycin, ciprofloxacin, Cy5-ciprofloxacin, MnSO4 and MgSO4 for wild type PAO1 was determined in triplicate using a 96-well microtiter plate. A log phase culture at 105 cells/ml was added to a range of antibiotic concentrations in LB broth. After a 24-h incubation at 37°C, bacterial growth was enumerated by spot dilution on LB agar.
Biofilm protection assay
Four-day-old biofilms were treated statically with 0, 10, 50, or 100 µg/ml tobramycin for 30 min and then transferred to 0 or 1 µg/ml tobramycin for 4 h. Biofilms was then stained with 30 µM propidium iodide (Sigma) to visualize the dead cells.
MBEC viability assay
Biofilms were cultivated in the Calgary Biofilm Device (MBEC™ Physiology and Genetics Assay, Innovotech Inc.) as described previously (Ceri et al., 1999; Harrison et al., 2010). Routine calibration and quality control of equipment setup was carried out according to the manufacturers’ directions (Harrison et al., 2010). Wells of the device were inoculated with 2 × 106 CFU in TSB, as verified by viable cell counting. The devices were sealed and incubated at 37°C for 24 h with shaking. Pegs of the device were then rinsed with phosphate buffered saline (PBS). Mean viable cell counts were determined for four pegs according to established methods (Harrison et al., 2010).
Checkerboard arrangements of MnSO4 (0 – 20 mM) and tobramycin (0 – 4 µg/ml) were made up in 96-well microtiter plates according to standard protocols (Moody, 2004). Biofilms on the pegs of the device were inserted into these plates. Following 24 h exposure at 37 °C under static conditions, biofilms were rinsed and cells were plated for viable cell counting as previously described (Harrison et al., 2010). Synergy was defined as a ≥100-fold (or 2-log10) decrease in viable cell count at 24 h by the antimicrobial combination compared with that of the most active single agent and as a ≥100-fold decrease in viable cell count compared with the starting cell number (White et al., 1996; Harrison et al., 2008).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are supported by the National Institutes of Heath (NIH R01AI081983). B.S.T., J.J.H, T.P.Q., and M.R.P. are also supported by the NIH (R01AI077628). Additionally, W.Z., J.L.S., and A.I.P. acknowledge support from the NIH (K25AI62977). B.S.T. is supported by the Cystic Fibrosis Foundation Postdoctoral Fellowship (TSENG11F0). J.J.H. was supported by a fellowship from the Canadian Institute for Health Research. The authors would like to thank Innovotech Inc. for donating the Calgary Biofilm Devices used in this research.
REFERENCES
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl JN, Zahller J, Roe F, Stewart PS. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47:1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GG, O'Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Hoiby N. Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2004;48:1168–1174. doi: 10.1128/AAC.48.4.1168-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay ML, Begg EJ, Chambers ST, Thornley PE, Pattemore PK, Grimwood K. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J Antimicrob Chemother. 1996;37:1155–1164. doi: 10.1093/jac/37.6.1155. [DOI] [PubMed] [Google Scholar]
- Beaudoin T, Zhang L, Hinz AJ, Parr CJ, Mah TF. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J Bacteriol. 2012;194:3128–3136. doi: 10.1128/JB.06178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/aac.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Allison DG, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013 doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa . Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- Christensen BB, Sternberg C, Andersen JB, Palmer RJ, Jr., Nielsen AT, Givskov M, Molin S. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa . PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dunne WM, Jr., Mason EO, Jr., Kaplan SL. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 1993;37:2522–2526. doi: 10.1128/aac.37.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa . Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis RJ, White KG, Choi KH, Wagner VE, Schweizer HP, Iglewski BH. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2005;49:3858–3867. doi: 10.1128/AAC.49.9.3858-3867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CA, Hodges NA, Marriott C. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa . J Antimicrob Chemother. 1988;22:667–674. doi: 10.1093/jac/22.5.667. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- Hansen MC, Palmer RJ, Jr., Udsen C, White DC, Molin S. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology. 2001;147:1383–1391. doi: 10.1099/00221287-147-5-1383. [DOI] [PubMed] [Google Scholar]
- Harrison JJ, Ceri H, Stremick CA, Turner RJ. Biofilm susceptibility to metal toxicity. Environ Microbiol. 2004;6:1220–1227. doi: 10.1111/j.1462-2920.2004.00656.x. [DOI] [PubMed] [Google Scholar]
- Harrison JJ, Turner RJ, Ceri H. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa . Environ Microbiol. 2005;7:981–994. doi: 10.1111/j.1462-2920.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- Harrison JJ, Turner RJ, Joo DA, Stan MA, Chan CS, Allan ND, et al. Copper and quaternary ammonium cations exert synergistic bactericidal and antibiofilm activity against Pseudomonas aeruginosa . Antimicrob Agents Chemother. 2008;52:2870–2881. doi: 10.1128/AAC.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JJ, Stremick CA, Turner RJ, Allan ND, Olson ME, Ceri H. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc. 2010;5:1236–1254. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- Hatch RA, Schiller NL. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa . Antimicrob Agents Chemother. 1998;42:974–977. doi: 10.1128/aac.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges NA, Gordon CA. Protection of Pseudomonas aeruginosa against ciprofloxacin and beta-lactams by homologous alginate. Antimicrob Agents Chemother. 1991;35:2450–2452. doi: 10.1128/aac.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Hoyle BD, Alcantara J, Costerton JW. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob Agents Chemother. 1992;36:2054–2056. doi: 10.1128/aac.36.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson KK, Goldmann DA, Pier GB. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2005;49:2467–2473. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky JA, Zelenitsky SA, Zhanel GG. Aminoglycoside adaptive resistance. Pharmacotherapy. 1997;17:549–555. [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Hahn JS, Franklin MJ, Stewart PS, Yoon J. Tolerance of dormant and active cells in Pseudomonas aeruginosa PA01 biofilm to antimicrobial agents. J Antimicrob Chemother. 2009;63:129–135. doi: 10.1093/jac/dkn462. [DOI] [PubMed] [Google Scholar]
- Kindrachuk KN, Fernandez L, Bains M, Hancock RE. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa . Antimicrob Agents Chemother. 2011;55:1874–1882. doi: 10.1128/AAC.00935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumon H, Tomochika K, Matunaga T, Ogawa M, Ohmori H. A sandwich cup method for the penetration assay of antimicrobial agents through Pseudomonas exopolysaccharides. Microbiol Immunol. 1994;38:615–619. doi: 10.1111/j.1348-0421.1994.tb01831.x. [DOI] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, et al. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci U S A. 2009;106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012 doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146(Pt 10):2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Makin SA, Beveridge TJ. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology. 1996;142(Pt 2):299–307. doi: 10.1099/13500872-142-2-299. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody JA. Synergism Testing: Broth Microdilution Checkerboard and Broth Macrodilution Methods. In: Isenberg HD, editor. In Clinical microbiology procedures handbook. Vol 2. Washington, D.C.: ASM; 2004. 5.12.11-15.12.23. [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WW, Dorrington SM, Slack MP, Walmsley HL. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–523. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WW, Evans MJ, Slack MP, Walmsley HL. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa . J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- Ortiz-Perez A, Martin-de-Hijas N, Alonso-Rodriguez N, Molina-Manso D, Fernandez-Roblas R, Esteban J. Importance of antibiotic penetration in the antimicrobial resistance of biofilm formed by non-pigmented rapidly growing mycobacteria against amikacin, ciprofloxacin and clarithromycin. Enferm Infecc Microbiol Clin. 2011;29:79–84. doi: 10.1016/j.eimc.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol. 2008;68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- Preston CA, Khoury AE, Reid G, Bruce AW, Costerton JW. Pseudomonas aeruginosa biofilms are more susceptible to ciprofloxacin than to tobramycin. Int J Antimicrob Agents. 1996;7:251–256. doi: 10.1016/s0924-8579(96)00330-5. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rocchetta HL, Burrows LL, Pacan JC, Lam JS. Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M, Gibson R, McNamara S, Emerson J, McCoyd KS, Shell R, et al. Serum and lower respiratory tract drug concentrations after tobramycin inhalation in young children with cystic fibrosis. J Pediatr. 2001;139:572–577. doi: 10.1067/mpd.2001.117785. [DOI] [PubMed] [Google Scholar]
- Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeta M, Tanaka G, Komatsuzawa H, Sugai M, Suginaka H, Usui T. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy. 1997;43:340–345. doi: 10.1159/000239587. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech. 1983;1:784–791. [Google Scholar]
- Singh R, Ray P, Das A, Sharma M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother. 2010;65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone G, Wood P, Dixon L, Keyhan M, Matin A. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob Agents Chemother. 2002;46:2458–2461. doi: 10.1128/AAC.46.8.2458-2461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suci PA, Mittelman MW, Yu FP, Geesey GG. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 1994;38:2125–2133. doi: 10.1128/aac.38.9.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szomolay B, Klapper I, Dockery J, Stewart PS. Adaptive responses to antimicrobial agents in biofilms. Environ Microbiol. 2005;7:1186–1191. doi: 10.1111/j.1462-2920.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- Vrany JD, Stewart PS, Suci PA. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa bofilms displaying rapid-transport characteristics. Antimicrob Agents Chemother. 1997;41:1352–1358. doi: 10.1128/aac.41.6.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2004;70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RL, Burgess DS, Manduru M, Bosso JA. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob Agents Chemother. 1996;40:1914–1918. doi: 10.1128/aac.40.8.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Ajiki Y, Koga T, Kawada H, Yokota T. Interaction between biofilms formed by Pseudomonas aeruginosa and clarithromycin. Antimicrob Agents Chemother. 1993;37:1749–1755. doi: 10.1128/aac.37.9.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mah TF. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol. 2008;190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Stewart PS. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2002;46:900–903. doi: 10.1128/AAC.46.3.900-903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.