Abstract
Given that most bone marrow cells are short-lived, the accumulation of multiple leukemogenic mutations in a single clonal lineage has been difficult to explain. Here, we propose that serial acquisition of mutations occurs in self-renewing hematopoietic stem cells (HSCs). We investigated this model through genomic analysis of HSCs from six patients with de novo acute myeloid leukemia (AML). Using exome sequencing, we identified mutations present in individual AML patients harboring the FLT3-ITD mutation. We then screened the residual HSCs and detected some of these mutations including mutations in the NPM1, TET2, and SMC1A genes. Finally, through single cell analysis, we determined that a clonal progression of multiple mutations occurred in the HSCs of some AML patients. These pre-leukemic HSCs suggest the clonal evolution of AML genomes from founder mutations, revealing a potential mechanism contributing to relapse. Such pre-leukemic HSCs may constitute a cellular reservoir that needs to be targeted therapeutically for more durable remissions.
INTRODUCTION
Acute myeloid leukemia (AML) is an aggressive malignancy of hematopoietic progenitor cells with a poor clinical outcome (1, 2). A number of cytogenetic and molecular abnormalities have been identified in AML, many of which have been found to be prognostic. For example, internal tandem duplications in the FLT3 tyrosine kinase (FLT3-ITD) occur frequently in AML and define a relatively high-risk subgroup (3). Recently, next-generation DNA sequencing has been used to describe AML genomes (4, 5) and to identify new recurrent mutations (6, 7). Despite these advances, our fundamental understanding of both the genomics of leukemogenesis and the genetic heterogeneity within the precursor cells is incomplete.
Initiating mutations in the majority of AML cases are largely unknown because pre-leukemic cells are clinically silent and are outcompeted by their malignant descendants (8). Our limited knowledge of initiating mutations comes from infrequent cases of AML arising secondary to antecedent clonal bone marrow disorders or rare instances of inherited syndromes, but this does not include the large majority of de novo AML cases.
Beyond the first mutation, the requirement for multiple coding mutations to generate AML raises the question of how these mutations can accumulate in a single clone given the low rate of spontaneous mutation in hematopoietic cells and the lack of global genome instability in leukemia. We propose a model in which serial mutations and/or epigenetic events must accumulate in self-renewing hematopoietic stem cells (HSCs), unless a mutation confers self-renewal ability on a downstream cell given that mutations occurring in non-self-renewing cells will be lost (9). Early evidence consistent with this model comes from our previous investigation of AML cases associated with the AML1-ETO translocation, where Lin−CD34+CD38−CD90+ HSCs isolated from patients in long-term remission up to 150 months after therapy, produced normal myeloid colonies in methylcellulose clonal in vitro progenitor assays and not leukemic blasts, yet contained detectable AML1-ETO transcripts (10). This was followed by our studies of chronic myeloid leukemia (CML) where t(9;22) resulting in the BCR-ABL translocation occurred in HSCs yielding chronic phase disease, yet progression to blast crisis was accompanied by additional mutations resulting in the formation of leukemia stem cells at the granulocyte-monocyte progenitor (GMP) stage (11, 12). Pre-leukemic HSCs were also observed in one twin pair with discordant TEL-AML1 pediatric acute lymphoblastic leukemia (ALL) (13), and additional studies of myelodysplastic syndrome (MDS) demonstrated sequential acquisition of chromosomal abnormalities in the HSC compartment of some patients (14). These studies investigated subtypes of AML/ALL/MDS defined by karyotypic abnormalities as clonal genomic events, and predate recent advances in cancer genome sequencing and mutation analysis. However, in the current era of cancer genome sequencing, the extent to which clonal evolution occurs in a pre-leukemic HSC population remains unknown.
Here, we investigated this model and the nature of initiating mutations through the genomic and functional analysis of de novo AML and patient-matched residual HSCs at the single cell level.
RESULTS
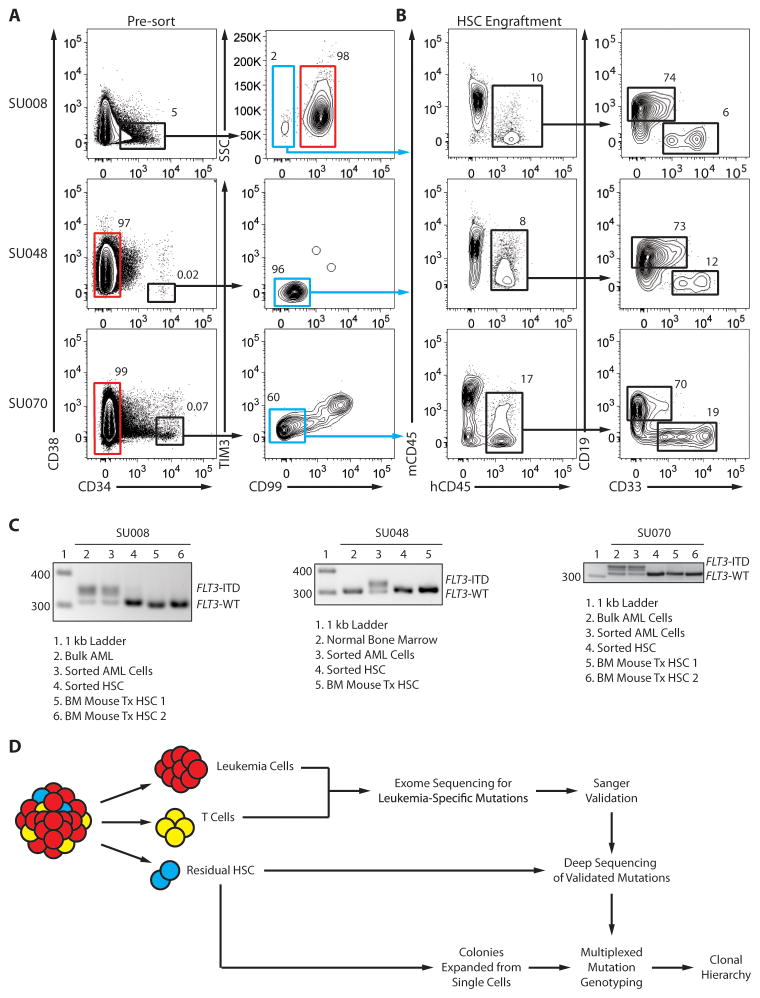
Advances in AML stem cell biology have enabled the prospective separation of residual hematopoietic stem cells (HSCs) from AML cells (15–19). We reported the prospective separation of residual HSCs from primary AML cells in diagnostic AML patient samples based on the differential expression of the cell surface proteins CD47 and TIM3 (15, 18). Six of these samples were normal karyotype AML harboring a FLT3-ITD mutation, a large relatively high-risk AML subgroup (clinical features are presented in the methods). Rare residual HSCs in the Lin−CD34+CD38− fraction were isolated from these samples by fluorescence-activated cell sorting (FACS) based on their lack of expression of the AML marker TIM3 and/or a new marker CD99 (Fig. 1A and fig. S1). The function of these cells was assessed by transplantation into immunodeficient non-obese diabetic scid gamma (NSG) mice, which resulted in long-term engraftment with both CD33+ myeloid and CD19+ lymphoid human cells, thereby establishing these cells as non-leukemic HSCs (Fig. 1B and fig. S2). Molecular analysis determined that these isolated cells, as well as their in vivo progeny, did not contain the FLT3-ITD mutation, further indicating that they were not part of the fully leukemic clone (Fig. 1C and fig. S3). However, even though they appeared to be functionally normal, we hypothesized that these residual HSCs might in fact constitute a reservoir of pre-leukemic HSCs harboring founder mutations, but lacking the complete complement of abnormalities required to generate AML.
Figure 1. Prospective separation of residual HSCs from leukemia cells.
(A) Flow cytometry analysis of samples from AML cases SU008 and SU048 indicating lineage negative cells (left panels) and Lin−CD34+CD38− subsets (right panels) further analyzed for expression of CD99 and/or TIM3. Leukemia cells (red gate) and putative residual HSCs (blue gate) were each purified via two rounds of FACS (Supplemental Figure 1). (B) Lin−CD34+CD38−CD99− cells (6000 cells) isolated from case SU008 or Lin−CD34+CD38−TIM3−CD99− cells (450 cells) isolated from case SU048 were transplanted into newborn NSG mice. 12 weeks later, bone marrow engraftment was analyzed by flow cytometry for the presence of human hematopoietic cells (hCD45+) (left panel), further subdivided into lymphoid (CD19+) and myeloid (CD33+) subsets (right panel). (C) The indicated cells were further analyzed for the presence of the FLT3-ITD mutation by PCR. Leukemia cells and HSCs from each case corresponded to the red and blue gates in panel A, respectively, and the analysis also included bone marrow from mice engrafted with the residual HSCs. (D) Experimental scheme for identification of pre-leukemic mutations and clonal evolution in de novo AML.
To identify mutations present in each AML sample, we used targeted exome sequencing (20, 21) of FACS-enriched AML cells and patient-matched CD3+ T-cells (see Fig. 1D for experimental scheme). Because the majority of T-cell development occurs in childhood, T-cells have previously been used as a germline control for the discovery of somatic mutations in adult hematologic malignancies (22, 23). AML and corresponding T cell exomes were sequenced using paired-end reads at a median depth of 67–239-fold coverage (fig. S4). The resulting sequencing data were analyzed for leukemia-specific mutations using single nucleotide polymorphism (SNP) variant calling algorithms from two independent sources and a separate algorithm for the identification of insertions or deletions (Indels). Analysis of these results identified high confidence somatic variations that were then validated by Sanger re-sequencing of genomic DNA. In addition, expression of mutant alleles was assessed by transcriptome sequencing of the leukemic cells. This analysis identified 3 to 19 variations in each of these samples that led to protein coding or 5′ or 3′ untranslated region (UTR) variants (table S1), similar numbers as reported for other AML genomes (4, 5). The 57 mutated genes of varied function and expression included FLT3, IDH1, NPM1, and TET2, which are known to be recurrently mutated in AML (table S1).
Notably, from these six patients, we identified two cases harboring mutations, R96H and R711G, in SMC1A, a gene that has recently been reported to be recurrently mutated in AML at a low frequency (24). Somatic mutations in SMC1A have been described in several cases of colorectal cancer (25). Germline mutations in SMC1A, including one reported case of R711 substitution, are causative in the congenital Cornelia de Lange Syndrome (CdLS) (26), indicating the functional role of SMC1A mutations in human disease. We conducted targeted sequencing of this locus in 120 additional AML cases and identified one further SMC1A mutation, identical to the R96H mutation detected in our original cohort. In total, we identified infrequent (3 out of 125 cases), but recurrent mutations in SMC1A (fig. S5). SMC1A is now the second cohesin complex component to be recurrently mutated in AML, as SMC3 mutations were reported in 6/200 cases elsewhere (27).
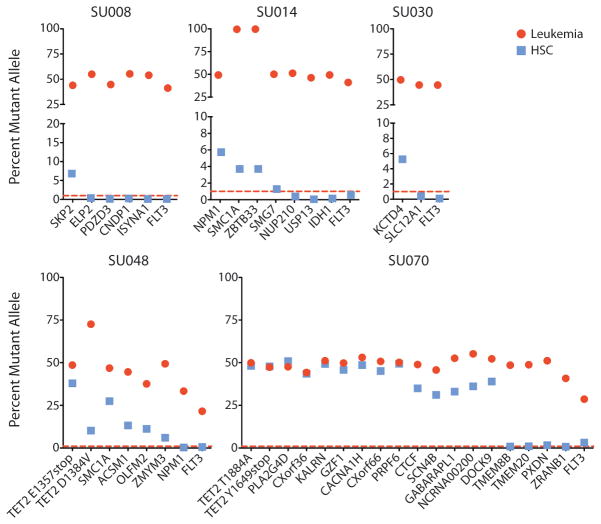
AML cells and the population of residual HSCs from each patient were analyzed for leukemia-associated mutations by targeted re-sequencing to a median depth of 24,470 reads. Consistent with the exome sequencing results, the mutant allele frequency was between 29% and 73% for most mutations in leukemia cells, suggesting that these mutations were heterozygous (Fig. 2 and table S2). The exceptions included 2 hemizygous mutations on the X chromosome in case SU014 that were present in nearly 100% of leukemic cell reads in this male patient. Sequencing of defined mixtures of normal and leukemic DNA determined that the lower threshold of sensitivity of our deep sequencing assay was less than 1% variant allele (fig. S6). Significantly, in 5 of the 6 cases (none in SU043), some, but not all, mutations were present in the population of residual HSCs, consistent with the presence of pre-leukemic cells in this population (Fig. 2 and table S2).
Figure 2. Targeted sequencing identifies pre-leukemic HSCs.
FACS-purified leukemia cells and residual HSCs from AML cases SU008, SU014, SU030, SU048, and SU070 were analyzed by targeted deep sequencing for the presence of patient-matched somatic mutations in genes with detectable mRNA expression identified in leukemia cells by exome and transcriptome sequencing. The percentage of mutant allele reads is indicated in each case. The dashed red line indicates the threshold of 1% for variant allele detection as determined by sequencing of defined mixtures of normal and leukemic DNA (fig. S6). Details of sequencing reads are presented in table S2.
In order to screen out likely passenger mutations, further investigation focused on mutations in genes found to be expressed by transcriptome analysis (Table 1 and table S1). Several mutations in genes recurrently mutated in AML were identified in residual HSCs, including the NPM1c mutation in SU014 (6% mutant allele frequency) (Fig. 2, table S2). Mutations of both alleles of TET2 were identified in residual HSCs from SU048 (40% and 10%) and SU070 (48% and 48%) (fig. S7). In addition, mutations in SMC1A were found in residual HSCs from both SU014 (4%) and SU048 (27%). In case SU008, 7% of sequenced alleles of SKP2, a known oncogene involved in regulating HSC quiescence (7, 28), were mutant. In case SU070, the R339Q mutation in CTCF is present in residual HSCs (35%). Mutation of this amino acid in the third DNA-binding zinc finger domain of CTCF has been previously described in Wilms’ tumor and alters DNA-binding specificity of the protein (29). In all 5 cases, the FLT3-ITD mutation was not detected in the residual HSCs, nor was IDH1 R132H detected in residual HSCs from SU014 (Fig. 2). In each case, the population of residual HSCs had varying allele frequencies for each mutation. However, relating the mutant allele frequency to the percentage of cells that contain each mutation may be complicated by copy number alterations (CNA) or loss of heterozygosity (LOH). To determine the percentage of cells that contain each mutation, single cell experiments were performed (see below). In summary, these results indicate that in 5 of the 6 cases of AML, a fraction of residual HSCs harbor some mutations found in the downstream leukemia cells, identifying some of these cells as pre-leukemic HSCs.
Table 1.
Somatic variations identified in five AML patients.
| Case | Somatic mutations present in leukemia and residual HSCs | Leukemia-specific somatic mutations |
|---|---|---|
| SU008 | SKP2, PDZD3, ELP2 | SEMA5A, OR4A47, CNDP1, ISYNA1, FLT3 |
| SU014 | NPM1, SMC1A, KAISO, SMG7, SLC22A10 | NUP210, USP13, SMPD3, IDH1, FLT3 |
| SU030 | KCTD4 | SLC12A1, FLT3 |
| SU048 | TET2 (biallelic), SMC1A, ACSM1, FKBP9L, GOLGA7B, NPHP4, OLFM2, ZMYM3 | RHCG, FLT3 |
| SU070 | TET2 (biallelic), CTCF, PLA2G4D, CXorf36, KALRN, GZF1, CACNA1H, CXorf66, PRPF6, SCN4B, GABARAPL1, NcRNA00200, DOCK9 | TMEM8B, TMEM20, PXDN, ZRANB1, FLT3 |
|
| ||
| Total | 32 (7) | 19 (6) |
Recurrent mutations.
Mutations in genes with RPKM gene expression <0.1.
Sanger re-sequencing of leukemia DNA and CD3+ T cell DNA confirmed all of the listed variations as somatic mutations. RNA sequencing was performed on bulk AML cells for leukemic samples SU008, SU014, SU030, and SU048 to determine which variations were expressed at a level of reads per kilobase of transcript per million mapped reads > 0.1.
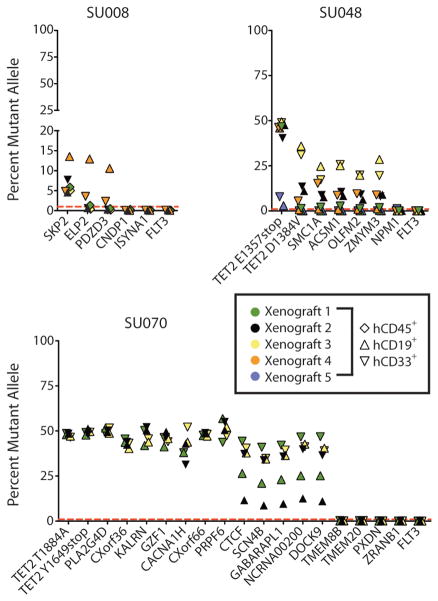
To formally demonstrate the presence of these mutations in functional HSCs, mutant allele frequencies were measured in human CD45+ leukocytes, including both CD33+ myeloid and CD19+ lymphoid cells, isolated from NSG mice transplanted with the FACS-purified residual HSCs from cases SU008, SU048, and SU070. In the case of SU008, human CD45+ cells isolated from the bone marrow of 3 mice 10–12 weeks after transplantation, representing progeny of engrafted HSCs (30), were found to contain the SKP2 mutant allele (Fig. 3). Moreover, in 1 additional mouse, SKP2, ELP2, and PDZD3 mutations were found in lymphoid and myeloid cells. In SU048, both lymphoid and myeloid cells isolated from 5 engrafted mice contained the TET2 E1357stop mutation at allele frequencies between 3% and 49% (Fig. 3). A second cluster of 5 mutations, including mutations in the remaining TET2 allele and SMC1A, was observed in myeloid cells from four engrafted mice and lymphoid cells from two of these mice. In SU070, lymphoid and myeloid cells from 3 engrafted mice contained a cluster of 14 mutations, including CTCF and both alleles of TET2 (Fig. 3).
Figure 3. Functional HSCs contain pre-leukemic mutations.
FACS-purified hCD45+, hCD45+CD19+, and/or hCD45+CD33+ cells isolated from the bone marrow of mice engrafted with residual HSCs from AML cases SU008 (n=4), SU048 (n=5), and SU070 (n=3) were analyzed by targeted deep sequencing for patient-matched somatic mutations. The percentage of mutant allele reads is indicated in each case. The dashed red line indicates the threshold of 1% for variant allele detection as determined by sequencing of defined mixtures of normal and leukemic DNA (fig. S6).
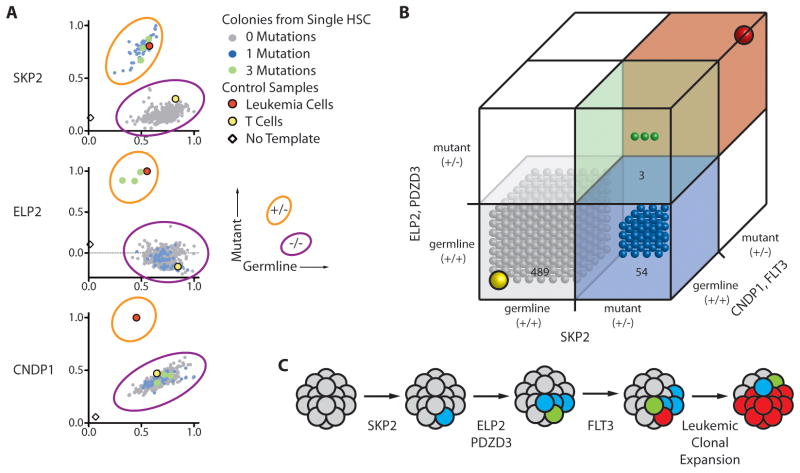
Next, we investigated the serial acquisition of mutations, and thereby clonal progression, in pre-leukemic HSCs by determining the presence of each mutation in single cells of this population. Single residual HSCs were clone sorted into 96-well plates containing methylcellulose capable of supporting myeloid colony formation. 14 days later, genomic DNA was prepped from each individual colony and analyzed for the presence of each mutation using an allele-specific SNP Taqman assay. In case SU008, 546 myeloid colonies were grown from single residual HSCs and genotyped for SKP2, ELP2, PDZD3, and CNDP1 mutations (Fig. 4A, B and fig. S8). Of these 546 colonies, 489 had none of the mutations, while 54 contained the SKP2 mutation alone. Three colonies contained SKP2, ELP2, and PDZD3 mutations, but were likely not contaminating leukemic colonies because they lacked the CNDP1 mutation. These mutation frequencies mirror those determined from targeted resequencing of this population (Fig. 2), suggesting that HSCs with or without mutations in SKP2, ELP2, and/or PDZD3 do not significantly differ in their colony formation potential, similar to the case for AML1-ETO pre-leukemic translocations (10). These results indicate that in the clonal evolution of SU008, the SKP2 mutation is a founding event, mutation of ELP2 and PDZD3 occurred next within this SKP2 mutant population, and the eventual dominant leukemic clone carried these pre-leukemic mutations, and further evolved through the mutation of ISYNA1 and FLT3 (Fig. 4C). Similarly, in case SU030, 165 myeloid colonies were grown from single residual HSCs and genotyped for KCTD4 and SLC12A1 mutations (fig. S9). Of these 165 colonies, 144 contained none of the mutations, while 21 contained KCTD4 mutations alone, indicating that the KCTD4 mutation preceded mutations in SLC12A1 and FLT3-ITD in this AML case.
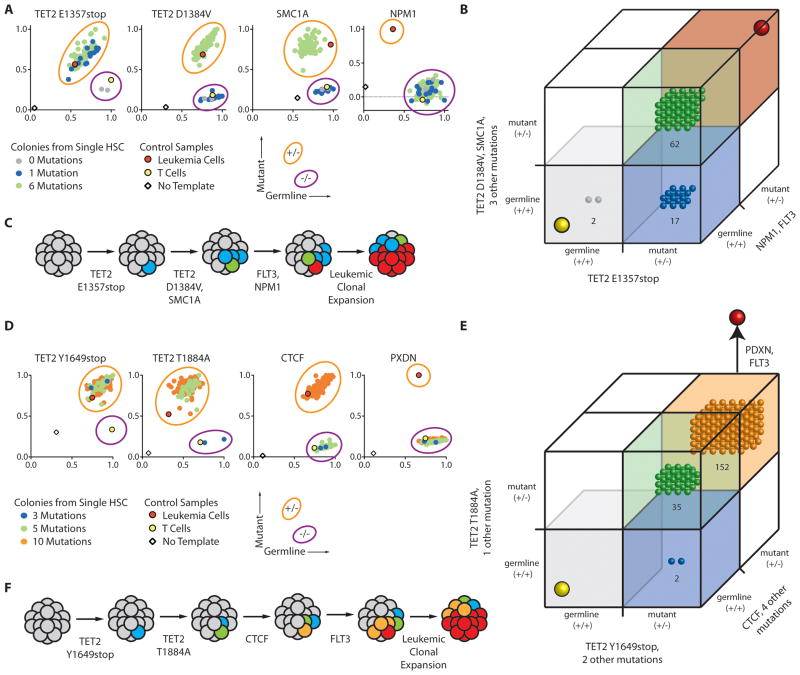
Figure 4. Single cell analysis identifies sequential mutation acquisition in pre-leukemic HSCs.
(A) The genotype of 546 myeloid colonies derived from clone sorted single residual HSCs from case SU008 were determined by multiplexed custom Taqman SNP assays for mutations identified by exome analysis to be present in each population. Selected assays are shown with the full data presented in fig. S8. Each colony is represented by a single dot in graphs for each mutation tested; colonies are colored according to genotype (see sure key). (B) 3-D plots illustrate the genotype of each colony. In both panels, the yellow dot indicates the genotype of patient T cells, and the red dot indicates the genotype of patient leukemia cells. (C) Shown is a model for the proposed clonal evolution of AML in patient SU008.
In SU008 and SU030, pre-leukemic mutations were identified in genes not previously known to be mutated in AML. In cases SU048 and SU070, pre-leukemic mutations were identified in a number of genes including TET2, a known recurrently mutated gene (31, 32), and SMC1A, which has recently been reported to be recurrently mutated in human AML (24). In SU048, we genotyped 81 myeloid colonies derived from single residual HSCs for 8 mutations identified in expressed genes (Fig. 5A, B and fig. S10). Of these 81 colonies, 17 contained only the nonsense mutation in TET2, 62 contained 6 mutations including both mutant alleles of TET2 and SMC1A, and only 2 were found to have no mutations. Thus, in this AML case, the TET2 nonsense mutation occurred first, followed by a dominant pre-leukemic clone with an additional 5 mutations including the second TET2 and the SMC1A mutations (Fig. 5C). These results mirror those observed in mouse models, where TET2-deficiency results in HSC expansion and a competitive clonal advantage (33, 34). Further clonal evolution through the acquisition of NPM1c and FLT3-ITD mutations occurred to generate the eventual AML (Fig. 5C).
Figure 5. Single cell analysis identifies sequential driver mutation acquisition in pre-leukemic HSCs.
(A, D) The genotype of 81 myeloid colonies derived from clone sorted single residual HSCs from AML case SU048 (A) or 189 colonies derived from single HSCs from AML case SU070 (D) were determined by multiplexed custom Taqman SNP assays for mutations identified by exome analysis to be present in each population. Selected assays are shown with the full data presented in fig. S10 (SU048) and fig. S11 (SU070). Each colony is represented by a single dot in graphs for each mutation tested; colonies are colored according to genotype (see figure key). (B, E) 3-D plots illustrating the genotype of each colony from SU048 (B) and SU070 (E). In both panels, the yellow dot indicates the genotype of patient T cells, and the red dot indicates the genotype of patient leukemia cells. (C, F) Models for the proposed clonal evolution of SU048 (C) and SU070 (F) are presented.
In case SU070, 189 myeloid colonies derived from single residual HSCs were genotyped for 13 mutations. None of these colonies contained 0 mutations, 2 colonies only contained 3 mutations including the nonsense mutation in TET2, 35 colonies additionally contained 2 mutations including TET2 T1884A, and 152 colonies contained the previous mutations as well as 5 others including CTCF (Fig. 5D, E and fig. S11). In this AML case, the TET2 nonsense mutation occurred first, followed by mutation of the second TET2 allele, followed by a dominant pre-leukemic clone with biallelic loss of TET2 and mutation of CTCF (Fig. 5F). After these mutations, the FLT3-ITD mutation occurred and is present in frank leukemia cells (Fig. 5F). As in case SU048, the majority of residual HSCs contained biallelic mutation of TET2. In all cases, FLT3-ITD is a late event, as this mutation is absent from the pre-leukemic compartment, which corroborates the clinical observation that FLT3-ITD can be a secondary mutation in leukemogenesis (35).
Using flow cytometric detection of residual HSCs and results of the single cell analysis, a subclonal portrait can be derived for AML cases SU008, SU048, and SU070 (fig. S12). In case SU008, residual HSC comprise 0.1% of total cells. Of these cells, 90% harbored no mutations, 10% contained the SKP2 mutation alone, and <1% were mutant for SKP2, ELP2, and PDZD3. In case SU048, residual HSCs comprise 0.02% of total cells, but unlike SU008, most of these cells were pre-leukemic and approximately 75% possessed multiple mutations including biallelic mutations in TET2. In case SU070, residual HSCs comprise 0.04% of total cells, of which all cells tested were pre-leukemic and 99% of which contained biallelic mutations in TET2.
DISCUSSION
We propose a model in which serial mutations and/or epigenetic events must accumulate in self-renewing HSCs unless a mutation confers self-renewal ability on a downstream cell, as mutations occurring in non-self-renewing cells will be lost (9). Here we provide evidence supporting this model in six cases of de novo AML through the use of advanced sequencing methods. Within the Lin−CD34+CD38− population of hematopoietic cells from de novo AML patients, we prospectively separated CD99-negative and/or TIM3-negative residual HSCs from leukemia cells, and we then identified pre-leukemic HSCs within this residual HSC population. We quantitatively assessed the fraction of total protein-altering mutations present in pre-leukemic HSCs compared to their leukemic descendants and further identified HSCs as a major target of somatic mutations antecedent to AML.
Specifically, we identified pre-leukemic mutations in residual HSCs in 5 out of 6 cases of normal karyotype AML harboring a FLT3-ITD mutation. In these 5 cases, 32 out of 51 total mutations found in leukemia cells were also present in the residual HSCs. Moreover, 7 out of 13 mutations in genes recurrently mutated in AML were also present in the residual HSC population, with the notable absence of FLT3-ITD in all 5 cases. Consequently, through deep sequencing and single cell genetic characterization of residual pre-leukemic HSCs, we established an order for the accumulation of mutations in AML. While it may turn out that FLT3-ITD mutations can occur closer to initiation in some instances, our data show that FLT3-ITD is a secondary mutation in all 5 cases in this study. Importantly, these data do not prove that the evolutionary path of AML includes pre-leukemic HSCs in all cases of de novo AML; instead we show that this model does occur in some patients. As the identification and prospective isolation of other tissue-specific stem cells occurs, for example in breast, colon, and brain, a similar approach may be used to search for premalignant progression in the tissue stem cell pool in other cancers. Such efforts would complement comparisons of primary and relapsed or metastatic tumors that have elucidated later events in the clonal progression of cancer (27, 36).
Through in vivo engraftment and in vitro single cell assays, we identified three cases with sequential pre-leukemic HSC subpopulations. According to the classic clonal evolution model of cancer (37), later and more genetically altered clones are increasingly dominant and more numerous than early, less altered clones. Our findings show that some AML cases follow this model, while others do not. In cases SU048 and SU070, pre-leukemic clones bearing biallelic mutations in TET2 were more numerous than clones carrying only one mutation in TET2, and this TET2 heterozygous population was in turn more numerous than HSCs lacking all pre-leukemic mutations. However, an opposite pattern was observed in SU008. In this case, SKP2-mutant cells expanded to occupy ~10% of the residual HSC pool, whereas descendent cells with the additional ELP2 and PDZD3 mutations only represented less than 1% of the residual HSC population. Several models may explain the relative size of these pre-leukemic subclones in cases that do not fit the classic clonal evolution model of cancer. First, some mutations may be passenger mutations. Second, some driver mutations may have functions independent of clonal advantage, such as impairment of differentiation. Third, as these mutations occurred at different points in time, the earlier pre-leukemic cells may simply have had more time to expand within the HSC pool than subsequent subclones. Consistent with the possibility that only some mutations confer a clonal advantage, large jumps in the number of mutations were observed, without identification of intermediate subclones. In cases SU048 and SU070, pre-leukemic subclones with 1 and 6 mutations and 3, 5, and 10 mutations, respectively, were found. Presumably, only the last mutation in each group confers a clonal advantage. Ultimately, the relationship between subclone size and clonal progression may be complex.
The methods used here allowed us to identify residual HSCs harboring some, but not all of the mutations found in the subsequent AML. We termed these cells pre-leukemic HSCs in that they represent progeny from a time point preceding full development of AML. Additional evidence that these cells are indeed pre-leukemic would come by demonstrating that they have an increased ability to form frank leukemia compared to HSCs lacking such mutations. The phylogenetic relationships identified here are most likely an underestimate of the true subclonal complexity of de novo AML. Although our investigation was limited to exome analysis, epigenetic, non-coding, and complex genomic events such as structural variations may contribute to pathogenesis and may help explain the relative dearth of mutations identified in case SU030. For example, we previously showed that increased expression of CD47 contributes to AML stem cell pathogenesis and clinical prognosis, but find no evidence of direct CD47 mutations (18). Moreover, increased expression of CD47 can distinguish AML leukemia stem cells (LSCs) from residual HSCs, indicating that it is a late-occurring event (18). Our study was not designed to assess clonal divergence among leukemia-initiating cells (LICs), as recently reported for acute lymphoblastic leukemia (ALL) (38, 39). Thus, our evidence for a linear clonal progression of pre-leukemic cells does not contradict evidence for complex branching phylogenetic relationships between subclones of ALL LICs (38, 39). Furthermore, we cannot detect pre-leukemic cells with mutations divergent from the same patient’s dominant presenting leukemic clone, which are a common cause of relapse in pediatric ALL (40). To identify divergent mutations within pre-leukemic subclones, techniques such as exome sequencing of single cells (41) are required. In this study, we have identified the cellular and genomic path from HSCs to the dominant presenting leukemic clone. It is reasonable to postulate that after the establishment of a frankly malignant leukemic stem cell clone, continued evolution occurs, resulting in the emergence of diverse subclones of increasing malignancy and refractoriness to therapy.
Although clonal antecedents of leukemia have been difficult to study, they may prove clinically important. Indeed, some cases of relapsed pediatric ALL arise from a clone ancestral to the presenting leukemia (40). The same may be true in AML, in which relapsed disease could develop from a pre-leukemic HSC clone that acquires additional new mutations resulting in a genetically divergent leukemic relapse. This possibility suggests that pre-leukemic HSCs constitute a cellular reservoir that may need to be targeted therapeutically for more durable remissions.
MATERIALS AND METHODS
Human Samples
Human AML samples were obtained from patients at the Stanford Medical Center with informed consent, according to Institutional Review Board (IRB)-approved protocols (Stanford IRB no. 76935 and 6453). In all cases, specimens were obtained prior to therapy. Mononuclear cells from each sample were isolated by Ficoll separation and cryopreserved in liquid nitrogen. All analyses conducted here utilized freshly thawed cells.
Animal Care
All mouse experiments were conducted according to an Institutional Animal Care and Use Committee-approved protocol and in adherence to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Flow Cytometry Analysis and Cell Sorting
A panel of antibodies was used for analysis and sorting of residual HSC within the Lin-CD34+CD38− compartment of AML samples as previously described (15, 18). CD99 antibody clone TÜ12 (BD Pharmingen) and TIM3 antibody clone 344823 (R&D Systems) were used to discriminate marker-positive leukemia cells from marker-negative HSC. In these cases, CD90+ cells were in the CD99- and TIM3- subsets, allowing exclusion of CD90 antibodies in these studies. The lineage consisted of CD3, CD19, and CD20. CD3 antibody clone SK7 was used to sort CD3+ T cells.
Exome Sequencing
Genomic DNA was purified from two sorted cell populations (reference from CD3+ T cells, leukemia from Lin-CD34+CD38-CD99+ or Lin-CD34+CD38-CD99+TIM3+ cells) and paired-end sequencing library preparation was carried out as recommended by Illumina. Targeted exome enrichment was then performed using the SeqCap EZ Exome SR kit per the manufacturer’s instructions (Roche). For additional details, see supplemental methods.
Targeted Resequencing of Leukemia-Associated Mutations
Amplicons spanning each mutation were amplified by PCR from an input of between 500 and 2000 cell-equivalents of gDNA from several different populations. Paired-end sequencing library preparation with barcoding was performed as recommended by Illumina. Reads that passed all filters were assayed for presence of the given SNP and tallied under the corresponding barcode into one of three categories: germline, “mutant” (the somatic variant discovered in leukemia cells), or “non-biological” (the remaining two possible nucleotides). INDELs were assayed for “germline” or “mutant” alleles. The mutant allele frequency was determined as follows: mutant read # / (germline read # + mutant read #). For additional details, see supplemental methods.
NSG Xenotransplantation Assay
FACS-purified cells were transplanted into newborn NOD/SCID/IL2R-gamma null (NSG) mice (Jackson Laboratory) conditioned with 100 rads of irradiation as described (30). After 12 weeks, mice were euthanized and bone marrow was analyzed for human engraftment (hCD45+) that was further characterized for lineage based on expression of myeloid (CD33+) and lymphoid (CD19+) cell surface markers. hCD45+, hCD45+CD33+, or hCD45+CD19+ cells were sorted for genetic analyses.
Single HSC-Derived Colony Genotyping Assay
Single cells were deposited into 96-well plates containing 100 μl complete methylcellulose (Methocult GF+ H4435; StemCell Technologies). For SU008, SU048, and SU070, single cells were clone sorted by FACS. For SU030, cells were diluted into media at a dilution of <1 CFU / ml. Colony formation was assayed after 14 days in culture by microscopy and scored based on morphology. Methylcellulose colony types were scored as follows: CFU-GEMM, colony forming unit –granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-E, colony forming unit-erythrocyte; CFU-GM, colony forming unit-granulocyte-macrophage. gDNA was isolated from each colony using TaqMan Sample-to-SNP kit (Applied Biosystems). Details of each Custom TaqMan SNP Genotyping Assay (Applied Biosystems) are available upon request. Multiplexed TaqMan SNP Genotyping Assays were conducted on each colony according to manufacturer’s specifications.
Supplementary Material
Fig. S1. Flow cytometry analysis of FACS-purified cell populations.
Fig. S2. Functional characterization of residual HSC.
Fig. S3. FLT3 genotyping of sorted and engrafted cells.
Fig. S4. Sequencing statistics and schematic of the data analysis pipeline used for variant identification.
Fig. S5. Schematic of SMC1A indicating identified mutations.
Fig. S6. Control sequencing to determine sensitivity of targeted re-sequencing.
Fig. S7. TET2 mutations in SU048 and SU070 occur on separate alleles.
Fig. S8. Single cell genotyping of residual HSCs from SU008.
Fig. S9. Single cell analysis identifies sequential mutation acquisition in pre-leukemic HSC from SU030.
Fig. S10. Single cell genotyping of residual HSC from SU048.
Fig. S11. Single cell genotyping of residual HSC from SU070.
Fig. S12. Methylcellulose colony formation from residual HSC and subclonal portraits.
Table S1. Summary of somatic mutations identified by targeted exome sequencing and validated by Sanger sequencing.
Table S2. Summary of deep sequencing of somatic mutations in FACS-purified cell populations.
Acknowledgments
We acknowledge Feifei Zhao and Serena Tseng for lab management, Norma Neff, Ben Passarelli, and Gary Mantalas for sequencing assistance and expertise and Emily Piccione for critical review of the manuscript. We acknowledge the Hematology Division Tissue Bank and the patients for donating their samples.
FUNDING:
M.J. is supported by the Lucille P. Markey Biomedical Research Fellowship and the National Science Foundation Graduate Research Fellowship. T.M.S. is supported by the Howard Hughes Medical Institute. M.R.C.Z. is supported by the Smith Fellowship and the National Science Foundation Graduate Research Fellowship. R.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. This research was supported by grants from the Stinehardt-Reed Foundation (to R.M.), Howard Hughes Medical Institute (to S.R.Q), NIH and Ludwig Foundation (to I.L.W.), and NIH grant U01HL099999 (to I.L.W., S.R.Q., and R.M.).
Footnotes
AUTHOR CONTRIBUTIONS:
M.J., T.M.S., and M.R.C.Z. contributed equally to this work. M.J., T.M.S., M.R.C.Z., I.L.W., S.R.Q., and R.M. designed research and wrote the paper; M.J., T.M.S., and M.R.C.Z. performed exome library preparation; T.M.S. performed transcriptome library preparation; M.J. and M.R.C.Z. performed single cell analyses; M.J., P.V., and M.R.C.Z. performed targeted resequencing of observed mutations; T.M.S. and M.R.C.Z. analyzed sequencing data.
COMPETING INTERESTS.
The authors declare no competing financial interests.
DATA AND MATERIALS:
The data for this study will be deposited in dbGaP. Requests for material should be addressed to R.M. (rmajeti@stanford.edu), S.R.Q. (quake@stanford.edu) or I.L.W. (irv@stanford.edu).
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 3.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Dohner H. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 4.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Han F, Wu J, Lee SW, Chan CH, Wu CY, Yang WL, Gao Y, Zhang X, Jeong YS, Moten A, Samaniego F, Huang P, Liu Q, Zeng YX, Lin HK. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. 2011;118:5429–5438. doi: 10.1182/blood-2010-10-312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves M. Darwin and evolutionary tales in leukemia. The Ham-Wasserman Lecture. Hematology Am Soc Hematol Educ Program. 2009:3–12. doi: 10.1182/asheducation-2009.1.3. [DOI] [PubMed] [Google Scholar]
- 9.Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. JAMA. 2005;294:1359–1366. doi: 10.1001/jama.294.11.1359. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci USA. 2000;97:7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, Jones C, Zehnder JL, Keating A, Negrin RS, Weissman IL, Jamieson CH. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci USA. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, Sawyers CL, Weissman IL. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 13.Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, Green J, Colman S, Piacibello W, Buckle V, Tsuzuki S, Greaves M, Enver T. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson L, Astrand-Grundstrom I, Anderson K, Arvidsson I, Hokland P, Bryder D, Kjeldsen L, Johansson B, Hellstrom-Lindberg E, Hast R, Jacobsen SE. Involvement and functional impairment of the CD34(+)CD38(−)Thy-1(+) hematopoietic stem cell pool in myelodysplastic syndromes with trisomy 8. Blood. 2002;100:259–267. doi: 10.1182/blood-2001-12-0188. [DOI] [PubMed] [Google Scholar]
- 15.Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, Majeti R. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki Y, Akashi K. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7:708–717. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci USA. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, Lillington D, Oakervee H, Cavenagh J, Agrawal SG, Lister TA, Gribben JG, Bonnet D. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(−) fraction. Blood. 2010;115:1976–1984. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 23.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 24.Welch JS, Larson D, Ding L, McLellan MD, Lamprecht T, Kandoth C, Payton JE, Baty J, Harris CC, Lichti CF, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Wylie T, Walker J, Westervelt P, Tomasson MH, Watson MA, Heath S, Shannon WD, Nagarajan R, Link DC, Graubert T, DiPersio JF, Mardis ER, Wilson RK, Ley TJ. Complete Sequencing and Comparison of 12 Normal Karyotype M1 AML Genomes with 12 t(15;17) Positive M3-APL Genomes. ASH Annual Meeting Abstracts. 2011;118:404. [Google Scholar]
- 25.Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannini L, Liu J, Krantz ID, Musio A. Spectrum and consequences of SMC1A mutations: the unexpected involvement of a core component of cohesin in human disease. Hum Mutat. 2010;31:5–10. doi: 10.1002/humu.21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez S, Wang L, Mumaw C, Srour EF, Lo Celso C, Nakayama K, Carlesso N. The SKP2 E3 ligase regulates basal homeostasis and stress-induced regeneration of HSCs. Blood. 2011;117:6509–6519. doi: 10.1182/blood-2010-11-321521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, Feinberg AP, Cleton-Jansen AM, Moerland EW, Cornelisse CJ, Suzuki H, Komiya A, Lindblom A, Dorion-Bonnet F, Neiman PE, Morse HC, 3rd, Collins SJ, Lobanenkov VV. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 30.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cloos J, Goemans BF, Hess CJ, van Oostveen JW, Waisfisz Q, Corthals S, de Lange D, Boeckx N, Hahlen K, Reinhardt D, Creutzig U, Schuurhuis GJ, Zwaan Ch M, Kaspers GJ. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006;20:1217–1220. doi: 10.1038/sj.leu.2404246. [DOI] [PubMed] [Google Scholar]
- 36.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 38.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 39.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, Kearney L, Enver T, Greaves M. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 40.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, Wu H, Ye X, Ye C, Wu R, Jian M, Chen Y, Xie W, Zhang R, Chen L, Liu X, Yao X, Zheng H, Yu C, Li Q, Gong Z, Mao M, Yang X, Yang L, Li J, Wang W, Lu Z, Gu N, Laurie G, Bolund L, Kristiansen K, Wang J, Yang H, Li Y, Zhang X. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Flow cytometry analysis of FACS-purified cell populations.
Fig. S2. Functional characterization of residual HSC.
Fig. S3. FLT3 genotyping of sorted and engrafted cells.
Fig. S4. Sequencing statistics and schematic of the data analysis pipeline used for variant identification.
Fig. S5. Schematic of SMC1A indicating identified mutations.
Fig. S6. Control sequencing to determine sensitivity of targeted re-sequencing.
Fig. S7. TET2 mutations in SU048 and SU070 occur on separate alleles.
Fig. S8. Single cell genotyping of residual HSCs from SU008.
Fig. S9. Single cell analysis identifies sequential mutation acquisition in pre-leukemic HSC from SU030.
Fig. S10. Single cell genotyping of residual HSC from SU048.
Fig. S11. Single cell genotyping of residual HSC from SU070.
Fig. S12. Methylcellulose colony formation from residual HSC and subclonal portraits.
Table S1. Summary of somatic mutations identified by targeted exome sequencing and validated by Sanger sequencing.
Table S2. Summary of deep sequencing of somatic mutations in FACS-purified cell populations.