Abstract
Purpose
Multimodality treatment for squamous cell carcinoma of the head and neck (SCCHN) often involves radiation (RT) and cisplatin-based therapy. Elevated activity of DNA repair mechanisms, such as the nucleotide excision repair (NER) pathway, of which ERCC1 is a rate-limiting element, are associated with cisplatin and possibly RT resistance. We have determined ERCC1 expression in HPV-negative SCCHN treated with surgery (+/− adjuvant RT/chemoradiation (CRT)).
Experimental Design
We assessed ERCC1 protein expression in archival tumors using automated, quantitative analysis (AQUA) immunohistochemistry (IHC) and three antibodies to ERCC1 (8F1 (2009, Lab Vision), FL297 (Santa Cruz) and HPA029773 (Sigma)). Analysis with Classification and Regression Tree Methods (CART) ascertained the cut-points between high/low ERCC1 expression. Multivariable analysis adjusted for age, T and N stage. Kaplan-Meier curves determined median survival. ERCC1 expression at initial tumor presentation and in recurrent disease were compared. Performance characteristics of antibodies were assessed.
Results
ERCC1 low/high groups were defined based on AQUA analysis with 8F1/2009, FL297 and HPA029773. Among patients treated with surgery plus adjuvant RT/CRT, longer median survival was observed in ERCC1 low tumors versus ERCC1 high (64 vs. 29 months, p=0.02 (HPA029773)). Data obtained with HPA029773 indicated no survival difference among patients treated only with surgery. Recurrent cancers had lower ERCC1 AQUA scores than tumors from initial presentation. Extensive characterization indicated optimal specificity and performance by the HPA029773 antibody.
Conclusions
Using AQUA, with the specific ERCC1 antibody HPA029773, we found a statistical difference in survival among high/low ERCC1 tumors from patients treated with surgery and adjuvant RT.
Keywords: ERCC1, radiation, head and neck cancer, immunohistochemistry
INTRODUCTION
Squamous cell carcinoma of the head and neck (SCCHN) is diagnosed in over 500,000 patients worldwide each year, accounting for 5% of all malignancies. In the United States some 52,000 new cases occur annually.(1) Risk factors for SCCHN include tobacco and alcohol use;(2) mounting evidence supports a pathogenic role of infection with the human papillomavirus (HPV), especially in patients lacking the usual habitual exposures.(3) p16 is a reliable surrogate biomarker for HPV-initiated oropharyngeal cancers, where p16 elevation is often associated with a favorable prognosis.(4–7) In contrast, tumors that arise from other head and neck sites such as the larynx and oral cavity are not associated with HPV infection, and have a poorer prognosis. Platinum chemotherapy using agents such as cisplatin is one important treatment for SCCHN.(8, 9), while chemoradiation is often used for SCCHN patients with high risk clinical features. (10, 11) In view of the significant morbidity of these treatments, it is important to ensure that they are administered only those patients who are likely to benefit.
Platinum-containing chemotherapies cause formation of platinum-DNA adducts, which interfere with DNA transcription and replication, and are typically controlled by activation of the Nucleotide Excision Repair (NER) pathway.(12, 13) Radiation typically induces double strand breaks (DSBs).(14) The Excision Repair Cross Complementing group 1 (ERCC1) enzyme has an essential role in the NER pathway, and also functions in the DSB pathway. ERCC1+ cell lines are more resistant to cisplatin and radiation than ERCC1- cell lines.(12, 15) These roles suggest ERCC1 expression is a potentially valuable predictor of response to chemotherapy and chemoradiation.
Scagliotti and colleagues have analyzed ERCC1 gene expression by RT-PCR in patients with advanced non-small cell lung cancer (NSCLC) treated with cisplatin and gemcitabine.(16) Among cisplatin-treated patients, those with low ERCC1 levels had increased survival of 23 versus 12.4 months (p=.001). Although these results are suggestive, RT-PCR reports mRNA rather than protein expression. Given additional factors including differential translation and stability, altered control of localization, and post-translational modifications that may affect enzymatic activity, results with protein may differ significantly from results with mRNA.(17, 18) Given these issues, we have used an immunohistochemistry (IHC) based platform in order to determine tissue ERCC1 levels.
A retrospective standard IHC analysis for ERCC1 protein expression has also been conducted on tumor specimens from the International Adjuvant Lung Trial (IALT), in which patients received cisplatin-based therapy. (19) In the original publication, the survival benefit from adjuvant chemotherapy was confined to the 56% of patients whose tumors were ERCC1 low. However, recent data from the same group have not reproduced these results in other adjuvant datasets.(20) Their report has also raised questions of antibody quality, and of whether IHC is a suitably precise tool for quantifying DNA repair biomarkers.(20)
In SCCHN, ERCC1 expression levels have been commonly studied with standard IHC using an H score scale with review from a pathologist, which renders ranking of ERCC1 expression subject to variation among pathologists. This prompted us to evaluate the ability of quantitative IHC analysis using AQUA (Automated Quantitative Analysis, HistoRx) to measure ERCC1 expression levels in archival tissue. AQUA provides a highly reproducible platform suitable for development as a clinical test.(21, 22) Further, due to the ability to localize signals associated with a tumor mask (which eliminates signal from adjacent stroma), or to the cytoplasmic or nuclear compartments of tumor cells, it is possible to quantify the marker of interest within specific subcellular regions of the most relevant cell type. This permits more accurate quantification of ERCC1 in the nuclear compartment, where it is known to localize.(23)
A critical concern in IHC studies is the quality of the antibody used. A growing literature surrounds the use of IHC analysis for ERCC1 as a biomarker in other malignancies.(19, 24–29) The Lab Vision 8F1 antibody has been most widely used in retrospective series from multiple tumor types, and has provided data with an ERCC1 cut point associated with survival differences in NSCLC and SCCHN, but recent batches of this antibody been criticized for lack of specificity (and for lot-to-lot variability), making its continued use as a reagent problematic, and calling into question the results of studies (including IALT) performed during the past decade, as the quality of this preparation deteriorated.(20, 30, 31) In this study, we have carefully compared early batches of Lab Vision 8F1 antibody (8F1/2009), but also with FL297 (Santa Cruz) (which is reported to be more specific for in vitro applications)(32), and a new reagent, HPA029773 (Sigma), that has not previously been characterized in SCCHN. We also supported this clinical study with parallel biochemical analysis of these antibodies in cell line models, to address the question of specificity for the ERCC1 protein. Our primary goal was to study the correlation of ERCC1 and survival in a retrospective tissue analysis of patients who were treated with either surgery or surgery plus adjuvant radiation.
MATERIALS/METHODS
Construction of tissue microarrays and annotation of clinical data
SCCHN surgical specimens from the Fox Chase Cancer Center Biosample Repository were used to construct tissue microarrays (TMA). Tissue from each tumor was placed in 2 unique spots on each TMA. Cases are a random representation of tumors resected in our facility from 1990 to 2007. All samples were obtained from primary tumors and/or nodal metastases at the time of initial resection. A small set of recurrent squamous cell cancers was also randomly sampled. Clinical information was available from the repository database and extracted from clinical databases in anonymized fashion. At the time of tissue acquisition patients provided IRB-approved informed consent for storing tissue and reviewing clinical data. Controls included SCCHN specimens with varying levels of ERCC1 expression.
Cell culture, siRNA, and quantitative RT-PCR
HeLa cervical adenocarcinoma cells and FaDu SCCHN cells (ATCC), and SV40-transformed XP2YO skin fibroblasts (Coriell Institute for Medical Research, Camden, NJ), were cultured as recommended by suppliers. Transfection with siRNAs employed RNAiMAX (Invitrogen, Carlsbad, CA). ERCC1 was depleted using Human ERCC1-ON-TARGET-plus SMARTpool (NM_001983) from Dharmacon (Lafayette, CO). The four siRNAs in this pool bind areas in common between all reported ERCC1 isoforms, and hence are predicted to deplete all ERCC1 species. Negative scrambled control was purchased from Dharmacon (Lafayette, CO). For evaluation of ERCC1 knockdown, total RNA was extracted using the RNeasy Mini Kit (Qiagen) 48 hours after transfection, reverse-transcribed using standard approaches, and analyzed by Taqman chemistry using Assay-on-Demand Hs01012158_m1 for ERCC1. Expression was normalized to housekeeping gene POLR2F, for which the primers and probe sequences were TGCCATGAAGGAACTCAAGG, TCATAGCTCCCATCTGGCAG and 6fam-CCCCATCATCATTCGCCGTTACC-bhq1. The assays were validated with a 4 fold 4-points dilution curve of cDNA.
Western blots and immunoprecipitation (IP)
Cells were lysed in CelLytic M lysis buffer from Sigma-Aldrich (St. Louis, MO) supplemented with Halt™ protease and phosphatase inhibitor cocktail from (Thermo Scientific, Rockford, IL). Tumor tissues were homogenized in T-PER Tissue protein extraction reagent from (ThermoScientific, Rockford, IL)) supplemented with Halt protease and phosphatase inhibitor cocktail on ice, then cleared by centrifugation. IP samples were incubated overnight with antibody at 4°C, the incubated for two hours with protein A/G-Sepharose (Thermo Scientific, Rockford, IL), washed, and resolved by SDS-PAGE. Western blotting used standard procedures and was developed by chemoluminescence using Luminata Western HRP substrates (Classico, Crescendo and Forte) from EMD Millipore (Billerica, MA). Primary antibodies included anti-ERCC1 (FL-297) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-ERCC1 (8F1) from Neomarker (Fremont, CA), anti-ERCC1 (HPA029773) from Sigma-Aldrich (St. Louis, MO), and mouse anti-β-actin conjugated to HRP (#ab49900) from Abcam (Cambridge, United Kingdom). Secondary anti-mouse and anti-rabbit HRP-conjugated antibodies from GE Healthcare (Little Chalfont, United Kingdom) were used at a dilution of 1:10,000. Image analysis was done using ImageJ (National Institute of Health, Bethesda, MD), with signal intensity normalized to β-actin.
Fluorescent immunohistochemistry (F-IHC)
Tissue sections were deparaffinized in xylene and rehydrated in a graded series of ethanol. The sections were subjected to antigen retrieval by boiling in 0.01 M sodium citrate buffer (pH 6.0) in a steamer for 20 minutes and allowed to cool in the buffer for 20 minutes. Endogenous peroxidase activity was blocked by incubating the slides in 3% hydrogen peroxide in absolute methanol for 15 min and washed once with distilled water and three times with phosphate buffered saline (PBS). After limiting nonspecific binding with blocking reagent (Protein Block Serum-Free, 162 DAKO) for 30 min, the sections were incubated overnight with ERCC1 antibody (1:100, 8F1/2009, Lab Vision; 1:400 FL297/2010, Santa Cruz; or 1:5000 HPA0297731/2011, Sigma) and pan-cytokeratin (tumor mask) in antibody diluent Da Vinci Green (Biocare Medical, PD900) at 4°C overnight. The pan-cytokeratin was probed with an Alexa Fluor 555 dye-labeled secondary antibody (Invitrogen). The primary antibody was targeted with Envision reagents (DAKO, Carpenteria, CA), which provides multiple horseradish peroxidase (HRP) moieties, intensifying signal. Target amplification and visualization was accomplished using a Cy-5-tyramide signal amplification system (TSA, PerkinElmer, Waltham, MA. Cat. AT705A): the Cy-5 (far-red) emission peak is well outside the green-orange spectrum of tissue autofluorescence. Prolong Gold mounting medium (P36931; Molecular Probes) containing 4,6-Diamidino-2-phenylindole (DAPI) was used to stain tissue nuclei. Positive and negative controls, discussed in Results, were stained simultaneously.
Image Acquisition and AQUA analysis
Automated image capture was performed by the HistoRx PM-2000 (HistoRx, New Haven, CT), using the AQUAsition software. High-resolution monochromatic digital images of the cytokeratin staining visualized with AF555, DAPI and target staining with Cy5 were captured and saved for each tumor histospot. Tumor mask was created from the cytokeratin image of each histospot, representing areas of epithelial tumor. Histospots were excluded if the tumor mask represented less than 5% of the total histospot area. DAPI immunoreactivity defined the nuclear compartment. The cytoplasmic compartment was defined by the tumor mask. Images were visually inspected and cropped for unfavorable factors such as “out of focus”, debris or damaged specimen before automatic analysis.(33) An AQUA score was generated by dividing the sum of target signals within the tumor mask. AQUA scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be compared directly. The nuclear scores from two nonoverlapping images were averaged for each case.
Standard Immunohistochemistry for ERCC1 and p16
Immunohistochemical staining was performed on 5-μm slides. Deparaffinization, rehydration, epitope retrieval, blocking of peroxidase activity and non-specific protein binding, and primary antibody incubation were as for F-IHC. Primary antibodies included ERCC1 (8F1/2009, Lab Vision, 1:100) and p16 (E6H4, MTM Laboratories, 1:3000), with mouse IgG as negative control. Immunodetection was performed with the Dako Envision+ system. The antigen-antibody immunoreaction was visualized using 3–3′-diaminobenzidine as the chromogen. The slides were washed, counterstained with hematoxylin, dehydrated with alcohol, cleared in xylene, and mounted. Patient samples that were shown previously to express high levels were used as positive controls.
Standard IHC Evaluation (H-score)
The slides were viewed and scored by a single pathologist. Immunoreactivity was semi-quantitatively scored using a well-established immunoreactivity score system (H-Score) by multiplying both the percentage of positive tumor cells (0~100%) and the intensity of staining (0–3). H-score range is 0~300.
Statistical analysis
Only HPV-negative primary tumors were included in the final statistical analysis (all oropharynx primary tumors in which p16 was positive or unknown were excluded). Nuclear ERCC1 AQUA scores were analyzed for all antibodies used. Survival time was measured as the number of months between diagnosis and death due from any cause. Individuals who were alive at time of last follow-up were considered censored. Overall survival time distributions were plotted by stage and level of nuclear ERCC1 expression using Kaplan-Meier curves and median survival times were estimated. Classification and Regression Tree (CART) methods for failure time data (34) were used to identify optimal cut points for nuclear ERCC1 expression levels with respect to the association with survival time within the population treated with adjuvant radiation therapy, separately for each antibody. Next, Cox proportional hazards models were used to compare survival times between nuclear ERCC1 expression level groups (above or below the optimal cut point) while adjusting for patient age, gender and T/N stage. These analyses were performed separately for surgery only and adjuvant radiation therapy groups using the same ERCC1 expression cut point. To assess the validity of our method, we randomly sampled 75% of the entire dataset as a training set and used the remaining 25% as a test set. Harrell’s C index was computed based on the multivariable Cox model on the test set.(37) Harrell’s C index measures the agreement of predictions with observed failure order. It is defined as the proportion of all usable subject pairs in which the predictions and outcomes are concordant. C-index ranges between 0 and 1 with a value of 0.5 indicating no predictive ability. We repeated this procedure 50 times and the C-index ranged between 0.70–0.89 with mean of 0.78, indicating good predictive ability of the CART method. These permutations were performed using HPA029773 (Sigma) expression data derived from patients treated with adjuvant radiation therapy. Spearman’s correlation coefficient was computed to assess the association between nuclear Aqua and H-scores measured on the same tissue sample. Wilcoxon ranked sum tests were used to compare ERCC1 expression values between primary and recurrent tissue samples. All test were two-sided with a 5% type-I error. Statistical analyses were conducted using Stata 12 (Stata Corporation, College Station, Texas).
RESULTS
Tissue Microarray Patient Characteristics
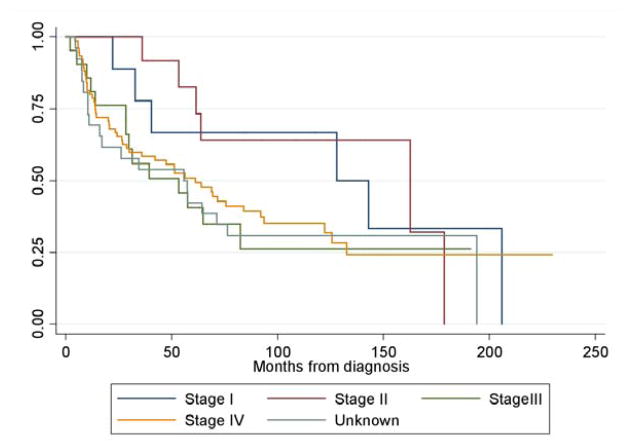
TMAs were initially constructed from tissue obtained from 105 patients at the time of surgery at FCCC for newly diagnosed SCCHN. Primary sites included tongue, tonsil, glottis, pyriform sinus, and non-tongue oral cavity (Table 1). The patient population is representative of SCCHN disease, as illustrated by the overall survival curves of the patients analyzed across all stages. (Figure 1) The limited number of oropharynx tumors reflects institutional practice in which such patients were predominantly treated with definitive chemoradiation. Thirty-three were treated with surgery alone and 72 received adjuvant RT; a minority (N=7) were treated with platinum-based chemoradiation (CRT). Standard indications for adjuvant therapy included multiple involved lymph nodes, close or positive margins or extracapsular spread of disease.
Table 1.
Patient characteristics of initial and expanded TMAs
| Initial TMA | Expanded TMA | |
|---|---|---|
| Primary Site | ||
| Oral Cavity | 46 | 51 |
| Tongue | 33 | 42 |
| Tonsil | 2 | 2 |
| Glottic | 15 | 13 |
| Pyriform Sinus | 3 | 4 |
| Other | 6 | 6 |
| Gender | ||
| Male | 66 | 76 |
| Female | 39 | 42 |
| Race | ||
| Caucasian | 95 | 107 |
| Non-caucasian | 10 | 11 |
| T stage | ||
| 1 | 14 | 15 |
| 2 | 23 | 33 |
| 3 | 14 | 14 |
| 4 | 32 | 38 |
| Unknown | 22 | 18 |
| N stage | ||
| 0 | 40 | 41 |
| 1 | 12 | 15 |
| 2 | 31 | 41 |
| Unknown | 22 | 18 |
| Treatment | ||
| Surgery alone | 33 | 38 |
| Radiation | 65 | 63 |
| Chemoradiation | 7 | 17 |
| Total | 105 | 118 |
Figure 1.
Overall survival by stage for total patient population. Median survival time Stage I: 128 months, Stage 2: 163 months, Stage 3: 53 months, Stage 4: 61 months, Unknown: 56 months.
Quantitative Immunohistochemistry Analysis
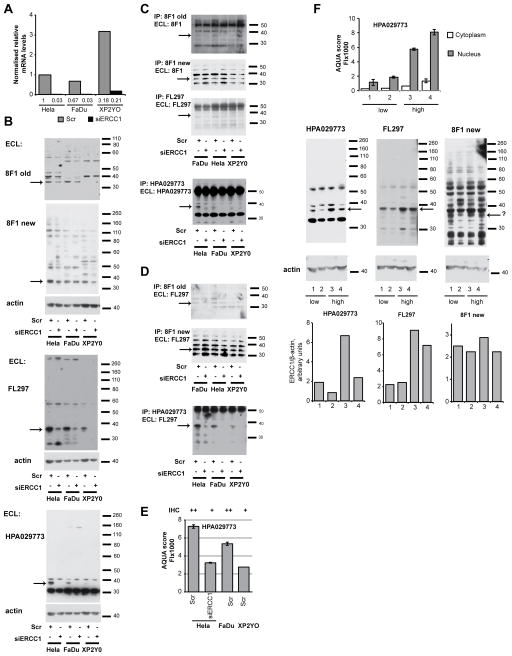
In order to optimize the AQUA-based assay with increased reliability, we analyzed these TMAs with three different ERCC1 antibodies: 8F1/2009, FL297, and HPA029773 (Supplemental Figure S1A). A 2009 lot of the antibody 8F1 (Lab Vision), produced prior to the time of reported deterioration in performance (20), represents a standard much reported in the literature. However, given increasing concerns about reproducibility and specificity with the 8F1 antibody (30), we evaluated expression of nuclear ERCC1 on these TMAs with additional antibodies. FL297 (32) is a rabbit polyclonal IgG developed against full length ERCC1. This reagent has been reported to be highly specific for ERCC1 by Western and immunoprecipitation analysis (32), although it has not been certified by its manufacturer for IHC. Finally, HPA029773 is a more recently developed polyclonal affinity-isolated antibody that targets ERCC1 with reported high specificity.(35) These three antibodies indicate a wide expression range of ERCC1 between different tumor specimens, indicating a potentially useful dynamic range. In most specimens, 8F1/2009 and HPA029773 staining was largely restricted to the nuclear compartment. In contrast, FL297 showed nuclear staining in some cases, but in other specimens, the staining was ambiguous with considerable cytoplasmic staining, raising some concerns. (Supplemental Figure S1A)
AQUA analysis
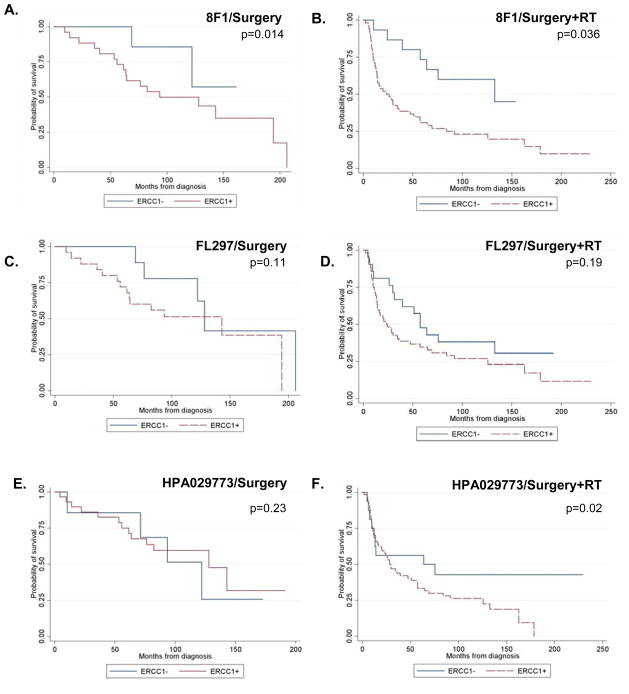
The cut point for nuclear ERCC1 expression (8F1/2009) based on CART was established at the 23rd percentile of AQUA scores from this cohort of tissue (172 in the nuclear compartment) (Figure 2A, 2B). With this value, in a multivariable analysis adjusted for age, gender, T and N stage, a higher median survival (133 months) was observed in ERCC1-low tumors compared to ERCC1-high (26 months) (p=0.036, Hazard Ratio (HR)=2.35) in SCCHN treated with adjuvant radiation therapy. At this cut point, there was a significantly increased survival among the patients who were treated with surgery alone as well (p=0.014). However, we note that in this cohort there were only seven patients in the ERCC1-low group, and merely two events. As an additional control, specimens stained with 8F1 antibody were also assigned H scores using standard IHC; these results indicated a strong correlation between AQUA and H scores (r=0.69), indicating robust performance of the AQUA technology (Supplemental Figure S1B)
Figure 2.
Kaplan-Meier curves showing overall survival based on nuclear ERCC1 level in patients treated with (A, C, E) surgery alone, or (B, D, F) surgery plus adjuvant radiation (RT) A, B. Specimens were stained with 8F1: The cut point used was the 23rd percentile (172) of nuclear staining. C, D. Specimens were stained with FL297: the cut point used was the 38th percentile (2492). E, F. Specimens were stained with HPA029773: the cut point used was the 28th percentile, 2136) N=33 for surgery and n=72 for surgery+RT for 8F1 and FL297; n=38 for surgery and n=80 for surgery+RT for HPA029773.
In comparison, FL297 did not provide a cut point for high/low ERCC1 expression that was associated with a significant survival difference (Figure 2C, 2D). With this antibody, a trend towards improved survival among the patients who had surgery plus radiation was observed at the 28th percentile cut point (2492 in the nuclear compartment). However, this was not statistically significant (p=0.19) after adjusting for patient age, gender and T/N stage.
We then used HPA029773 to stain 97 of the original specimens, augmented with 21 additional clinical samples obtained from the Fox Chase Cancer Center Biosample Repository, for a total of 118 cases (see Table 1). Thirty-eight patients were treated with surgery alone; 80 received adjuvant treatment, either with radiation or chemoradiation. At a cut point of 2136, which was the 28th percentile, there were 24 ERCC1 low cases and 94 ERCC1 high cases (Figure 2E, 2F). For this new reagent, in additional assessment, we found that the ERCC1-low specimens showed the same level of cytokeratin and DAPI staining as the ERCC1-high specimens, indicating that the difference of ERCC1 expression is intrinsic and not due to differences in tissue preservation (Supplemental Figure S2). Using the HPA029773 antibody on the AQUA platform, there was a statistically significant difference in survival (p=0.02, HR=2.72) after adjusting for patient age, gender and T/N stage, with an overall median survival of 64 months in the low ERCC1 group and 29 months in the high ERCC1 group among patients receiving adjuvant radiation/chemoradiation therapy. Among patients treated with surgery alone, there was no difference in overall survival between the two ERCC1 groups (p=0.23).
Based on AQUA nuclear scores, there was weak to moderate relationship of ERCC1 high/low assignment using the CART-based cutpoints between all three antibodies, with 82% concordance between 8F1 and FL297, 75% agreement between 8F1 and HPA029773, and 73% between FL297 and HPA209773.
Recurrent Disease
Tissues were available from a cohort of SCCHN patients with recurrent cancer. In further analysis with HPA029773, we sought to determine if there was a difference in nuclear ERCC1 levels among previously untreated cancers and tumor samples from sites of recurrent disease (Figure 3). Specimens from recurrences were available from oral cavity, oropharynx, and laryngeal tumors. Initial treatment was heterogeneous and included radiation, chemoradiation, surgery, and surgery plus radiation (Supplemental Table 1). This series includes 15 non-recurrent nodal metastases which were removed at the time of the original presentation of malignancy, 11 recurrent nodal metastases, 20 recurrences at the primary site, and 43 untreated primary cancers (which were randomly selected from the original TMA). There was an insufficient number of matched previously untreated and recurrent specimens from the same patient for direct intra-patient comparison. Overall, the ERCC1 AQUA values were lower in recurrent tumors (p<0.01) with a mean score of 1243 compared to 2879 for previously untreated tumors. Non-recurrent nodes, recurrent nodes and primary tumors all had similar values.
Figure 3.
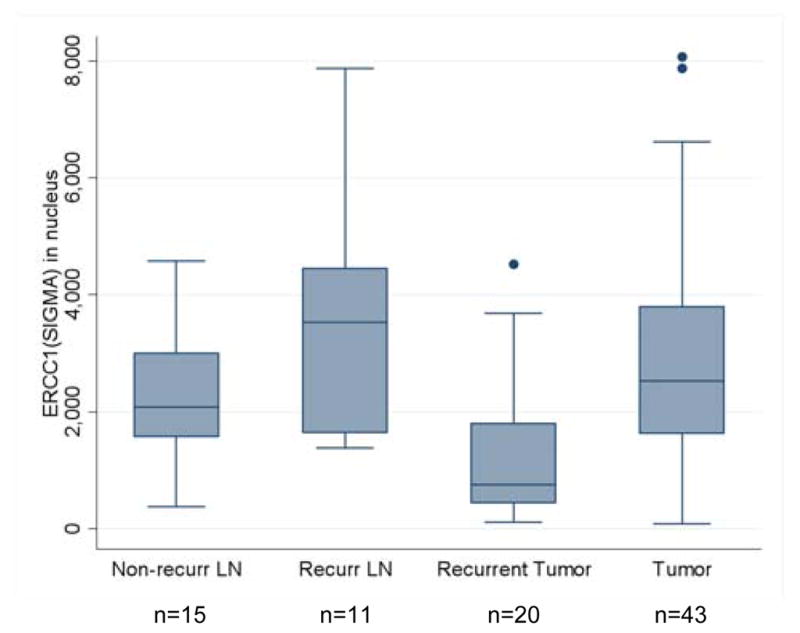
Comparison of ERCC1 values with HPA029773 between primary and recurrent tumor. Overall, values for the recurrence tumor are smaller (p<.01). Non-recurrent lymph nodes, recurrent lymph nodes and primary tumors have similar values.
Comparison of Antibodies by Western and Immunoprecipitation Analysis
Our data demonstrated non-equivalent behavior of three different commercial reagents for detection of ERCC1 in predicting clinical outcome. Several recent studies have emphasized changes in behavior in the erstwhile gold standard 8F1 affecting antibody lots produced in recent years(20) (31), while FL297 and HPA029773 have not been as extensively characterized. To address this issue, we compared 1) the early batch of 8F1/2009 used here for AQUA and IHC on tumors, 2) a more recent batch of 8F1 (from 2010), 3) FL297, and 4) HPA029773, using Western and immunoprecipitation analysis (Figure 4). As a test set for evaluation of antibody specificity, we used the HeLa, FaDu, and XP2YO cell lines (in this last cell line, a mutation eliminating the ERCC1 partner XPF destabilizes ERCC1 protein)(36), transfected with an siRNA to deplete ERCC1 or a negative control scrambled siRNA. Control assessment using quantitative RT-PCR confirmed very effective depletion of ERCC1 mRNA using the ERCC1 siRNA (Figure 4A). Western analysis indicated that 8F1/2009 and FL297 both identified a single species with a molecular weight corresponding to the reported gel mobility of ERCC1 (~35–38 kDa) whether analyzed by Western (Figure 4B) or immunoprecipitation (IP) (Figures 5C, D). HPA029773 unambiguously identified a single specific ERCC1 species, but additionally identified a prominent cross-reacting species (Figures 4B, C). In contrast, the recent batch lot of 8F1 did not detect a specific species either by Western or by IP analysis (Figure 4B–D), agreeing with other recent studies indicating a loss of performance. (20, 31)
Figure 4. qRT-PCR, Western, and IP analysis of ERCC1 in cell lines and tumor lysates.
A. Expression of ERCC1 following transfection with siRNA to ERCC1 (siERCC1) or scrambled (Scr) non-specific siRNA in HeLa, FaDu and XP2YO cell lines. Data are normalized to the HeLa-Scr combination; numbers below bars represent quantification. Note, basal mRNA levels are elevated in the XP2YO cell line, likely in compensation for the destabilization of the protein associated with absence of XPF/ERCC4. For each sample, the values are average and standard deviation of data from two PCR reactions performed with the cDNAs from two RT reactions. B. Western detection of ERCC1 protein in HeLa, FaDu and XP2YO cells transfected with siERCC1 or Scr, using old and new batches of 8F1 antibody, FL297 antibody and HPA029773 antibody as indicated. C. ERCC1 immunoprecipitated and visualized with old and new batches of 8F1, FL297, or HPA029773 antibody from indicated cell lines transfected with siERCC1 or Scr. D. ERCC1 immunoprecipitated with old and new batches of 8F1 antibody and HPA029773 antibody indicated cell lines, detected by FL297 antibody. E. AQUA and IHC scores generated following analysis of FFPE cell pellets from HeLa, FaDu and XP2YO cells transfected with the indicated siRNAs. F. Graph indicates expression levels of ERCC1 in 4 tumor samples characterized by AQUA. Below, Western blots indicate ERCC1 levels detected in these tumor lysates with low (lanes 1 and 2) and high (lanes 3 and 4) expression levels of ERCC1 as characterized by AQUA.
In AQUA analysis, low ERCC1 expression as measured with FL297 did not predict an increased survival benefit, in contrast to data generated with 8F1/2009 or with HPA029773. One potential explanation suggests that FL297 does not recognize ERCC1 following preparation of FFPE tissue, and hence, AQUA data with this antibody reflect non-specific cross-reactivity, while early batch 8F1 and HPA029773, despite having partial cross-reactivity with non-ERCC1 proteins detectable by Western, predominantly react with ERCC1 in IHC and AQUA analysis. If this hypothesis is correct, ERCC1 is predictive of overall survival. An alternative possibility is that FL297 accurately and specifically identifies ERCC1 in IHC and AQUA analysis, while early batch 8F1 and HPA029773 do not. In that scenario, ERCC1 expression does not predict overall survival. To discriminate between these models, we used AQUA and IHC to analyze FFPE cell pellets prepared from some of the cell lines used for Western and IP analysis (Figure 4E). In this setting, the reduced expression of ERCC1 associated with siRNA depletion or with the XP2YO genotype was reflected by reduced HPA029773 signal levels, consistent with a specific signal. In reciprocal analysis, in Western blotting and IP of tumor lysates from tumors characterized by AQUA using the HPA029773 antibody as high or low for ERCC1, signal variance detected by the FL297 and HPA029773 reflected ERCC1, rather than the non-specific cross-reacting band, again supporting the idea of a specific signal for HPA029773 in AQUA analysis (Figure 4F). Taken in sum, these data are most simply interpreted as indicating the presence of a common, prognostic protein that is part of a larger pool of cross-reacting species detected by 8F1 and HPA029773.
DISCUSSION
In summary, our AQUA data indicate that low nuclear ERCC1 expression detected by either 8F1/2009 or current HPA029773 antibodies is associated with a survival benefit among HPV-negative SCCHN patients who received adjuvant radiation after surgery. Since HPV-negative tumors are known to be less sensitive to chemoradiation, this association provides a potential mechanistic explanation for this well-established clinical finding. Importantly this work includes a cohort of patients treated with surgery alone; ERCC1 biomarker data have not been studied previously in such patients, because patients in all prior reports received chemoradiation. With HPA029773, there was no appreciable difference in survival associated with ERCC1 expression among these patients with a more favorable prognosis, who needed no adjuvant therapy and were treated with resection alone. Our finding that low ERCC1 expression correlates with improved treatment outcomes is consistent with previous reports.(24, 25, 37) The finding that recurrent disease shows lower ERCC1 expression levels was unexpected and potentially reflects altered DNA repair capacity in response to prior therapy; this warrants study in a larger series. Our retrospective, non-randomized series suggests, but does not prove, the hypothesis that ERCC1 is a predictive marker for radiation sensitivity in SCCHN. Similarly, it suggests, but does not prove the hypothesis that ERCC1 expression has no influence on the prognosis of SCCHN, a factor independent of treatment.
Other retrospective studies of ERCC1 levels and correlation to chemotherapy response and survival in SCCHN have been reported.(24, 25, 38) In one of the larger series, Handra-Luca and colleagues studied specimens from 107 patients with SCCHN who had received cisplatin based induction therapy. In this report, which was published in 2007, ERCC1 expression was also analyzed with an older batch of 8F1 and assigned an H sore with standard IHC. 71% of tumors had high level expression, while 29% had low level expression. This later group derived a fourfold greater response and benefit to cisplatin based induction therapy, as well as an increase in median survival.(24) However, additional series yielded conflicting results, thus engendering controversy whether ERCC1 is truly a marker for platinum resistance and if IHC is the best method to interrogate this marker. (38, 39)
Many previous reports employed the 8F1 ERCC1 antibody, with specific batch undefined; most reports in the past two years probably involve 8F1 batches that have lost specificity for ERCC1.(24, 25, 37) In a 2012 ASCO report, Austin et al. analyzed tissue from 84 platinum-treated patients who were enrolled in a clinical trial for locally advanced SCCHN. In standard IHC reported as an H score, and cut point established at the median, 8F1-based determination of high ERCC1 expression (46%) was not associated with an inferior response rate or progression free survival (PFS), while in FL297-based determination high ERCC1 levels were associated with a significantly inferior PFS.(40) Similarly, in 2012 Hao et al. also reported a retrospective AQUA IHC using 8F1 and FL297 to analyze 55 SCCHN tumors from patients who underwent chemoradiation.(38) Results with 8F1 indicated no difference in survival, while with FL297, there was a significant relationship between low ERCC1 expression and increased overall survival (p=0.004). This relationship remained significant among the HPV-negative tumors, but not the HPV-positive group, which comprised half of their study population. This work in general supports the results of our study, conducted entirely in an HPV-negative group. However, while our studies with FL297 suggest a survival difference predicted by this reagent, statistical significance was not reached. Why FL297 would be prognostic in the Hao et al. paper but not our series is unclear. In prior analyses, and consistent with our experience with automated platforms, FL297 nuclear staining was weak and cytoplasmic staining strong, suggesting cross-reactivity in tissues fixed for IHC which might account for the varying results observed with FL297.(41, 42)
Accurate and reproducible methods are essential when assaying clinical biomarkers. The recent report by Fibroulet et al (20) in lung cancer emphasizes the importance of integrating careful controls into the performance of IHC-based biomarker studies, particularly when interrogating ERCC1. Their report is consistent with our conclusion that currently available lots of 8F1 should not be used in future biomarker studies. While Fibroulet et al. conclusively showed that, they did not re-evaluate the lung tumors in their cohort with additional antibodies against ERCC1. Based on Western blot analysis, our data indicate that HPA029773 is a reasonable replacement for the early, “specific” batches of 8F1 in IHC/AQUA applications for research applications. Fibroulet also noted that all commercially available antibodies to ERCC1 react with multiple isoforms, some of which are catalytically inactive, which may obscure interpretations. However, there are limited data regarding the role of ERCC1 isoforms in DNA repair24, and our own Western analyses (Figure 4) typically detected only a single species of correct molecular weight in analyzed specimens, reducing this concern.
ERCC1 association with XPF (ERCC4) is essential for both NER and DSB repair, and the expression levels of these enzymes are closely linked.(15, 32, 43) Fibroblasts deficient in the ERCC1-XPF (ERCC4) complex are sensitive to radiation, with coincident defects in DSB repair.(15) Decreased ERCC1-XPF activity has also been implicated in increased hypoxic radiosensitivity, which is of particular interest in SCCHN.(44, 45) Thus, improved survival among ERCC1-low patients treated with radiation has a sound biologic rationale. Further, a retrospective series reported by Vaezi et al.(51), used a quantitative IHC platform to measure XPF expression in 80 SCCHN tumors. Their finding that low XPF expression is statistically associated with a prolonged PFS thus corroborates our findings.(46) In that study, the primary site of disease was heterogeneous, and treatments consisting of chemoradiation, radiation alone, or surgery plus adjuvant radiation, were not considered separately. In spite of this, the authors showed a statistically significant increase in PFS among tumors with low XPF expression. Nonetheless, the DNA repair mechanisms associated with radio-resistance are complex and will warrant evaluation of interactions between ERCC1, XPF and other DNA repair pathways.
In view of the magnitude of the survival disadvantage associated with high ERCC1 expression in resected patients requiring radiation or chemoradiation, future studies of adjuvant therapy, especially among HPV-negative patients, may benefit from incorporation of ERCC1 status as a stratification variable to ensure balance across treatment arms. Currently, there are limited non-platinum chemotherapy options for SCCHN patients. Future work should focus on developing optimal therapies for HPV-negative, ERCC1-high patients, and might also explore methods to target and inhibit ERCC1-XPF in order to restore sensitivity to cisplatin and radiation.
In conclusion, although limited, the results of our retrospective review are intriguing. While the lack of randomized data hinders our ability to make definite conclusions as to whether ERCC1 represents a predictive biomarker for treatment selection, the data presented here are strongly suggestive. A prospective, randomized trial design will be necessary in order to define this with complete confidence. The current work additionally supports the significance of such a study and illustrates the importance of selecting the appropriate antibody and assay conditions for all such endeavors. Much of the debate surrounding the utility of ERCC1 IHC would be addressed by the availability of a clinical-grade reagent. We are now working under the auspices of the NCI Clinical Assay Development Program in order to develop a standard clinical assay that uses an antibody adequate for this purpose.
Supplementary Material
Statement of Translational Relevance.
Human papillomavirus (HPV)-negative squamous cell carcinoma of the head and neck (SCCHN) is comparatively insensitive to cisplatin and radiation. Cisplatin resistance is associated with increased activity of the excision repair cross complementing group 1 (ERCC1) enzyme, a rate-limiting element in the NER pathway, and also acts in repair of double stranded breaks (DSB). Using immunofluorescence staining and automated quantitative analysis (AQUA) to probe ERCC1 expression in archival surgical specimens from patients who received adjuvant radiation, as compared to those submitted only to resection, this work suggests that SCCHN patients with high ERCC1 levels may not receive a survival benefit from adjuvant radiotherapy. Identification of patients with this form of intrinsic treatment resistance will help guide new treatment strategies in the high risk HPV-negative population, and allow more effective stratification of patients in future trials.
Acknowledgments
Financial support: This work and the authors were supported by the CA06927 NIH core grant (to Fox Chase Cancer Center), CA63366 (to EAG), and the Fox Chase Cancer Center Head and Neck Keystone.
We would like to thank Eric Ross, Ph.D (Director, Biostatistics and Bioinformatics Facility), Emmanuelle Nicolas (Genomics Facility), and Mary Donovan (Data Manager, Biosample Repository), for their assistance with this work.
Funding Sources: We would like to acknowledge the following funding sources which contributed to our work: support from CA06927 NIH core grant and the Pew Charitable Trust (to Fox Chase Cancer Center) and CA63366 (to EAG), and support from the Fox Chase Cancer Center Head and Neck Keystone.
Footnotes
Conflict of Interest:
Mehra: No potential conflicts
Zhu: No potential conflicts
Yang: No potential conflicts
Cai: No potential conflicts
Weaver: No potential conflicts
Singh: No potential conflicts
Nikonova: No potential conflicts
Golemis: No potential conflicts
Flieder: No potential conflicts
Cooper: No potential conflicts
Lango: No potential conflicts
Ridge: No potential conflicts
Burtness: No potential conflicts
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010 Sep-Oct;60(5):277–300. doi: 10.3322/caac.20073. eng. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg D, Lee J, Koch WM, et al. Habitual risk factors for head and neck cancer. Otolaryngal Head Neck Surg. 2004;131(6):986–93. doi: 10.1016/j.otohns.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 3.Herrero R, Castellsague X, Pawlita M. Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J Natl Cancer Inst. 2003;95:1772–83. doi: 10.1093/jnci/djg107. [DOI] [PubMed] [Google Scholar]
- 4.Spector NL, Xia W, Burris H, 3rd, Hurwitz H, Dees EC, Dowlati A, et al. Study of the biologic effects of lapatinib, a reversible inhibitor of ErbB1 and ErbB2 tyrosine kinases, on tumor growth and survival pathways in patients with advanced malignancies. J Clin Oncol. 2005 Apr 10;23(11):2502–12. doi: 10.1200/JCO.2005.12.157. eng. [DOI] [PubMed] [Google Scholar]
- 5.Rischin D, Young R, Fisher R, et al., editors. ASCO. 2009. Prognostic significance of HPV and p16 status in patients with oropharyngeal cancer treated on a large international phase III trial. [Google Scholar]
- 6.Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Sasaki C, et al. Prognostic significance of p16 protein levels in oropharyngeal squamous cell cancer. Clin Cancer Res. 2004 Sep 1;10(17):5684–91. doi: 10.1158/1078-0432.CCR-04-0448. eng. [DOI] [PubMed] [Google Scholar]
- 7.Lingen MW, Xiao W, Schmitt A, Jiang B, Pickard R, Kreinbrink P, et al. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral oncology. 2013 Jan;49(1):1–8. doi: 10.1016/j.oraloncology.2012.07.002. eng. [DOI] [PubMed] [Google Scholar]
- 8.Forastiere AA, Metch B, Schuller DE, Ensley JF, Hutchins LF, Triozzi P, et al. Randomized comparison of cisplatin plus fluorouracil and carboplatin plus fluorouracil versus methotrexate in advanced squamous-cell carcinoma of the head and neck: a Southwest Oncology Group study. J Clin Oncol. 1992 Aug;10(8):1245–51. doi: 10.1200/JCO.1992.10.8.1245. eng. [DOI] [PubMed] [Google Scholar]
- 9.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. The New England journal of medicine. 2008 Sep 11;359(11):1116–27. doi: 10.1056/NEJMoa0802656. eng. [DOI] [PubMed] [Google Scholar]
- 10.Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, Greiner RH, et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004 May 6;350(19):1945–52. doi: 10.1056/NEJMoa032641. eng. [DOI] [PubMed] [Google Scholar]
- 11.Pignon JP, Bourhis J, Domenge C, Designe L. Chemotherapy added to locoregional treatment for head and neck squamous-cell carcinoma: three meta-analyses of updated individual data. MACH-NC Collaborative Group. Meta-Analysis of Chemotherapy on Head and Neck Cancer. Lancet. 2000 Mar 18;355(9208):949–55. eng. [PubMed] [Google Scholar]
- 12.Lee KB, Parker RJ, Bohr V, Cornelison T, Reed E. Cisplatin sensitivity/resistance in UV repair-deficient Chinese hamster ovary cells of complementation groups 1 and 3. Carcinogenesis. 1993 Oct;14(10):2177–80. doi: 10.1093/carcin/14.10.2177. eng. [DOI] [PubMed] [Google Scholar]
- 13.Dabholkar M, Vionnet J, Bostick-Bruton F, Yu JJ, Reed E. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. The Journal of clinical investigation. 1994 Aug;94(2):703–8. doi: 10.1172/JCI117388. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moeller BJ, Yordy JS, Williams MD, Giri U, Raju U, Molkentine DP, et al. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clin Cancer Res. 2011 Apr 1;17(7):2035–43. doi: 10.1158/1078-0432.CCR-10-2641. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Molecular and cellular biology. 2008 Aug;28(16):5082–92. doi: 10.1128/MCB.00293-08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006 Dec;17(12):1818–25. doi: 10.1093/annonc/mdl300. eng. [DOI] [PubMed] [Google Scholar]
- 17.Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. The Journal of biological chemistry. 1997 Feb 7;272(6):3833–7. doi: 10.1074/jbc.272.6.3833. eng. [DOI] [PubMed] [Google Scholar]
- 18.Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Scharer OD. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. The Journal of biological chemistry. 2009 Feb 5;285(6):3705–12. doi: 10.1074/jbc.M109.067538. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. The New England journal of medicine. 2006 Sep 7;355(10):983–91. doi: 10.1056/NEJMoa060570. eng. [DOI] [PubMed] [Google Scholar]
- 20.Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. The New England journal of medicine. 2013 Mar 21;368(12):1101–10. doi: 10.1056/NEJMoa1214271. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustavson MD, Bourke-Martin B, Reilly DM, Cregger M, Williams C, Tedeschi G, et al. Development of an Unsupervised Pixel-based Clustering Algorithm for Compartmentalization of Immunohistochemical Expression Using Automated QUantitative Analysis. Appl Immunohistochem Mol Morphol. 2009 Jul;17(4):329–37. doi: 10.1097/PAI.0b013e318195ecaa. eng. [DOI] [PubMed] [Google Scholar]
- 22.Moeder CB, Giltnane JM, Moulis SP, Rimm DL. Quantitative, fluorescence-based in-situ assessment of protein expression. Methods in molecular biology (Clifton, NJ. 2009;520:163–75. doi: 10.1007/978-1-60327-811-9_12. eng. [DOI] [PubMed] [Google Scholar]
- 23.Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science (New York, NY. 1999 May 7;284(5416):958–61. doi: 10.1126/science.284.5416.958. eng. [DOI] [PubMed] [Google Scholar]
- 24.Handra-Luca A, Hernandez J, Mountzios G, Taranchon E, Lacau-St-Guily J, Soria JC, et al. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin Cancer Res. 2007 Jul 1;13(13):3855–9. doi: 10.1158/1078-0432.CCR-07-0252. eng. [DOI] [PubMed] [Google Scholar]
- 25.Jun HJ, Ahn MJ, Kim HS, Yi SY, Han J, Lee SK, et al. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. British journal of cancer. 2008 Jul 8;99(1):167–72. doi: 10.1038/sj.bjc.6604464. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MK, Cho KJ, Kwon GY, Park SI, Kim YH, Kim JH, et al. Patients with ERCC1-negative locally advanced esophageal cancers may benefit from preoperative chemoradiotherapy. Clin Cancer Res. 2008 Jul 1;14(13):4225–31. doi: 10.1158/1078-0432.CCR-07-4848. eng. [DOI] [PubMed] [Google Scholar]
- 27.Kim MK, Cho KJ, Kwon GY, Park SI, Kim YH, Kim JH, et al. ERCC1 predicting chemoradiation resistance and poor outcome in oesophageal cancer. Eur J Cancer. 2008 Jan;44(1):54–60. doi: 10.1016/j.ejca.2007.09.006. eng. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. The New England journal of medicine. 2007 Feb 22;356(8):800–8. doi: 10.1056/NEJMoa065411. eng. [DOI] [PubMed] [Google Scholar]
- 29.Akita H, Zheng Z, Takeda Y, Kim C, Kittaka N, Kobayashi S, et al. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009 Jun 22; doi: 10.1038/onc.2009.158. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. The New England journal of medicine. 2007 Jun 14;356(24):2538–40. doi: 10.1056/NEJMc070742. author reply 40–1. eng. [DOI] [PubMed] [Google Scholar]
- 31.Besse B, Olaussen KA, Soria JC. ERCC1 and RRM1: ready for prime time? J Clin Oncol. 2013 Mar 10;31(8):1050–60. doi: 10.1200/JCO.2012.43.0900. eng. [DOI] [PubMed] [Google Scholar]
- 32.Bhagwat NR, Roginskaya VY, Acquafondata MB, Dhir R, Wood RD, Niedernhofer LJ. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer research. 2009 Sep 1;69(17):6831–8. doi: 10.1158/0008-5472.CAN-09-1237. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. Journal of the National Cancer Institute. 2005 Dec 21;97(24):1808–15. doi: 10.1093/jnci/dji427. eng. [DOI] [PubMed] [Google Scholar]
- 34.Van Putten W. CART: Stata module to perform Classification and Regression Tree analysis. Boston College Department of Economics; 2006. [Google Scholar]
- 35.Mooradian AD. Biomarkers of aging: do we know what to look for? J Geront. 1990;45:B183–6. doi: 10.1093/geronj/45.6.b183. [DOI] [PubMed] [Google Scholar]
- 36.Miura M, Nakamura S, Sasaki T, Takasaki Y, Shiomi T, Yamaizumi M. Roles of XPG and XPF/ERCC1 endonucleases in UV-induced immunostaining of PCNA in fibroblasts. Experimental cell research. 1996 Jul 10;226(1):126–32. doi: 10.1006/excr.1996.0210. eng. [DOI] [PubMed] [Google Scholar]
- 37.Fountzilas G, Bamias A, Kalogera-Fountzila A, Karayannopoulou G, Bobos M, Athanassiou E, et al. Induction chemotherapy with docetaxel and cisplatin followed by concomitant chemoradiotherapy in patients with inoperable non-nasopharyngeal carcinoma of the head and neck. Anticancer research. 2009 Feb;29(2):529–38. eng. [PubMed] [Google Scholar]
- 38.Hao D, Lau HY, Eliasziw M, Box A, Diaz R, Klimowicz AC, et al. Comparing ERCC1 protein expression, mRNA levels, and genotype in squamous cell carcinomas of the head and neck treated with concurrent chemoradiation stratified by HPV status. Head & neck. 2011 Jun;34(6):785–91. doi: 10.1002/hed.21817. eng. [DOI] [PubMed] [Google Scholar]
- 39.Seiwert TY, Cohen EE, Wang X, Kocherginsky M, Bhayani M, Stenson K, et al., editors. ASCO. 2008. Use of systematic analysis of DNA repair pathways in head and neck cancer (HNC) to identify XPF as a novel predictor of induction response, and pMK2 relationship to chemoradiotherapy. [Google Scholar]
- 40.Austin M, Schmidt R, Parvathaneni U, Bauman J, Hayes D, Papagikos M, et al., editors. ASCO. 2012. Expression of p16, ERCC1, and EGFR amplification as predictors of responsiveness of locally advanced squamous cell carcinomas of head and neck (SCCHN) to cisplatin, radiotherapy, and erlotinib: A phase II randomized trial. [Google Scholar]
- 41.Olaussen KA, Soria JC. Validation of ERCC1-XPF immunodetection--letter. Cancer research. 2010 May 1;70(9):3851–2. doi: 10.1158/0008-5472.CAN-09-4352. author reply 2. eng. [DOI] [PubMed] [Google Scholar]
- 42.Arbogast S, Behnke S, Opitz I, Stahel RA, Seifert B, Weder W, et al. Automated ERCC1 immunohistochemistry in non-small cell lung cancer: comparison of anti-ERCC1 antibodies 8F1, D-10, and FL-297. Appl Immunohistochem Mol Morphol. 2011 Mar;19(2):99–105. doi: 10.1097/PAI.0b013e3181f1feeb. eng. [DOI] [PubMed] [Google Scholar]
- 43.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, et al. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Molecular and cellular biology. 2009 Dec;29(24):6427–37. doi: 10.1128/MCB.00086-09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray D, Macann A, Hanson J, Rosenberg E. ERCC1/ERCC4 5′-endonuclease activity as a determinant of hypoxic cell radiosensitivity. International journal of radiation biology. 1996 Mar;69(3):319–27. doi: 10.1080/095530096145878. eng. [DOI] [PubMed] [Google Scholar]
- 45.Murray D, Rosenberg E. The importance of the ERCC1/ERCC4[XPF] complex for hypoxic-cell radioresistance does not appear to derive from its participation in the nucleotide excision repair pathway. Mutation research. 1996 Dec 2;364(3):217–26. doi: 10.1016/s0921-8777(96)00036-5. eng. [DOI] [PubMed] [Google Scholar]
- 46.Vaezi A, Wang X, Buch S, Gooding W, Wang L, Seethala RR, et al. XPF expression correlates with clinical outcome in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2011 Aug 15;17(16):5513–22. doi: 10.1158/1078-0432.CCR-11-0086. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.