Abstract
Here we report the convergent total synthesis of two proteins: DARPin pE59 and RNase B. a. (Barnase). Leveraging our recently developed fast flow peptide synthesis platform, we rapidly explored numerous conditions for the assembly of long polypeptides and were able to mitigate common side reactions including deletion and aspartimide products. We report general strategies for improving the synthetic quality of difficult peptide sequences with our system. High-quality protein fragments produced under optimal synthetic conditions were subjected to convergent native chemical ligation, which afforded native full-length proteins after a final desulfurization step. Both DARPin and Barnase were folded and found to be as active as their recombinant analogues.
Keywords: Fast flow, Native chemical ligation, Peptides, Protein synthesis, Solid-phase synthesis
Introduction
Chemically modified proteins play a vital role in modern science, especially in the pharmaceutical sector and chemical biology research.[1] However, our ability to selectively modify the properties of proteins is often restricted by the use of biological expression systems. Although impressive advances have been made towards the routine incorporation of select non-proteogenic amino acids with in vitro[2] and in vivo[3] expression systems, only a limited number of unnatural moieties can be introduced. In contrast, total synthesis enables complete chemical control and the uncompromised ability to tailor a protein's properties as desired. Using solid phase peptide synthesis (SPPS) to assemble smaller fragments, it is possible to construct moderately sized proteins through the use of native chemical ligation (NCL).[4]
Since Kent's first report of NCL, much work has been done to expand the scope of the reaction through auxiliary thiols,[5] desulfurization,[6] and other strategies.[7] These efforts have resulted in robust, flexible ligation chemistry. However, obtaining the high quality peptide fragments required for ligation can prove to be difficult and time consuming. Fmoc-based SPPS, often preferred for its safety and the low cost of reagents, typically takes 60 minutes or more to incorporate each amino acid residue.[8] Furthermore, sequence-specific optimization is commonly required, thus necessitating repeated synthesis of fragments under different conditions.
In order to accelerate this process, we have recently developed a flow based peptide synthesizer that incorporates an amino acid residue every 3 minutes (see the accompanying manuscript). With our fast flow peptide synthesis methodology well validated for shorter peptides, we undertook the total synthesis of two proteins, a 130 residue Designed Ankyrin Repeat Protein (DARPin) and the 113 residue RNase B. a. (Barnase, EC 3.1.27). The syntheses of fragments for these two proteins serve as case studies for the application of our fast flow synthesizer methodology towards longer and difficult peptides.
We chose to synthesize a DARPin because of its established versatility as a protein-binding scaffold.[9] Inspired by nature, DARPins consist of several individual modules on the order of 30 residues each. Each module is comprised of a β turn and two α helices. Variable regions within the helices and loops may be engineered to bind a diverse array of therapeutically relevant targets, and multiple modules can be stacked to expand the binding surface. The DARPin chosen for synthesis was pE59, a DARPin with high nanomolar affinity for phosphorylated ERK2.[9b]
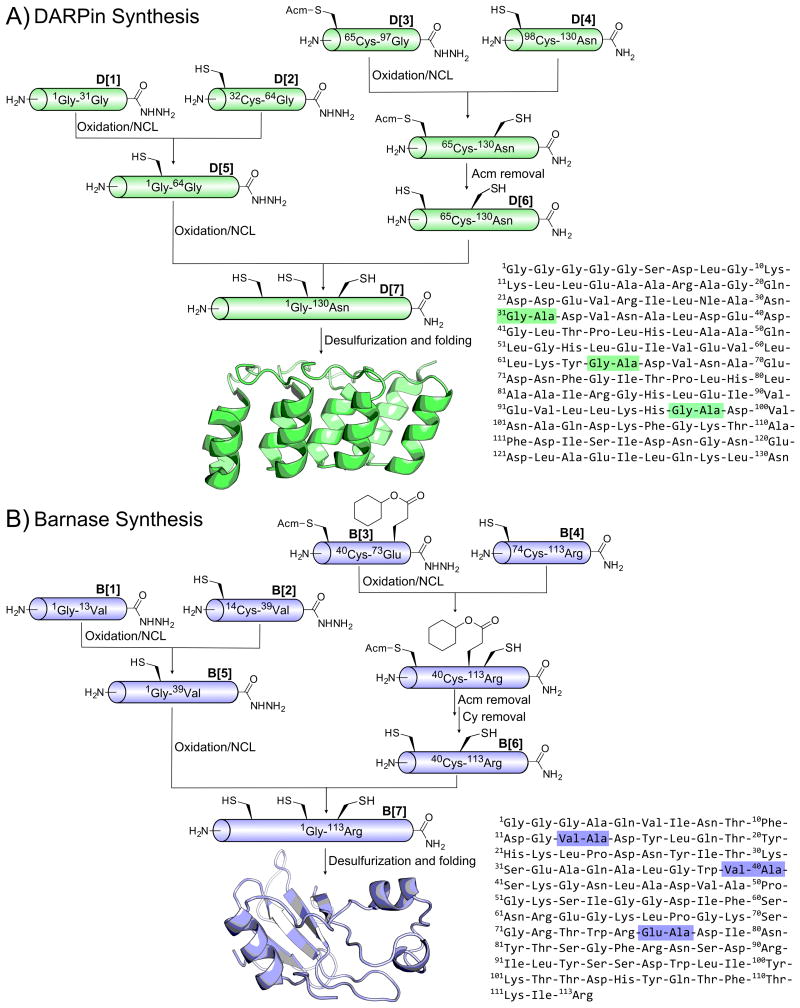
The synthetic route for DARPin pE59, shown in Figure 1a, was devised to preserve the modular assembly of a DARPin, with the recurring glycine-alanine junctions serving as ligation sites. A pentaglycine motif was appended to the N-terminus in order to facilitate future enzymatic ligation by the transpeptidase, Sortase A. Because DARPins are based on a consensus sequence[10] and this synthetic strategy reflects their modularity, we envision that this synthetic route can be adapted to produce chemically enhanced DARPins of varying lengths.
Figure 1.
a) Synthetic scheme of DARPin pE59 with the protein sequence on the right. Ligation sites are highlighted in green. b) Synthetic scheme of barnase with the protein sequence on the right. Ligation sites are highlighted in blue.
Barnase is a small 12 kDa protein produced and secreted by Bacillus amyloliquefaciens.[11] Barnase is a potent RNase with endonuclease activity and a prime requirement for GpN nucleotides with the selectivity A > G > C > U for N.[12] Barnase is composed of one polypeptide chain, free of disulfide bonds, and does not require cofactors or metal ions for its folding and catalytic activity.[13] These factors make Barnase an appealing candidate for future in vivo model studies.
Our initial strategy was to assemble Barnase from three peptide fragments of equal length, but we ultimately opted for convergent synthesis from four fragments, as shown in Figure 1b. This change resulted in a strategy nearly identical to the DARPin except 76Glu was protected with the non-standard cyclohexyl ester in order to prevent possible isomerization during NCL.[14]
Results
Initial Peptide Synthesis
The DARPin was assembled from four fragments:
D[1] (H2N-[1Gly-31Gly]-CONHNH2),
D[2] (H2N-[32Cys-64Gly]-CONHNH2),
D[3] (H2N-[65Cys-97Gly]-CONHNH2), and
D[4] (H2N-[98Cys-130Asn]-CONH2).
Barnase was initially split into three fragments:
B[1+2] (H2N-[4Ala-39Val]-CONHNH2, B[1+2]),
B[3] (H2N-[40Cys-76Glu]-CONHNH2) and
B[4] (H2N-[74Cys-113Arg]-CONH2).
Ultimately, B[1+2] was divided into:
B[1] (NH2-[1Gly-13Val]-CONHNH2) and
B[2] (NH2-[14Cys–39Val]-CONHNH2).
The initial crude DARPin and Barnase peptides synthesized on a standard three minute cycle are shown in Figures 2 and 3, respectively. D[1], D[2], D[3], B[1+2], B[1], B[2], and B[3] were synthesized as peptide hydrazides on 2-chlorotrityl hydrazine polystyrene resin. D[4] was synthesized as a C-terminal carboxamide using a Rink-MBHA polystyrene resin, and B[4] was synthesized as a C-terminal carboxamide on Rink-aminomethyl polystyrene resin.
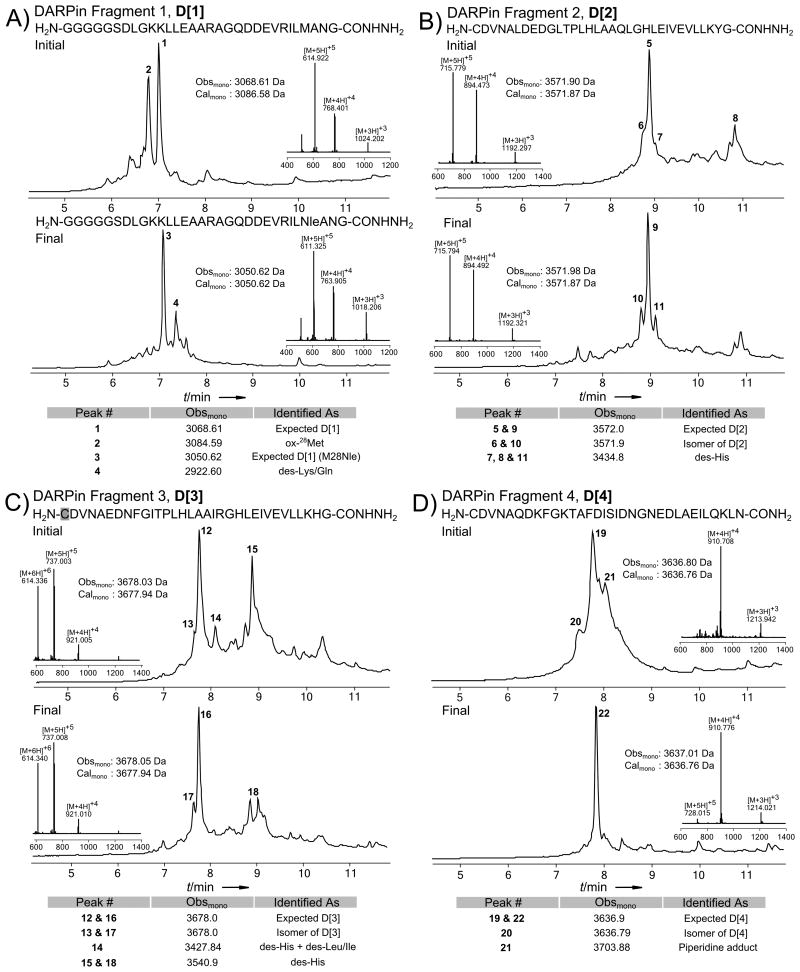
Figure 2.
LC-MS data (total ion current vs. time) of crude DARPin fragments. The initial as well as the final syntheses are shown for every peptide. Each panel also displays peptide sequences, MS insets of the major peak, comparison of monoisotopic calculated and observed molecular masses for the expected product, and a table with identification of the major impurities. The charge state series on the inset mass spectra display the most abundant ions; observed and calculated masses are monoisotopic. The data for a) DARPin fragment 1; b) DARPin fragment 2; c) DARPin fragment 3; cysteine highlighted in grey was incorporated as Cys(Acm); d) DARPin fragment 4.
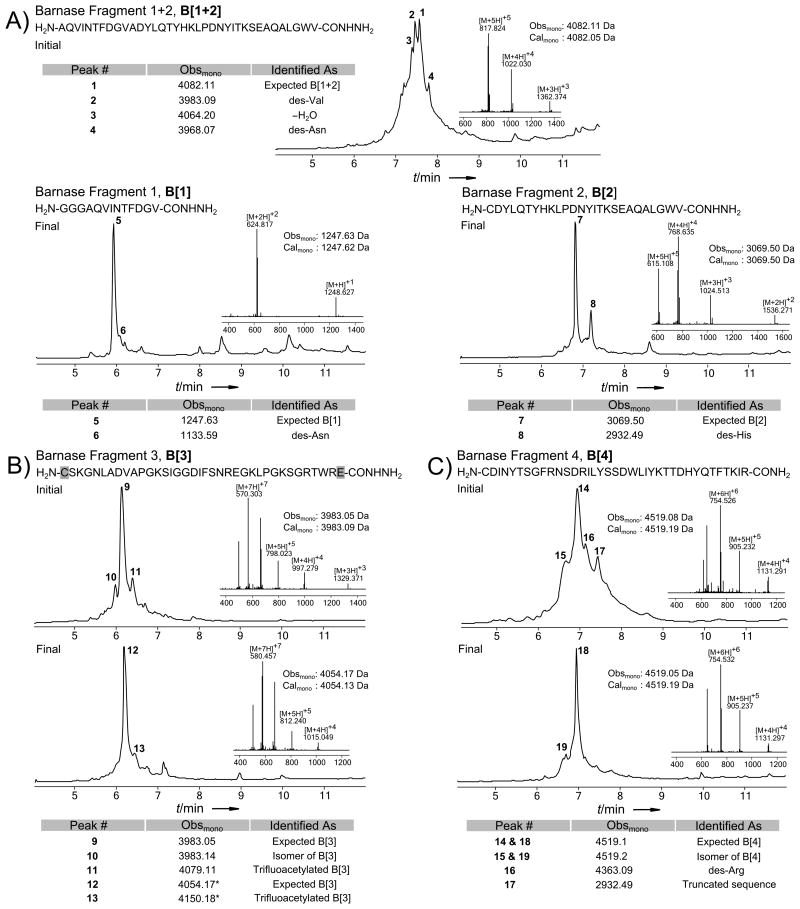
Figure 3.
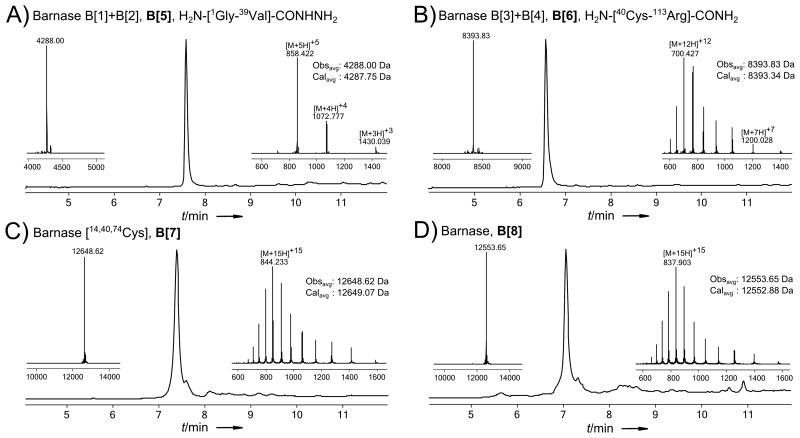
LC-MS data (total ion current vs. time) of crude Barnase fragments. The initial as well as the final syntheses are shown for every peptide. Each panel also displays peptide sequences, MS insets of the major peak, comparison of monoisotopic calculated and observed molecular masses for the expected product, and a table with identification of the major impurities. The charge state series on the inset mass spectra display the most abundant ions; observed and calculated masses are monoisotopic. a) Barnase fragments 1 and 2. b) Barnase fragment 3. Residues highlighted in grey were incorporated with non-standard side-chain protection; Cys(Trt) is utilized in the initial synthesis and Cys(Acm) in the final synthesis; Glu is protected as cyclohexyl ester in both cases. c) Barnase fragment 4.
While crude D[1], D[2], D[3], and B[3] were readily purifiable, D[4], B[1+2], and B[4] required improvements in the synthetic conditions prior to RP-HPLC. Additionally, we attempted to refine synthetic conditions for peptides D[1], D[2], D[3], and B[2] in order to increase yields and explore factors which contribute to peptide quality with our platform. Because our fast flow synthesis platform allowed us to obtain long polypeptides in hours rather than days, we systematically studied numerous synthetic conditions. Ultimately, we made only a few changes to the standard synthetic strategy to achieve significant improvement in crude peptide quality.
Improvement of DARPin Fragment Synthesis
Although the crude products of the initial D[1], D[2], and D[3] syntheses were readily purified by RP-HPLC, we attempted to improve synthetic conditions to increase the yield. As shown in Figure 2a, the major impurity in the initial synthesis of D[1] was a product 15.98 Da more massive than the desired product, which we hypothesized to be oxidation of 28Met. An anti-oxidative cleavage solution reported by Rabenstein and co-workers[15] effectively eliminated this side product, but the sulfoxide re-formed during workup and purification. Thus, we mutated the native 28Met to norleucine (Nle); this substitution significantly simplified synthesis and handling by eliminating oxidation. Based on the analysis of the DARPin crystal structure we concluded that this mutation should not affect the activity of the protein.
In the initial syntheses of D[2] and D[3], the major impurities were single and double deletion products with elution times from 9 to 11 minutes and 8 to 10 minutes on the chromatograms in Figures 2b and 2c respectively. To improve the quality of these polypeptides, which differ by only seven internal residues, we first ensured complete removal of Fmoc during each deblocking step. To accomplish this, we monitored the formation of the Fmoc-dibenzofulvene adduct via UV absorbance of the reactor effluent at 304 nm, and every residue was double deprotected. If the second deprotection step produced a second UV absorbance peak, a third deprotection was employed, and so forth, until no UV active deprotection product was observed. In both D[2] and D[3], we found the initial deprotection step to be inadequate for the 9th to 17th residues coupled. However, exhaustive Fmoc had negligible effect on the quality of the crude product (Figures S4).
Next, in addition to complete deprotection, we coupled each of the identified residues for 10 minutes following the hypothesis that acylation is difficult in the same regions where Fmoc removal is difficult, due to chain aggregation.[16] This was accomplished by pausing the flow of reagent through the synthesis vessel for 10 minutes after delivering the activated amino acids, for an effective coupling time of 30 s + 10 min. We assumed that a twentyfold increase in reaction time would be sufficient to ensure complete acylation, if it was possible under the given conditions. While in the case of D[2], the quality of the peptide was only marginally improved (Figure 2b), for D[3] one significant des-His was nearly eliminated, and the intensity of the other deletion products were slightly reduced. Although only one deletion product was significantly reduced, these peptides were purified by RP-HPLC without further optimization.
Finally, we turned our attention to improving the quality of the most difficult DARPin fragment, D[4]. As shown in Figure 2d, the initial synthesis of this fragment resulted in material which was heavily contaminated by many low intensity deletions and prominent aspartimide-derived products. These compounds were poorly resolved from the desired compound and we were unable to readily purify this material by RP-HPLC. Therefore, we changed the synthetic conditions to obtain a purifiable product. First, we repeated the synthesis reducing the duration of the deblocking step from 20 s to 5 s, in an effort to reduce the levels of aspartimide-derived products. The abbreviated deprotection step was somewhat successful at reducing the intensities of the prominent early eluting isomer and the late eluting piperidine adduct (Figure S9), without considerably increasing levels of the deletion products. Retaining the 5 s deprotection, we next attempted to reduce the levels of deletion products. First, we switched to a more potent HATU activator,[17] but a direct substitution of 0.4 m HATU for 0.4 m HBTU did not appreciably alter the quality of the crude peptide. We then increased the temperature of the synthesis to 70 °C, which led to an increase in aspartimide derived products without a concomitant decrease in deletion products. Therefore, we abandoned both of these strategies. Next we assembled D[4] on four different resins, under otherwise identical conditions.
The initial synthesis of D[4] was carried out on Rink-MBHA polystyrene resin, and produced the peptide shown in Figure 2d. Assembly on Rink-polyethylene glycol-polystyrene hybrid resin (PEG-PS) produced a peptide of even lower crude purity, but syntheses on Rink-aminomethyl polystyrene prepared in-house (AM) and Rink-polyethylene glycol (PEG) resins resulted in markedly refined crude material (Figures S10-S11). AM resin was selected for subsequent experiments for its higher loading and lower cost. Although improved, peptide D[4] still contained poorly resolved deletion products and aspartimide derived peptides.
The crude quality of D[4] was further improved through the use of an Nα‐backbone protected amino acid. In order to identify sites for possible backbone protection we performed intermediate test-scale cleavages during chain assembly. We avoided the multiple deprotection strategy used for D[2] and D[3] as we assumed that prolonged exposure of the protected polypeptide to piperidine would promote based-catalyzed formation of aspartimide. Analysis of consecutive segments of D[4] allowed us to conclude that chain assembly became ineffective in a region between 114Ser and 110Ala. During the next synthesis of D[4], we incorporated Fmoc-Ile-Ser(ψMe,MePro)-OH in place of 114Ser and 113Ile. This substitution led to a drastic increase in the crude quality of D[4], suppressing nearly all deletion products (Figure S14). However, RP-HPLC failed to resolve the desired product from two major co-eluting impurities with masses -18.02 and +14.02 Da relative to desired product (Figure S13). These were presumed to be aspartimide derived products (condensation and methanol adduct, respectively).
To address aspartimide formation, the last major problem with the synthesis of D[4], we first changed the deprotection protocol from 5 seconds with 50% (v/v) piperidine in DMF to 20 seconds with 20% (v/v) piperidine and 0.1 m HOBt in DMF, a deprotection reagent reported to suppress aspartimide formation.[18] This new deblocking solution somewhat reduced the content of aspartimide products, but did not eliminate them. In a separate experiment, we incorporated 116Asp, the aspartic acid suspected to form aspartimide, as the methyl pentyl ester, Fmoc-Asp(OMpe)-OH. This side chain protecting group is reported to alleviate aspartimide formation.[19] To test the effect of Asp(OMpe) alone, it was incorporated using 5 second deprotection steps with 50% (v/v) piperidine. Under these conditions, the use of Asp(OMpe) slightly mitigated the formation of aspartimide-derived products. Combining the incorporation of Asp(OMpe) with the improved deprotection conditions (20 s, 20% (v/v) piperidine and 0.1 m HOBt in DMF), afforded crude D[4] which was nearly free of aspartimide products (Figures 2d and S14). Preparative RP-HPLC of this material readily afforded D[4] which was homogenous by LC-MS (Figure S19).
Improvement of Barnase Fragment Synthesis
Our initial synthetic strategy for Barnase included assembly of fragments B[1] and B[2] as a single polypeptide, H2N-[4Ala-39Val]-CONHNH2 (B[1+2]), by stepwise SPPS. The first synthesis of this peptide using HBTU as an activating agent yielded crude material of unsatisfactory quality. The major impurities were aspartimide derived compounds, and deletion products, including 50% des-Valine (Figure 3a). We hypothesized that the des-Val product may have resulted from incomplete coupling of the C-terminal 39Val to the resin, and, thus, we coupled 39Val for 10 minutes during the next synthesis. Additionally, in an attempt to prevent the formation of other deletion products, we employed HATU activation throughout the sequence. These measures generally increased the quality of the crude peptide (Figure S29), including elimination of the des-Val product, but an isomer and several deletion products prevented facile RP-HPLC purification. Impressed with the results of HATU in this synthesis, we used it for all other Barnase fragments.
Striving to suppress incomplete couplings throughout the synthesis, we doubled the amount of activated amino acid for every coupling step, while maintaining 30 s coupling times by doubling the flow rate. However, this did not improve synthetic quality. In a separate experiment, reducing the amount of DIEA used to activate the amino acids did not improve the peptide quality (Figures S30-S31). At this point, we placed the synthesis of B[1+2] on hold and turned our attention to fragments B[3] and B[4].
Synthesis of the 37 residue B[3] under standard conditions resulted in the high quality crude product shown in Figure 3b. The first synthesis of B[3] did not employ the orthogonally protected cysteine ultimately required by our synthetic strategy, so it was resynthesized with Acm protection. Additionally, the resynthesis used 20% (v/v) piperidine as the deprotection reagent, which somewhat suppressed formation of an early eluting isomer. The late eluting triflouroacetylation product was also less prevalent, for poorly understood reasons.
Preliminary experiments suggested that B[4] would be difficult to synthesize, and prone to aspartimide formation. For these reasons, we conducted the initial synthesis with 20% (v/v) piperidine and 0.1 m HOBt in DMF as the deblocking solution, and used the Rink-AM resin that was effective for D[4]. Unfortunately, the resulting peptide, shown in Figure 3c, contained numerous deletion products, a prominent isomer, and a significant premature chain termination product. Because the resin may affect the quality of synthesized peptides, we next assembled B[4] under the same conditions but on PEG resin, which resulted in a significant improvement in the quality of the peptide. Under these conditions, the two major impurities were a well resolved isomer and a product of premature chain termination at 90Arg (Figure S47). Deletion products were nearly eradicated. Although the peptide was of acceptable quality, we attempted to further improve the yield. In a successful attempt to suppress the persistent sequence truncation which had a mass 42.05 Da greater than H2N-[91Ile-113Arg]-CONH2, we performed the synthesis of B[4] again, but used DCC/HOBt activation of 90Arg, instead of HATU. Additionally, we removed HOBt from the deprotection solution throughout the synthesis. As a result, the formation of truncated peptide was alleviated, while removal of HOBt from the deprotection solution had little to no effect on the formation of the isomer (Figure 3c). Based on these findings, we selected 20% (v/v) piperidine in DMF as a universal deprotection agent for Barnase fragment assembly.
Finally, we revisited synthesis of B[1+2]. Based on the data obtained earlier, we assembled B[1+2] as a C-terminal carboxamide on Rink-PEG resin using 20% (v/v) piperidine for deprotection. Unfortunately, these measures did not improve the quality of the crude product. To identify regions of incomplete Fmoc removal and slow coupling, we employed UV monitoring of the reactor effluent and intermediate cleavage of the polypeptide chain, as described above. Real time monitoring of the deprotection step identified incomplete Fmoc removal in the region 29Thr – 27Tyr, and the data obtained from intermediate cleavage at 20Tyr supported this finding.
At this point, we divided B[1+2] into two peptides: B[1] (NH2-[1Gly-13Val]-CONHNH2) and B[2] (NH2-[14Cys–39Val]-CONHNH2), concurrently appending an olygoglycine motif to the N-terminus of B[1]. We assembled B[1] and B[2] as C-terminal peptide-hydrazides on 2-chlorotrityl hydrazine polystyrene resin under standard conditions, except 20% (v/v) piperidine was used as a deprotection reagent and C-terminal Val residues were coupled for 10 minutes. Synthesis of B[1] produced very high quality crude peptide, as shown in Figure 3a, and RP-HPLC afforded pure B[1] in good yield. Although crude B[2] was of acceptable quality, we sought to reduce the proportions of several prominent deletion products (Figure S39). As with DARPin fragments D[2] and D[3], we hypothesized that prolonged coupling times may suppress deletion products. During the next synthesis of B[2], we deprotected 27Tyr – 25Asp for one minute with 20% (v/v) piperidine and coupled 28Ile for 10 minutes and 27Tyr –25Asp for 20 minutes. This synthesis failed to improve the crude peptide quality (Figure S40), and thus we abandoned the strategy of extended coupling times.
Next, we turned to the 2-hydroxy-4-methoxybenzyl (Hmb) protecting group, a well-known Nα-protecting group which is supposed to disrupt peptide chain aggregation on resin. We identified 33Ala, the closest alanine to the problematic region, as a suitable site for incorporation of Fmoc-(Hmb)-Ala-OH. Model coupling studies with Fmoc-(Hmb)-Ala-OH on our synthetic platform led us to activate this and the subsequent residue with DCC/HOBt rather than HATU or HBTU, to avoid premature termination of the peptide chain. Further, at 60 °C, coupling of the Hmb backbone protected amino acid required 25 minutes, presumably due to intramolecular formation of the less active Hmb-ester, and coupling to the resulting secondary amine required half an hour. Otherwise standard conditions were used. Incorporation of the Nα-protected residue near the region of incomplete coupling drastically improved the quality of the crude peptide, leaving the His deletion as the only significant impurity (Figure 3a). RP-HPLC readily afforded the last Barnase fragment, B[2], as a homogeneous peptide.
Native Chemical Ligation and Synthesis of Proteins
Following facile RP-HPLC purification of the crude peptides, we turned our attention to NCL. A one-pot oxidation/NCL procedure reported by Lei Lu and coworkers[20] was employed with only minor modifications. This procedure relies on oxidation of C-terminal hydrazides to the azide, followed by addition of thiol to generate thioesters for NCL. Thus, the N-terminal fragments (D[1] or B[1]) were oxidized with NaNO2, and 4-mercaptophenyl acetic acid (MPAA) was added to yield C-terminal thioesters.[21] The second peptide (D[2] or B[2], respectively) was added to afford ligated fragments D[5], H2N-[1Gly-64Gly]-CONHNH2, and B[5], H2N-[1Gly-39Val]-CONHNH2. The C-terminal segments (H2N-[65(Acm)Cys-130Asn]-CONH2) and (H2N-[40(Acm)Cys-113Arg]-CONH2) were prepared in an analogous manner from peptides D[3] and D[4], and B[3] and B[4], respectively. A typical ligation reaction was allowed to proceed overnight at 1.5 mM peptide concentrations and at pH 7.0. For detailed information, refer to the SI. All ligated products were isolated in high yield as pure compounds after RP-HPLC (Figures 4 and 5).
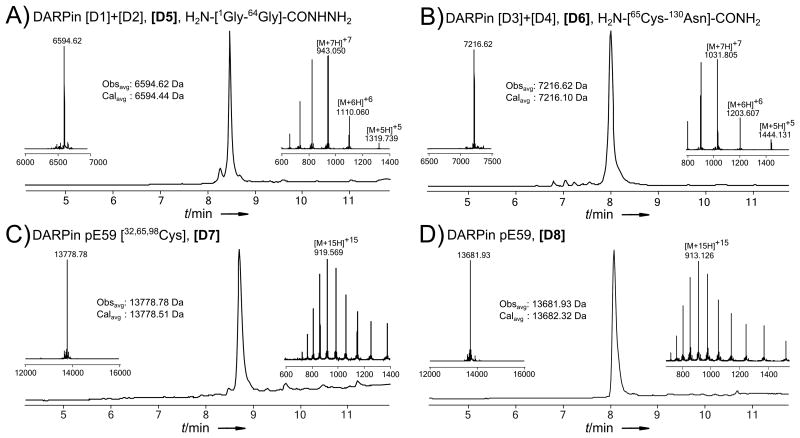
Figure 4.
LC-MS data (total ion current vs time) of purified DARPin ligation products. Each panel also displays MS insets of the major peak, comparison of average calculated and observed molecular masses for the expected product, and a deconvolution result. The charge state series on the inset mass spectra display the most abundant ions; observed and calculated masses are average. a) Purified N-terminal polypeptide H2N-[1Gly-64Gly]-CONHNH2. b) Purified C-terminal polypeptide H2N-[65Cys-130Asn]-CONH2. c) Purified full length DARPin [32,65,98Cys]. d) Purified full length, native, desulfurized DARPin pE59.
Figure 5.
LC-MS data (total ion current vs time) of purified Barnase ligation products. Each panel also displays MS insets of the major peak, comparison of average calculated and observed molecular masses for the expected product, and a deconvolution result. The charge state series on the inset mass spectra display the most abundant ions; observed and calculated masses are average. a) Purified N-terminal polypeptide H2N-[1Gly-39Val]-CONHNH2. b) Purified C-terminal polypeptide H2N-[40Cys-113Arg]-CONH2. c) Purified full length Barnase [14,40,77Cys]. d) Purified full length, native, desulfurized Barnase.
In order to prepare peptides for the final ligation, DARPin segment (H2N-[65(Acm)Cys-130Asn]-CONH2) was subjected to silver (I) acetate in 50% aqueous acetic acid to remove Acm protection on the N-terminal cysteine,[22] affording D[6] for ligation. Barnase segment (H2N-[40(Acm)Cys-113Arg]-CONH2) was similarly treated to afford a free N-terminal cysteine, and the cyclohexyl ester of 76Glu was removed using 10% v/v p-cresol in HF to prepare protecting group free peptide B[6] for ligation. NaNO2 oxidation, MPAA thioesterefication, and subsequent ligation of peptides D[5] and B[5] with D[6] and B[6], respectively, afforded the full length proteins.
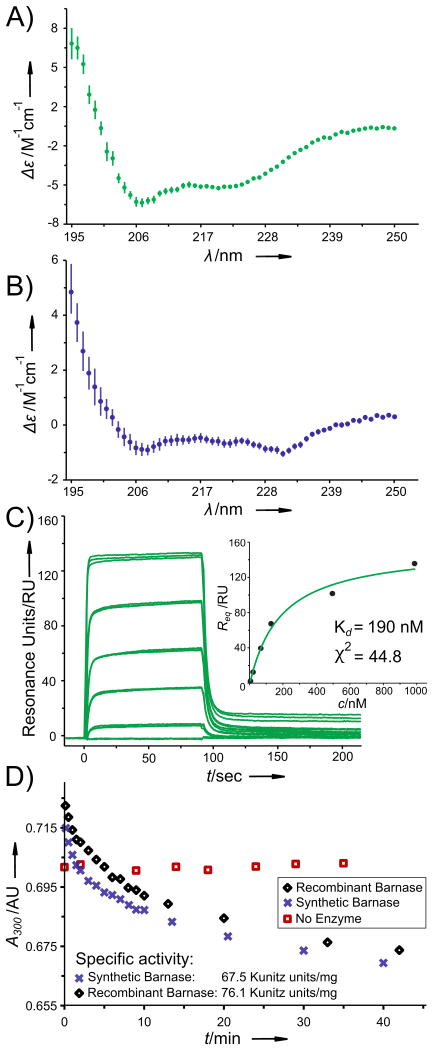
Finally, to obtain native sequences, the desulfurization protocol developed by Danishefsky and coworkers was used to globally convert Cys to Ala.[6a] The DARPin [32,65,98Cys] and Barnase [14,40,77Cys] were reacted with excess TCEP and t-BuSH or Mesna in the presence of VA-044 radical initiator, affording the native proteins (Figures 4d and 5d). After both the DARPin and Barnase were successfully refolded from a 6M Gn·HCl solution (characterized by circular dichroism spectroscopy), we characterized the bioactivity of the synthetic proteins and determined them to be comparable to recombinant analogs.
DARPin pE59 was characterized by binding to phosphorylated extracellular regulated kinase 2 (pERK2), for which it was evolved to have high specificity and nanomolar affinity.[9b] Using surface plasmon resonance and a steady-state approximation, the dissociation constant for our synthetic DARPin, KD = 190 nM (χ2 = 44.8, Figure 6c), was in close agreement to previously reported values of KD = 117 nM[9b] and the values we measured for the recombinant material.
Figure 6.
Characterization of the synthesized proteins. a) CD spectrum of the synthesized DARPin pE59. b) CD spectrum of the synthesized Barnase. c) SPR binding assay showing the synthetic DARPin pE59 binding to pERK2; KD of the complex was found to be 190 nM. d) Barnase RNase activity assay; specific activity was calculated using additional data points (see the SI).
Barnase is a potent RNase, and its activity was assayed by measuring the catalytic hydrolysis of Yeast Torula RNA according to an RNAse hydrolytic assay described by Kunitz et al.[23] We found that that the specific activity of the synthetic Barnase was 67.5 ± 10.5 units/mg (Figure 6d), which corresponds to the values we obtained for the recombinant analogue (76.1 ± 9.2 units/mg).
Discussion
The total convergent syntheses of the DARPin pE59 and Barnase serve as case studies in the application of our fast flow peptide synthesizer to the rapid and facile assembly of longer and difficult sequences via Fmoc SPPS. Initial syntheses of DARPin and Barnase peptide fragments on our system identified two major classes of impurities: point deletions, and aspartimide-derived products. Additionally, in isolated instances, we observed carboxylation, premature chain termination, and methionine oxidation. First, the two major classes of impurities are addressed, and then miscellaneous problems are discussed. Finally, major side reactions encountered during oxidation and thioesterefication of C-terminal peptide hydrazides are described.
Deletion Products
Impurities arising from incomplete acylation are the most common side products in peptide synthesis.[24] As the polypeptide grows, these products constitute a greater fraction of the crude material and are less resolved by RP-HPLC. This fact limits the maximum length of a polypeptide prepared by stepwise SPPS, if homogenous material is desired. Many strategies are employed to reduce deletion side products and in the following sections we discuss some common approaches in the context of our fast flow platform.
Changing the Activating Agent
As we report in the accompanying manuscript, the use of HATU over HBTU as an activating agent may improve synthetic outcomes with our flow synthesizer. Because of this, it was often our first option to suppress deletion products. In the synthesis of B[1+2] we observed a substantial improvement in the crude peptide quality with HATU instead of HBTU activation, and HATU was generally adopted for synthesis of Barnase fragments. In contrast, fragments D[3] and D[4] showed little or no improvement when individual amino acids or the entire peptide was assembled with HATU activation. With our flow synthesizer, HATU activation does not always improve synthetic outcomes, but changing the activating agent is an easy strategy to implement and a good response to the poor synthetic quality of a peptide.
Increased Coupling Time
One of the most common protocols to overcome difficult couplings is to increase the coupling time, with or without adding fresh reagents (i.e double coupling).[8] To examine the efficacy of these strategies in the context of our rapid synthesis platform, we synthesized peptides D[2], D[3], and B[1+2] with extended coupling times on stretches of problematic residues, and on individual residues in D[4], B[1], and B[2].
The results were mixed. In B[1+2], B[1], and B[2], coupling of the first residue, a C-terminal Val, to the resin proceeded to completion within 10 minutes, eliminating a 50% valine deletion product. Additionally the C-terminal des-Asn of D[4] was successfully mitigated by coupling it for 10 minutes. Unfortunately, increased coupling time only substantially reduced one non-C-terminal deletion, the des-His product in D[3]. All other deletions were essentially unaffected by increased coupling time. Although extended couplings are time consuming and typically ineffective, they are extremely easy to implement, and a reasonable early response to deletion products with our flow system.
Increased Deprotection Time
Another commonly used procedure to reduce deletion products, often used in conjunction with extended acylation times, is multiple or extended deprotection steps.[16b] To examine the utility of this strategy with our rapid synthesis protocol, we monitored the formation of Fmoc-dibenzofulvene via the UV absorbance of the reactor effluent at 304 nm and exhaustively deprotected each residue of peptides D[2] and D[3]. We successfully identified regions where Fmoc removal was slower than usual, but no change in synthetic quality was observed. This strategy is unlikely to improve synthetic outcomes with our platform, although it may provide valuable data about the location of deletion prone residues. This information may not be clear from LC-MS analysis alone if there are several copies of a problematic amino acid in a sequence.
Resins
The nature of the solid support may have a drastic effect on the outcome of peptide synthesis.[25] While working with D[4], B[1+2], and B[4], we screened several resins, including three polystyrene resins with various functionalities, a polyethylene glycol resin, and a hybrid polyethylene glycol-polystyrene resin. We found that the synthesis of D[4] on PEG or AM-PS resins radically improved the crude quality compared to other resins. While PEG resin was also highly effective for B[4] it did not alter the outcome of B[1+2] synthesis. In general, we expect the success of this strategy to be highly sequence dependent, but changing resins can be strikingly effective.
Backbone Protection
In two cases, D[4] and B[2], the above measures did not produce crude peptides of the desired purity. For this reason we explored the use of Nα-protected amino acids: Fmoc-Ile-Ser(ψMe,Mepro)-OH and Fmoc-(Hmb)-Ala-OH. These residues are known to drastically improve the synthesis of difficult sequences, presumably by disrupting chain aggregation.[26] Using procedures outlined above, we identified 114Ser and 110Ala in peptide D[4] and 29Thr – 27Tyr in B[2] as difficult stretches. We installed Fmoc-Ile-Ser(ψMe,Mepro)-OH at 114Ser of D[4]and Fmoc-(Hmb)-Ala-OH at 33Ala of B[2]. The quality of the synthesis vastly improved in both cases. We reserved Nα‐protected amino acids for special cases because they are relatively difficult to prepare, and quite expensive when obtained commercially. When used, however, these amino acid derivatives were highly effective.
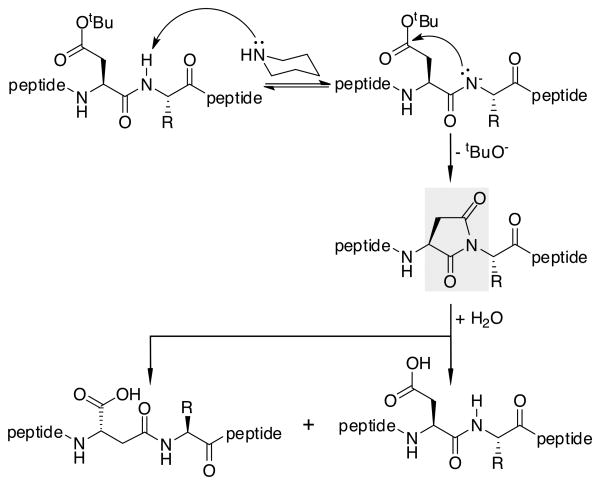
Aspartimide Formation
Another common side reaction is formation of a five membered imide ring with the aspartic acid side chain and adjacent Nα‐nitrogen as shown in Scheme 1.[19,27] The 5-membered ring can then be opened by a variety of nucleophiles, such as water, piperidine, or methanol. Ring opening can occur at either of two electrophilic sites, producing two isomers from each reaction. As a result, aspartimide leads to multiple products in the crude peptide. Formation of the imide is reported to be more common at sterically less hindered Asp-Ala, Asp-Gly, and Asp-Asn sites.[19] Below, several known strategies to reduce formation of this side product are evaluated for use in our fast flow system.
Scheme 1.
Base-promoted formation of aspartimide and its hydrolysis during Fmoc-based SPPS. Key aspartimide is highlighted in red.
Reduced Piperidine
To reduce highly undesirable aspartimide formation, we first turned to the most straightforward solutions: lower piperidine concentrations, shorter deprotection steps, and addition of HOBt to the deprotection reagent. In B[3] and B[4], reducing the concentration of piperidine from 50% to 20% (v/v in DMF) significantly reduced the formation of isomers, without otherwise affecting the synthesis. For this reason, 20% piperidine in DMF was adopted as the standard deprotection reagent for synthesis of Barnase fragments. In the synthesis of D[4], we decreased the deprotection time with 50% piperidine from 20 s to 5 s which mitigated, but did not eliminate, formation of aspartimide. Further reduction in aspartimide products was obtained by deprotecting for 20 s with 20% v/v piperidine and 0.1 m HOBt in DMF, a reagent known to suppress aspartimide formation.[28] We expect the use of less piperidine, with or without added HOBt, to be generally effective against aspartimide formation in our flow synthesizer.
3-Methylpentyl ester of aspartic acid
After the above optimization of deprotection conditions, D[4] still contained inseparable aspartimide derived impurities. The problem was partially resolved by using methylpentyl ester side chain protected aspartic acid, Fmoc-Asp(OMpe)-OH, in conjunction with the above deprotection techniques. OMpe ester of aspartic acid is more sterically hindered than the common tert-butyl ester, and is reported to effectively reduce aspartimide formation during Fmoc synthesis.[19] The use of Asp(OMpe) may generally reduce aspartimide formation with our system, as well. Like Nα-protected amino acids, however, Asp(OMpe) is not readily available, so we reserved its use for cases where other solutions were inadequate.
Alternate Deprotection Base
Another reported solution to the aspartimide problem is the use of secondary amines, such as piperazine and morpholine, which are less basic than piperidine but still sufficiently strong to remove Fmoc.[18,29] Unfortunately, piperazine is only soluble to 5% (w/v) in DMF. The use of volatile DCM is incompatible with our flow system as this solvent cannot be effectively drawn into the pump, and would boil in the heated synthesis vessel. Using 5% piperazine solution in DMF resulted in slow (over 60 sec) Fmoc removal, so we decided that piperazine was unsuitable for use in our rapid flow synthesizer. Next, we turned to morpholine, which is less basic than piperazine, but miscible with DMF. Predictably, Fmoc removal with any proportion of morpholine in DMF was extremely slow and this deprotection strategy was abandoned. We were unable to implement alternate bases for Fmoc removal without substantially increasing cycle times; piperazine and morpholine are unlikely to be viable with our rapid synthesis platform.
Miscellaneous Problems
Reversible Carboxylation
In Barnase fragments, we observed addition of CO2 (+43.99 Da relative to the expected product) as an intense, close eluting impurity. This impurity likely arose from the carboxylation of tryptophan and was found to disappear upon incubation in acetonitrile/water with 0.1% TFA as reported.[30] No other measures against it were explored.
Methionine Oxidation
The first synthesis of D[1] resulted in a large amount of methionine oxidation product (+15.98 Da) This impurity was effectively suppressed by using the anti-oxidative cleavage cocktail reported by Huang et al,[15] which includes dimethylsulfide and ammonium iodide. Unfortunately, this fragment re-oxidized during subsequent handling, so we mutated 28Met to Nle, eliminating the problem. If special care is taken in handling the peptide, methionine oxidation might be obviated.
Premature Chain Termination
We identified a truncated impurity in the crude product of B[4] synthesis. The molecular weight of the impurity indicated that the chain was abruptly capped with a 42.05 Da moiety immediately before 90Arg. Although we could not detect any acetic acid in the reagents upon MS and NMR analysis, changing activating reagent mitigated the problem. Thus, when we activated 90Arg with DCC/HOBt instead of HATU, the truncated product disappeared. As we only observed one major truncation product during our studies, we refrain from commenting on the general utility of this strategy to suppress chain termination with our flow system.
Trifluoroacetylation
We observed triflouroacetylation (+95.98 Da) in several peptide fragments. It is reported to be a prominent side product of Fmoc synthesis, formed during cleavage and global side chain deprotection in TFA.[31] The levels of this side product were highly variable, even amongst almost identical syntheses, and we were unable to eliminate it by incubating the peptide at pH 10 overnight. The impurity elutes very late during RP-HPLC and is easily removed during purification.
Oxidation of peptide hydrazides and NCL
During the ligation steps of this work, we identified several side products necessitating minor optimization of Lu's oxidation/thioesterification protocol (Scheme 2). The most severe was the almost quantitative formation of a side product with molecular mass 15.00 Da greater than the expected thioester upon oxidation/MPAA thioesterification of B[1]. After additional experimentation (See SI 8.2.1), we concluded that this side-product resulted from a Curtius rearrangement of the C-terminal azide to an isocyanate, followed by addition of MPAA to yield undesired carbamothioate, as shown in Scheme 2. We screened the temperature, pH and duration of the reaction to no avail. Fortuitously, we discovered that the formation of carbamothioate can be avoided if the azide is allowed to stand at room temperature with MPAA for 20 minutes before adding the second peptide and adjusting the pH from 3 to 7.
Scheme 2.
Transformations of C-terminal peptide hydrazides during oxidation with sodium nitrite. Major identified side products are highlighted in grey.
Additionally, we identified several less prominent side reactions, including formation of C-terminal carboxamide, hydrolysis of the azide to yield a C-terminal carboxylate, and products of incomplete oxidation. Formation of these species is well-documented in acyl azide chemistry.[32]
Carboxamide and carboxylate products were especially pronounced in the case of B[5] oxidation. Following suggestions from the literature,[32c] we reduced the temperature and the pH. While reduction of the pH was unsuccessful, lowering the temperature to −30 °C alleviated side reactions and afforded MPAA thioesters of Barnase fragments suitable for subsequent ligation. Incomplete oxidation was a prominent impurity in many DARPin oxidation reactions. We mitigated this problem by increasing the reaction time to 30 minutes.
Conclusion
In summary, we have successfully synthesized the native DARPin pE59 and RNase B. a. proteins by Fmoc chemistry on a novel fast flow peptide synthesizer. The rate at which amino acids could be incorporated allowed us to explore numerous conditions for peptide synthesis, dramatically improve the synthetic quality of our peptides, and systematically study important factors contributing to the successful synthesis of difficult sequences using this platform. Using identified optimal synthetic conditions, high-quality crude peptide fragments were produced in a rapid manner. Convergent NCL via peptide hydrazides afforded full-length proteins, which, after desulfurization and refolding, were characterized and found to be biologically active.
This work highlights the capacity of the fast flow peptide synthesis methodology to quickly and reliably make high quality peptide fragments, and serves as a guide to sequence specific optimization of rapid peptide synthesis. We present an extensive analysis of the utility of various strategies to improve synthetic outcomes with our fast flow platform and report conditions that effectively mitigate or eliminate deletion and aspartimide-derived products, while incorporating most amino acid residues in three minutes.
We believe this work represents a major step towards routine total synthesis of proteins. Our fast flow platform overcomes the primary bottleneck in protein synthesis — the rate at which pure peptides can be obtained. During the course of this study, we were able to routinely assemble ∼30 residue polypeptides in a matter of hours, rather than days. We coupled approximately 1000 amino acids, which translates to about 60 hours of linear synthetic time; using traditional Fmoc SPPS this task would require about 1000 hours (125 working days) of continuous effort. Such rapid synthesis allows peptides to be made multiple times, under different conditions, to obtain crude material of exceptional quality, simplifying purification and increasing yields. Using standard Fmoc chemistry, we were able to assemble peptides of excellent crude quality as long as 37 residues, and it seems that even longer fragments, often necessary for protein synthesis, could be routinely obtained.
We believe this work will make the total chemical synthesis of proteins significantly faster and more appealing to a broad scientific community.
Experimental Section
Synthetic Platform
All peptide synthesis was performed using our flow synthesizer and a second generation synthesis vessel (see accompanying manuscript). As a starting point for peptide assembly, we incorporated an amino acid residue every three minutes using a synthetic cycle consisting of four steps at 60 °C: a 30 second amide bond forming step (coupling), a 60 second DMF wash, a 20 second deprotection, and another 60 second wash. Ten seconds were reserved for manual manipulation of the device. Typically, 1 mmol of amino acids were activated with either 1 equivalent of HBTU (for DARPin and Barnase H2N-[4Ala-39Val]-CONHNH2) or HATU (all other Barnase peptides). In initial syntheses, 50% (v/v) piperidine in DMF was used as the deprotection agent. Syntheses were performed on a 0.20 mmol or smaller scale, dictated by the loading and swelling properties of the resin. The resin swells continuously as the peptide is elongated, and no more than 2 mL of swollen resin could be accommodated by the synthesis vessels that were used. No overpressure problems were encountered when assembling peptides as long as 37 residues. The detailed conditions for the synthesis of all peptides described in the manuscript are included in the SI.
Final Synthetic Conditions for DARPin Fragments
The final crude DARPin fragments are shown in Figure 2. No global changes were made to the synthetic procedure, but each peptide had sequence specific changes. D[1], D[2], and D[3] were synthesized on 2-chlorotrityl hydrazine polystyrene resin, and D[4] was assembled on Rink-AM resin.
D[1] was assembled on a two minute, rather than three minute, cycle. This required 20 mL of wash solvent delivered over thirty seconds, rather than one minute. This resulted in a 30 second coupling, a 20 second deprotection, two 30 second washes, and 10 seconds reserved for manual actions. Additionally, 28Met was mutated to Nle. Although the initial syntheses produced acceptable material D[2] and D[3] were ultimately assembled using double or triple deprotection (3 × 20 seconds) and 10 minute extended coupling for the 9th through 17th residues coupled. Additionally, after incorporation of 40Asp and 71Asp in D[2] and D[3] respectively, deprotection times were shortened to 10 and 6 seconds in order to mitigate potential aspartimide formation. Additionally, 116Asp was incorporated with methyl pentyl ester protection and coupled for 10 minutes, 114Ser and 113Ile were incorporated as the Fmoc-Ile-Ser(ψMe,Mepro)-OH dipeptide and coupled for 10 minutes, and the C-terminal Asn was coupled to the resin for 10 minutes.
Final Synthetic Conditions for Barnase Fragments
The final crude Barnase peptides are shown in Figure 3. All final Barnase fragments were produced with a standard three minute cycle using HATU activation and 20% piperidine in DMF (v/v) replacing the initial 50% (v/v) piperidine deprotection reagent. B[1], B[2], and B[3] were produced on 2-chlorotrityl hydrazine resin, and B[4] was produced on Rink-PEG resin.
Additionally, several peptide specific modifications were made to the synthesis. For B[1] and B[2], the C-terminal Val were coupled to the resin for 10 minutes. For B[2] 33Ala, incorporated with Hmb backbone-protection, was coupled for 25 minutes, and the subsequent residue, 32Glu, was coupled for 30 minutes. Both 33Ala and 32Glu were activated with HOBt/DCC, rather than HATU. For B[3], no additional modifications were made. For B[4], 90Arg was activated with HOBt/DCC. The masses and RP-HPLC purification yields of all the peptides assembled above, as well as overall yields of synthetic proteins, are summarized in Table 1.
Table 1.
Yields of peptides and overall yield of synthetic proteins.
| Fragments | Crude peptide, mg | Purification Yield, % |
|---|---|---|
| D[1] | 250 | 27 |
| D[2] | 270 | 13 |
| D[3] | 270 | 8 |
| D[4] | 430 | 19 |
| B[1] | 183 | 39 |
| B[2] | 340 | 30 |
| B[3] | 308 | 36 |
| B[4] | 115 | 28 |
| DARPin | N/A | 9 (1)a |
| Barnase | N/A | 12 (3)a |
Yields are calculated from purified starting fragments and exclude the purification yield of the peptides. Yields in parenthesis are calculated from crude peptides and include the purification yield of the peptide.
Supplementary Material
Acknowledgments
This research was supported by the MIT startup funds for B.L.P., MIT Reed Fund, MIT Deshpande Center, Damon Runyon-Rachleff Innovation Award, Sontag Foundation Distinguished Scientist Award, AstraZeneca Distinguished Graduate Student Fellowship for A.A.V., and Daniel S. Kemp Summer Fellowship and NIGMS/MIT Biotechnology Training Program (5T32GM008334-25) for S.K.M. We thank R. John Collier for providing some of the laboratory equipment used to carry out the experiments. We also thank Dr. Xiaoli Liao, Dr. Alexander Spokoyny, Ms. Amy Rabideau, Mr. Richard Chang and Ms. Tatiana Berger for expert technical assistance.
Abbreviations
- RP-HPLC
Reverse Phase High Performance Liquid Chromatography
- LC-MS
Liquid Chromatography-Mass Spectroscopy
- DMF
dimethylformamide
- Fmoc
Fluorenylme-hyloxycarbonyl
- HBTU
N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl)-uronium hexafluor-phosphate
- HATU
o-(7-Azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate
- TFA
trifluoroacetic acid
- HOBt
hy-droxybenzotriazole
- DCC
N,N′-Dicyclohexylcarbodiimide
- Gn•HCl
guanidinium chloride
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org.
References
- 1.(a) Kochendoerfer G, Chen S, Mao F, Cressman S, Traviglia S, Shao H, Hunter C, Low D, Cagle E, Carnevali M, Gueriguian V, Keogh P, Porter H, Stratton S, Wiedeke M, Wilken J, Tang J, Levy J, Miranda L, Crnogorac M, Kalbag S, Botti P, Schindler-Horvat J, Savatski L, Adamson J, Kung A, Kent S, Bradburne J. Science. 2003;299:884–887. doi: 10.1126/science.1079085. [DOI] [PubMed] [Google Scholar]; (b) Chalker J, Lercher L, Rose N, Schofield C, Davis B. Angew Chem, Int Ed. 2012;51:1835–1839. doi: 10.1002/anie.201106432. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2012;124:1871–1875. [Google Scholar]; (c) Roberts M, Bentley M, Harris J. Adv Drug Deliver Rev. 2012;64:116–127. [Google Scholar]; (d) Wu A, Senter P. Nat Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]; (e) Alley S, Okeley N, Senter P. Curr Opin Chem Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]; (f) Prescher J, Bertozzi C. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]; (g) Macmillan D, Bill R, Sage K, Fern D, Flitsch S. Chem Biol. 2001;8:133–145. doi: 10.1016/s1074-5521(00)90065-6. [DOI] [PubMed] [Google Scholar]; (h) Hackenberger C, Friel C, Radford S, Imperiali B. J Am Chem Soc. 2005;127:12882–12889. doi: 10.1021/ja051855k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Armishaw C, Daly N, Nevin S, Adams D, Craik D, Alewood P. J Biol Chem. 2006;281:14136–14143. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]; (j) Kumar A, Bavikar S, Spasser L, Moyal T, Ohayon S, Brik A. Angew Chem, Int Ed. 2011;50:6137–6141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2011;123:6261–6265. [Google Scholar]; (k) Kotch F, Raines R. Proc Natl Acad Sci, USA. 2006;103:3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goto Y, Katoh T, Suga H. Nat Protoc. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 3.(a) Xie J, Schultz P. Curr Opin Chem Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]; (b) Chin J, Cropp T, Anderson J, Mukherji M, Zhang Z, Schultz P. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]; (c) Döring V, Mootz H, Nangle L, Hendrickson T, de Crécy-Lagard V, Schimmel P, Marlière P. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 4.(a) Dawson P, Muir T, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]; (b) Dawson P, Kent SB. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 5.(a) Ollivier N, Behr J, El-Mahdi O, Blanpain A, Melnyk O. Org Lett. 2005;7:2647–2650. doi: 10.1021/ol050776a. [DOI] [PubMed] [Google Scholar]; (b) Chen J, Wan Q, Yuan Y, Zhu J, Danishefsky SJ. Angew Chem, Int Ed. 2008;120:8649–8652. [Google Scholar]; (c) Crich D, Banerjee A. A J Am Chem Soc. 2007;129:10064–10065. doi: 10.1021/ja072804l. [DOI] [PubMed] [Google Scholar]; (d) Haase C, Rohde H, Seitz O. Angew Chem, Int Ed. 2008;47:6807–6810. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2008;120:6912–6915. [Google Scholar]; (e) Yan L, Dawson P. J Am Chem Soc. 2001;123:526–533. doi: 10.1021/ja003265m. [DOI] [PubMed] [Google Scholar]
- 6.(a) Wan Q, Danishefsky S. Angew Chem Int Ed. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:9408–9412. [Google Scholar]; (b) Pentelute B, Kent S. Org Lett. 2007;9:687–690. doi: 10.1021/ol0630144. [DOI] [PubMed] [Google Scholar]
- 7.(a) Bang D, Kent S. Angew Chem Int Ed. 2004;43:2534–2538. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2004;116:2588–2592. [Google Scholar]; (b) Bang D, Pentelute B, Kent S. Angew Chem Int Ed. 2006;45:3985–3988. doi: 10.1002/anie.200600702. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2006;118:4089–4092. [Google Scholar]; (c) Okamoto R, Izumi M, Kajihara Y. Int J Pept Res Ther. 2010;16:191–198. [Google Scholar]; (d) Mende F, Seitz O. Angew Chem Int Ed. 2007;46:4577–4582. doi: 10.1002/anie.200700356. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:4661–4665. [Google Scholar]; (e) Li X, Kawakami T, Aimoto S. Tetrahedron Letters. 1998;39:8669–8672. [Google Scholar]; (f) Muir T, Sondhi D, Cole P. Proc Nat Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coin I, Beyermann M, Bienert M. Nat Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 9.(a) Binz H, Amstutz P, Kohl A, Stumpp M, Briand C, Forrer P, Grutter M, Pluckthun A. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]; (b) Kummer L, Parizek P, Rube P, Millgramm B, Prinz A, Mittl P, Kaufholz M, Zimmermann B, Herberg F, Pluckthun A. Proc Nat Acad Sci USA. 2012;109:E2248–E2257. doi: 10.1073/pnas.1205399109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zahnd C, Wyler E, Schwenk J, Steiner D, Lawrence M, McKern N, Pecorari F, Ward C, Joos T, Pluckthun A. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]; (d) Parizek P, Kummer L, Rube P, Prinz A, Herberg F, Pluckthun A. ACS Chem Biol. 2012;7:1356–1366. doi: 10.1021/cb3001167. [DOI] [PubMed] [Google Scholar]; (e) Schroeder T, Barandun J, Flutsch A, Briand C, Mittl P, Grutter M. Structure. 2013;21:277–289. doi: 10.1016/j.str.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Binz H, Stumpp M, Forrer P, Amstutz P, Pluckthun A. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 11.(a) Hartley R, Barker E, E Nature: New Biol. 1972;235:15–16. doi: 10.1038/newbio235015a0. [DOI] [PubMed] [Google Scholar]; (b) Dawson P, Churchill M, Ghadiri R, Kent S. J Am Chem Soc. 1997;119(19):4325–4329. [Google Scholar]
- 12.Mossakowska D, Nyberg K, Fersht A. Biochemistry. 1989;28:3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- 13.Hartley R. Trends Biochem Sci. 1989;14:450–454. doi: 10.1016/0968-0004(89)90104-7. [DOI] [PubMed] [Google Scholar]
- 14.(a) Dawson P, Churchill M, Ghadiri M, Kent S. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]; (b) Villain M, Gaertner H, Botti P. Eur J Org Chem. 2003;2003:3267–3272. [Google Scholar]
- 15.Huang H, Rabenstein D. J Pept Res. 1999;53:548–553. doi: 10.1034/j.1399-3011.1999.00059.x. [DOI] [PubMed] [Google Scholar]
- 16.(a) Chan W, White P, P . Fmoc Solid Phase Peptide Synthesis: A practical approach. Oxford University Press; New York: 2000. 346 pp. [Google Scholar]; (b) Quibell M, Turnell W, Johnson T. J Org Chem. 1994;59:1745–1750. [Google Scholar]
- 17.Carpino L, Imazumi H, El-Faham A, Zhang C, Lee Y, Foxman B, Henklein P, Hanay C, Mügge C, Wenschuh H, Klose J, Beyerman M, Bienert M. Angew Chem Int Ed. 2002;41:441–445. doi: 10.1002/1521-3773(20020201)41:3<441::aid-anie441>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2002;114:457–461. [Google Scholar]
- 18.Palasek S, Cox Z, Collins J. J Pept Sci. 2007;13:143–148. doi: 10.1002/psc.804. [DOI] [PubMed] [Google Scholar]
- 19.Mergler M, Dick F, Sax B, Weiler P, Vorherr T. J Pept Sci. 2003;9:36–46. doi: 10.1002/psc.430. [DOI] [PubMed] [Google Scholar]
- 20.Fang G, Li Y, Shen F, Huang Y, Li J, Lin Y, Cui H, Liu L. Angew Chem Int Ed. 2011;50:7645–7649. doi: 10.1002/anie.201100996. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2011;123:7787–7791. [Google Scholar]
- 21.Johnson E, Kent S. J Am Chem Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Pentelute B, Kent S, S Angew Chem Int Ed. 2012;51:993–999. doi: 10.1002/anie.201106060. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2012;124:1017–1023. [Google Scholar]
- 23.Kunitz M. J Biol Chem. 1946;164:563–568. [PubMed] [Google Scholar]
- 24.Hyde C, Johnson T, Owen D, Quibell M, Sheppard R. Int J Pept Prot Res. 1994;43:431–440. [PubMed] [Google Scholar]
- 25.Garcia-Martin F, Quintanar-Audelo M, Garcia-Ramos Y, Cruz L, Gravel C, Furic R, Cote S, Tulla-Puche J, Albericio F. J Comb Chem. 2006;8:213–220. doi: 10.1021/cc0600019. [DOI] [PubMed] [Google Scholar]
- 26.(a) Johnson T, Quibell M, Owen D, Sheppard R. J Chem Soc Chem Comm. 1993:369–372. [Google Scholar]; (b) Nicolas E, Pujades M, Bacardit J, Giralt E, Albericio F. Tetrahedron Lett. 1997;38:2317–2320. [Google Scholar]; (c) Wöhr T, Wahl F, Nefzi A, Rohwedder B, Sato T, Sun X, Mutter M. J Am Chem Soc. 1996;118:9218–9227. [Google Scholar]
- 27.(a) Mergler M, Dick F, Sax B, Stahelin C, Vorherr T. J Pept Sci. 2003;9:518–526. doi: 10.1002/psc.473. [DOI] [PubMed] [Google Scholar]; (b) Mergler M, Dick F. J Pept Sci. 2005;11:650–657. doi: 10.1002/psc.668. [DOI] [PubMed] [Google Scholar]
- 28.Dolling R, Beyermann M, Haenel J, Kernchen F, Krause E, Franke P, Brudel M, Bienert M. J Chem Soc Chem Comm. 1994:853–854. [Google Scholar]
- 29.Chang C, Waki M, Ahmad M, Meienhofer J, Lundell E, Haug J. Int J Pept Prot Res. 1980;15:59–66. doi: 10.1111/j.1399-3011.1980.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 30.Franzen H, Grehn L, Ragnarsson U. J Chem Soc, Chem Commun. 1984:1699–1700. [Google Scholar]
- 31.(a) Kocsis L, Ruff F, Orosz G. J Pept Sci. 2006;12:428–436. doi: 10.1002/psc.745. [DOI] [PubMed] [Google Scholar]; (b) Haase C, Burton M, Agten S, Brunsveld L. Tetrahedron Lett. 2012;53:4763–4765. [Google Scholar]
- 32.(a) Prelog V, Wieland P. Helv Chim Acta. 1946;29:1128–1132. [Google Scholar]; (b) Hofmann K, Thompson T, Yajima H, Schwartz E, Inouye H. J Am Chem Soc. 1960;82:3715–3720. [Google Scholar]; (c) Honzl J, Rudinger J. Collect Czech Chem C. 1961;26:2333–2344. [Google Scholar]; (d) Schwyzer R, Kappeler H. Helv Chim Acta. 1961;44:1991–2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.