Abstract
Newcastle disease is an avian pathogen causing severe economic losses to the Indian poultry industry due to recurring outbreaks in vaccinated and unvaccinated flocks. India being an endemic country, advocates vaccination against the virus using lentogenic and mesogenic strains. Two virus strains which are commonly used for vaccination are strain F (a lentogenic virus) and strain R2B (a mesogenic virus). Strain F is given to 0–7 days old chicks and R2B is given to older birds which are around 6–8 weeks old. To understand the genetic makeup of these two strains, a complete genome study and phylogenetic analysis of the F, HN genes of these vaccine strains were carried out. Both the viral strains had a genome length of 15,186 nucleotides and consisted of six genes with conserved complimentary 3' leader and 5' trailer regions. The fusion protein cleavage site of strain F is GGRQGRL and strain R2B is RRQKRF. Although both the viral strains had different virulence attributes, the length of the HN protein was similar with 577 amino acids. Phylogenetic analysis of F, HN and complete genome sequences grouped these two strains in genotype II category which are considered as early genotypes and corroborated with their years of isolation.
Introduction
Newcastle disease virus (NDV), the prototype of paramyxovirus, causes the highly contagious Newcastle disease (ND) in many avian species, resulting in substantial economic losses in the poultry industry worldwide. Strains of NDV are classified into three main pathogens as highly virulent (velogenic), intermediate (mesogenic) and non-virulent (lentogenic) on the basis of their pathogenicity for chickens. NDV is a member of the Avulavirus genus in the Paramyxovirus family [1]. NDV genome is approximately 15 kb long, non-segmented, single-stranded, negative-sense RNA that codes for six proteins: nucleoprotein (NP), phosphoprotein (P), matrix (M) protein, fusion (F) protein, haemagglutinin-neuraminidase (HN) protein, and polymerase (L) protein [2]. Although NDV is monotypic in nature, both antigenic and genetic diversities are recognised among NDV isolates. Two different systems classifying NDV are currently used worldwide. One of the systems classifies NDV into two major divisions represented by Class I and Class II, with Class I being further divided into nine genotypes and Class II into ten, when comparing the sequences isolated [3], [4], [5]. Class II viruses have been studied in more detail and the genotypes that are considered ‘early’ (1930–1960) I, II, III, IV and IX contain 15,186 nucleotides [4]. Viruses that emerged ‘late’ (after 1960) V, VI, VII, VIII and X contain 15,192 nucleotides. Vaccination of commercially reared birds is the only way to reduce disease and the losses resulting from infection. India, being an endemic country for NDV, outbreaks still occurs in spite of regular vaccination programmes. Though many reasons could be attributed to this scenario, presence of the etiological agent in the vicinity may always pose a severe threat even to vaccinated population. This gains importance by the fact that many of the free-roaming local birds, water fowls and wild birds are reported to harbour velogenic NDV without manifesting clinical signs [6].
Currently, lentogenic NDV strains Hitchner B1, La Sota, Fuller (F), and mesogenic strain R2B are widely used as live vaccines in India. Strain F NDV is a virus of low virulence originally reported by Asplin [7] in England. Since then, in several countries in Europe, Africa and Asia, the use of the virus as an immunizing agent in the form of a live vaccine has been studied [8]. Similarly the vaccine strain R2B used in the Indian subcontinent has given excellent results in older birds (>6 to 8 weeks old) with long lasting immunity but has been proven to be severely pathogenic for baby chicks. The virus strain had originated by passaging three Indian field isolates in embryonated chicken eggs, with one of the lines showing signs of attenuation after 19 passages [9]. Recently the complete genome sequence of NDV mesogenic strain R2B was elucidated [10]. In this paper, we elucidate the complete genome of the hitherto lentogenic NDV strain ‘F’ and studied the genotype characterization of the commonly used vaccine strains in India, namely ‘F’ and ‘R2B’.
Materials and Methods
Virus strains
NDV strain F (referred to as Dobson's ‘F’ strain) seed virus was obtained from the viral repository maintained at the Indian Veterinary Research Institute. This virus had undergone 8 serial passages in embryonated eggs in April 1953 when it was received at this Institute. It was further propagated to 39 serial passages in embryonated eggs and the seed virus was prepared [11]. The R2B strain of NDV was also obtained from the viral repository at the Indian Veterinary Research Institute. Both the seed viruses were plaque purified for further work.
Virus propagation, RNA isolation and genome sequencing
The virus was propagated in eleven days-old embryonated chicken eggs and purified according to previously established procedures [12]. Total viral RNA was extracted using Trizol (Sigma, USA), according to manufacturer's instructions. Reverse transcription was carried out using the Thermoscript RT kit (Invitrogen, USA) to synthesize the first strand cDNA. Oligonucleotide primers were synthesized for amplifying the entire genome of F and R2B viral strains as overlapping fragments. The sequences of primers used in the study are given in Table 1. The leader and trailer regions were generated by rapid amplification of cDNA end (RACE) protocol, as described elsewhere [13]. Three recombinant plasmids containing the amplified product from each overlapping fragment were purified and sequenced in ABI 3730 DNA analyzer (Applied Biosystems, USA) at the DNA sequencing facility, University of Delhi, South Campus, New Delhi.
Table 1. Primers used in this study to amplify R2B and F strains of NDV.
| Name | Primer sequence (5′–3′) | Genomic locationab | Expected size |
| Strain R2B | |||
| 1Fc | ACCAAACAGAGAATCCGTGAA | 1–21 | 1900 bp |
| 1Rd | GTGAAGGTGGCCATCTTCACT | 1880–1900 | |
| 2F | ACAACACGGGCACAACTCGAC | 1741–1761 | 1559 bp |
| 2R | GATGAGTCCATCCTGGCACAA | 3280–3300 | |
| 3F | TTCCATCCCACTGAATGATCG | 3191–3211 | 2171 bp |
| 3R | AGGGTTACCGGTGATCAAGCC | 5342–5362 | |
| 4F | GGCTCTTTACAATCTGGCTGG | 5251–5271 | 2230 bp |
| 4R | GACTTAGCCATCCGAATCTGG | 7461–7481 | |
| 5F | GGTTGCGATTGCGTGAGTCAC | 7356–7376 | 2044 bp |
| 5R | GGTTGCGATTGCGTGAGTCAC | 9380–9400 | |
| 6F | CTAATGGAGGGATTCGCATAC | 9242–9262 | 2046 bp |
| 6R | CCCAATCTCCATTTCCAGGCG | 11268–11288 | |
| 7F | GTGCACTCATATGTCCTGACC | 11093–11113 | 1951 bp |
| 7R | CGTCGAGTGCAAGAGACTAGC | 13024–13044 | |
| 8F | CACCCTGTCATCCATTCAAGG | 12923–12943 | 2263 bp |
| 8R | CCCACCAAACAAAGATTTGGTGAATG | 15164–15186 | |
| Strain F | |||
| 1Fc | ACCAAACAGAGAATCCGTAAGTTA | 1–24 | 2363 bp |
| 1Rd | GTTTCCGCGGCTGGGTTGACTCCCCT | 2338–2363 | |
| 2F | AGTCAACCCAGCCGCGGAAACAG | 2343–2365 | 2621 bp |
| 2R | GAGCTGCGGCCGCTGTTATTTG | 4943–4964 | |
| 3F | AACAGCGGCCGCAGCTCTGAT | 4948–4968 | 1353 bp |
| 3R | TACAACGCGTAGTTTTTTCTTAACTC | 6276–6301 | |
| 4F | AACTACGCGTTGTAGATGACCAAAG | 6288–6312 | 2078 bp |
| 4R | GCACTACGTATTTTGCCTTGTATCTC | 8351–8366 | |
| 5F | CAAAATACGTAATGGTAAATAATACGGGTAGGACATG | 8358–8383 | 1694 bp |
| 5R | TCAGCTTAGCGAAGATCCGTCCATTAACT | 10024–10052 | |
| 6F | CTTCGCTAAGCTGACAAAGAAGTTAAGGAACTG | 10039–10071 | 2805 bp |
| 6R | GTCTAGGCCTCTTACTCTCAGGTAATAG | 12817–12844 | |
| 7F | AGAGGCCTAGACAATATTGTCT | 12833–12854 | 2353 bp |
| 7R | ACCAAACAAAGATTTGGTGAATGACGAG | 15159–15186 |
Genomic location of strain R2B is based on the complete genomic sequence of NDV strain Mukteswar.
Genomic location of strain F is based on the complete genomic sequence of NDV strain La Sota.
F stands for forward primer.
R stands for reverse primer.
Sequence and phylogenetic analysis
The complete genome sequence analysis of NDV vaccine strains ‘F’ and ‘R2B’ were carried out using the Seqman, Editseq and Megalign modules of the Lasergene software package. Sequences representing genotypes of NDV were aligned using Muscle [14] with default settings implemented in MEGA 5. MEGA 5 was used to select best fit model of nucleotide substitution and phylogenetic analysis was conducted using the nucleotide substitution model under maximum likelihood with boot strap values for 1000 replicates. Markov Chain Monte Carlo method implemented in BEAST was used to generate Bayesian evolutionary analysis. Three independent Markov Chains were run for 70 million generations for complete genome and 10 million for individual genes datasets and first 10% of samples were discarded as burn in. Logs were combined using Logcombiner V 1.7.5. Stationarity was assessed as effective sample size >200 using Tracer Maximum clade credibility tree generated using Tree Annotator V.1.7.5. The support for nodes in Bayesian tree deduction was assessed using posterior probabilities values calculated in BEAST. Trees were visualized using FigTree V 1.4.0. The assignment of genotypes to the sequences used in this study was in accordance to the criteria described by Diel et al. [15].
Pathogenicity test
NDV vaccine strains F and R2B were subjected to pathogenicity tests that included mean death time (MDT) analysis in 9 days-old specific pathogen free (SPF) embryonated chicken eggs, intra-cerebral pathogenicity index (ICPI) test in one-day old SPF chickens and intra-venous pathogenicity index (IVPI) test in six-weeks old SPF chickens using standard procedures [16].
Results
Complete sequences analysis of NDV strains F and R2B
On alignment of the various overlapping fragments generated in this study, it was found that the complete length of the genome of both the vaccine strains of NDV, namely strains ‘F’ and ‘R2B’, is 15,186 nucleotides (nts), which is considered to be of early genotypes [4]. The complete sequence of strain F and strain R2B is available in the GenBank under the accession numbers KC987036.1 and JX316216.1 respectively. The various defining features of the genome of these viral strains are given in Table 2. The 3′ leader sequence of strains F and R2B consists of 55 nts, a length present in all NDV strains [17]. The 5′ trailer sequence of both the strains was 114 nts. It was also found that the 3′- and 5′- termini were highly conserved; especially the first 12 nts in the 3′-terminus and 8 nts in the 5′-terminus were identical. The P gene contains a putative editing site 477AAAAAGGG484 (mRNA sense) that is identical in position to other NDV strains. The gene order of 3′-N-P-M-F-HN-L-5′ coding from six open reading frames was also similar to other NDV strains.
Table 2. Genomic features and protein characteristics of NDV strains R2B and F.
| Gene | Hexamer Phasing Position | Gene Length | ORF | IGS | 5'UTR | 3'UTR | Amino acids | MW (kDa) | pI |
| Strain R2B | |||||||||
| N | 2 | 1746 | 1470 | 2 | 66 | 210 | 490 | 53.2 | 5.51 |
| P | 4 | 1451 | 1188 | 1 | 83 | 180 | 396 | 41.84 | 6.21 |
| P/V | 4 | 1452 | 720 | - | 83 | 649 | 240 | 25.25 | 5.63 |
| P/W | 4 | 1453 | 684 | - | 83 | 686 | 228 | 24.49 | 10.11 |
| M | 4 | 1241 | 1095 | 1 | 34 | 112 | 365 | 39.63 | 9.64 |
| F | 4 | 1792 | 1662 | 31 | 46 | 84 | 554 | 58.8 | 8.56 |
| HN | 3 | 2002 | 1734 | 47 | 91 | 177 | 578 | 63.17 | 7.88 |
| L | 6 | 6703 | 6615 | - | 11 | 77 | 2205 | 248.44 | 6.53 |
| Strain F | |||||||||
| N | 2 | 1746 | 1470 | 2 | 66 | 210 | 490 | 53.27 | 5.35 |
| P | 4 | 1451 | 1188 | 1 | 83 | 180 | 396 | 42.38 | 6.19 |
| P/V | 4 | 1452 | 720 | - | 83 | 649 | 240 | 25.53 | 5.23 |
| P/W | 4 | 1453 | 546 | - | 83 | 824 | 182 | 19.48 | 9.07 |
| M | 4 | 1241 | 1095 | 1 | 34 | 112 | 365 | 39.49 | 9.59 |
| F | 4 | 1792 | 1662 | 31 | 46 | 84 | 554 | 58.81 | 8.17 |
| HN | 3 | 2002 | 1734 | 47 | 91 | 177 | 578 | 63.18 | 7.55 |
| L | 6 | 6703 | 6615 | - | 11 | 77 | 2205 | 248.4 | 7.05 |
A comparison of NP gene of F and R2B with other vaccine strains of genotype II viruses showed that the amino acid sequence of NP was conserved (the divergence between representative genotype II strains is 0.052±0.004 S.E.) while the C-terminal sequence of NP was relatively variable compared to other regions of NP protein. One of the characteristics of M protein of NDV is nuclear localization, and it is provided by the nuclear localization signals which are due to clusters of basic amino acids in the M protein. There are two highly basic clusters in the position of amino acid sequence from 246 to 263 [18]. The amino acid sequence of both F and R2B, with respect to M protein, is 246DRKGKKVTFDKLEKKIR263.
Transcription starts in the 3′ leader region, and the genes are transcribed into separate mRNAs in a start-stop-restart mechanism mediated by the conserved transcriptional control sequences at the beginning (Gene Start, GS) and end (Gene end, GE) of each gene. As in other NDV strains, the non-coding intergenic sequences (IGS) lie between GE and GS. The GS, GE and IGS of strains F and R2B for NP, P, M, F, HN and L genes are given in Table 3.
Table 3. Sequences of gene start, gene end and intergenic regions of NDV strains R2B and F.
| Gene | Gene start sequence | Start position (nt) | Gene end sequence | Intergenic sequences |
| Strain R2B | ||||
| NP | ACGGGTAGAA | 56 | TTAGAAAAAA | AT |
| P | ACGGGTAGAA | 1804 | TAAGAAAAAA | T |
| M | ACGGGTAGAA | 3256 | TTAGAAAAAA | C |
| F | ACGGGTAGAA | 4498 | TAAGAAAAAA | CTACCGGATGTAGGTGAACAAAGGCAATAT |
| HN | ACGGGTAGAA | 6321 | TAAGAAAAAA | TGTAAGTGGCAATGAGATACAAGGCAAAACAGCTCATGGTAAATAGT |
| L | ACGGGTAGGA | 8370 | TTAGAAAAAA | - |
| Strain F | ||||
| NP | ACGGGTAGAA | 56 | TTAGAAAAAA | GT |
| P | ACGGGTAGAA | 1804 | TAAGAAAAAA | T |
| M | ACGGGTAGAA | 3256 | TTAGAAAAAA | C |
| F | ACGGGTAGAA | 4498 | TTAGAAAAAA | CTACGCGTTGTAGATGACCAAAGGACGATAT |
| HN | ACGGGTAGAA | 6321 | TAAGAAAAAA | TGTAAGTGGCAATGAGATACAAGGCAAAATACGTAATGGTAAATAAT |
| L | ACGGGTAGGA | 8370 | TTAGAAAAAA | - |
F and HN gene analysis
The F protein is considered as a key determinant in virulence of NDV [19]. However, recent analysis of the complete genome sequences of viral isolates and by reverse genetics, it has been proven that the fusion protein cleavage site (FPCS), spanning amino acids 112 to 117, of the F protein alone cannot confer virulence to an otherwise avirulent strain [20], [21]. In this context, the FPCS of strain F is GGRQG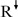 L and that of R2B is RRQK
L and that of R2B is RRQK F. Virulent NDV strains typically contain a polybasic cleavage site (R-X-K/R-
F. Virulent NDV strains typically contain a polybasic cleavage site (R-X-K/R- F), which is recognized by most cells. In addition, the presence of phenylalanine (F) residue at position 117 has been described as being possible contributor to neurological effects [22]. In contrast, the lentogenic strains are being encoded by a single pair of basic amino acids and F at position 117 being replaced by L. This further substantiates the fact that strain F is lentogenic while R2B is mesogenic.
F), which is recognized by most cells. In addition, the presence of phenylalanine (F) residue at position 117 has been described as being possible contributor to neurological effects [22]. In contrast, the lentogenic strains are being encoded by a single pair of basic amino acids and F at position 117 being replaced by L. This further substantiates the fact that strain F is lentogenic while R2B is mesogenic.
HN is a type II homotetrameric glycoprotein with a monomer length of 577 amino acids for most NDV strains [23]. The ability to bind a sialic acid containing receptor is one of the functions of the HN protein and plays a key role in the initial steps of the NDV life cycle [24]. Several amino acid residues, R 174, Y 526, E 401 and R 416 in HN protein were reported to be involved in the activity of sialic acid binding [25]. The salient features of HN protein of strains F and R2B include the following conserved amino acids: R 174, I 175, D 198, K 236, R 416, R 498, Y 526 and E 547; potential glycosylation sites being G1-119, G2-341, G3-433, G4-481 and G6-538, which is similar to genotype II vaccine strains Beaudette C, B1 and Ulster. The glycosylation site G5 present in other NDV strains has been replaced by serine (G5-N508S). Further, there is a conserved amino acid E 347 present both in F and R2B, a feature common to all vaccine strains of genotype II ND viruses. The percent similarity in relation to amino acids between F and R2B is 91% and that of R2B and Mukteswar is 96%. The most variable portion of NDV HN protein is present in the N-terminal 78 amino acids that included the transmembrane domain. The predicted amino acids present in the transmembrane domain of strain R2B and Mukteswar is 27IAALLLMVITLAVSAVALAYSME49, which is different from that of strain F –27IAILLLTIVTLAISVISLVYIMG49.
Phylogenetic analysis of complete genome, complete F and HN genes of strains F and R2B
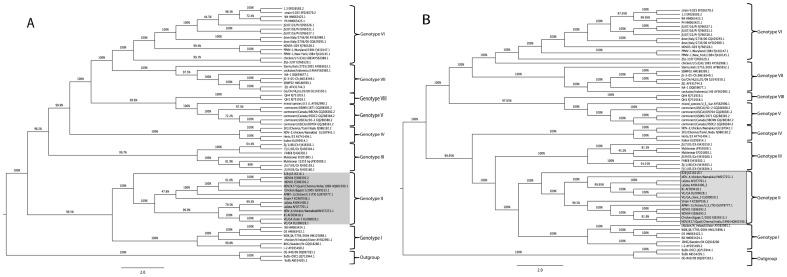
The commonly used Indian vaccine strains of NDV grouped along with genotype II viruses that also contained viral strains exhibiting different virulence attributes, such as La Sota, B1, VG/GA that are lentogenic, Beaudette C that is mesogenic and Texas GB (TX/GB) being velogenic. The vaccine strain F was more closely related to Beaudette C strain than to La Sota or B1. The vaccine strain R2B branched out separately from the two sub-clusters within genotype II viruses, one cluster involving the lentogenic vaccine strains La Sota, B1, VG/GA and the other with virulent virus strains Beaudette C, Texas GB and Egypt/2005 (Figure 1).
Figure 1. Phylogenetic analysis of complete genome of vaccine strains R2B and F.
The whole genome sequence of NDV strain R2B and F was aligned with other NDV strain sequences from GenBank representing different genotypes using Muscle algorithm in MEGA5.0. A. A rooted consensus tree drawn to scale was obtained using Maximum Likelihood method employed with discrete gamma distribution model with evolutionary invariable (GTR+G+I) using MEGA5.0. B. A Bayesian tree analysis of complete genome analysis of Indian vaccine strains. The scale indicates the number of substitutions per site. Posterior probabilities are indicated above the branches.
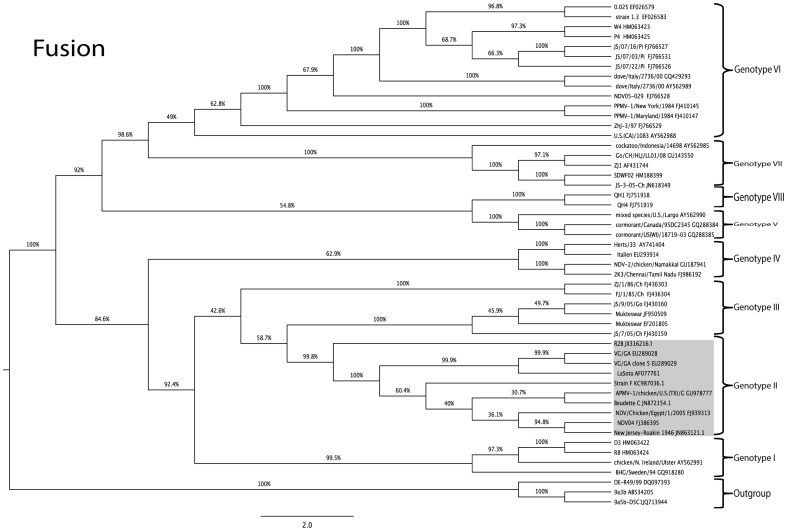
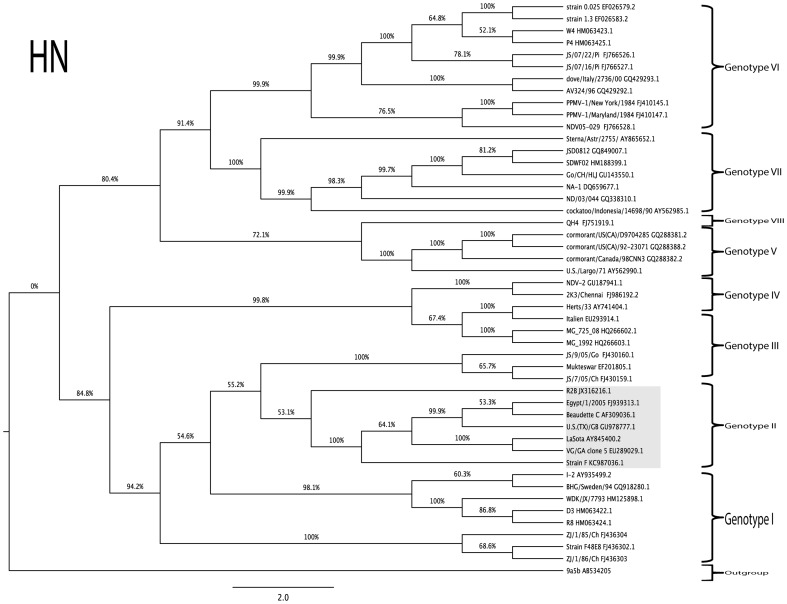
Further, the topology of the tree did not have any significant differences when the complete fusion and HN genes of the viruses were used for comparison in this study (Figure 2 and Figure 3).
Figure 2. Phylogenetic relationship of vaccine strains R2B and strain F (boxed) based on Fusion protein gene.
The phylogenetic tree was drawn using maximum likelihood method employed in MEGA program (Version 5). The percentage of replicate trees in which associate taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Trees are drawn to scale, with branch length in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed and are in the units of the number of base substitutions per site.
Figure 3. Phylogenetic relationship of vaccine strains R2B and strain F (boxed) based on HN protein gene.
The phylogenetic tree was drawn using maximum likelihood method employed in MEGA program (Version 5). The percentage of replicate trees in which associate taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Trees are drawn to scale, with branch length in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed and are in the units of the number of base substitutions per site.
Pathogenicity
The NDV vaccine strains F and R2B had a MDT of 184 and 63 hrs, ICPI value of 0.1 and 1.45 and IVPI value of 0.0 and 0.55 respectively, confirming the lentogenicity and mesogenicity of the strains.
Discussion
The backyard poultry sector in India is increasingly being recognized as an important area of intervention for poverty alleviation. Because rearing is based on traditional practices with no focus on veterinary and health sciences, there is high mortality caused by diseases. Newcastle disease is identified as the most fatal disease wiping out entire flocks, severely constraining the growth of this sector. Vaccination is the principal method of controlling the losses caused by ND. For prophylactic use, the lentogenic strains of NDV of chick embryo origin, such as B1, La Sota and F are likely used as vaccines in most countries of the world. In addition, mesogenic strains of NDV, Komarov and R2B are still used extensively in many Asian countries, including India for its very strong immune response evoked in susceptible birds even up to 4 years [26]. The puzzling persistence of virulent NDV in poultry despite intensive vaccination efforts has been a recurrent phenomenon in endemic countries of Asia, Africa and Central America [27], [4]. Almost exclusive predominance of low virulence class I and mesogenic virus of class II [4] suggests that the immune pressure from vaccination may be selecting variant form of virulent NDV.
Both the vaccine strains used in this study have a genome length of 15,186 nts, suggesting that these belong to the ‘early’ (1930–1960) genotypes among the class II ND viruses [4]. This could be further corroborated with their years of isolation being 1950 for strain F in England, and 1945 for strain R2B in India. Phylogenetic analysis of complete genome of these vaccine strains aligned them with class II genotype II viruses, which also harbour some of the commonly used vaccine strains across the world including B1, La Sota, VG/GA and Beaudette C. Interestingly, this genotype also includes the neurotropic virulent chicken/U.S (TX) GB/1948 (TXGB) isolate which was isolated in 1948, and is used in the USA as a challenge virus to show efficacy of ND commercial vaccines before production [5]. Within the genotype II viruses, two distinct separate groups were delineated in which lentogenic strains, such as B1, La Sota, VG/GA formed a group and mesogenic/velogenic strains, such as Beaudette C, TXGB and Egypt/2005 formed another group. The vaccine strain F was related to the mesogenic group of viruses, while R2B strain forming a separate clade branching out from both these groups but within genotype II. This is in concurrence with the earlier findings [28], [29], wherein the FPCS region of the virus was used to study the relationship among different viruses. The topology of the tree and the grouping of Indian vaccine strains did not change when the full-length F and HN genes were compared with the sequences belonging to different genotypes from the GenBank.
Both NDV surface glycoproteins interact during infection of cells [30]. The principal function of paramyxovirus F protein is pH independent cell fusion, following cleavage activation by cellular proteases [31]. All five potential glycosylation sites were conserved among NDV isolates and at least four of them are utilized in the ectodomain of the mature protein [32]. The F glycoprotein of strains R2B and F contains six potential N-linked glycosylation acceptor sites at residues 85, 191, 366, 447, 471, and 541 that are all conserved in other NDV strains. Cysteines are important for disulphide bond formation in the F protein [33], [34]. R2B strain has 13 cysteine residues, whereas strain F carries 12, which is similar to another vaccine strain Ulster.
Virulence of NDV isolates is primarily determined by sequence at the F cleavage site from positions 112 to 116 [35]. The difference in the cleavage site of both the vaccine strains used in this study is indicative of the difference in the virulence attribute of these strains, namely lentogenic for strain F and mesogenic for strain R2B in corroboration with the pathogenicity test data.
An interesting feature, especially with the genome of R2B in relation to the polymerase gene, is that this genome is closely related to the genome of Egypt/2005 which has been designated as a velogenic virus [36]. This further substantiates to the fact that there are other virulence factors such as polymerase gene, which also play a major role in determining the virulence of NDV [21].
With reference to the HN protein, different amino acid sequence lengths of 571, 577, 581 and 616 have been reported for different NDV strains [37]. Analyzing the HN sequences of different NDV strains, a 571 amino acid HN protein, which is the shortest of all, was found in genotypes III–VIII. In this context, the length of HN protein of strain Mukteswar also has 571 amino acids (consisting exclusively of viscerotropic, velogenic strains). Genotype II strains had 6 amino acids extension regardless of whether the strain was lentogenic, mesogenic or velogenic [38]. Strains F and R2B with different virulence attributes have 577 amino acids that concurred with this fact. This is in contradiction to the findings of Zanetti et al. [39] who showed that the NDV pathogenicity increased through serial viral passages by introduction of point mutation at the carboxy terminus region of HN to shorten this protein. However, based on experimental evidence it has been proven that the length of HN protein has no role in NDV pathogenicity [19].
India being an endemic country for NDV, outbreaks of the disease occur every passing year. It has also been recently reported about the persistence of genotype IV strains in India [40]. The results of the study reported herein indicate that the conventional vaccines like strain F and R2B to belong to genotype II. There have been reports suggesting that vaccination against NDV although protects against clinical disease, it fails to protect against virus shedding when challenged with a different genotype virus [41], [42]. Considering these facts, it can be concluded that the commonly used vaccine strains in India need better jurisprudence in its usage as a prophylactic agent.
Acknowledgments
We thank the Director, Indian Veterinary Research Institute for extending the facilities to carry out the work.
Funding Statement
This work was supported by a research grant from the National Fund for Basic, Strategic and Frontier Application Research in Agriculture, Indian Council of Agricultural Research to S.D. (NFBSFARA/BS-3010), and a grant from Department of Biotechnology, Ministry of Science and Technology to M.M.C. (BT/PR15373/AAQ/57/116/2011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. de Leeuw O, Peeters B (1999) Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J Gen Virol 80: 131–136. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DJ, Senne DA (2008) Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. In: Saif YM, Fadly AM, Glisson JR, Mc Dougald LR, Nolan LK, Swayne DE, Editors. Diseases of poultry. Iowa State University Press. pp. 75–116.
- 3. Ballagi-Pordány A, Wehmann E, Herczeg J, Belák S, Lomniczi B (1996) Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch Virol 141: 243–261. [DOI] [PubMed] [Google Scholar]
- 4. Czeglédi A, Ujvári D, Somogyi E, Wehmann E, Werner O, et al. (2006) Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res 120: 36–48. [DOI] [PubMed] [Google Scholar]
- 5. Miller PJ, Decanini EL, Afonso CL (2010) Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol 10: 26–35. [DOI] [PubMed] [Google Scholar]
- 6. Raghavan VS, Kumanan K, Nachimuthu K (1998) Use of MDBK cells in characterizing Newcastle disease virus isolates from desi chicken. Indian Vet J 75: 1079–1082. [DOI] [PubMed] [Google Scholar]
- 7. Asplin FD (1952) Immunisation against Newcastle disease with a virus of low virulence (Strain F) and observations on sub-clinical infection in partially resistant fowls. Vet Rec 64: 245. [Google Scholar]
- 8. Lancaster JE (1962) The world distribution of Newcastle Disease-1961. Can J Comp Med Vet Sci 26: 244–245. [PMC free article] [PubMed] [Google Scholar]
- 9. Iyer GS, Hashmi ZA (1945) Studies on Newcastle (Ranikhet) disease virus strain differences in amenability to attenuations. Indian J Vet Sci 15: 155–157. [Google Scholar]
- 10. Chellappa MM, Dey S, Gaikwad S, Kataria JM, Vakharia VN (2012) Complete genome sequence of Newcastle disease virus mesogenic vaccine strain R2B from India. J Virol 86: 13814–13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IVRI Report (1954) Annual Report of the Indian Veterinary Research Institute Izatnagar and Mukteswar, Division of Pathology and Bacteriology 15 p. [Google Scholar]
- 12. Panda A, Huang Z, Elankumaran S, Rockermann DD, Samal SK (2004) Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb Pathog 36: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z, Yu M, Zhang H, Wang HY, Wang LF (2005) Improved rapid amplification of cDNA ends (RACE) for mapping both the 5' and 3' terminal sequences of paramyxovirus genomes. J Virol Methods 130: 154–156. [DOI] [PubMed] [Google Scholar]
- 14. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, et al. (2012) Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol 12: 1770–1779. [DOI] [PubMed] [Google Scholar]
- 16.Alexander DJ (1989). Newcastle disease. In: Purchase HG, Arp LH, Domermuth CH, Pearson JE, editors. A laboratory manual for the isolation and identification of avian pathogens, Pennysylvania: American Association of Avian Pathologists, pp. 114–120.
- 17. Krishnamurthy S, Samal SK (1998) Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J Gen Virol 79: 2419–2424. [DOI] [PubMed] [Google Scholar]
- 18. Coleman NA, Peeples ME (1993) The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology 195: 596–607. [DOI] [PubMed] [Google Scholar]
- 19. Römer-Oberdörfer A, Werner O, Veits J, Mebatsion T, Mettenleiter TC (2003) Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. J Gen Virol 84: 3121–3129. [DOI] [PubMed] [Google Scholar]
- 20. Khattar SK, Yan Y, Panda A, Collins PL, Samal SK (2009) A Y526Q mutation in the Newcastle disease virus HN protein reduces its functional activities and attenuates virus replication and pathogenicity. J Virol 83: 7779–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rout SN, Samal SK (2008) The large polymerase protein is associated with the virulence of Newcastle disease virus. J Virol 82: 7828–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kattenbelt JA, Stevens MP, Gould AR (2006) Sequence variation in the Newcastle disease virus genome. Virus Res 116: 168–184. [DOI] [PubMed] [Google Scholar]
- 23. Schuy W, Garten W, Linder D, Klenk HD (1984) The carboxy-terminus of the hemagglutinin-neuraminidase of Newcastle disease virus is exposed at the surface of the viral envelope. Virus Res 1: 415–426. [DOI] [PubMed] [Google Scholar]
- 24. Huang Z, Panda A, Elankumaran S, Govindarajan D, Rockemann DD, et al. (2004) The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. J Virol 78: 4176–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Connaris H, Takimoto T, Russell R, Crennell S, Moustafa I, et al. (2002) Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J Virol 76: 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seetharaman C (1951) Immunity in fowls following vaccination against Ranikhet disease. Indian Vet J 28: 13–16. [Google Scholar]
- 27. Alexander DJ (2001) Gordon Memorial Lecture. Newcastle disease. Br Poult Sci 42: 5–22. [DOI] [PubMed] [Google Scholar]
- 28. Nanthakumar T, Tiwari AK, Kataria RS, Butchaiah G, Kataria JM, et al. (2000) Sequence analysis of the cleavage site-encoding region of the fusion protein gene of Newcastle disease viruses from India and Nepal. Avian Pathol 29: 603–607. [DOI] [PubMed] [Google Scholar]
- 29. Czeglédi A, Wehmann E, Lomniczi B (2003) On the origins and relationships of Newcastle disease virus vaccine strains Hertfordshire and Mukteswar, and virulent strain Herts'33. Avian Pathol 32: 271–276. [DOI] [PubMed] [Google Scholar]
- 30. Stone-Hulslander J, Morrison TG (1997) Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J Virol 71: 6287–6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morrison TG (2003) Structure and function of a paramyxovirus fusion protein. Biochim Biophys Acta 1614: 73–84. [DOI] [PubMed] [Google Scholar]
- 32. McGinnes LW, Serge T, Chen H, Hamo L, Schwertz S, et al. (2001) Mutational analysis of the membrane proximal heptad repeat of the Newcastle disease virus fusion protein. Virology 289: 343–352. [DOI] [PubMed] [Google Scholar]
- 33. Chen L, Colman PM, Cosgrove LJ, Lawrence MC, Lawrence LJ, et al. (2001a) Cloning, expression and crystallization of the fusion protein of Newcastle disease virus. Virology 290: 290–299. [DOI] [PubMed] [Google Scholar]
- 34. Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, et al. (2001b) The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9: 255–266. [DOI] [PubMed] [Google Scholar]
- 35. Nagai Y, Klenk HD, Rott R (1976) Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology 72: 494–508. [DOI] [PubMed] [Google Scholar]
- 36. Mohamed Mahmoud HA, Kumar S, Paldurai A, Megahed MM, Ghanem IA, et al. (2009) Complete genome sequence of a virulent Newcastle disease virus isolated from an outbreak in chickens in Egypt. Virus Genes 39: 234–237. [DOI] [PubMed] [Google Scholar]
- 37. Römer-Oberdörfer A, Veits J, Werner O, Mettenleiter TC (2006) Enhancement of pathogenicity of Newcastle disease virus by alteration of specific amino acid residues in the surface glycoproteins F and HN. Avian Dis 50: 259–263. [DOI] [PubMed] [Google Scholar]
- 38. Sakaguchi T, Toyoda T, Gotoh B, Inocencio NM, Kuma K, et al. (1989) Newcastle disease virus evolution. I. Multiple lineages defined by sequence variability of the hemagglutinin-neuraminidase gene. Virology 169: 260–72. [DOI] [PubMed] [Google Scholar]
- 39. Zanetti F, Berinstein A, Carrillo E (2008) Effect of host selective pressure on Newcastle disease virus virulence. Microb Pathog 44: 135–140. [DOI] [PubMed] [Google Scholar]
- 40. Tirumurugaan KG, Kapgate S, Vinupriya MK, Vijayarani K, Kumanan K, et al. (2011) Genotypic and pathotypic characterization of Newcastle disease viruses from India. PLoS One 6: e28414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller PJ, King DJ, Afonso CL, Suarez DL (2007) Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine 25: 7238–7246. [DOI] [PubMed] [Google Scholar]
- 42. Kapczynski DR, King DJ (2005) Protection of chickens against overt clinical disease and determination of viral shedding following vaccination with commercially available Newcastle disease virus vaccines upon challenge with highly virulent virus from the California 2002 exotic Newcastle disease outbreak. Vaccine 23: 3424–3433. [DOI] [PubMed] [Google Scholar]