Abstract
Porcine reproductive and respiratory syndrome (PRRS) continues to be one of the most significant diseases of swine. IDEXX HerdChek PRRS, a commercially available enzyme-linked immunosorbent assay (ELISA), has become the industry standard for the detection of antibodies against PRRS virus (PRRSV). The need to accurately determine the PRRSV serostatus of herds and individual animals has prompted the development of several follow-up assay methods. A highly specific and repeatable blocking ELISA (bELISA) was developed on the basis of the use of an expressed PRRSV nucleocapsid (N) protein as the antigen and a biotinylated monoclonal antibody. Validation of the bELISA used sera from 316 animals experimentally and naturally infected with North American PRRSV and sera from 370 uninfected animals. Receiver operating characteristic analysis of the data calculated a diagnostic sensitivity of 97.8% and a diagnostic specificity of 100%. The between-run coefficient of variation of an internal quality control serum was 4.24%. The bELISA was able to detect seroconversion as well as the IDEXX ELISA and the indirect fluorescent antibody (IFA) assay; kappa values were 0.94 and 0.96, respectively. A collection of 133 serum samples with unexpected positive IDEXX ELISA results was obtained from 4,038 diagnostic samples submitted from farms from which PRRS-negative results were expected. The bELISA identified 97% of the samples as PRRS seronegative, while the IFA identified 100% as seronegative. The anticipated use of the bELISA is as a follow-up test to the IDEXX ELISA for determining the PRRSV serostatus of individual animals with unexpected positive test results from swine herds from which negative results are expected.
Porcine reproductive and respiratory syndrome (PRRS) continues to be one of the most devastating diseases of swine throughout the world. The syndrome is characterized by respiratory disease leading to increased mortality in young pigs, respiratory disease leading to decreased performance in adults, and infertility or late-term abortions in sows. Two distinct strains of PRRS virus (PRRSV) cause the disease in Europe and North America (7, 32). PRRSV was classified with three other viruses (equine arteritis virus, lactate dehydrogenase-elevating virus, and simian hemorrhagic fever virus) in genus Arterivirus, family Arteriviridae, order Nidovirales (27).
The PRRSV is a small, enveloped virus with a 15-kb positive-sense RNA genome composed of nine open reading frames (ORF). Most of the genome is comprised of ORF 1a and ORF 1b that code for RNA replicases. ORF 2a, ORF 3, and ORF 4 encode minor glycoproteins of the PRRSV particle (18). ORF 2b encodes a minor nonglycosylated protein (35). The major structural proteins are encoded by ORF 5, ORF 6, and ORF 7 and are called glycoprotein 5 (GP5), nonglycosylated membrane protein (M), and nucleocapsid (N), respectively. GP5 is a glycosylated, 25-kDa membrane-associated protein, while M is a nonglycosylated, 19-kDa structural matrix protein for the membrane. The N protein is a basic, 15-kDa protein that forms an icosahedral shell around the RNA genome (18). It is expressed abundantly in infected cells and constitutes 20 to 40% of the viral protein (22). Most of the antibodies in the immune response of an infected animal are directed against the N and M proteins. Anti-N antibodies can be detected as early as 7 days postinfection (dpi), while anti-M antibodies can be detected by 14 dpi (19). The N proteins of the European and North American strains of PRRSV have both conserved and divergent epitopes that are recognized by monoclonal antibodies (MAbs) (22). Because of these properties, the N protein has been chosen as the antigen for several diagnostic tests (8, 9, 25, 33).
Serological testing to determine the PRRS status of herds and individual animals is often included in management strategies for monitoring and controlling PRRS. An indirect enzyme-linked immunosorbent assay (ELISA) was first described by Albina et al. in 1992 (1). The IDEXX HerdChek 2XR PRRS ELISA, a commercially available indirect ELISA, has become the industry standard for monitoring the serostatus of swine herds. IDEXX reports excellent sensitivity and specificity values for the assay (97.4 and 99.6%, respectively) (HerdChek PRRS ELISA package insert; IDEXX Laboratories, Westbrook, Maine). The IDEXX ELISA detected the presence of antibodies in serum samples from 14 to 105 dpi (11). Although results for other indirect ELISAs have been reported previously (4, 9, 25, 29, 33), the IDEXX ELISA has gained wide acceptance by the diagnostic community as a screening test because of the good repeatability, good reproducibility, fast turnaround time, and relatively low cost of the assay.
Even though many veterinarians are aware that the IDEXX ELISA was developed as a herd-screening tool, individual unexpected positive IDEXX ELISA results in otherwise seronegative herds have caused great concern. This has led to a perception among some veterinarians that there might be an unacceptable level of false-positive results with the assay. For example, a false-positive IDEXX ELISA result was demonstrated in a controlled experiment by Horter et al. (11). Sera from multiple collection dates and several tissue samples were available to investigate the false-positive results in that experiment, but this is not typically the case in the field. Often, several diagnostic methods are applied to resolve the question of the serostatus of the sample. Virus isolation (VI), PCR, serum virus neutralization (SVN), indirect fluorescent antibody assay (IFA), and ELISA have been used. VI and PCR indicate the presence of the virus in serum samples, and positive results from these methods may not always correlate with antibody levels. SVN, IFA, and ELISA are serological methods that have been used to detect PRRSV-specific antibodies in serum samples.
The viremic stage of the PRRSV infection is generally considered to be from 1 to 6 weeks after infection, but VI has been used to confirm the presence of infectious virus as early as 12 h postexposure (23). The success of VI from serum decreases as the number of dpi increases (5). The PRRSV is temperature sensitive (3) and may not survive shipping to a diagnostic laboratory if precautions are not taken. Also, the cell type used for VI can affect the success of virus recovery (2). Therefore, an IDEXX ELISA-positive and a VI-negative result combination is possible when testing an infected animal. This potential lack of correlation between the IDEXX ELISA and VI limits the use of VI as a follow-up test. The relatively long turnaround time of VI is also a negative aspect of the assay, since the isolation of virus from cell culture may take several days.
PCR is an extremely sensitive assay to detect the presence of the virus (6). Due to the sensitivity of the assay, false-positive results can be a concern. Infectious virus is not required for detection by PCR. The turnaround time of the assay is faster than that of VI, but the assay is often more expensive than ELISA. PCR typically will detect virus for a longer period of time than VI, but PCR would also have a potential for lack of correlation with the IDEXX ELISA. Therefore, the use of PCR as a follow-up test for confirming antibody status is also limited.
SVN is considered less sensitive than other methods because it detects the presence of neutralizing antibodies that appear at 30 to 60 dpi (3). Antigenic differences between the virus isolate used in the test and the PRRSV in the herd can cause decreased sensitivity (31, 37). Therefore, the SVN results do not correlate well with the ELISA results; this decreases the method's effectiveness as a follow-up test.
Many diagnostic laboratories provide IFA tests as a follow up to the IDEXX ELISA. Titer values higher than 1:16 or 1:20 are generally considered to represent positive test results. The IFA detects antibodies to PRRSV at 7 to 90 dpi (36). This detection window is similar to that of the IDEXX ELISA. There are inherent limitations in the IFA as well. Cell culture techniques are required that may not be available in all labs. If the PRRSV isolate used in the assay is antigenically different from the virus in the herd, false-negative results may be generated. Also, the IFA results are more subjective because the assay is read manually using a microscope with a UV light source (2, 37). The IFA subjectivity can be caused by method variation between labs, technician interpretation, nonspecific fluorescence, and increased background staining with some samples. Due to these inherent differences, decreased IFA reproducibility can be expected between laboratories.
To address the limitations of these follow-up methods, several different competitive or blocking ELISAs (bELISAs) have been developed to detect antibodies against PRRSV (8, 12, 28, 33). The intent of these published competitive or bELISA methods was to provide a more specific, repeatable, and reproducible method that can be used to investigate unexpected positive IDEXX ELISA results by confirming the presence of PRRSV antibodies in individual animals. Houben et al. used a cell culture-derived antigen with a horseradish peroxidase-conjugated polyclonal antibody (12). Sorensen et al. used a cell culture-propagated antigen with a biotinylated polyclonal antibody (28). Dea et al. used a recombinant PRRSV N protein as the antigen, an anti-N MAb as the primary antibody, and a horseradish peroxidase-conjugated anti-mouse immunoglobulin G antibody as the secondary antibody (8). These bELISAs use different antibodies and have been validated in different ways. However, none of the methods have found wide acceptance by the diagnostic community. The goal of this project was to validate a highly specific, highly repeatable bELISA designed on the basis of the use of a purified recombinant nucleocapsid antigen and the well-characterized SDOW17 MAb (22) with the validation methods described by Jacobson for the Office International des Epizooties, the world organization for animal health and testing standards (13).
MATERIALS AND METHODS
Antigen production.
The antigen used for the bELISA was a recombinant ORF 7 nucleocapsid protein of the ATCC VR-2332 isolate of PRRSV. The ORF 7 gene was inserted into the pET 24b plasmid (Novagen, Madison, Wis.) and transformed into BL21-CodonPlus (DE3)-RP-competent cells (Stratagene, La Jolla, Calif.) with the method recommended in the Stratagene package insert. The transformed cells were plated onto Luria-Bertani agar plates containing 30 μg of kanamycin/ml and 50 μg of chloramphenicol/ml. The ORF 7 protein expression was induced using isopropyl β-d-1-thiogalactopyranoside (IPTG). Single colonies were grown in cultures overnight at 37°C in 1 ml of 2× yeast extract tryptone (YT) culture medium with 30 μg of kanamycin/ml and 50 μg of chloramphenicol/ml. The 1-ml culture was added to 50 ml of 2× YT medium with 30 μg of kanamycin/ml and 50 μg of chloramphenicol/ml and grown in cultures at 37°C for 2 h. The 50-ml culture was added to 1 liter of 2× YT culture medium with 30 μg of kanamycin/ml and 50 μg of chloramphenicol/ml and grown in cultures at 37°C for 2 h. IPTG was added to achieve a final concentration of 0.4 mM and grown in cultures for an additional 4 h. The bacteria were pelleted by centrifugation at 12,500 × g for 15 min at 4°C. The pellet was resuspended in 97 ml of lysis solution containing Tris-EDTA buffer (TE buffer) (25 mM Tris, 10 mM EDTA, pH 8.0), 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 μM leupeptin, 1 μM pepstatin, 0.5% (vol/vol) Triton X-100, 0.05% (vol/vol) Tween 20, and 10 mg of lysozyme (Sigma Chemical Company, St. Louis, Mo.). The suspension was incubated overnight at 4°C and then centrifuged at 17,000 × g for 20 min at 4°C. The pellet was resuspended in 25 ml of TE buffer-1 mM PMSF-1 μM pepstatin-1 μM leupeptin, sonicated on ice at 70% output with a 50% duty cycle and with pulses in bursts of 30 s, and centrifuged at 17,000 × g for 20 min at 4°C. The supernatant was added to 12.5 ml of 5 M LiCl and 50 ml of ice-cold absolute ethanol. The suspension was centrifuged at 17,000 × g for 20 min at 4°C. The LiCl-ethanol supernatant was collected, and 350 ml of acetone was added. The suspension was centrifuged at 17,000 × g for 20 min at 4°C. The protein pellet was resuspended in 10 ml of TE buffer (pH 8.0)-1 mM PMSF-1 μM leupeptin-1 μM pepstatin and stored at −80°C.
MAb production and biotinylation.
The MAb SDOW17 recognizes a highly conserved epitope on the PRRSV N protein (22). SDOW17 mouse ascites fluid was partially purified by ammonium sulfate precipitation. A solution of 1 mg of biotin-N-hydroxysuccinimide (biotin-NHS)-100 μl of dimethyl sulfoxide was added to purified MAb for a final concentration of 15% (vol/vol). The mixture was incubated at room temperature for 4 h with gentle stirring. To neutralize the biotin-NHS, 80 μl of 1 M NH4Cl was added for every milligram of biotin-NHS. The solution was dialyzed twice using Spectra/Por dialysis tubing (MWCO 6-8000; Spectrum Laboratories, Rancho Dominguez, Calif.) in 4 liters of PBS for more than 8 h. Bovine serum albumin (BSA) (1%) was added for stability, and aliquots of the biotinylated MAb were frozen at −80°C until use.
bELISA.
The bELISA was performed using Immulon 2 HB 96-well microtiter plates (Thermo Labsystems, Franklin, Mass.). The optimal dilution of N protein antigen was experimentally determined so that the MAb generated an optical density of approximately 2 in the absence of a competitor. The recombinant, expressed N protein was diluted 1:2,000 with 15 mM sodium carbonate-35 mM sodium bicarbonate (ACB), pH 9.6. The plates were coated with 100 μl of the diluted N protein in columns 1, 3, 5, 7, 9, and 11. Columns 2, 4, 6, 8, 10, and 12 were treated with 100 μl of ACB as a background control. The plates were incubated for 1 h at 37°C and then held at 4°C overnight. The following day, 200 μl of PBS containing 2% (wt/vol) BSA was pipetted into all wells of the microtiter plate and the plates were incubated for 1 h at 37°C. The plates were washed six times using an Immuno Wash eight-channel manifold (Nalgene Nunc International, Rochester, N.Y.) and more than 300 μl of PBS containing 0.05% Tween 20 (PBS-T20). Test and control sera were diluted 1:4 with PBS-T20 containing 0.1% BSA, and 100 μl of the dilution was pipetted into an N protein-coated well and a background well. The plates were incubated for 1 h at 37°C. Following the sample incubation, 100 μl of a 1:200 dilution of biotinylated SDOW17 MAb was added to all wells. The plates were gently swirled to mix the antibody and then incubated for an additional 30 min at 37°C. The plates were washed six times using a Nunc-Immuno Wash eight-channel manifold and more than 300 μl of PBS-T20. Streptavidin-horseradish peroxidase conjugate (Zymed, South San Francisco, Calif.) was added to all wells as 100 μl of a 1:8,000 dilution, and plates were incubated for 1 h at 37°C. Plates were washed six times using a Nunc-Immuno Wash eight channel manifold and more than 300 μl of PBS-T20, and 100 μl of ABTS peroxidase substrate (KPL, Gaithersburg, Md.) was added to all wells. The plates were gently swirled and then incubated in a 25°C incubator. At 15 min, color development was stopped by the addition of 100 μl of ABTS stop solution (KPL) to all wells. The color development was quantified by reading at 405 nm with an EL800 microplate reader (BioTek Instruments Inc., Winooski, Vt.) controlled by XChek Software (IDEXX Laboratories). The raw plate data were copied to an Excel spreadsheet to calculate the percent inhibition (PI) using the following formula:
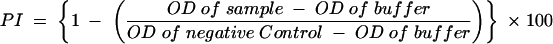 |
where OD represents optical density.
A single lot of porcine serum (which was obtained prior to the emergence of PRRSV and tested seronegative by all available assays) was used as the negative-control serum and assayed in quadruplicate on each plate. Three internal quality control serum samples (also used for the IDEXX ELISA and the IFA) were assayed on each plate. A well without any test or control serum was used as an MAb control.
IFA.
The IFA procedure has been described previously (21). Briefly, MARC-145 cells (15) were grown in cultures for 3 to 4 days to confluence on 96-well Falcon cell culture plates (BD Biosciences, San Jose, Calif.). Every other column was infected with the SD23983 isolate of PRRSV (5 × 103 50% tissue culture infective doses/ml), and the plates were incubated for an additional 18 to 24 h. The plates were then fixed with 300 μl of 80% (vol/vol) acetone/well for 15 min at room temperature and then air dried and frozen with a desiccant at −20°C until use. Serum samples to be assayed were diluted 1:20 and 1:40 with PBS, and 100 μl of each dilution was transferred to paired wells of PRRSV-infected and uninfected MARC-145 cells. The plates were incubated at 37°C for 30 min and then washed three times with at least 300 μl of PBS. A 30-μl volume of 41.7 μg of fluorescein isothiocyanate-labeled goat anti-swine immunoglobulin G (KPL)/ml was added to each well. The plates were gently tapped to allow the fluorescein isothiocyanate conjugate to cover the bottom of the well entirely. The plates were incubated at 37°C for 30 min. The plates were washed again with at least 300 μl of PBS three times and patted dry between the washes. A fourth wash of 150 μl was left on the cells. The wells were examined for specific fluorescence with an inverted microscope and a UV light source.
Method validation. (i) Analytical sensitivity.
To establish the smallest detectable unit of the assay, six 1:2 serial dilutions of a high-level positive- and a negative-internal-control serum were analyzed in triplicate by the bELISA. Analysis of variance (ANOVA) was performed using Minitab Inc. software (release 13) to evaluate differences between the dilutions of the two control sera.
(ii) Cutoff determination.
To accurately assess the diagnostic sensitivity and diagnostic specificity of the bELISA, 686 serum samples from individual animals with established PRRSV status were analyzed using the bELISA, the IDEXX ELISA, and the IFA. Since there is no universally accepted serological method to determine the PRRS status of an animal, great care was taken to establish the PRRSV serostatus of the individual animals before inclusion into the validation populations. Results of assays of individual animal serum samples from challenge studies were considered to be negative (uninfected) when collected prior to inoculation with the virus. No negative-testing (uninfected) field samples were included in the validation population due to the high-level prevalence of PRRS.
The positive-testing (infected) validation population was composed of samples from animal challenge studies and known positive-testing field samples. The animal challenge study samples were collected at 21 dpi. A collection of serum samples from a university challenge study was supplied by Jeffery Zimmerman, Department of Veterinary Diagnostic and Production Animal Medicine, Iowa State University. The animals were from PRRS-negative sites and were 3 weeks old at the start of the experiment (11). Another collection of serum samples from a challenged-animal experiment was supplied by Mike Roof, Research and Development, Boehringer Ingelheim Vetmedica, Ames, Iowa. The animals were 4 to 6 weeks of age at the start of the experiment. Field samples were included in the positive-testing validation population when the case was submitted from a farm with a history of PRRSV-positive status and the IDEXX ELISA and IFA results of the entire case were positive. Field serum samples were supplied by the Health Management Center of Boehringer Ingelheim Vetmedica. The field serum samples were from animals at various stages of production, including isolation gilts, sows, nursery pigs, and finishing pigs. Also, four different geographic regions of the United States (the upper Midwest, the lower Midwest, and the eastern and western regions) were represented. Receiver Operating Characteristic (ROC) analysis methodology assessment was performed using GRAPH ROC software (14) (Version 2.0; [http://members.tripod.com/refstat/GraphROC.htm]).
(iii) Measurement of repeatability.
The repeatability of the bELISA was assessed by running the same lot of internal quality control sera 20 times on one plate, once per plate on 10 plates in 1 run, and at least once in 10 different runs. The same lot of internal-quality-control serum has been used with the IDEXX ELISA, IFA, and bELISA at the South Dakota Animal Disease Research and Diagnostic Laboratory (SD ADRDL) since 2001. Means, standard deviations (sd), percent coefficient of variation (%CV) values, and Levey-Jennings control charts were calculated using ControlChart!Pro Plus software (version 7.12.24; ChemSW).
(iv) Detection of seroconversion.
To compare the detection of seroconversion by the bELISA to detection by the IDEXX ELISA and IFA, serum from a vaccination and virus challenge study was supplied by Mike Roof. A total of 30 animals were vaccinated with a commercially available vaccine, and then the total group was split into three groups of 10 animals that were challenged with three different isolates of PRRSV (ATCC VR-2385, field isolate 1-8-4, and field isolate 1-4-4). Three groups of unvaccinated animals (n = 9, 10, and 11) were inoculated with the three different isolates of PRRSV. One group of 11 was not vaccinated or inoculated. One group of three animals received the vaccine only. Serum samples were collected approximately weekly over a 52-day period. All samples were assayed with the bELISA, the IDEXX ELISA, and the IFA. The ANOVA statistical comparison was made using Minitab software. The agreement between the three methods was assessed using kappa analysis and MedCalc software (version 7.2.0.2).
(v) Evaluation of IDEXX ELISA unexpected positive results.
To compare the abilities of the IFA and the bELISA to resolve the PRRS status of serum samples demonstrating unexpected positive IDEXX ELISA results, 4,038 samples from sites expected to be PRRS negative were analyzed. The samples comprised 70 different cases submitted to the University of Minnesota Veterinary Diagnostic Laboratory (MVDL). Following analysis using the IDEXX ELISA, samples with unexpected positive results were split for follow-up investigation. The MVDL repeated the IDEXX ELISA and performed the IFA on the first subsample. The second subsample was sent to the SD ADRDL for PRRS bELISA. The true PRRS status of the sample was determined retrospectively on the basis of subsequent monthly serological testing of the herd according to established protocols of the producer. The sampling protocols were designed to find at least one positive sample at a 95% confidence interval in a herd with a PRRS prevalence of 10%. The amounts of monthly samples per farm differed but were approximately 30 animals per site per month. At 6 months after the conclusion of the study, all the sites remained PRRS negative.
RESULTS
Antigen production.
The expressed protein was evaluated for purity using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As shown in Fig. 1, a dilution series of the N protein preparation (lanes 1 to 4) revealed two bands. The larger 15- to 20-kDa band contained approximately 90% of the protein preparation and was the recombinant N protein. The smaller 12- to 13-kDa band was lysozyme that copurified with N protein. Western blot analysis indicated the MAb SDOW17 specifically recognized the larger 15- to 20-kDa N protein (data not shown). Lysozyme did not interfere in the ELISA and showed no ELISA reactivity to PRRSV-negative or PRRSV-positive pig sera at any dilution (data not shown).
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of PRRSV N protein preparation. Lanes 1, 2, 3, and 4 represent 10, 5, 2.5, and 1 μl of the N protein preparation. Lane 5 represents 4 μl of the concentration standard (1 mg of BSA/ml and 1 mg of lysozyme/ml). Lane 6 represents 1 μl of the concentration standard (1 mg of BSA/ml and 1 mg of lysozyme/ml). Lane 7 shows the molecular size standard. Protein concentration was densitometrically estimated at 0.52 mg/ml (with about 90% due to the presence of PRRSV N protein).
Analytical sensitivity.
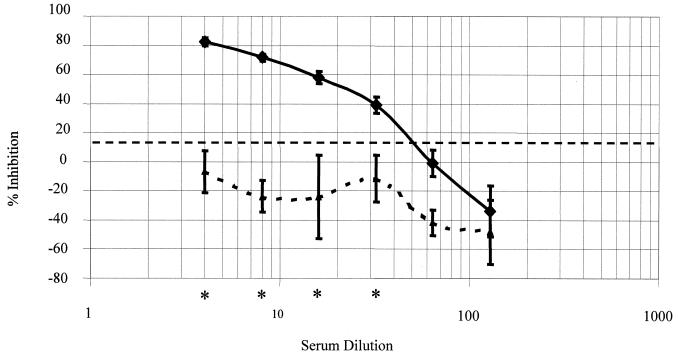
The serial dilutions of a high-level positive-internal-control serum and a negative-control serum were assayed under the optimized conditions. A 1:32 dilution of the sample was the highest dilution that was significantly different (P = 0.001) from that of the negative control (Fig. 2).
FIG. 2.
Analytical sensitivity of the bELISA. Six twofold serial dilutions of a high-level positive-internal-control serum (solid line) and a negative-internal-control serum (curved dashed line) were tested. The analytical sensitivity is the largest dilution of a high-level positive serum in which antibody is no longer detected. A 1:32 dilution of the high-level positive internal control was the limit of the analytical sensitivity. Bars, ± 2 standard deviations. The horizontal dashed line represents the optimized cutoff. Asterisks indicate dilutions at which the PI of the positive-internal-control serum was statistically different from the PI of the negative-internal-control serum.
Cutoff determination, diagnostic sensitivity, and diagnostic specificity.
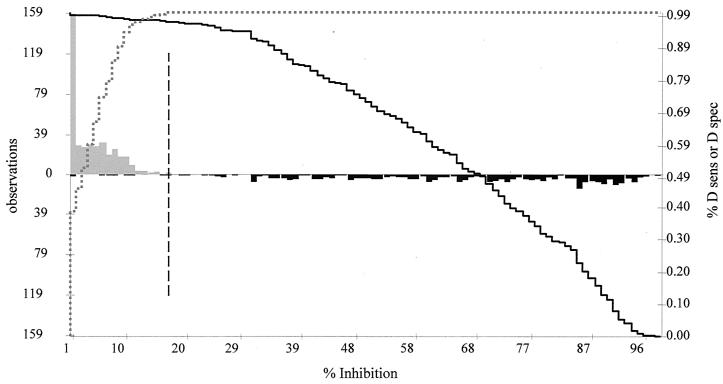
Serum samples from a positive-testing (North American PRRSV-infected) population of 316 animals and 370 serum samples from a negative-testing (PRRSV-uninfected) population were analyzed with the bELISA, the IDEXX ELISA, and the IFA at the SD ADRDL (Table 1). GRAPHROC software was used for ROC analysis of all three methods to compare histograms of the results obtained with the uninfected and infected populations to determine an optimized cutoff that maximizes both the diagnostic specificity and diagnostic sensitivity of the assays (Table 2). A two-graph ROC plot for the bELISA was generated (Fig. 3). An optimized cutoff that maximized the efficiency of the assay was calculated at 17 PI for the bELISA. A diagnostic sensitivity of 97.8% with a 95% confidence interval of 95.5 to 99.1% and a corresponding diagnostic specificity of 100% with a 95% confidence interval of 99.1 to 100% were calculated for the bELISA. Using a cutoff of greater than or equal to 1:20, the IFA displayed a diagnostic sensitivity of 99.4% with a 95% confidence interval of 97.7 to 99.9%. The diagnostic specificity of the IFA was the same as that of the bELISA (100%, with a 95% confidence interval of 99.1 to 100%). When the 0.4 sample to positive ratio (S/P) cutoff determined by IDEXX was used, the diagnostic sensitivity was 97.2 with a 95% confidence interval of 94.7 to 98.7%; the diagnostic specificity of the IDEXX ELISA was also the same as that of the bELISA (100%, with a 95% confidence interval of 99.1 to 100%). The positive and negative predictive values of the three methods vary with the expected herd prevalence. The calculated values are displayed in Table 2 for 1, 10, and 40% prevalence.
TABLE 1.
Validation data
| Groupa | No. of pigs
|
|
|---|---|---|
| Infected (n = 316)b | Healthy (n = 370)c | |
| bELISA positive | 309 | 0 |
| bELISA negative | 7 | 370 |
| IDEXX ELISA positive | 308 | 0 |
| IDEXX ELISA negative | 8 | 370 |
| IFA positive | 314 | 0 |
| IFA negative | 2 | 370 |
Diagnostic sensitivity and specificity for bELISA-positive and -negative groups, 97.8 and 100%, respectively. Diagnostic sensitivity and specificity for IDEXX ELISA-positive and -negative groups, 97.2 and 100%, respectively. Diagnostic sensitivity and specificity for IFA-positive and -negative groups, 99.4 and 100%, respectively.
Individual animals at dpi 21 of challenge study or field animals with positive results by IDEXX, with positive results by IFA, and farm with history of PRRS.
Individual pigs never exposed to PRRSV.
TABLE 2.
ROC data
| Characteristic | Value for bELISA | Value for IDEXX | Value for IFA |
|---|---|---|---|
| Optimized cutoff | 17.0 PI | 0.400 S/P | ≥1:20 |
| Diagnostic sensitivity (%) | 97.8 | 97.2 | 99.4 |
| 95% confidence interval | 95.5-99.1 | 94.7-98.7 | 97.7-99.9 |
| Diagnostic specificity (%) | 1.000 | 1.000 | 1.000 |
| 95% confidence interval | 99.1-100 | 99.1-100 | 99.1-100 |
| AUC | 0.993 | 1.000 | 0.997 |
| 95% confidence interval | 0.979-0.996 | 0.995-1.000 | 0.989-0.999 |
| Positive predictive value (%) | |||
| Estimated herd prevalence | |||
| 1% | 100 | 100 | 100 |
| 10% | 100 | 100 | 100 |
| 40% | 100 | 100 | 100 |
| Negative predictive value (%) | |||
| Estimated herd prevalence | |||
| 1% | 100 | 100 | 100 |
| 10% | 99.8 | 99.7 | 99.7 |
| 40% | 98.5 | 98.1 | 98.1 |
FIG. 3.
Two-graph ROC plot of the bELISA. The graph was calculated using the 686 individual animal serum samples and GraphROC software. The upward-pointing gray histogram on the left side of the figure represents the uninfected animals. The downward-pointing black histogram on the right represents the PRRSV-infected animals. The solid black line represents the diagnostic sensitivity (D sens) of the assay as the cutoff is moved from 1 to 100 PI. The dotted gray line represents the changes in the diagnostic specificity (D spec) values of the assay as the cutoff is moved from 1 to 100 PI. The black dashed vertical line represents the optimized cutoff value of 17 (the PI that corresponds to the maximum diagnostic sensitivity and diagnostic specificity).
All three methods were able to detect all of the uninfected pigs in the validation population as giving negative results. No false-positive results were observed.
Each of the three methods demonstrated a slight decrease in sensitivity. False-negative results from all the methods were observed in the positive-testing validation population. Seven (2.2%) of the animals from the infected population had negative bELISA results. The average PI of these seven samples was 3.54. Four of the seven tested positive by both IFA and IDEXX ELISA. Three of the seven had positive IFA results but IDEXX ELISA values that were less than 0.400 (0.263, 0.327, and 0.341). Eight (2.5%) infected animals had negative IDEXX ELISA results when analyzed at the SD ADRDL. The S/P of these eight samples ranged from 0.260 to 0.352, with an average of 0.308. All of the eight had corresponding positive IFA results, and each was the only sample in its respective case with an S/P value of less than 0.400. Two (0.6%) infected animals had negative IFA results. Both of these samples had corresponding positive bELISA results (22.80 and 49.50 PI) and IDEXX ELISA results (0.901 and 0.552). These two samples were the only samples with negative IFA results in their respective cases as well.
A single-graph ROC plot (data not shown) is a representation of test performance comparing true-positive results (diagnostic sensitivity) to false-positive results (1 − diagnostic specificity). The plots are used to assess the overall accuracy of the assay by calculating the area under the curve (AUC). An AUC of more than 0.800 is considered highly accurate. The AUCs of the bELISA, IDEXX ELISA, and IFA were calculated to be 0.993, 1.00, and 0.997, respectively (Table 2).
Repeatability of the bELISA.
The levels of precision of the IDEXX ELISA and the bELISA were compared using an SD ADRDL internal-control serum. The %CV was calculated using the protocol described earlier. The IDEXX ELISA within-plate %CV was 6.5, the %CV between plates in one run was 9.92, and the %CV between runs was 18.1. The bELISA within-plate %CV was 3.2, the %CV between plates in one run was 3.96, and the %CV between runs was 4.24 (Table 3).
TABLE 3.
Assay repeatability
| Assay | Repeatability result (% CV)a
|
||
|---|---|---|---|
| Within plate | Within run | Between runs | |
| IDEXX ELISA | 6.25 | 9.92 | 18.1 |
| bELISA | 3.22 | 3.96 | 4.24 |
Values listed are %CV of high-level positive-internal-control serum. Within-plate precision was calculated from 20 replicates on one plate. Within-run precision was calculated using one serum on 10 plates in one run. Between-run precision was calculated from at least one serum in 10 different runs.
Detection of seroconversion.
A vaccination-inoculation challenge study was used to compare the detection of seroconversion by the bELISA, IDEXX ELISA, and IFA (Table 4). A total of 30 animals were vaccinated with a commercially available PRRS vaccine (Ingelvac ATP; Boehringer Ingelheim Vetmedica). At 14 days later, 29 of the 30 vaccinated animals had seroconverted, as revealed by all three methods. The 30th animal was positive by IFA and negative by bELISA and IDEXX ELISA but had seroconverted by all methods by 28 dpi.
TABLE 4.
Seroconversion detection
| Method | No. of seropositive animals detected by indicated method on indicated dpi/total no. of animalsa
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccinated animals
|
Animals inoculated with indicated PRRSV isolate
|
|||||||||||||||
| 10
|
13
|
17
|
24
|
|||||||||||||
| 7 | 14 | 21 | 28 | 1-4-4 | VR-2385 | 1-8-4 | 1-4-4 | VR-2385 | 1-8-4 | 1-4-4 | VR-2385 | 1-8-4 | 1-4-4 | VR-2385 | 1-8-4 | |
| bELISA | 0/30 | 29/30 | 29/30 | 30/30 | 0/10 | 0/9 | 0/11 | 0/10 | 0/9 | 0/11 | 10/10 | 8/9 | 10/11 | 9/10 | 9/9 | 11/11 |
| IDEXX | 0/30 | 29/30 | 28/30 | 29/30 | 0/10 | 0/9 | 0/11 | 0/10 | 1/9 | 0/11 | 9/10 | 3/9 | 7/11 | 10/10 | 9/9 | 11/11 |
| IFA | 0/30 | 30/30 | 30/30 | 30/30 | 0/10 | 0/9 | 0/11 | 0/10 | 0/9 | 0/11 | 10/10 | 3/9 | 9/11 | 10/10 | 9/9 | 11/11 |
Results of seroconversion detection by bELISA, IDEXX ELISA, and IFA were compared using samples from a vaccination-inoculation study. Overall, the methods agreed well. The kappa value for the comparison of bELISA to IDEXX was 0.94. The kappa value for the comparison of IDEXX to IFA was 0.95. The kappa value for the comparison of IFA to bELISA was 0.96. There were differences at certain dpi (depending on the virus isolate examined).
When seroconversion after infection with field isolates was detected, differences were observed among the methods. Three groups of nonvaccinated animals (n = 9, 10, and 11) were inoculated with three different field isolates of PRRSV. At 13 dpi, all samples tested negative by all methods (with the exception of one weak positive IDEXX ELISA result when ATCC VR-2385 PRRSV was the inoculum). At 17 dpi, greater differences were associated with the isolate of virus used in the inoculum. When PRRSV field isolate 1-4-4 was used as the inoculum, the bELISA and the IFA detected 10 positive results with 10 animals whereas the IDEXX ELISA detected 9 positive results with 10 animals. When PRRSV field isolate 1-8-4 was used as the inoculum, the bELISA detected 10 positive results with 11 animals, the IFA detected 9 positive results with 11 animals, and the IDEXX ELISA detected 7 positive results with 11 animals. When the ATCC VR-2385 PRRSV isolate was used as the inoculum, the bELISA detected eight positive results with nine animals, the IFA detected three positive results with nine animals, and the IDEXX ELISA detected three positive results with nine animals. By 24 dpi, all three methods detected seroconversion in all the samples regardless of the isolate of virus used for inoculation (Table 4).
Evaluation of IDEXX ELISA unexpected positive results.
Of the total of 4,038 serum samples, 3,471 samples from 64 cases were analyzed using the older antigen format of the IDEXX ELISA at the MVDL. The older antigen format was a cell culture-derived North American antigen with a recombinant European antigen and was available prior to May of 2002. Unexpected positive results were obtained from 104 serum samples (3.0% of the total). The data could be evaluated as three resultant groups. Group 1 consisted of 15 of the 104 samples that had only one positive IDEXX ELISA result and negative results by both the IFA and the bELISA. The average S/P value for the first IDEXX ELISA results for these 15 samples was 0.519. The average S/P value for the repeated IDEXX ELISA was 0.208. Group 2 consisted of 85 samples that had repeatable positive IDEXX ELISA results, negative IFA results, and negative bELISA results. The average IDEXX ELISA S/P of group 2 was 0.858. Group 3 consisted of four samples that had repeatable positive IDEXX ELISA results and negative IFA results but were positive by the bELISA. The average IDEXX ELISA S/P of group 3 was 1.086.
The MVDL used the new 2XR antigen format of the IDEXX ELISA that became available in May of 2002 to analyze 567 serum samples from six cases. Both of the PRRSV antigens are recombinant proteins in the 2XR format. Unexpected positive results were obtained from 29 serum samples (5.1% of the samples). The data from the 2XR assay could be divided into two additional resultant groups, because none of the 29 samples had positive bELISA results. Group 4 consisted of 8 of the 29 samples that had only one positive IDEXX ELISA result and negative results by both the IFA and the bELISA. The average S/P value for the first IDEXX ELISA results of these 8 samples was 0.538. The average S/P value for the repeated IDEXX ELISA was 0.293. Group 5 consisted of 21 samples that had repeatable positive IDEXX ELISA results, negative IFA results, and negative bELISA results. The average IDEXX ELISA S/P value for group 5 was 0.893. The combined results of the two kit formats are displayed in Table 5.
TABLE 5.
Unexpected IDEXX positive result follow-up testing
| Format and group | Resulta | n | IDEXX
|
IFA
|
bELISA
|
||
|---|---|---|---|---|---|---|---|
| Avg (S/P) | SD | Titer | Avg (S/P) | SD | |||
| Old IDEXX | |||||||
| 1 | 1 IDEXX Pos, IFA Neg, bELISA Neg | 15 | 0.519 | 0.152 | <1:16 | 0.97 | 5.69 |
| 2 | >1 IDEXX Pos, IFA Neg, bELISA Neg | 85 | 0.858 | 0.594 | <1:16 | −0.17 | 5.67 |
| 3 | >1 IDEXX Pos, IFA Neg, bELISA Pos | 4 | 1.086 | 0.479 | <1:16 | 32.64 | 11.19 |
| IDEXX 2XR | |||||||
| 4 | 1 IDEXX Pos, IFA Neg, bELISA Neg | 8 | 0.538 | 0.131 | <1:16 | −35.4 | 18.5 |
| 5 | >1 IDEXX Pos, IFA Neg, bELISA Neg | 21 | 0.893 | 0.358 | <1:16 | −39.89 | 19.5 |
Pos, positive; Neg, negative.
ANOVA results indicated that the IDEXX S/P values of groups 1 and 4 were not significantly different from each other. The values for groups 2, 3, and 5 were not significantly different from each other but were different from those of groups 1 and 4 (P < 0.001). A dot plot (Fig. 4) demonstrates the relationships among the five groups.
FIG. 4.
Dot plot of S/P values of unexpected positive IDEXX ELISA results in herds expected to give negative results. The IDEXX ELISA, IFA, and bELISA were used to analyze 4,038 serum samples from herds expected to give negative results. Groups 1 and 4 had only one IDEXX ELISA-positive result (with negative IFA and bELISA results). Groups 2 and 5 had repeatable IDEXX ELISA-positive results (with negative IFA and bELISA results). Group 3 had repeatable IDEXX ELISA-positive results and IFA-negative results but bELISA-positive results. ANOVA was performed using MedCalc. The results seen with groups with superscripts sharing the same letter were not significantly different from each other (P < 0.001).
A comparison of the percentages of the unexpected positive results of the two antigen formats was made. The old IDEXX ELISA antigen format yielded 104 of 3,471 (3.0%) unexpected positive results. The new 2XR antigen format yielded 29 of 567 (5.1%) unexpected positive results. A chi-square test for the comparison of the two proportions calculated a P value of 0.0135. The data indicate that a significantly higher percentage of unexpected positive results was observed with the 2XR antigen format.
DISCUSSION
Monitoring the serostatus of PRRSV-negative or low-prevalence herds is an issue of great importance to the swine industry. When the IDEXX ELISA is used as a screening tool, unexpected positive results from samples in negative-testing or low-prevalence herds may require additional testing to resolve the issue. Several methods (including VI, PCR, SVN, and IFA) are used to clarify the PRRSV status of individual animals. The inherent limitations that make each of these common methods less desirable as a follow-up test have been presented. Several other competitive or bELISA methods have been published previously to address these limitations (8, 12, 28). Each bELISA has slightly different assay methods and different validation designs. Houben et al. (12) and Sorensen et al. (28) both used cell culture-propagated antigens with polyclonal antibodies. This may have led to a lower-than-desired level of specificity. ROC analysis was not done by either group. Dea et al. (8) used a recombinant protein paired with a MAb. They reported a sensitivity of 80% and a specificity of 98% when the results were compared to those obtained with the IDEXX ELISA and the IFA. Kappa values resulting from comparisons of their bELISA to the IDEXX ELISA and IFA were 0.76 and 0.803. ROC analysis was not done. Dea's estimates of the sensitivity and specificity were more accurate than those of previous researchers but still not as accurate as desired. A more thoroughly validated bELISA with accurate and high-level estimates of sensitivity and specificity was still needed.
Jacobson described the validation of an ELISA as a five-stage ongoing process: selection of the antigen, antibody, and control sera; optimization of the assay conditions; determination of the sensitivity and specificity; assessment of predictive value; and monitoring of the repeatability of the assay (13). The Office International des Epizooties supports this method to validate ELISAs. This study uses the method described by Jacobson to validate the use of a bELISA designed on the basis of the use of a recombinant nucleocapsid antigen paired with biotinylated SDOW17 MAb for the detection of antibodies to PRRSV.
Stage 1 of the validation process is selection of the ELISA antigen, method, and control sera. ELISA antigens have been previously prepared from a cell culture preparation of the whole virus (12). The nucleocapsid protein was chosen as the antigen for this assay, because a high proportion of the antibody response in an early infection is directed against it (19, 28). To increase the specificity of the ELISA, a purified recombinant antigen preparation of the nucleocapsid protein was used. Use of a purified antigen is expected to significantly decrease false-positive reactions to other proteins in the antigen preparation (as may occur in cell culture-prepared antigens).
The choice of the method of the ELISA can affect the overall specificity, and each step of that method contributes to the performance of the assay. A competitive or blocking method was chosen, because that type of ELISA is inherently more specific than an indirect ELISA (24). A biotinylated competing MAb and a streptavidin conjugate were intended to maximize the sensitivity of the ELISA at subsequent steps.
The internal control sera used with the bELISA are also used with the IDEXX ELISA and the IFA at the SD ADRDL for routine diagnostic testing. The sera were collected from individual animals and have been well characterized over a period of several years. This facilitates comparison of results among the three methods.
Stage 2 is the optimization of all the assay conditions. This was accomplished by checkerboard titration of the nucleocapsid antigen, MAb, streptavidin conjugate, and other reagents. Once the assay conditions were optimized, the analytical sensitivity was determined to be a 1:32 dilution of the high-level positive-internal-control serum. This information indicates that a 1:4 dilution of the sample and control serum ensures the maximum signal difference between positive and negative results without increasing the background.
The analytical specificity of the bELISA is determined on the basis of the MAb SDOW17. SDOW17 is directed against a highly conserved epitope on the nucleocapsid protein (34). SDOW17 binds to essentially all isolates of the PRRSV except for isolates of a vaccine no longer on the market (16, 17, 22).
Stage 3 is used to determine the accuracy parameters of the assay. Jacobson stated that 700 uninfected and 300 infected animals should be used to validate the method but that lesser numbers could be used as available (13). ROC analysis was performed using serum samples from 370 known uninfected animals and 316 known North American PRRSV-infected animals analyzed by the bELISA, IDEXX ELISA, and IFA to determine the diagnostic sensitivity and diagnostic specificity of the methods (10). In this study, a cutoff of 17 PI for the bELISA gave the optimum diagnostic sensitivity of 97.8% (95.5 to 99.1%; 95% confidence interval) and diagnostic specificity of 100% (99.1 to 100%; 95% confidence interval). The two-graph ROC plot of the bELISA displays the histograms of the uninfected and PRRSV-infected populations and demonstrates minimal overlap of the two populations (Fig. 3). None of the animals from the uninfected population had PI values of more than 17. The overlap between the two populations can be attributed to seven samples from the PRRSV infected population that had PI values of less than 17. Closer examination of the seven samples revealed that all were field samples from farms with a history of PRRSV infections. The PI values of the seven samples ranged from −4.72 to 16.09, with an average PI of 7.0. The average IDEXX ELISA value of the seven samples was 0.627, with three of the samples having S/P values of less than 0.400 at the SD ADRDL. The geometric mean of the IFA data for the seven samples was 44.2. The farm history, initial Boehringer Ingelheim Vetmedica IDEXX data, and IFA data indicate that the serum samples were indeed positive for antibodies to PRRSV. This decrease in sensitivity of the bELISA indicates that the antibody responses of those animals were not able to compete effectively against the SDOW17 MAb for binding to the N protein. Variations in the individual immune responses, the number of dpi that the sample was collected, and sample quality and antigenic differences of PRRSV infecting the herd are all possible explanations.
With the IDEXX ELISA cutoff of 0.400 S/P, the diagnostic sensitivity was 97.2% (94.7 to 98.7%; 95% confidence interval) and diagnostic specificity was 100% (99.1 to 100%; 95% confidence interval). None of the animals from the uninfected population had S/P values of more than 0.200. Eight field samples of the positive population had S/P values of less 0.400 and therefore had negative results at the SD ADRDL. The S/P values ranged from 0.260 to 0.352. All of the eight had positive corresponding IFA results and were the only samples in their respective cases with an S/P value of less than 0.400. Only three of the eight samples also had negative results by the bELISA. The average bELISA value of the eight samples was 31.9 PI, and the geometric mean of the IFA data was 40.0. These samples were from a herd with a history of PRRSV infection and had positive IDEXX ELISA values at Boehringer Ingelheim Vetmedica. The decrease in sensitivity with respect to these samples could be attributed to IDEXX ELISA serial lot differences, ELISA variation around the cutoff, or sample degradation. Interestingly, none of the infected population had S/P values of less than 0.260. This observation may support the practice by some veterinarians of using follow-up testing for any sample from a herd expected to give negative results that has an S/P value of more than 0.200. ROC analysis demonstrated that an optimized cutoff of 0.200 by the IDEXX ELISA would have a diagnostic sensitivity of 100% while still maintaining the diagnostic specificity at 100%.
The IFA had a diagnostic sensitivity of 99.4% (97.7 to 99.9%; 95% confidence interval) and diagnostic specificity of 100% (99.1 to 100%; 95% confidence interval). Two field samples from the infected population yielded negative IFA results. The average bELISA value was 36.13 PI, and the average IDEXX ELISA S/P value was 0.727. This observation could also be attributed to sample quality or assay variations.
A single-curve ROC plot (data not shown) compares the true positive results (sensitivity) to the false-positive results (1 − specificity) and is a representation of the overall accuracy of the assay. A very accurate assay will have an AUC of more than 0.90. The AUCs of the bELISA, IDEXX ELISA, and IFA were 0.993 (0.979 to 0.996; 95% confidence interval), 1.000 (0.995 to 1.000; 95% confidence interval), and 0.997 (0.989 to 0.999; 95% confidence interval), respectively. There was also a high level of agreement between the bELISA and the IDEXX ELISA, between the IDEXX ELISA and the IFA, and between the IFA and the bELISA, as demonstrated by kappa values of 0.973, 0.974, and 0.971, respectively. The ROC data for the IDEXX ELISA and the IFA may be skewed higher than could be observed in the field, since positive IDEXX ELISA and IFA results were required for inclusion into the infected population. The comparisons are relevant as a method of evaluating the bELISA in strictly defined populations.
Stage 4 is the assessment of the clinical accuracy by calculation of the positive and negative predictive values. When GraphROC software was used, the clinical accuracy at 10% prevalence of PRRSV in a herd was 1.00 for the positive predictive value and 0.997 for the negative predictive value. These values indicate that a high degree on confidence can be placed in the bELISA results.
Stage 5 is the assessment of repeatability. The other methods used for follow-up testing have inherent qualities that can affect repeatability and reproducibility. The initial repeatability of the bELISA was evaluated using SD ADRDL internal-quality-control sera. The within-plate, within-run, and between-run %CV values of the assay were 3.22, 3.96, and 4.24, respectively. These %CV values are very small compared to those of other ELISAs performed at the SD ADRDL and indicate that the bELISA is very repeatable.
The main anticipated use of this assay is as a follow-up test to the IDEXX ELISA when investigations of unexpected positive results in herds expected to give negative results are conducted. The results of detection of seroconversion by the different methods must have a high level of agreement. The detection of seroconversion by the bELISA was compared to the results seen with the IDEXX ELISA and the IFA by assaying samples from a controlled vaccination and inoculation study. A total of 30 animals were vaccinated with a commercially available vaccine. All three methods detected seroconversion by vaccination by 14 dpi equally well. When animals were inoculated with different isolates of PRRSV, slight differences in the serodetection abilities of the methods at 17 dpi were observed.
Overall, the agreement between the methods was excellent. The entire serodetection experiment contained 590 samples taken from 7 to 52 dpi. The kappa value obtained in the comparison of bELISA to IDEXX was 0.94. The kappa value obtained in the comparison of IDEXX to IFA was 0.95. The kappa value obtained in the comparison of IFA to bELISA was 0.96. The data indicate that the bELISA has a window of detection very similar to those of the IDEXX ELISA and the IFA. No false-positive results were demonstrated with the uninfected population in the assay validation. Since most of the uninfected population was composed of animals from university challenge studies, those animals may have been healthier or had less previous exposure to other pathogens than the general swine population. To compare the follow-up methods with respect to the ability to correctly identify the PRRS status of unexpected positive IDEXX ELISA results, 4,038 serum samples were collected from sites expected to give negative results. The negative-testing status of the site was determined retrospectively. At 6 months after the conclusion of the study, all sites remained PRRS negative. The percentages of unexpected positive results were 3.0% for the older IDEXX antigen format and 5.1% for the newer 2XR format, indicating a significantly larger percentage of unexpected positive results obtained using the 2XR antigen format. An increase in the percentage of unexpected positive results from the IDEXX ELISA would emphasize the need for more follow-up testing alternatives.
When the S/P values for the unexpected positive results in group 1 were examined closely, the single IDEXX-positive S/P values (average, 0.519) were not significantly different than the S/P values of the repeated samples (average, 0.208). This may indicate a variation of the assay about the cutoff point of 0.400 S/P. Although the differences between the first and repeated IDEXX S/P values of group 1 are not statistically significant, they are diagnostically significant (since the variation includes the assay cutoff value). Efforts to maximize the repeatability of the IDEXX ELISA could reduce the occurrence of this type of unexpected positive result. The serum samples from groups 2 and 3 with repeatable positive IDEXX ELISA results (averages, 0.858 and 1.086) were significantly different than the group 1 samples but not significantly different from each other. This would indicate that when an unexpected IDEXX value is repeatable, it is significantly different from a negative result and that further investigation is warranted. Also, the data demonstrate that truly negative-testing samples can have very high and repeatable S/P values. The S/P values of the groups were not significantly different between the two antigen formats.
Of all the 133 total unexpected positive results, the IFA was able to detect all of the sera as negative testing whereas the bELISA returned 129 negative results (a 97% agreement between the IFA and the bELISA). The four samples with bELISA-positive results had lower PI values (average PI = 32.64) and were from four different farms. Three of the four samples were also positive for antibodies to Mycoplasma hyopneumoniae. Thacker et al. showed a potentiation of PRRSV infections by M. hyopneumoniae (30). Other researchers have demonstrated that mycoplasmas can stimulate B cells nonspecifically (20, 26). When the M. hyopneumoniae antibody data available from other diagnostic cases were reviewed in the study, however, no correlation between the percentages of IDEXX ELISA unexpected positive results and M. hyopneumoniae antibody levels could be confirmed (data not shown). The process responsible for the IDEXX ELISA and bELISA false-positive reactions remains unknown.
In another interesting case, four samples initially in the data set had repeatable positive results by the IDEXX ELISA and three of the four had bELISA-positive results. However, all were IFA negative. Further evaluation revealed that the animals had previously been PRRSV seropositive; therefore, the IFA results were considered to be false-negative results. The antibody levels appeared to have declined below the level detectable by the IFA. This case demonstrates that the IFA can be less sensitive than other available methods for some types of samples.
In conclusion, the IDEXX HerdChek ELISA is a well-accepted assay, but unexpected positive results in herds expected to give negative results have been regularly documented, which makes determination of the serostatus of individual animals and herds unclear. Several other follow-up tests are used, but the inherent limitations of these methods have not allowed resolution of the issue. The bELISA (designed on the basis of the use of a recombinant expressed nucleocapsid protein with the biotinylated SDOW17 MAb) addresses these limitations. The data demonstrate that the bELISA is a highly repeatable assay that shows strong agreement with the IDEXX ELISA and the IFA with respect to detection of seroconversion. The bELISA also demonstrates high-level agreement with the IFA when resolving unexpected IDEXX ELISA positive results. The diagnostic sensitivity, diagnostic specificity, and accuracy of the bELISA make it a useful follow-up test when unexpected positive results in a herd expected to give negative results are investigated.
Acknowledgments
Funding and resources in support of this project were provided by the National Pork Board, PIC USA, Boehringer Ingelheim Vetmedica, the Department of Pathobiology, University of Minnesota, the South Dakota Agricultural Experiment Station, and the South Dakota Animal Disease Research and Diagnostic Laboratory.
We thank Jeffery Zimmerman, Mike Roof, Dennis Horter, and Wayne Chittick for providing and coordinating serum samples and technical assistance. We thank Christopher Chase, Jane Christopher-Hennings, and David Francis for valuable suggestions with the review of the manuscript. The staff of the South Dakota Animal Disease Research and Diagnostic Laboratory provided invaluable assistance and support.
Footnotes
This is South Dakota Experiment Station paper 3394.
REFERENCES
- 1.Albina, E., Y. Leforban, T. Baron, J. P. Plana Duran, and P. Vannier. 1992. An enzyme linked immunosorbent assay (ELISA) for the detection of antibodies to the porcine reproductive and respiratory syndrome (PRRS) virus. Ann. Rech. Vet. 23:167-176. [PubMed] [Google Scholar]
- 2.Bautista, E. M., S. M. Goyal, I. J. Yoon, H. S. Joo, and J. E. Collins. 1993. Comparison of porcine alveolar macrophages and CL 2621 for the detection of porcine reproductive and respiratory syndrome (PRRS) virus and anti-PRRS antibody. J. Vet. Diagn. Investig. 5:163-165. [DOI] [PubMed] [Google Scholar]
- 3.Benfield, D. A., E. A. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robison, W. T. Christianson, R. B. Morrison, D. Gorcyca, and D. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 4:127-133. [DOI] [PubMed] [Google Scholar]
- 4.Cho, H. J., D. Deregt, and H. S. Joo. 1996. An ELISA for porcine reproductive and respiratory syndrome: production of antigen of high quality. Can. J. Vet. Res. 60:89-93. [PMC free article] [PubMed] [Google Scholar]
- 5.Christopher-Hennings, J., L. D. Holler, D. A. Benfield, and E. A. Nelson. 2001. Detection and duration of porcine reproductive and respiratory syndrome virus in semen, serum, peripheral blood mononuclear cells, and tissues from Yorkshire, Hampshire, and Landrace boars. J. Vet. Diagn. Investig. 13:133-142. [DOI] [PubMed] [Google Scholar]
- 6.Christopher-Hennings, J., E. A. Nelson, J. K. Nelson, R. J. Hines, S. L. Swenson, H. T. Hill, J. J. Zimmerman, J. B. Katz, M. J. Yaeger, and C. C. Chase. 1995. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J. Clin. Microbiol. 33:1730-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, J. E., D. A. Benfield, W. T. Christianson, L. Harris, J. C. Hennings, D. P. Shaw, S. M. Goyal, S. McCullough, R. B. Morrison, H. S. Joo, et al. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 8.Dea, S., L. Wilson, D. Therrien, and E. Cornaglia. 2000. Competitive ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus using recombinant E. coli-expressed nucleocapsid protein as antigen. J. Virol. Methods 87:109-122. [DOI] [PubMed] [Google Scholar]
- 9.Denac, H., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65:169-181. [DOI] [PubMed] [Google Scholar]
- 10.Greiner, M., D. Pfeiffer, and R. D. Smith. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 45:23-41. [DOI] [PubMed] [Google Scholar]
- 11.Horter, D. C., R. M. Pogranichniy, C. C. Chang, R. B. Evans, K. J. Yoon, and J. J. Zimmerman. 2002. Characterization of the carrier state in porcine reproductive and respiratory syndrome virus infection. Vet. Microbiol. 86:213-228. [DOI] [PubMed] [Google Scholar]
- 12.Houben, S., P. Callebaut, and M. B. Pensaert. 1995. Comparative study of a blocking enzyme-linked immunosorbent assay and the immunoperoxidase monolayer assay for the detection of antibodies to the porcine reproductive and respiratory syndrome virus in pigs. J. Virol. Methods 51:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson, R. H. 1998. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. Off. Int. Epizoot. 17:469-526. [DOI] [PubMed] [Google Scholar]
- 14.Kairisto, V., and A. Poola. 1995. Software for illustrative presentation of basic clinical characteristics of laboratory tests—GraphROC for Windows. Scand. J. Clin. Lab. Investig. Suppl. 222:43-60. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 16.Magar, R., R. Larochelle, S. Dea, C. A. Gagnon, E. A. Nelson, J. Christopher-Hennings, and D. A. Benfield. 1995. Antigenic comparison of Canadian and US isolates of porcine reproductive and respiratory syndrome virus using monoclonal antibodies to the nucleocapsid protein. Can. J. Vet. Res. 59:232-234. [PMC free article] [PubMed] [Google Scholar]
- 17.Magar, R., R. Larochelle, E. A. Nelson, and C. Charreyre. 1997. Differential reactivity of a monoclonal antibody directed to the membrane protein of porcine reproductive and respiratory syndrome virus. Can. J. Vet. Res. 61:69-71. [PMC free article] [PubMed] [Google Scholar]
- 18.Meulenberg, J. J., A. Petersen-den Besten, E. P. De Kluyver, R. J. Moormann, W. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murtaugh, M. P., Z. Xiao, and F. Zuckermann. 2002. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 15:533-547. [DOI] [PubMed] [Google Scholar]
- 20.Naot, Y. 1982. In vitro studies on the mitogenic activity of mycoplasmal species toward lymphocytes. Rev Infect. Dis. 4(Suppl.):S205-S209. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, E. A., J. Christopher-Hennings, and D. A. Benfield. 1994. Serum immune responses to the proteins of porcine reproductive and respiratory syndrome (PRRS) virus. J. Vet. Diagn. Investig. 6:410-415. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossow, K. D., J. E. Collins, S. M. Goyal, E. A. Nelson, J. Christopher-Hennings, and D. A. Benfield. 1995. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet. Pathol. 32:361-373. [DOI] [PubMed] [Google Scholar]
- 24.Schrijver, R. S., and J. A. Kramps. 1998. Critical factors affecting the diagnostic reliability of enzyme-linked immunosorbent assay formats. Rev. Sci. Tech. Off. Int. Epizoot. 17:550-561. [DOI] [PubMed] [Google Scholar]
- 25.Seuberlich, T., J. D. Tratschin, B. Thur, and M. A. Hofmann. 2002. Nucleocapsid protein-based enzyme-linked immunosorbent assay for detection and differentiation of antibodies against European and North American porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 9:1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simecka, J. W., S. E. Ross, G. H. Cassell, and J. K. Davis. 1993. Interactions of mycoplasmas with B cells: antibody production and nonspecific effects. Clin. Infect. Dis. 17(Suppl. 1):S176-S182. [DOI] [PubMed] [Google Scholar]
- 27.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79(Pt. 5):961-979. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen, K. J., A. Botner, E. S. Madsen, B. Strandbygaard, and J. Nielsen. 1997. Evaluation of a blocking ELISA for screening of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 56:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Takikawa, N., S. Kobayashi, S. Ide, Y. Yamane, Y. Tanaka, and H. Yamagishi. 1996. Detection of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus in swine sera by enzyme-linked immunosorbent assay. J. Vet. Med. Sci. 58:355-357. [DOI] [PubMed] [Google Scholar]
- 30.Thacker, E. L., P. G. Halbur, R. F. Ross, R. Thanawongnuwech, and B. J. Thacker. 1999. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J. Clin. Microbiol. 37:620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wensvoort, G., E. P. de Kluyver, E. A. Luijtze, A. den Besten, L. Harris, J. E. Collins, W. T. Christianson, and D. Chladek. 1992. Antigenic comparison of Lelystad virus and swine infertility and respiratory syndrome (SIRS) virus. J. Vet. Diagn. Investig. 4:134-138. [DOI] [PubMed] [Google Scholar]
- 32.Wensvoort, G., C. Terpstra, J. M. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, F. Wagenaar, and a. l. et. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 33.Witte, S. B., C. Chard-Bergstrom, T. A. Loughin, and S. Kapil. 2000. Development of a recombinant nucleoprotein-based enzyme-linked immunosorbent assay for quantification of antibodies against porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 7:700-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wootton, S. K., E. A. Nelson, and D. Yoo. 1998. Antigenic structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 5:773-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]
- 36.Yoon, I. J., H. S. Joo, W. T. Christianson, H. S. Kim, J. E. Collins, R. B. Morrison, and G. D. Dial. 1992. An indirect fluorescent antibody test for the detection of antibody to swine infertility and respiratory syndrome virus in swine sera. J. Vet. Diagn. Investig. 4:144-147. [DOI] [PubMed] [Google Scholar]
- 37.Yoon, K. J., J. J. Zimmerman, M. J. McGinley, J. Landgraf, M. L. Frey, H. T. Hill, and K. B. Platt. 1995. Failure to consider the antigenic diversity of porcine reproductive and respiratory syndrome (PRRS) virus isolates may lead to misdiagnosis. J. Vet. Diagn. Investig. 7:386-387. [DOI] [PubMed] [Google Scholar]