Abstract
Mechanistic insights regarding the activity enhancement of dehydrogenase by metal ion substitution were investigated by a simple method using a kinetic and thermodynamic analysis. By profiling the binding energy of both the substrate and product, the metal ion's role in catalysis enhancement was revealed. Glycerol dehydrogenase (GDH) from Klebsiella pneumoniae sp., which demonstrated an improvement in activity by the substitution of a zinc ion with a manganese ion, was used as a model for the mechanistic study of metal ion substitution. A kinetic model based on an ordered Bi-Bi mechanism was proposed considering the noncompetitive product inhibition of dihydroxyacetone (DHA) and the competitive product inhibition of NADH. By obtaining preliminary kinetic parameters of substrate and product inhibition, the number of estimated parameters was reduced from 10 to 4 for a nonlinear regression-based kinetic parameter estimation. The simulated values of time-concentration curves fit the experimental values well, with an average relative error of 11.5% and 12.7% for Mn-GDH and GDH, respectively. A comparison of the binding energy of enzyme ternary complex for Mn-GDH and GDH derived from kinetic parameters indicated that metal ion substitution accelerated the release of dioxyacetone. The metal ion's role in catalysis enhancement was explicated.
Introduction
Various metalloenzymes act in fundamental biological processes found in nature. The metal ion of most metalloenzymes participates in the catalytic process involved in the function of the polarization of chemical bonds, nucleophile activation, and substrate or product coordination [1]. Substitution of metal ions in metalloenzymes is a mild and effective modification that is used in structure-function relationship studies [2]. Metal ion substitutions have been reported to change the catalytic activity [3], substrate specificity [4], and stability [5], [6] of metalloenzymes and have stimulated much research interest. The mechanism of metal ion substitution has been mainly studied by molecular simulation and the analysis of substituted enzyme crystals. Sparta [7] studied the metal ion substitution effects of catechol-O-Methyltransferase activity by a quantum mechanical/molecular mechanical dynamics method. Metal ion substitution affected the rate-limiting step, which was explicated as the methyl transfer that occurred with a significant increase in the activation barrier. D'Antonio investigated the structure of cobalt-reconstituted human arginase I, revealing the change of the catalytic mechanism upon metal ion substitution [8]. However, the molecular simulation of the enzyme structure and the generation of metal ion substituted enzyme crystals are lengthy and expensive processes.
Compared with dynamic simulation and structure analysis, thermodynamic and kinetic analysis is a simple, quick and valid strategy aimed at explaining the activity and functional properties of biocatalysis systems [9]. The microenvironment of the active site of the metalloenzyme involved in the reaction mechanism undergoes dynamic and structural changes upon metal ion substitution [10]. Free energy binding profiles of substrates and products with modified enzymes and enzyme-substrate complexes, which demonstrate the catalytic requirement for transition state stabilization and ground state stabilization, offer an alternative method for studying the influence of metal ion substitution on catalytic reactions [11]. A kinetic and thermodynamic analysis of each binding step of the substrate and product can be used for profiling the significant changes caused by metal ion substitution and can provide valuable information for further study.
Glycerol dehydrogenases play crucial roles enzymes in the pathway of glycerol metabolism, industrial applications and even pathogenicity [12], [13]. The NAD+-linked GDHs are members of the medium-chain alcohol dehydrogenase family, most of which are metalloenzymes [14]. They dehydrogenate glycerol to dihydroxyacetone and lead to the production of value-added products, namely, DHA, butanol, succinic acid and citric acid [15], [16]. GDHs are also widely used for the enzymatic determination of glycerol for medical diagnoses and fermentation process analyses [17], [18]. Coupling with other oxidoreductases, GDH is part of a multi-enzyme system for the biosynthesis of chiral intermediates with cofactor regeneration [19]. Therefore, glycerol dehydrogenase was selected as a model enzyme for this metal ion substitution study.
Glycerol dehydrogenase from Klebsiella pneumoniae sp. is a zinc-dependent metalloenzyme [20]. It has been previously shown that Mn2+ substituted GDH exhibits improved activity and thermostability [21]. In this paper, mechanistic insights of activity improvement are studied based on kinetic and thermodynamic analysis. A kinetic model based on an ordered Bi-Bi mechanism with substrate and product inhibition is proposed. The equilibrium constants for each ligand-binding are calculated by using the forward and reverse rate constants. By profiling the binding rate and energy for substrate and product with enzyme, the rate accelerating step is determined. The metal ion's role in catalysis enhancement is investigated.
Results and Discussion
The influence of substrate concentration
The influence of substrate concentration of both substrates, NAD+ and glycerol, on GDH (Fig. 1a) and Mn-GDH (Fig. 1b) were studied. Double reciprocal plots of six NAD+ concentrations versus reaction rates at six fixed glycerol concentrations were drawn [22]. Earlier studies that were conducted on glycerol dehydrogenase from Klebsiella pneumonia [23] and from other microorganisms [24], [25] reported the GDHs follow an ordered Bi-Bi sequential mechanism. Therefore, it was reasonable to assume that GDH from Klebsiella pneumonia and Mn-GDH obey the ordered Bi-Bi sequential mechanism. Kinetic parameters were determined from Lineweaver-Burke plots. The kinetic parameters for GDH and Mn-GDH are listed in Table 1.
Figure 1. The influence of glycerol concentration.
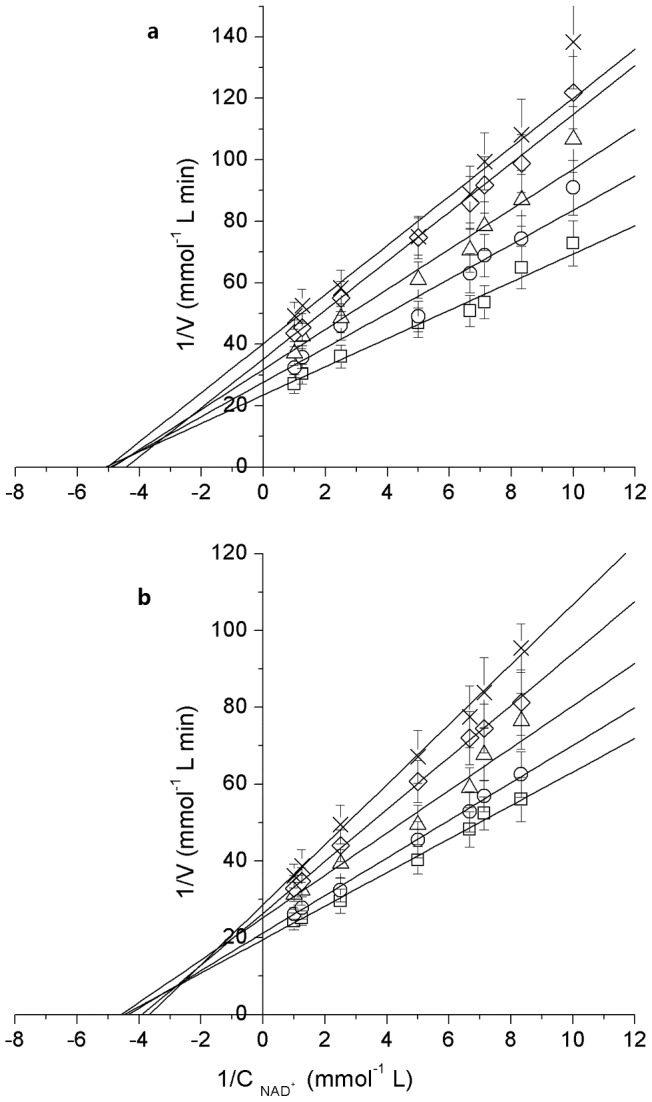
a. The influence of glycerol concentration for GDH; b. The influence of glycerol concentration for Mn-GDH. Reaction conditions: Glycerol concentration (×: 0.010 mol/L, 0.015 mol/L, Δ: 0.025 mol/L, O: 0.100 mol/L, □: 0.200 mol/L); enzyme, 1 mg/L; pH 12.0; temperature, 45°C.
Table 1. Kinetic parameters of Mn-GDH and GDH.
| Kinetic parameters | Mn-GDH | GDH |
| KmA * (mmol/L) | 0.008±0.001 | 0.005±0.001 |
| KmB *(mmol/L) | 0.226±0.013 | 0.262±0.014 |
| KiA (mmol/L) | 0. 012±0.002 | 0.008±0.001 |
*KmA, the Michaelis-Menten constant of NAD+; KmB, the Michaelis-Menten constant of glycerol.
Reaction conditions: Glycerol concentration (0.010–0.400 mmol/L); enzyme, 1 mg/L; pH 12.0; temperature, 45°C.
The influence of product concentration
The product inhibition of Mn-GDH was studied by varying the concentration of each product, DHA (0–0.40 mmol/L) and NADH (0–0.20 mmol/L). NADH competitively inhibited the enzyme at a constant concentration of glycerol (Fig. 2a), which was consistent with a compulsory ordered Bi-Bi reaction mechanism. The inhibition constant of NADH, K iQ, which was derived by secondary plots (Fig.2b) of the slopes determined from the primary double-reciprocal plots [22] against each fixed NADH concentration, was calculated as 0.02 mmol/L.
Figure 2. Product inhibition of NADH for Mn-GDH.
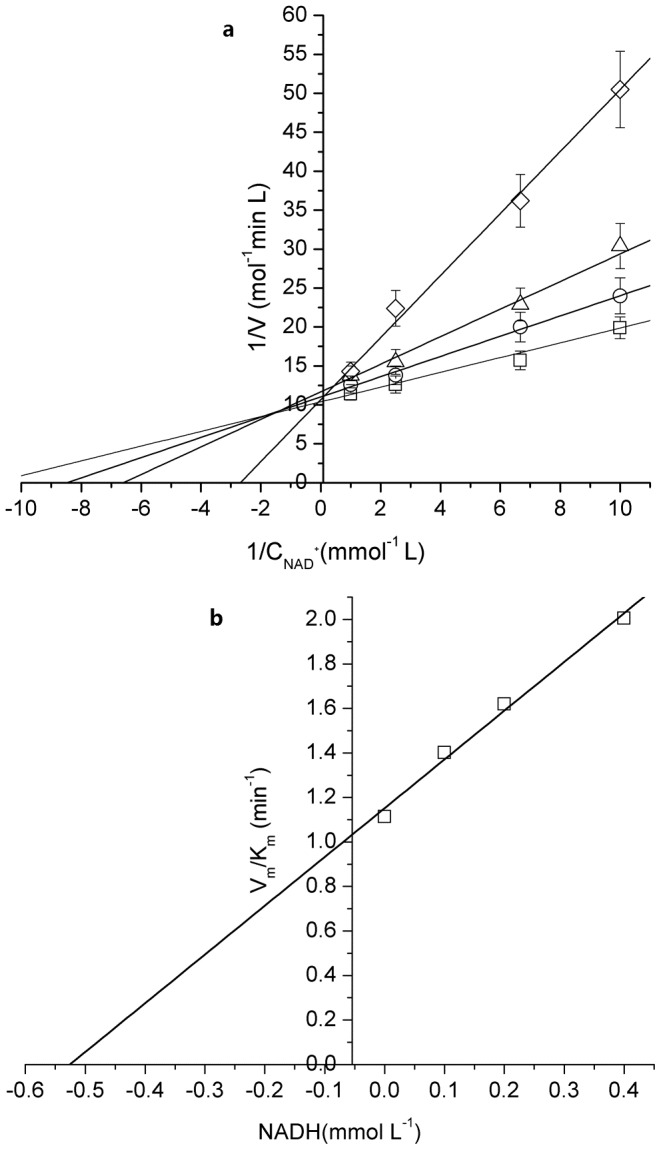
a. Double reciprocal plots of NADH concentrations versus reaction rates b. The secondary plots of the slopes for product inhibition of NADH for Mn-GDH. Reaction conditions: NADH concentration (□: 0 mmol/L, Ο: 0.05 mmol/L, Δ: 0.10 mmol/L, ◊: 0.20 mmol/L); glycerol 0.40 mol/L; Mn-GDH, 1 mg/L; pH 12.0; temperature, 45°C.
DHA inhibited the enzyme noncompetitively with respect to NAD+ at a constant concentration of glycerol (Fig. 3a). The inhibition constant of DHA, KiP, derived by secondary plots of the slopes determined from the primary double-reciprocal plots (Fig.3b), was calculated as 0.52 mmol/L. Product inhibition indicated that Mn-GDH obeyed a compulsory ordered-Bi-Bi mechanism. The inhibition constant of DHA and NADH for GDH were calculated. KiP, and KiQ were 0.45 mmoL/L and 0.015 mmoL/L, respectively.
Figure 3. Product inhibition of DHA for Mn-GDH.
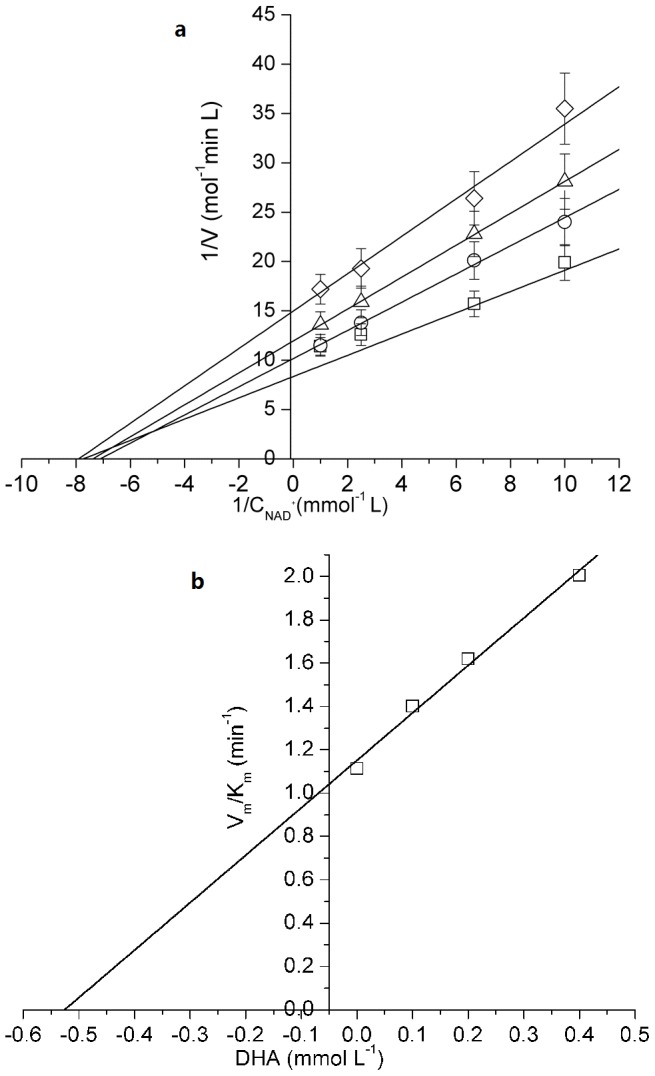
a. Double reciprocal plots of NADH concentrations versus reaction rates. b. The secondary plots of the slopes for product inhibition of DHA for Mn-GDH. Reaction conditions: DHA (□: 0 mmol/L, Ο: 0.10 mmol/L, Δ: 0.20 mmol/L, ◊: 0.40 mmol/L); glycerol 0.40 mol/L; Mn-GDH, 1 mg/L; pH 12.0; temperature, 45°C.
Kinetic model development
An ordered Bi-Bi mechanism kinetic model was proposed. The King-Altman plot of this model is shown in Fig. 4. In this model, NAD+ (A) binds first to the free enzyme (E). The second substrate, glycerol, binds subsequently, forming the ternary complex (EAB). Upon isomerization, the product-bound complex (EPQ) is formed. After the release of the first product, DHA (P), the second product, NADH (Q), dissociates, leaving the free enzyme.
Figure 4. Compulsory ordered Bi-Bi reaction mechanism.
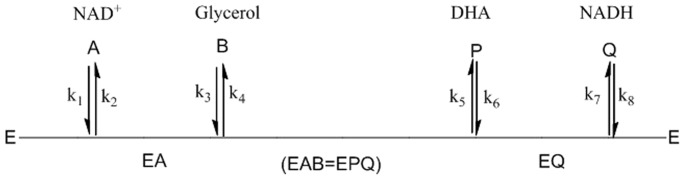
The corresponding volumetric rate of NADH, derived from King-Altman method and transformed using Cleland's coefficient form, is given by Eq.1.
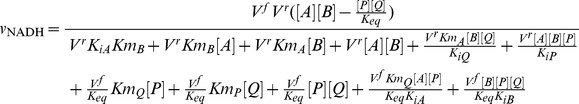 |
(1) |
Eq.2 and Eq.3, which describe the relevance between kinetic constants, were obtained by applying Haldane equations. Keq and KiB were replaced with Eq.2, Eq.3, respectively. The value of KmA, KmB, KiA, KiP and KiQ were derived from the above experiments.
| (2) |
| (3) |
Parameter estimation
The values of the kinetic parameters, namely KmA, KmB, KiA, KiB, KiP, KiQ, was obtained previously, which were used for the final parameter estimation via nonlinear regression [26]. Keq is a dependent parameter. Therefore, the number of estimated parameters was reduced from 10 to 4. Kinetic parameters for estimation can be remarkably decreased. The concentration-time curve data with various substrate concentrations were used for the simulation of the remaining four parameters, namely, Vf, Vr, KmP, and KmQ, by the MATLAB program. Parameter estimation was carried out by a combination of fourth- and fifth-order Runge kutta method using ode45 module in MATLAB software. All datasets were fitted at once. These parameters are listed in Table 2. The comparisons of simulated values with the experimental data for Mn-GDH and GDH are shown in Fig. 5 and Fig. 6. There was a good agreement between the experimental and simulated values, with 11.5% and 12.7% average relative error for Mn-GDH and GDH, respectively.
Table 2. Estimated values of kinetic parameters.
| Kinetic parameters | Mn-GDH | GDH |
| Vf (mmol/L/min) | 0.080 | 0.068 |
| Vr (mmol/L/min) | 0.0104 | 0.0074 |
| KmA *(mmol/L) | 0.008 | 0.262 |
| KmB * (mmol/L) | 0.005 | 0.226 |
| KiA * (mmol/L) | 0.012 | 0.008 |
| KiB ** (mmol/L) | 0.301 | 0.66 |
| KmP (mmol/L) | 0.451 | 0.28 |
| KmQ (mmol/L) | 0.002 | 0.0016 |
| KiP * (mmol/L) | 0.52 | 0.45 |
| KiQ * (mmol/L) | 0.02 | 0.015 |
| Keq ** | 25.6 | 18.4 |
*: the fixed parameters.
**: the dependent parameters.
Figure 5. Comparison of the simulated values with the experimental data for Mn-GDH reaction conditions: NAD+ (□: 1.0 mmol/L; Δ: 0.8 mmol/L, Ο: 0.4 mmol/L, +: 0.2 mmol/L, ×: 0.12 mmol/L), simulated (lines, —); glycerol, 0.1 mol/L; Mn-GDH, 1 mg/L; pH, 12.0; temperature, 45°C.
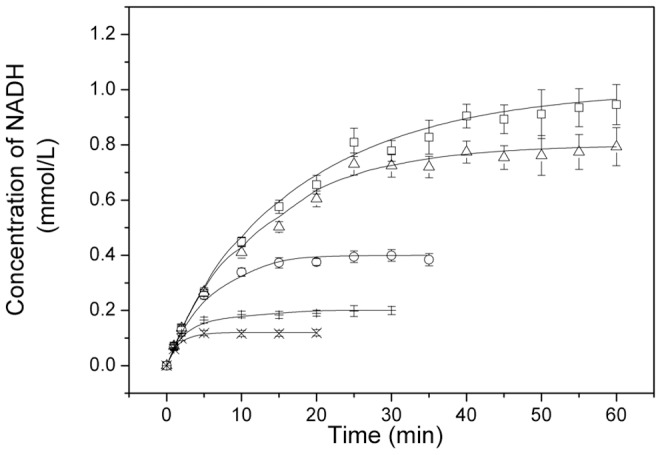
Figure 6. Comparison of the simulated values with the experimental data for GDH.
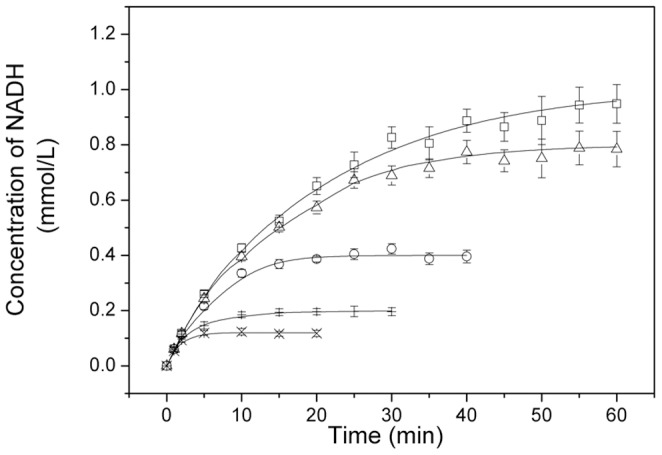
reaction conditions: NAD+ (□: 1.0 mmol/L; Δ: 0.8 mmol/L, Ο: 0.4 mmol/L, +: 0.2 mmol/L, ×: 0.12 mmol/L), simulated (lines, —); glycerol, 0.1 mol/L; GDH, 1 mg/L; pH, 12.0; temperature, 45°C.
Thermodynamic study
The thermodynamic parameters, which are highly complex and interdependent, can be derived from kinetic parameters. Most ligand-binding reactions are studied by calculating the values of the binding equilibrium constant and binding energy of each step [11]. Parameters for each step provide further insights into the binding process, which reveals the effect of metal ion substitution.
The rate constants for each binding step (k1-k8), which are shown in Table 3, were calculated according to the definition of rate constants in terms of kinetic constants [22]. KEA, KEAB, KEPQ and KEQ were assigned as the equilibrium constants for each ligand-binding event [11]. The binding equilibrium constants are listed in Table 4. The Mn-GDH and GDH have nearly the same KEAB value, while a higher KEPQ value for Mn-GDH.
Table 3. The rate constants.
| Rate constants | Mn-GDH | GDH |
| k1 (L/min/mmol) | 10000 | 13600 |
| k2 (/min) | 120 | 109 |
| k3 (L/min/mmol) | 366 | 260 |
| k4 (/min) | 11 | 8 |
| k5 (/min) | 347 | 3431 |
| k6 (L/min/mmol) | 725 | 11446 |
| k7 (/min) | 104 | 69 |
| k8 (L/min/mmol) | 5200 | 4625 |
Table 4. The binding equilibrium constants of enzyme-substrate complex.
| Binding equilibrium constants | Mn-GDH | GDH |
| KEA | 0.012 | 0.008 |
| KEAB | 0.030 | 0.030 |
| KEPQ | 0.48 | 0.30 |
| KEQ | 0.02 | 0.015 |
Binding energy of the enzyme ternary complex EPQ (enzyme-DHA-NADH complex), which were defined as -RTlnKEPQ, for Mn-GDH and GDH were 1.95 and 3.18 KJ/mol, respectively. This revealed that the substitution of manganese mainly depended on accelerating the release of dioxyacetone [27], [28].
Manganese has the similarity with the other divalent cations, which are Lewis acids and electrostatic stabilizers. Manganese can be replaceable with other metals, namely, magnesium and zinc. This depends on the intermediate properties of Mn2+ relative to these other ions, including its radius length and borderline hard–soft character [29]. Metal ions with important biological function can be classified as hard and soft, while hard acids and bases are weak polar, have small ionic radius and high oxidation state. The soft species are on the contrary [30]. Replacing the catalytic zinc with manganese revealed electronic requirements for the specific geometry of the catalytic site of GDH.
Conclusion
A simple, quick and valid strategy based on kinetic and thermodynamic analysis for understanding the significant mechanistic changes induced by metal ion substitution was proposed. By profiling the binding rate and energy for the substrate and product of the enzyme, the rate accelerating step was determined. The mechanism of increasing the catalytic activity of glycerol dehydrogenase by manganese substitution was studied. A kinetic model based on the ordered Bi-Bi mechanism with both product and substrate inhibition was proposed. Further mechanistic insights regarding the role of the metal ion in catalysis enhancement were investigated by characterizing the binding kinetics and thermodynamics of each step. The kinetic parameters were simulated by the MATLAB program. The thermodynamic parameters were derived from the kinetic parameters, which indicated that the metal ion substitution facilitated the release of dioxyacetone.
The kinetic and thermodynamic analysis contributed further insights regarding the prediction and optimization of polyol dehydrogenases that are widely used for chiral alcohol production. This study also provided valuable information for further investigation by molecular simulation. Metal coordination is a key structural and functional component of enzyme. Given this dual role, metal coordination plays a template role in folded and functional protein domains and complexes. The study of metal ions coordination effect based on kinetic and thermodynamic study will provided a promising method metal induced multi-enzyme assembly study [31].
Materials and Methods
Materials
Glycerol, NAD+, NADH, and ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma Chem. Co. (Beijing, China). The media (Tryptone, yeast extract, nutrient broth) were purchased from Sangon Biotech Co. Ltd. (Shanghai, China). All other chemicals used were analytical grade and were purchased from either Sigma or Merck (Beijing, China).
Preparation of the apoenzyme and metal substitution
The preparation of purified GDH followed a previously published method [19]. Recombinant GDH was expressed by transforming plasmids containing the gldA gene (GenBank: AKAM01000021.1), which codes for glycerol dehydrogenase, into E. coli BL21 (DE3). Metal substitution was accomplished using purified GDH using the following steps: chelation of GDH's catalytic zinc ion using EDTA (1.0 mmol/L) for 4 h at 28°C and removal of EDTA-Zn2+ from the GDH solution by dialysis in binding buffer (pH 7.4, changed every 8 h) at 4°C for 53 h. The manganese ion was introduced by the co-incubation of the GDH apoenzyme with MnCl2 (10.0 mmol/L) for 1 h at 37°C. A reference experiment was performed by directly adding Mn2+ (10 mmol/L) to native GDH to investigate the activation effects of bivalent ions. The results indicated that the addition of Mn2+ to the reaction solution decreased the activity of GDH. The activity of the apoenzyme after EDTA treatment was nearly undetectable, which confirmed the activity enhancement by metal ion substitution [21].
Enzyme assay and kinetic study
The activity of glycerol dehydrogenase was measured by following the increase of NADH concentration using a Spectra Max M5 Microplate Reader (California, United States). Measurements were taken at 340 nm, and a molar extinction coefficient of 6.22/mM/cm was used. One unit of GDH activity was defined as the amount of enzyme necessary to oxidize 1 µmol of NADH per minute under the following conditions (45°C, 0.1 mol/L potassium carbonate buffer, pH 12.0). The assay mixture contained 0.4 mol/L glycerol, 0.1–1.0 mmol/L NAD+, and 0.1 mol/L potassium carbonate buffer (pH 12.0). The volume of the reaction mixture was 200 µL in all cases. Reactions were started by the addition of the enzyme solution. Enzyme activities were determined in triplicate. Three blank controls were used as a reaction mixture with the apoenzyme, without the enzyme, and without NAD+. For the product inhibition study, the reactions were carried out with different DHA or NADH concentrations.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 41176111, No. 41306124), the Key Program of National Natural Science Foundation of China (No. 21336009), the Natural Science Foundation of Fujian Province of China (No. 2012J01049), and the Fundamental Research Funds for the Central Universities (No. 2013121029). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13: 1205–1218. [DOI] [PubMed] [Google Scholar]
- 2. Kleifeld O, Frenkel A, Martin JML, Sagi I (2003) Active site electronic structure and dynamics during metalloenzyme catalysis. Nat Struct Biol 10: 98–103. [DOI] [PubMed] [Google Scholar]
- 3. Cappiello M, Alterio V, Amodeo P, Del Corso A, Scaloni A, et al. (2006) Metal ion substitution in the catalytic site greatly affects the binding of sulfhydryl-containing compounds to leucyl aminopeptidase. Biochem 45: 3226–3234. [DOI] [PubMed] [Google Scholar]
- 4. Arima J, Uesugi Y, Hatanaka T (2009) Bacillus D-stereospecific metallo-amidohydrolase: Active-site metal-ion substitution changes substrate specificity. Biochim 91: 568–576. [DOI] [PubMed] [Google Scholar]
- 5. Bogin O, Peretz M, Burstein Y (1997) Thermoanaerobacter brockii alcohol dehydrogenase: Characterization of the active site metal and its ligand amino acids. Protein Sci 6: 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rochu D, Viguie N, Renault F, Crouzier D, Froment MT, et al. (2004) Contribution of the active-site metal cation to the catalytic activity and to the conformational stability of phosphotriesterase: temperature- and pH-dependence. Biochem J 380: 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sparta M, Alexandrova AN (2012) How metal substitution affects the enzymatic activity of catechol-O-nethyltransferase. PLoS ONE 10: e47172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. D'Antonio EL, Christianson DW (2011) Crystal structures of complexes with cobalt-reconstituted human arginase I. Biochemistry 50: 8018–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alberty RA (2010) Biochemical thermodynamics and rapid-equilibrium enzyme kinetics. J Phys Chem B 114: 17003–17012. [DOI] [PubMed] [Google Scholar]
- 10. Bohacek RS, McMartin C (1994) Multiple highly diverse structures complementary to enzyme binding sites: results of extensive application of a de Novo design method incorporating combinatorial growth. J Am Chem Soc 116: 5560–5571. [Google Scholar]
- 11. Fisher HF (2010) Protein–ligand interactions: thermodynamic basis and mechanistic consequences. Encyclopedia of life sciences, 1–10. DOI: 10.1002/9780470015902.a0001341.pub [Google Scholar]
- 12. Wang Y, Tao F, Xu P (2014) Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2, 3-butanediol formation in Klebsiella pneumoniae . J Biol Chem 113: 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, Zhao WD, Li Q, Fang WG, Zhu L, et al. (2009) Tentative identification of glycerol dehydrogenase as Escherichia coli K1 virulence factor cglD and its involvement in the pathogenesis of experimental neonatal meningitis. Med. Microbiol. Immunol. 198: , 195–204 [DOI] [PubMed] [Google Scholar]
- 14. Ruzheinikov SN, Burke J, Sedelnikova S, Baker PJ, Taylor R, et al. (2001) Glycerol dehydrogenase: Structure, specificity, and mechanism of a family III polyol dehydrogenase. Structure 9: 789–802. [DOI] [PubMed] [Google Scholar]
- 15. Dobson R, Gray V, Rumbold K (2012) Microbial utilization of crude glycerol for the production of value-added products. J Ind Microbiol Biotechnol 39: 217–226. [DOI] [PubMed] [Google Scholar]
- 16. Enders D, Voith M, Lenzen A (2005) The dihydroxyacetone unit - A versatile C-3 building block in organic synthesis. Angew Chem Int Ed 44: 1304–1325. [DOI] [PubMed] [Google Scholar]
- 17. Sokic-Lazic D, Arechederra RL, Treu BL, Minteer SD (2010) Oxidation of biofuels: fuel diversity and effectiveness of fuel oxidation through multiple enzyme cascades. Electroanal 22: 757–764. [Google Scholar]
- 18. Lapenaite I, Ramanaviciene A, Ramanavicius A (2006) Current trends in enzymatic determination of glycerol. Crit Rev in Anal Chem 36: 13–25. [Google Scholar]
- 19. Zhang Y, Gao F, Zhang SP, Su ZG, Ma GH, et al. (2011) Simultaneous production of 1,3-dihydroxyacetone and xylitol from glycerol and xylose using a nanoparticle-supported multi-enzyme system with in situ cofactor regeneration. Bioresource Technol 102: 1837–1843. [DOI] [PubMed] [Google Scholar]
- 20. Tang JCT, Forage RG, Lin ECC (1982) Immunochemical properties of NAD+-linked glycerol dehydrogenases from Escherichia coli and Klebsiella pneumonia . J Bacteriol 152: 1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang SZ, Wang J, Zhou XF, Guo YX, Fang BS (2013) The improvement of stability, activity and substrate promiscuity of glycerol dehydrogenase substituted by divalent metal ions. Biotechnol Bioproc E 18: 796–800. [Google Scholar]
- 22.Leskovac V (2004) Comprehensive enzyme kinetics. NewYorkKluwer Academic/Plenum Publishers, 141–147.
- 23. Chen HW, Nie JF, Chen G, Fang BS (2010) Kinetic mechanisms of glycerol dehydrogenase and 1,3-propanediol oxidoreductase from Klebsiella pneumonia . Chin J Biotech 26: 177–182. [PubMed] [Google Scholar]
- 24. Zheng MQ, Zhang SP (2011) Immobilization of glycerol dehydrogenase on magnetic silica nanoparticles for conversion of glycerol to value-added 1,3-dihydroxyacetone. Biocatal Biotransfor 29: 278–287. [Google Scholar]
- 25. Nishise H, Nagao A, Tani Y, Yamada H (1984) Further characterization of glycerol dehydrogenase from Cellulomonas sp. NT3060. Agr Biol Chem 48: 1603–1609. [Google Scholar]
- 26. Chen BH, Hibbert EG, Dalby PA, Woodley JM (2008) A new approach to bioconversion reaction kinetic parameter identification. AIChE J 54: 2155–2163. [Google Scholar]
- 27. Siddiqui KS, Cavicchioli R (2006) Cold-adapted enzymes. Annu Rev Biochem 75: 403–433. [DOI] [PubMed] [Google Scholar]
- 28. Fisher HF (2001) Protein ligand interactions: Molecular basis. Encyclopedia of life sciences, 1–9 DOI:10.1038/npg.els.0001341 [Google Scholar]
- 29. Martinez-Rodriguez S, Encinar JA, Hurtado-Gomez E, Prieto J, Clemente-Jimenez JM, et al. (2009) Metal-triggered changes in the stability and secondary structure of a tetrameric dihydropyrimidinase: A biophysical characterization. Biophys Chem 139: 42–52. [DOI] [PubMed] [Google Scholar]
- 30. Lemire JA, Harrison JJ, Turner RJ (2013) Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 11: 371–384. [DOI] [PubMed] [Google Scholar]
- 31. Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, et al. (2010) Metal templated design of protein interfaces. P Natl Acad Sci USA 107: 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]


