Abstract
Background
Furfural is a major growth inhibitor in lignocellulosic hydrolysates and improving furfural tolerance of microorganisms is critical for rapid and efficient fermentation of lignocellulosic biomass. In this study, we used the RNAi-Assisted Genome Evolution (RAGE) method to select for furfural resistant mutants of Saccharomyces cerevisiae, and identified a new determinant of furfural tolerance.
Results
By using a genome-wide RNAi (RNA-interference) screen in S. cerevisiae for genes involved in furfural tolerance, we identified SIZ1, a gene encoding an E3 SUMO-protein ligase. Disruption of SIZ1 gene function by knockdown or deletion conferred significantly higher furfural tolerance compared to other previously reported metabolic engineering strategies in S. cerevisiae. This improved furfural tolerance of siz1Δ cells is accompanied by rapid furfural reduction to furfuryl alcohol and leads to higher ethanol productivity in the presence of furfural. In addition, the siz1Δ mutant also exhibited tolerance towards oxidative stress, suggesting that oxidative stress tolerance related proteins may be under the SUMO regulation of SIZ1p and responsible for furfural tolerance.
Conclusions
Using a genome-wide approach, we identified a novel determinant for furfural tolerance, providing valuable insights into the design of recombinant microbes for efficient lignocellulose fermentation.
Keywords: Furfural tolerance, RAGE, Saccharomyces cerevisiae, SIZ1, SUMO E3 ligase
Background
There is a growing interest worldwide in using lignocellulose, the most abundant renewable biomass, to replace cereal substrates in the production of biofuels and biochemicals [1,2]. However, efficient fermentation of lignocellulosic hydrolysates is limited by inhibitors that are inevitably released during pretreatment and hydrolysis of lignocellulosic substrates [3]. Formed by dehydration of pentoses during dilute acid pretreatment of lignocelluloses, furfural is one of the major inhibitors present in lignocellulosic hydrolysates [4]. The toxicity of hydrolysates correlates with furfural concentration, with 1 to 5 g/L of furfural leading to complete growth inhibition of Escherichia coli, Zymomonas mobilis and Saccharomyces cerevisiae, significantly reducing the yield and productivity of desired products [5-8]. Although physical or chemical strategies for furfural detoxification can be adopted during fermentation, the additional equipment and time required increase the production costs [9]. Thus, improving furfural tolerance in microorganisms would provide a cost-effective means for lignocellulose fermentation.
S. cerevisiae is the most widely studied model organism for furfural tolerance and has higher furfural tolerance compared to other potential biofuel and biochemical production hosts [10-12]. Furfural modulates expression of genes involved in a variety of general stress responses in S. cerevisiae, including oxidative stress, nutrient starvation, DNA damage, unfolded protein response, as well as osmotic and salt stress [11]. However, whether and how these genes contribute to furfural tolerance is unknown [13]. To date, the known mechanism of furfural detoxification is its reduction into the less toxic furfuryl alcohol through reduced nicotinamide adenine dinucleotide phosphate(NADPH)-dependent enzymes [13,14]. Attributed to the significant increase in mRNA abundance and protein expression level observed in adapted S. cerevisiae under stress challenge, the NADPH-dependent oxidoreductases ADH7p and YKL071Wp were found to be responsible for furfural detoxification [15,16]. Due to limited knowledge on the mechanisms of furfural toxicity towards cells, strategies for improving furfural tolerance focus mainly on overexpression of the enzymes that convert furfural to furfuryl alcohol [12,17]. To fill this knowledge gap and at the same time develop strains with strong furfural resistance, genetic determinants of furfural tolerance need to be identified.
Genes associated with furfural tolerance have been identified by comparative analyses of wild-type strains with furfural tolerant mutants generated via random mutation, directed evolution or adaptation strategies. However, the existence of multiple simultaneous mutations in these tolerant strains often complicates the analyses [18]. Therefore, dissecting the functional contribution of each gene towards furfural tolerance remains a significant challenge. RAGE (RNAi-assisted genome evolution) is a recently developed genome engineering method that can continuously improve a desired trait by allowing the sequential introduction of tractable reduction-of-function modifications to the genome [19]. In this study, we used RAGE to select for clones with increased furfural tolerance. The genome-wide RNAi library of S. cerevisiae BY4741 was selected for clones with increased furfural tolerance to discover determinants of furfural resistance. Deletion of the gene SIZ1, which encodes an E3 SUMO-protein ligase, was found to play an important role in tolerance to furfural and general oxidative stress in S. cerevisiae.
Results
RAGE screen and isolation of furfural resistant strains
To uncover new genetic determinants of furfural tolerance, we sought to determine if furfural tolerance can be enhanced through reduction of gene function by using RAGE to select for furfural resistant mutants (Figure 1A). Reconstitution of the RNAi machinery in S. cerevisiae BY4741 was carried out as previously reported [19] to yield the BAD strain. The genomic DNA derived RNAi library was constructed with additional modifications to prevent self-ligation of vectors and fragments [20]. DNA sequencing of 17 randomly picked plasmids from the RNAi library showed that only one locus was targeted by each RNAi construct (Additional file 1: Figure S1). The number of Sau3AI digested fragments (35,837) from S. cerevisiae genomic DNA [21] was considered as the number of possible equiprobable variants. With a library size of 3.4 × 105, more than 99% coverage of the yeast genome was achieved [22].
Figure 1.
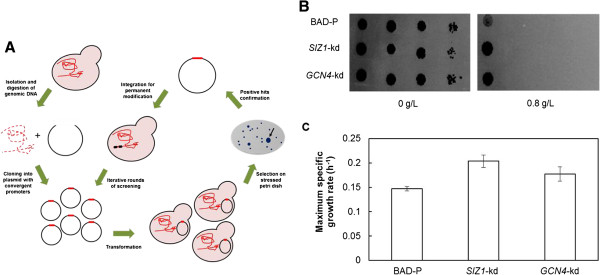
RNAi-assisted genome evolution (RAGE) screen and isolation of furfural resistant S. cerevisiae strains. (A) Schematic representation of RAGE [19]. (B) Isolation of furfural tolerant strains. Ten-fold dilutions of the indicated strains were spotted on synthetic complete (SC) plates containing 0 or 0.8 g/L furfural and incubated at 30°C for 2 and 5 days, respectively. BAD-P is the parent wild-type strain. Strains SIZ1-kd and GCN4-kd are furfural resistant strains isolated from the RAGE screen. (C) Maximum specific growth rates of furfural tolerant strains in the presence of 0.8 g/L furfural. Error bars represent SD (n = 3).
By selecting mutant colonies that grew larger than that of strain BAD-P (strain BAD with plasmid backbone alone) on synthetic complete medium deficient in uracil (SC-URA) plates containing 0.8 g/L furfural, we isolated and confirmed four clones with increased furfural tolerance. Sequencing revealed that three out of the four RNAi constructs from these furfural resistant clones contained the same fragment of the SIZ1 gene, which encodes an E3 small ubiquitin-like modifier (SUMO)-protein ligase (Additional file 1: Figure S2). The RNAi construct isolated from the fourth clone contained a fragment of the GCN4 gene encoding a basic leucine zipper transcriptional activator of amino acid biosynthesis (Additional file 1: Figure S3). Strains expressing the RNAi cassettes targeting genes SIZ1 (SIZ1-kd) and GCN4 (GCN4-kd) exhibited significant improvement in furfural tolerance compared to the control BAD-P strain (Figure 1B and C). Notably, SIZ1-kd and GCN4-kd strains showed no growth advantage over the control strain in the absence of furfural (Additional file 1: Table S3).
Disruption of SIZ1 function increased furfural tolerance in S. cerevisiae
To determine if the reduction or loss of SIZ1 and GCN4 gene function contributes to furfural resistance, furfural tolerance of the respective knockout mutants was tested. The maximum specific growth rate in the presence of 0.8 g/L furfural of the siz1Δ strain was 73% higher than wild type. Increased furfural tolerance was also found in the siz1Δ strain when higher furfural concentrations were adopted (Additional file 1: Table S4). Complementation of the siz1Δ strain with a plasmid-borne copy of gene SIZ1 but not with plasmid alone (siz1Δ-P) restored furfural sensitivity (Table 1). On the other hand, deletion of GCN4 did not phenocopy the improved tolerance observed for the GCN4-kd strain, suggesting that gene dosage is important for GCN4 associated furfural tolerance (Table 1). We chose to focus on SIZ1 as SIZ1-kd and siz1Δ have a more significant effect on furfural tolerance compared to GCN4-kd.
Table 1.
Furfural tolerance assay of siz1Δ mutant and its complementary derivatives
| Strain | Maximum specific growth rate (h -1 ) |
|---|---|
| BAD |
0.15 ± 0.00 |
|
siz1Δ |
0.26 ± 0.00 |
|
siz1Δ-P |
0.26 ± 0.01 |
|
siz1Δ-SIZ1 |
0.19 ± 0.00 |
| gcn4Δ | 0.16 ± 0.01 |
Strains were grown in the presence of 0.8 g/L furfural. Results are presented as mean ± SD (n = 3).
To investigate whether increased furfural tolerance via deletion of SIZ1 is a strain-specific or general attribute, SIZ1 was also deleted in two other S. cerevisiae strains: HZ848 [23] and W303a [24]. Furfural tolerance of these mutants was tested in the presence of 0.8 g/L furfural. Strains HZ848-siz1Δ and W303a-siz1Δ exhibited 27% and 58% higher maximum specific growth rates respectively, as compared to their respective parent strains (Table 2). These results showed that SIZ1 was indeed an important determinant for furfural resistance in S. cerevisiae.
Table 2.
Furfural tolerance assay of S. cerevisiae siz1Δ mutants in SC medium containing 0.8 g/L furfural
| Strain | Maximum specific growth rate (h -1 ) |
|---|---|
| HZ848 |
0.15 ± 0.01 |
| HZ848-siz1Δ |
0.19 ± 0.00 |
| W303a |
0.19 ± 0.01 |
| W303a-siz1Δ | 0.30 ± 0.01 |
The maximum specific growth rates of different S. cerevisiae siz1Δ mutants are statistically significant over their corresponding wild-type (P <0.05) as determined by the Student t-test. Results are presented as mean ± SD (n = 3).
Increased rate of furfural reduction and ethanol productivity by siz1Δ strain
Having demonstrated that disruption of SIZ1 gene function greatly increases furfural tolerance, we sought to determine the effect and utility of the enhanced furfural tolerance observed for the siz1Δ strain. Batch fermentation containing 20 g/L glucose and 0.8 g/L furfural was conducted using the siz1Δ and wild type (BAD) strains. While both control and siz1Δ strains experience a delay in entering exponential growth in the presence of furfural, a shorter initial lag was observed in the siz1Δ strain (Figure 2A). Finally, furfural was consumed and converted to the less toxic furfuryl alcohol at a rate that was 48% faster in the siz1Δ strain compared to that of the wild type (0.031 g/(L · h) versus 0.021 g/(L · h)) (Figure 2B). Strain siz1Δ consumed all glucose in 30 h, which was 18 h faster than that of the wild-type strain BAD (Figure 2B and C). As a result, strain siz1Δ was able to produce 9.0 g/L ethanol after 30 h, resulting in 275% higher productivity and 254% higher ethanol yield than that observed for strain BAD (Figure 2C, Additional file 1: Table S5). The molar ratios of carbon used for ethanol production were comparable between strain BAD and ∆siz1, indicating the improved furfural tolerance in strain ∆siz1 was not at the cost of ethanol yield (Additional file 1: Table S5). Overall, these results demonstrate that the increased furfural tolerance observed with disruption of SIZ1 function was accompanied by faster furfural reduction and this improved trait has clear utility in improving the efficiency of lignocellulose fermentation containing furfural.
Figure 2.
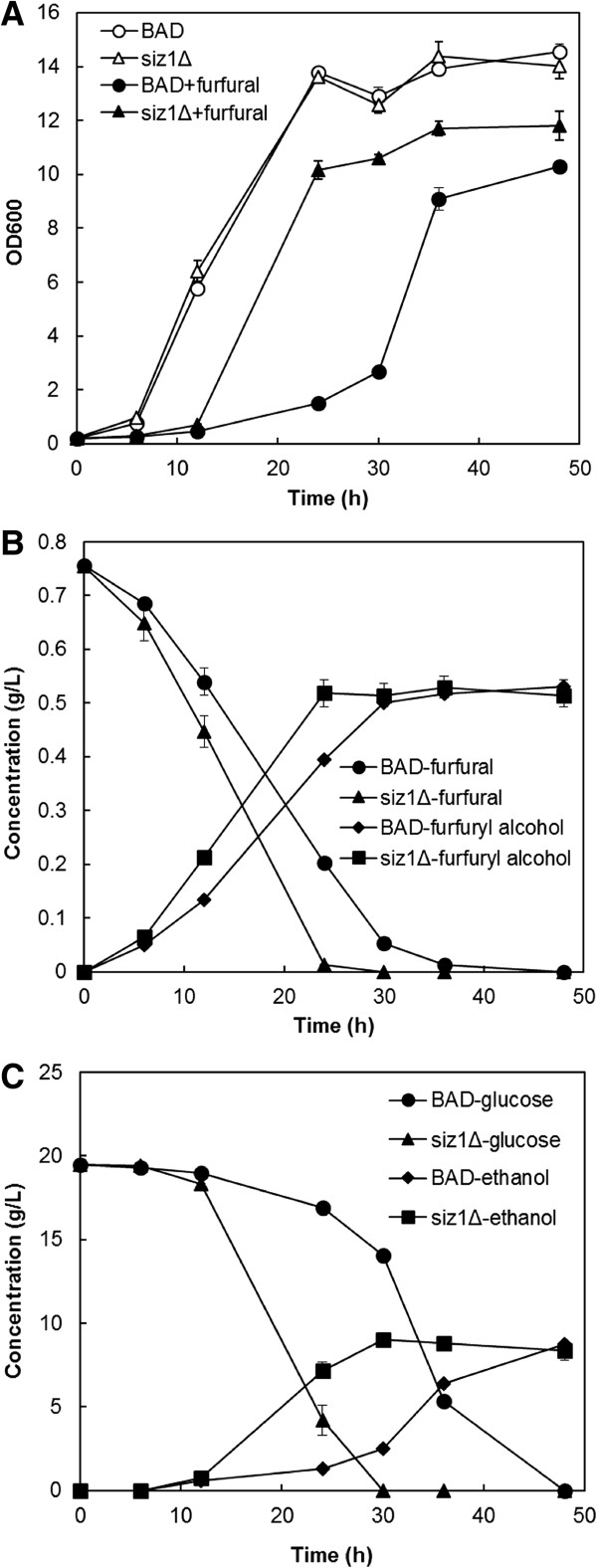
Growth and metabolite profiles in batch fermentation of parent BAD and siz1Δ strains. Strains were grown in SC medium with 20 g/L glucose in the presence and absence of 0.8 g/L furfural. (A) Cell growth as measured by optical density (OD)600. (B) Furfural consumption and furfuryl alcohol production. (C) Glucose consumption and ethanol production. Error bars represent SD of the mean (n = 3).
Comparison of SIZ1 deletion with other previously reported strategies for improving furfural tolerance in S. cerevisiae
The rapid furfural reduction observed for siz1Δ cells is reminiscent of furfural detoxification by enzymes that catalyze aldehyde reduction coupled with cofactors NADPH and/or NADH [13]. Indeed, overexpression of various aldehyde reduction enzymes encoded by genes YKL071W, ALD6, ADH7 and ARI1 have been demonstrated to be strongly associated with furfural resistance in yeast [8,12,17]. In addition, overexpression of glucose-6-phosphate dehydrogenase encoding gene ZWF1 and transcriptional activator encoding gene MSN2, which are involved in regeneration of NAD(P)H and stress response, respectively, have also been confirmed to increase furfural tolerance in S. cerevisiae[25,26]. To compare these reported targets with siz1Δ-associated furfural tolerance, individual overexpression of each gene was performed in strain BAD. Unexpectedly, only overexpression of gene ADH7 and ARI1 resulted in increased furfural tolerance (Figure 3), which may be attributed to the different promoters adopted for over-expression, different growth media tested for furfural tolerance and/or different strains used. Among the engineered strains tested, the siz1Δ mutant exhibited the highest maximum specific growth rate in the presence of 0.8 g/L furfural (Figure 3).
Figure 3.
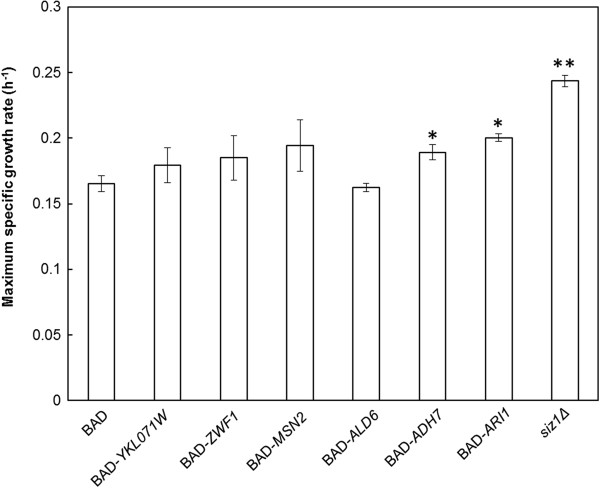
Maximum specific growth rates to furfural tolerance assay of parent strain BAD and its indicated derivatives in SC medium containing 0.8 g/L furfural. The Student t-test was performed to determine whether the specific growth rates of indicated derivatives was statistically significant over that of the parent strain BAD. *P <0.05; **P <0.01. Error bars represent the SD of the mean (n = 3).
Furfural tolerance is specific to SIZ1p and not to other SUMO E3 ligases
Protein sumoylation, an important post-translational modification in various cellular processes, involves the covalent attachment of the SUMO polypeptide to specific lysine residues of target proteins [27]. The E3 SUMO-protein ligase facilitates the transfer of SUMO to the substrate proteins [28]. To investigate whether furfural tolerance is specific to SIZ1p, genes SIZ2, MMS21 and CST9 that encode for the other three E3 SUMO-protein ligases in yeast [29] were individually deleted but did not affect furfural tolerance of the cells (Figure 4).
Figure 4.
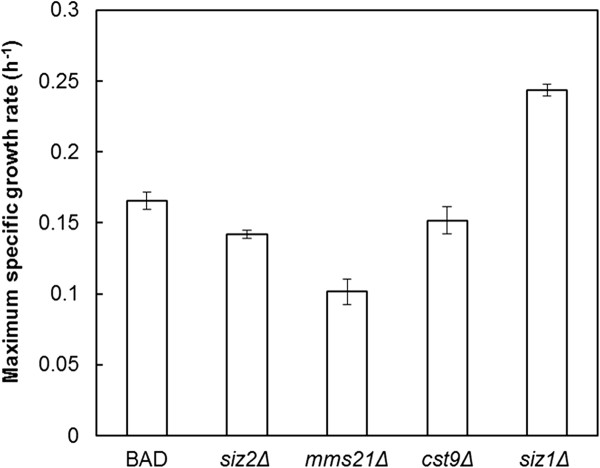
Furfural tolerance assay of indicated E3 small ubiquitin-like modifier (SUMO)-protein ligase gene deletion mutants in SC medium containing 0.8 g/L furfural. Error bars represent SD of the mean (n = 3).
As downregulation of gene GCN4 increased the furfural tolerance of strain BAD (Figure 1), this strategy was evaluated in siz1Δ strain. No significant difference in the maximum specific growth rates was found between strain siz1Δ-GCN4-kd and the siz1Δ strain in the presence of 0.8 g/L furfural. This observation suggests that downregulation of GCN4 may act in the same pathway as SIZ1 deletion (Figure 5).
Figure 5.
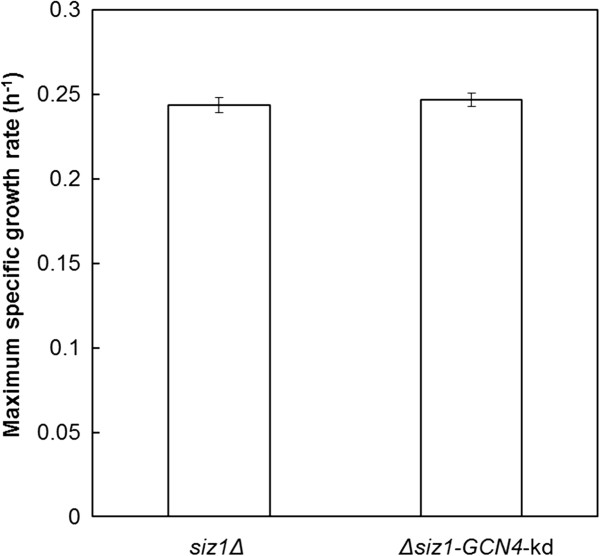
Furfural tolerance assay of siz1Δstrain and its derivatives in SC medium containing 0.8 g/L furfural. Error bars represent the SD of the mean (n = 3).
Disruption of SIZ1 function increases tolerance to oxidative stresses
According to a previous study, downregulation of GCN4 increases tolerance of a furfural-like chemical 5-hydroxymethylfurfural (5-HMF), which is another major inhibitor in lignocellulose hydrolysates that is derived from dehydration of hexoses in lignocellulosic hydrolysates [15,30]. The siz1Δ mutant also exhibits higher maximum specific growth rate in the presence of 1.26 g/L 5-HMF compared to the control strain (Figure 6A), while the maximum specific growth rates of both strains were similar in the absence of 5-HMF (Additional file 1: Table S3). Given the higher tolerance achieved by SIZ1 deletion as compared to downregulation of GCN4 in the presence of either furfural or 5-HMF, other proteins that are under the SUMO regulation of SIZ1p may also be involved in furfural tolerance besides GCN4p (Figure 1C, Table 1, Figure 6A and Figure 5). Furfural induces accumulation of reactive oxygen species (ROS), the toxicity of which is greatly attenuated in hosts with strong oxidative stress tolerance [10]. To investigate whether siz1Δ strain has a detoxification effect on ROS, the oxidative stress tolerance of the siz1Δ mutant was further tested in the presence of 1.72 mg/L menadione, which is known to generate ROS in vivo[31]. As shown in Figure 6B, the maximum specific growth rate of the siz1Δ mutant was 28% higher than that of the wild type. This result indicated that proteins that are related to oxidative stress tolerance may be under the SUMO regulation of SIZ1 and responsible for furfural tolerance.
Figure 6.
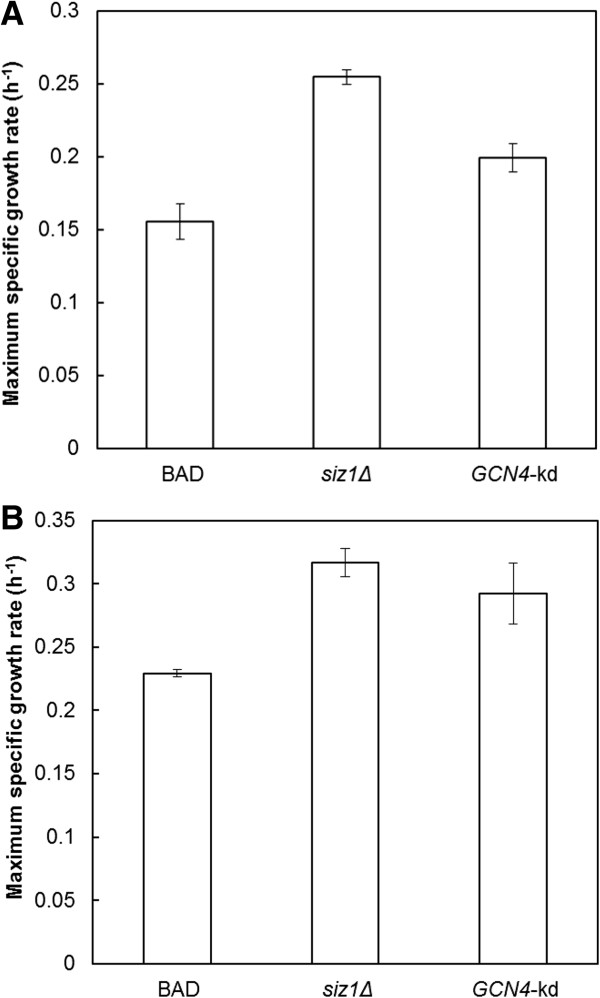
Tolerance assay of siz1Δ strain in SC medium containing (A) 1.26 g/L HMF or (B) 1.72 mg/L menadione. Error bars represent the SD of the mean (n = 3).
Discussion
Furfural toxicity is a major hurdle in the economical fermentative processes for biofuel and biochemical production using lignocellulosic hydrolysates as substrate [32]. Given the incomplete knowledge on furfural tolerance mechanisms, discovering new targets of furfural resistance would facilitate development of new metabolic engineering strategies for improving furfural tolerance. Whereas significant attention has been paid to the induced genes in furfural tolerance studies, the importance of the repressed genes is often neglected [13]. In this study, RAGE was used for selection of furfural resistant mutants, in order to identify those genes with previously undiscovered roles in furfural tolerance. Genes with downregulation and/or loss of function can be selected out in our case, which distinguishes RAGE from other screening methods (for example, gain-of-function-based screening and S. cerevisiae single gene-knockout collection-based screening). In a previous study, RAGE was demonstrated to continuously improve acetic acid tolerance by accumulating reduction-of-function modifications in the genome [19]. In this study, however, no further improvement of furfural tolerance was observed after the second round of selection in the siz1Δ strain by RAGE (data not shown).
RNAi cassettes targeting genes SIZ1 and GCN4 were recovered during selection for furfural resistance and were shown to increase furfural tolerance (Figure 1). Downregulation of GCN4 also increased 5-HMF tolerance according to a previous study [15]. GCN4p is a transcriptional activator of gene expression related to amino acid biosynthesis during amino acid starvation in yeast [33,34]. Downregulated expression of GCN4 could be an efficient means of energy utilization for economic pathway development [15]. However, deletion of GCN4 did not show increased furfural tolerance in our work, suggesting that an appropriate expression level of amino acid biosynthetic genes may facilitate cell survival under stress challenge.
Replicated discovery of SIZ1 by RAGE in the presence of furfural, along with the ability of siz1Δ to increase furfural tolerance in different S. cerevisiae strains, showed the important role of SIZ1 in furfural resistance (Figure 1, Table 2, Additional file 1: Figure S2). To our knowledge, this is a novel determinant of furfural tolerance. SUMO-modified proteins participate in transcription, nuclear transport, cell cycle, DNA repair and signal transduction [35]. The vast majority of sumoylation in yeast is mediated by SUMO E3 ligase [36]. SIZ1p, together with another SUMO E3 ligase SIZ2p, accounts for 90% of the total sumoylation in yeast [37,38]. As furfural tolerance is siz1Δ-dependent (Figure 4), targets that improve furfural tolerance may be exclusively under the SUMO regulation of SIZ1p. Alternatively, these targets may also be under SUMO regulation of other E3 SUMO-protein ligases, but the effect of furfural tolerance from deletion of other E3 SUMO-protein ligases was masked by interactions with other targets which increased susceptibility to furfural.
Downregulation of GCN4 and deletion of SIZ1 increased furfural tolerance (Figure 1 and Figure 2). However, no increase of furfural tolerance was found when GCN4 expression was reduced in the siz1Δ strain (Figure 5), indicating these two genes may act in the same pathway. GCN4p stability was demonstrated to be regulated by sumoylation, which occurs after it binds to target promoters and facilitates the subsequent removal of GCN4p from these promoters to ensure accurate transcription of its target genes [39,40]. A possible mechanism for furfural tolerance in the siz1Δ strain was speculated upon here. In the siz1Δ strain, non-sumoylated GCN4p cannot dissociate from target promoters after recruitment of RNA polymerase II, which may decrease the transcriptional efficiency of the target genes and facilitate cell survival in the presence of furfural.
SIZ1-kd or deletion strains exhibited higher furfural tolerance as compared to the GCN4-kd strain, indicating more SUMO targets of SIZ1p are likely to be involved in furfural resistance besides GCN4p (Figure 1 and Table 1). A newly identified SUMO substrate of SIZ1p is the NADHX dehydratase YKL151Cp, which converts (S)-NADHX to NADH [41,42]. More reducing power for furfural detoxification may be generated through this reaction. In addition, the siz1Δ strain also exhibited increased oxidative stress tolerance, suggesting that determinants of oxidative stress tolerance may be protein substrates of SIZ1p responsible for furfural tolerance (Figure 6B). A total of 159 proteins were identified to be sumoylated in a proteomics study [40], among which the 6-phosphogluconate dehydrogenase GND1p, the basic leucine zipper transcription factor SKO1p and the redoxin peroxidase TSA1p are implicated in oxidative stress response in yeast [43-45]. These candidates could be possible determinants of furfural resistance under SUMO regulation of SIZ1p. Identification of protein substrates that are differentially sumoylated in wild type and siz1Δ cells in the presence of furfural will further reveal the detailed molecular mechanism of furfural resistance in the mutant.
Conclusions
In this study, RNAi knockdown of genes SIZ1 and GCN4 was demonstrated to improve furfural tolerance in S. cerevisiae. The siz1Δ mutant was further found to exhibit superior performance with cell growth, glucose consumption, furfural consumption and ethanol productivity as compared to the parent strain, while the gcn4Δ strain did not exhibit improved furfural tolerance. Deletion of SIZ1 also resulted in higher furfural tolerance in different S. cerevisiae strains, indicating SIZ1 deletion may play an important role in furfural resistance in S. cerevisiae strains. To our knowledge, this is a novel determinant of furfural resistance. Preliminary exploration of furfural tolerance in the siz1Δ mutant showed that the proteins responsible for furfural tolerance, among which GCN4p is a possible candidate, may be exclusively under the SUMO regulation by SIZ1p. Besides furfural tolerance, the siz1Δ mutant also exhibited tolerance towards oxidative stress, suggesting that proteins that are related to oxidative stress tolerance may be under the SUMO regulation of SIZ1p and responsible for furfural tolerance. These findings provide valuable insights into the engineering of furfural resistant microbes for efficient lignocellulose-based fermentation.
Methods
Strains and growth media
The strains and plasmids used in this study are listed in Table 3. Cells were grown in liquid SC [46], SC-URA or synthetic complete medium deficient in leucine (SC-LEU) supplemented with 20 g/L glucose as the carbon source or on solid 1% yeast extract, 2% peptone, 0.01% adenine hemisulfate, 2% glucose and 2% agar (YPAD) medium unless otherwise noted. The initial pH value of SC medium was adjusted to 5.6 using 12 M NaOH.
Table 3.
Strains and plasmids used in this study
| Strains or plasmids | Characteristics | Reference or source |
|---|---|---|
| Strains |
|
|
|
S. cerevisiae |
|
|
| BY4741 |
MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 |
[47] |
| BAD |
BY4741/δ::TEF1p-ago1-TPI1p-dcr1 |
This study |
| BAD-P |
BAD/pRS416-TTrcx |
This study |
|
SIZ1-kd |
BAD/pRS416-TTrcx-siz1 |
This study |
|
GCN4-kd |
BAD/pRS416-TTrcx-gcn4 |
This study |
|
siz1Δ |
BAD/siz1Δ::leu2 |
This study |
|
siz1Δ-P |
siz1Δ/pRS416e |
This study |
|
siz1Δ-SIZ1 |
siz1Δ/pRS416e-siz1 |
This study |
|
gcn4Δ |
BAD/gcn4Δ::leu2 |
This study |
|
siz2Δ |
BAD/siz2Δ::leu2 |
This study |
|
mms21Δ |
BAD/siz1Δ::leu2 |
This study |
|
cst9Δ |
BAD/cst9Δ::leu2 |
This study |
| BAD-YKL071W |
BAD/pRS416e-ykl071w |
This study |
| BAD-ZWF1 |
BAD/pRS416e-zwf1 |
This study |
| BAD-MSN2 |
BAD/pRS416e-msn2 |
This study |
| BAD-ALD6 |
BAD/pRS416e-ald6 |
This study |
| BAD-ADH7 |
BAD/pRS416e-adh7 |
This study |
| BAD-ARI1 |
BAD/pRS416e-ari1 |
This study |
|
siz1Δ-GCN4-kd |
siz1Δ/pRS416-TTrcx-gcn4 |
This study |
| HZ848 |
MATα, ade2-1, Δura3, his3-11, 15, trp1-1, leu2-3, 112, and can1-100 |
[23] |
| HZ848-siz1Δ |
HZ848/siz1Δ::ura3 |
This study |
| W303a |
MATa; ura3-1; trp1Δ 2; leu2-3,112; his3-11,15; ade2-1; can1-100 |
[24] |
| W303a-siz1Δ |
W303a/siz1Δ::hygromycin B |
This study |
|
E. coli |
|
|
| DH5α |
General cloning host |
Takara |
| WM1788 |
Cloning host |
Provided by Professor William Metcalf |
| Plasmids |
|
|
| pRS416 |
Yeast centromere with URA3 marker |
[48] |
| pRS425-TEF1p-PmeI-PGK1t |
Yeast gene expression vector |
[19] |
| pRS416e |
Derived from pRS416, with TEF1 promoter and PGK1 terminator added |
This study |
| pRS-delta-KanMX-LoxP-TEF1p-AGO1-PGK1t-TPI1p-DCR1-GPD1t |
Helper plasmid for integration of S. castellii RNAi pathway into delta-site |
[19] |
| pRS416-TTrc |
Derived from pRS416, with convergent promoters to produce dsRNA |
[19] |
| pRS416-TTrcx |
Derived from pRS416-TTrc, with XhoI restriction recognition sequence instead of BamHI |
This study |
| pRS416-TTrcx-siz1 |
Derived from pRS416-TTrcx, with gene SIZ1 fragment added |
This study |
| pRS416-TTrcx-gcn4 |
Derived from pRS416-TTrcx, with gene GCN4 fragment added |
This study |
| pRS415 |
Yeast centromere with LEU2 marker |
[48] |
| pUG6 |
The loxP-KanMX-loxP disruption module |
Euroscarf |
| pUG72 |
The loxP-URA3-loxP disruption module |
Euroscarf |
| pLHCX |
Template for amplification of hygromycin B resistance gene |
Clontech |
| pXZ5 |
Derived from pUG72, with hygromycin B resistance gene expression cassette instead of ura3 |
This study |
| pRS416e-siz1 |
Derived from pRS416e, with SIZ1 gene cassette added |
This study |
| pRS416e-ykl071w |
Derived from pRS416e, with YKL071W gene cassette added |
This study |
| pRS416e-zwf1 |
Derived from pRS416e, with ZWF1 gene cassette added |
This study |
| pRS416e-msn2 |
Derived from pRS416e, with MSN2 gene cassette added |
This study |
| pRS416e-ald6 |
Derived from pRS416e, with ALD6 gene cassette added |
This study |
| pRS416e-adh7 |
Derived from pRS416e, with ADH7 gene cassette added |
This study |
| pRS416e-ari1 | Derived from pRS416e, with ARI1 gene cassette added | This study |
Construction of plasmids, genome-wide RNAi library and reconstitution of RNAi machinery in S. cerevisiae BY4741
The primers used in this study are listed in Additional file 1: Table S1. Plasmid constructions are summarized in Additional file 1: Table S2. All plasmid construction was performed by In-fusion HD cloning (Clontech Laboratories, Inc., Mountain View, CA, USA) following the manufacturer’s instructions, or by the DNA assembler method [23]. Construction of the genomic library of S. cerevisiae BY4741 was carried out as previously described [19] with modifications to prevent self-ligation of vectors and fragments [20]. Finally, a library size of 2.6 × 106 transformants was obtained, while the control reaction with only linearized plasmid gave 4 × 104 transformants. The plasmid library was isolated from an overnight E. coli culture.
DNA transformation of S. cerevisiae strains
DNA transformation of S. cerevisiae strains was carried out using the method developed by Gietz and Schiestl [49].
RAGE screen for increased furfural tolerance
The RNAi library (20 μg) or control plasmid pRS416-TTrcx was transformed into the BAD strain harboring the RNAi machinery. A library size of 3.4 × 105 was achieved, ensuring >99% coverage of the yeast genome [22]. Following transformation, yeast cells were recovered in 1 mL YPAD medium for 4 h, washed with ddH2O and plated onto solid SC-URA medium containing 0.8 g/L furfural. The library and control plates were incubated at 30°C for 3 to 5 days. Thirty-three colonies of sizes bigger than the largest colonies on the control plates were picked from the library plates into SC-URA liquid medium. The growth performance of the selected colonies and control strain were compared in the presence of 0.8 g/L furfural. The initial OD600 for all the strains was 0.2, and the growth rate was measured after 24 h. The RNAi plasmids from the top 14 strains with OD600 values at least 20% higher than the control strain were isolated and amplified in E. coli. The selected plasmids were then individually retransformed, of which four were able to retain the enhanced furfural tolerance in a fresh genetic background with three biological replicates. The four plasmids were sequenced with the primer pRS416-TTrc-S (Additional file 1: Table S1).
Spot assay
Yeast cells in the stationary phase were transferred into 5 mL of SC media in a 15-mL round-bottom Falcon tube at an initial OD600 of 0.2 and grown to an OD600 of 0.7 (30°C, 250 rpm). The cells were serially diluted 10-fold with sterile water and 5 μl of each dilution was spotted onto furfural-free SC agar (control) and SC agar medium containing 0.8 g/L furfural. The plates were incubated at 30°C for 2 to 5 days.
Tolerance assay - calculation of maximum specific growth rates
Maximum specific growth rate was used as an indicator for the cellular tolerance towards various inhibitors [12,50,51]. For calculating the maximum specific growth rates of S. cerevisiae strains, stationary-phase cells grown in SC medium were transferred into 5 mL of SC medium containing a specific inhibitor (0.8 g/L furfural, 1.2 g/L furfural, 2.0 g/L furfural, 1.26 g/L HMF or 1.72 mg/L menadione) in a 15-mL round-bottom Falcon tube (30°C, 250 rpm). The initial OD600 was 0.2. The maximum specific growth rate was determined from the maximum slope of the OD600 values over time.
Fermentation
Batch fermentations were carried out as follows: a single colony grown on a YPAD plate was inoculated into 3 mL of SC medium containing 20 g/L glucose in a 15-mL round-bottom Falcon tube and grown until saturation (30°C, 250 rpm). About 400 μL of the stationary-phase cells were transferred into 25 mL of fresh SC media containing 0.8 g/L furfural in 250 mL non-baffled shake flasks. Cells were grown under oxygen-limited conditions (30°C, 100 rpm) as previously reported [46]. The initial OD600 was 0.2.
HPLC analysis
The samples were centrifuged and the supernatants were diluted five to ten times before HPLC analysis. An Agilent 1100 series HPLC (Agilent Technologies, Palo Alto, CA, USA) coupled with an Agilent ZORBAX 80A Extend-C18 column was used for detection of furfural and furfuryl alcohol. HPLC parameters were as follows: solvent A, water; solvent B, acetonitrile; 5% B for 15 minutes, then 100% B for 5 minutes, followed by 5% B for 5 minutes; flow rate 1 mL/minute; detection by UV spectroscopy at 277 nm (furfural) or 210 nm (furfuryl alcohol). Under such conditions, furfural and furfuryl alcohol were eluted at 6.7 minutes and 5.6 minutes, respectively. An HPLC system equipped with a refractive index detector (Shimadzu Scientific Instruments, Columbia, MD, USA) was used to analyze the concentrations of glucose and ethanol in the broth. To separate glucose and ethanol, an HPX-87H column (BioRad, Hercules, CA, USA) was used as described [46].
Abbreviations
5-HMF: 5-hydroxymethylfurfural; OD: optical density; NAPDH: reduced nicotinamide adenine dinucleotide phosphate; RAGE: RNAi-assisted genome evolution; RNAi: RNA interference; ROS: reactive oxygen species; SC: synthetic complete; SC-LEU: synthetic complete medium deficient in leucine; SC-URA: synthetic complete medium deficient in uracil; SUMO: small ubiquitin-like modifier.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All the experiments were performed by HX. Both authors contributed to designing the experiments, writing the manuscript and have approved the final manuscript
Supplementary Material
Sequencing of 17 randomly picked plasmids from the RNAi library. Locations have been mapped to the S. cerevisiae genome. Each column represents one chromosome, the height of which is proportional to the size of the indicated chromosome. Each horizontal bar indicates the location of a fragment. Figure S2. Sequencing result of pRS416-TTrcx-siz1, which contains a fragment of gene SIZ1 (underlined). Figure S3. Sequencing result of pRS416-TTrcx-gcn4, which contains a fragment of gene GCN4 (underlined). Table S1. Primers used in this study. Table S2. Construction of plasmids. Table S3. Maximum specific growth rates of strain BAD and its derivatives cultured in SC medium containing 20 g/L glucose. Table S4. Maximum specific growth rates of strain BAD and its derivatives cultured in SC medium containing different concentrations of furfural. Table S5. Fermentation parameters and estimation of carbon balance in strain BAD and siz1Δ after 30 h in SC medium containing 20 g/L glucose and 0.8 g/L furfural.
Contributor Information
Han Xiao, Email: hanxiao@illinois.edu.
Huimin Zhao, Email: zhao5@illinois.edu.
Acknowledgements
The project was supported by the Center for Industrial Biotechnology at the University of Illinois at Urbana-Champaign. We thank Dr William Metcalf (Department of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, USA) for providing E. coli strain WM1788, Dr Mingzi M Zhang (Metabolic Engineering Research Laboratory, Institute of Chemical and Engineering Sciences, Agency for Science, Technology and Research, Singapore), Mr Tong Si, Mr Ryan E Cobb, Dr. Jing Liang (Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, Urbana, USA), Dr Xueyang Feng, Mr Jiazhang Lian and Ms Sijin Li (Department of Chemical and Biomolecular Engineering, Institute for Genomic Biology, University of Illinois at Urbana-Champaign) for helpful suggestions and thoughtful discussions.
References
- Han K-H, Ko J-H, Yang SH. Optimizing lignocellulosic feedstock for improved biofuel productivity and processing. Biofuel Bioprod Bior. 2007;1:135–146. doi: 10.1002/bbb.14. [DOI] [Google Scholar]
- Kumar R, Singh S, Singh OV. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 2008;35:377–391. doi: 10.1007/s10295-008-0327-8. [DOI] [PubMed] [Google Scholar]
- Pienkos PT, Zhang M. Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose. 2009;16:743–762. doi: 10.1007/s10570-009-9309-x. [DOI] [Google Scholar]
- Jeong TS, Um BH, Kim JS, Oh KK. Optimizing dilute-acid pretreatment of rapeseed straw for extraction of hemicellulose. Appl Biochem Biotech. 2010;161:22–33. doi: 10.1007/s12010-009-8898-z. [DOI] [PubMed] [Google Scholar]
- Delgenes JP, Moletta R, Navarro JM. Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzyme Microb Technol. 1996;19:220–225. doi: 10.1016/0141-0229(95)00237-5. [DOI] [Google Scholar]
- Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol. 2009;75:6132–6141. doi: 10.1128/AEM.01187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist E, Grage H, Meinander NQ, Hahn-Hagerdal B. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng. 1999;63:46–55. doi: 10.1002/(SICI)1097-0290(19990405)63:1<46::AID-BIT5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Heer D, Sauer U. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol. 2008;1:497–506. doi: 10.1111/j.1751-7915.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussatto SI, Roberto IC. Alternatives for detoxification of diluted-acid lignocellulosic hydrolyzates for use in fermentative processes: a review. Bioresour Technol. 2004;93:1–10. doi: 10.1016/j.biortech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels. 2010;3:2. doi: 10.1186/1754-6834-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FM, Qiao B, Yuan YJ. Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol. 2009;75:3765–3776. doi: 10.1128/AEM.02594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SE, Koo HM, Park YK, Park SM, Park JC, Lee OK, Park YC, Seo JH. Expression of aldehyde dehydrogenase 6 reduces inhibitory effect of furan derivatives on cell growth and ethanol production in Saccharomyces cerevisiae. Bioresour Technol. 2011;102:6033–6038. doi: 10.1016/j.biortech.2011.02.101. [DOI] [PubMed] [Google Scholar]
- Liu ZL. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol. 2011;90:809–825. doi: 10.1007/s00253-011-3167-9. [DOI] [PubMed] [Google Scholar]
- Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels. 2009;2:26. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Liu ZL. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics. 2010;11:660. doi: 10.1186/1471-2164-11-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer D, Heine D, Sauer U. Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl Environ Microbiol. 2009;75:7631–7638. doi: 10.1128/AEM.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZL, Moon J. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene. 2009;446:1–10. doi: 10.1016/j.gene.2009.06.018. [DOI] [PubMed] [Google Scholar]
- Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol. 2009;75:4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si T, Luo Y, Bao Z, Zhao H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Syn Bio. 2014. doi:10.1021/sb500074a. [DOI] [PubMed]
- Liu HP. Constructing yeast libraries. Method Enzymol. 2002;350:72–86. doi: 10.1016/s0076-6879(02)50956-3. [DOI] [PubMed] [Google Scholar]
- Online Sequence Analysis Tools. http://tools.neb.com/~posfai/project/Yeast_Digests.html.
- Statistics of Randomized Library Construction. http://guinevere.otago.ac.nz/mlrgd/STATS/
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H, Takagi H. Overexpression of the yeast transcription activator MSN2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng. 2012;113:451–455. doi: 10.1016/j.jbiosc.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2006;71:339–349. doi: 10.1007/s00253-005-0142-3. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes CC, Peterson JJ, Roslander C, Zacchi G, Mullinnix MT, Shanmugam KT, Ingram LO. Optimizing the saccharification of sugar cane bagasse using dilute phosphoric acid followed by fungal cellulases. Bioresource Technol. 2010;101:1851–1857. doi: 10.1016/j.biortech.2009.09.070. [DOI] [PubMed] [Google Scholar]
- Kim IS, Sohn HY, Jin I. Adaptive stress response to menadione-induced oxidative stress in Saccharomyces cerevisiae KNU5377. J Microbiol. 2011;49:816–823. doi: 10.1007/s12275-011-1154-6. [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng. 1999;65:24–33. doi: 10.1002/(SICI)1097-0290(19991005)65:1<24::AID-BIT4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that GCN4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/S0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T, Mullen JR, Slagle CE, Brill SJ. Stimulation of in vitro sumoylation by SLX5-SLX8: evidence for a functional interaction with the SUMO pathway. DNA Repair. 2007;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL. Sumoylation of transcription factor GCN4 facilitates its SRB10-mediated clearance from promoters in yeast. Genes Dev. 2012;26:350–355. doi: 10.1101/gad.184689.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- Srikumar T, Lewicki MC, Raught B. A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol Syst Biol. 2013;9:668. doi: 10.1038/msb.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger JM, Schmitt AP, Simon JR, McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:26875–26879. doi: 10.1074/jbc.273.41.26875. [DOI] [PubMed] [Google Scholar]
- Wong CM, Zhou Y, Ng RWM, Kung HF, Jin DY. Cooperation of yeast peroxiredoxins TSA1p and TSA2p in the cellular defense against oxidative and nitrosative stress. J Biol Chem. 2002;277:5385–5394. doi: 10.1074/jbc.M106846200. [DOI] [PubMed] [Google Scholar]
- Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae SKO1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- Izawa S, Maeda K, Miki T, Mano J, Inoue Y, Kimura A. Importance of glucose-6-phosphate dehydrogenase in the adaptive response to hydrogen peroxide in Saccharomyces cerevisiae. Biochemical J. 1998;330(Pt 2):811–817. doi: 10.1042/bj3300811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yuan Y, Si T, Lian J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40:e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Kelly C, Jones O, Barnhart C, Lajoie C. Effect of furfural, vanillin and syringaldehyde on Candida guilliermondii growth and xylitol biosynthesis. Appl Biochem Biotechnol. 2008;148:97–108. doi: 10.1007/s12010-007-8103-1. [DOI] [PubMed] [Google Scholar]
- Otero JM, Cimini D, Patil KR, Poulsen SG, Olsson L, Nielsen J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. Plos One. 2013;8:e54144. doi: 10.1371/journal.pone.0054144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing of 17 randomly picked plasmids from the RNAi library. Locations have been mapped to the S. cerevisiae genome. Each column represents one chromosome, the height of which is proportional to the size of the indicated chromosome. Each horizontal bar indicates the location of a fragment. Figure S2. Sequencing result of pRS416-TTrcx-siz1, which contains a fragment of gene SIZ1 (underlined). Figure S3. Sequencing result of pRS416-TTrcx-gcn4, which contains a fragment of gene GCN4 (underlined). Table S1. Primers used in this study. Table S2. Construction of plasmids. Table S3. Maximum specific growth rates of strain BAD and its derivatives cultured in SC medium containing 20 g/L glucose. Table S4. Maximum specific growth rates of strain BAD and its derivatives cultured in SC medium containing different concentrations of furfural. Table S5. Fermentation parameters and estimation of carbon balance in strain BAD and siz1Δ after 30 h in SC medium containing 20 g/L glucose and 0.8 g/L furfural.


