Abstract
Background
Mutations in epigenetic modifiers were reported in patients with acute myeloid leukaemia (AML) including mutations in DNA methyltransferase 3A gene (DNMT3A) in 20%-30% patients and mutations in isocitrate dehydrogenase 1/2 gene (IDH1/2) in 5%-15% patients. Novel studies have shown that mutations in DNMT3A and IDH1/2 influence prognosis, indicating an increasing need to detect these mutations during routine laboratory analysis. DNA sequencing for the identification of these mutations is time-consuming and cost-intensive. This study aimed to establish rapid screening tests to identify mutations in DNMT3A and IDH1/2 that could be applied in routine laboratory procedures and that could influence initial patient management.
Methods
In this study we developed an endonuclease restriction method to identify the most common DNMT3A mutation (R882H) and an amplification-refractory mutation system (ARMS) to analyse IDH2 R140Q mutations. Furthermore, we compared these methods with HRM analysis and evaluated the latter for the detection of IDH1 mutations.
Results
Of 230 samples from patients with AML 30 (13%) samples had DNMT3A mutations, 16 (7%) samples had IDH2 R140Q mutations and 36 (16%) samples had IDH1 mutations. Sensitivity assays performed using serial dilutions of mutated DNA showed that ARMS analysis had a sensitivity of 4.5%, endonuclease restriction had a sensitivity of 0.05% and HRM analysis had a sensitivity of 5.9%–7.8% for detecting different mutations. HRM analysis was the best screening method to determine the heterogeneity of IDH1 mutations. Furthermore, for the identification of mutations in IDH2 and DNMT3A, endonuclease restriction and ARMS methods showed a perfect concordance (100%) with Sanger sequencing while HRM analysis showed a near-perfect concordance (approximately 98%).
Conclusion
Our study suggested that all the developed methods were rapid, specific and easy to use and interpret. HRM analysis is the most timesaving and cost-efficient method to rapidly screen all the 3 genes at diagnosis in samples obtained from patients with AML. Endonuclease restriction and ARMS assays can be used separately or in combination with HRM analysis to obtain more reliable results. We propose that early screening of mutations in patients with AML having normal karyotype could facilitate risk stratification and improve treatment options.
Keywords: AML, DNMT3A, IDH1/2 mutations, DNA sequencing, HRM analysis, ARMS PCR, Endonuclease restriction
Background
Acute myeloid leukaemia (AML) is a clonal disorder characterised by the accumulation of myeloid cells and impairment of normal haematopoiesis [1]. The recent large-scale sequencing of AML genomes is now providing opportunities for patient stratification and personalised approaches to treatments that are based on an individual’s mutation profiles [1-3]. A few recurring gene mutations and overexpressed genes having prognostic relevance in AML have been identified and incorporated in the current prognostication models.
Recently, a new class of mutations affecting genes for DNA methylation and post-translational histone modification was identified in AML. These mutations frequently occur in the DNA nucleotide methyltransferase 3A gene (DNMT3A) [4-8] and isocitrate dehydrogenase 1/2 gene (IDH1/2) (isocitrat dehydrogenase 1/2) [9-13]. DNMT3A belongs to the mammalian methyltransferase gene family, which also includes DNTM1, DNMT3B and DNMT3L. Methyltransferases modify methylation patterns by enzymatically adding a methyl group to cytosine residues in CpG islands and are involved in tissue-specific gene expression [4,14]. Studies in different AML cohorts have reported the incidence of DNMT3A mutations in up to 22% de novo AML and 36% cytogenetically normal AML samples [5,6]. Nonsense, frameshift and missense mutations commonly occur in DNMT3A; however a point mutation at position R882 is the most frequently (40%–60%) observed mutation [7]. In vitro studies suggest that mutations at this position disturb the formation of heterodimers with DNMT3L, thereby preventing the catalytic activity of DNMT3A. Different studies have shown a negative impact of DNMT3A mutation on outcomes in patients with AML [3,15-19]. Prognostic effect is known to depend on certain biological factors as well as a combination of cytogenetics and other mutations such as those in FLT3 and NPM1[3,6,8].
Somatic mutations in IDH1/2 occur in 5–30% patients with AML and are commonly associated with nucleophosmin 1 (NPM1) mutations [9,10]. Both the genes play a critical role in the citric acid cycle IDH1 in the cytoplasm and peroxisome and IDH2 in the mitochondria. Both IDH1 and IDH2 promote the conversion of isocitrate to α-ketoglutarate (α-KG) that is associated with the reduction of nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH [8,11,20]. Mutations in IDH1 and IDH2 are heterozygous and occur in highly conserved arginine residues (IDH1 R132 and IDH2 R140/R172). Mutations at IDH2 R140 always result in the conversion of arginine to glutamine, whereas substitutions at IDH1 R132 and IDH2 R172 result in a wide range of amino acid replacements [12]. All point mutations in IDH1/2 lead to a gain of function, enabling the conversion of α-KG to 2-hydroxyglutarate (2-HG) and oxidation of NADPH to NADP+. Furthermore, an increase in 2-HG-levels leads to the functional impairment of α-KG-dependent enzymes through competitive inhibition [13].
In contrast to the impact of DNMT3A mutations, the impact of IDH1/2 mutations on prognosis is not completely understood. It appears that prognosis may depend on specific patient populations and a combination with NPM1 mutations [21-23].
The increasing evidence of high incidence particularly in cytogenetically normal AML and prognostic pertinence of DNMT3A and IDH1/2 mutations support the need to identify these mutations in routine diagnostic screening. Importantly, the presence of DNMT3A and IDH1/2 mutations may confer sensitivity to novel therapeutic approaches, including demethylating agents [24,25].
The current available methods like direct sequencing are informative but time consuming and cost intensive. In this study, we validated the polymerase chain reaction (PCR)-based high resolution melt (HRM) assay for screening DNMT3A, IDH1 and IDH2 mutations in samples obtained from patients with AML at diagnosis and developed 2 rapid methods for detecting more common mutations, DNMT3A R882H and IDH2 R140Q. We evaluated the utility of endonuclease restriction-based detection method to identify mutations in DNMT3A and designed an amplification-refractory mutation system (ARMS) to detect mutations in IDH2. In addition we compared both the systems with the HRM assay for all the studied mutations.
Methods
Patient characteristics
Bone marrow (BM) samples from 230 patients with newly diagnosed AML were included in the study. All patients were treated at the University Clinic Charité, Campus Benjamin Franklin, from May 2000 to July 2013. Patient’s characteristics are summarised in the Additional file 1: Table S1. The male/female ratio of the study population was 116/114, and the median age was 57 years (range, 16–94 years). Diagnoses were established according to the WHO criteria [26]. Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki and the ethical guidelines of the Charite University School of Medicine, which approved this study.
DNA extraction
Mononuclear cells from BM aspirates were isolated using Ficoll density gradient centrifugation as described [27]. DNA was extracted using Allprep DNA/RNA mini kit (Qiagen) as per the manufacturer’s instructions.
ARMS analysis of IDH2-R140Q mutations
All primers were designed using Primer 3 Software (Additional file 2: Table S2). ARMS analysis was performed using 2 control primers flanking exon 23 and 2 allele-specific primers IDH2-RI and IDH2-FI that are complementary to the wild-type (wt) and mutated alleles, respectively. To enhance specificity, both the primers had an additional medium mismatch at the preliminary base. The PCR mixture and reaction conditions are specified in the Additional file 3: PCR reaction mixtures and conditions. The generated PCR products were analysed on a 1.5% agarose gel.
Endonuclease restriction analysis of DNMT3A-R882H mutations
PCR amplification for endonuclease restriction analysis was conducted using primers DNMT3A-ResF/R (Additional file 2: Table S2). PCR reaction mixture was prepared as that described for ARMS assay. The reaction conditions are specified in the Additional file 3. In all, 10 μl of the PCR product was directly applied for endonuclease treatment with 1 μl Fnu4HI and 5 μl of CutSmart Buffer (New England Biolabs). After incubation at 37°C for 15 min products were analysed on a 1.5% agarose gel containing 10% ethidium bromide (voltage 150 V).
HRM assay
The reaction mixture and HRM conditions are specified in the Additional file 3. The analysis was performed in a Rotor Gene 6000 Real-Time PCR Cycler (Qiagen). Samples, including a control sample for each mutation and wt allele, were analysed in duplicates. For DNMT3A and IDH2, the wt allele was used for normalisation, while for IDH1 R132C mutation control was used as the baseline. Normalisation regions for the optimal detection of DNMT3A were 82°C-83°C (leading range) and 87°C-88°C (trailing range), for the optimal detection of IDH1 were 73°C-74°C (leading range) and 82°C-83°C (trailing range) and for the optimal detection of IDH2 were 77°C-78°C (leading range) and 87°C-88°C (trailing range). Confidence threshold was set to 70% for all the genes.
DNA sequencing
All the primers used for sequencing are listed in the Additional file 2: Table S2. All PCR reaction conditions are specified in the Additional file 3. The obtained products were purified using the PCR Purification Kit (Qiagen), as described in the manual. Sequencing reaction was performed using Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequencing products were purified using DyeEx 2.0 Spin Kit (Qiagen) according to the manufacturer’s instructions. The purified products were diluted with 18 μl HiDi-Formamid (Applied Biosystems), incubated at 95°C for 3 min and chilled on ice for 3 min. Sequencing was performed using ABI310 Genetic Analyser (Applied Biosystems), and data were collected using ABI Prism 310 Data Collection Software.
Results and discussion
All the positive and negative controls used in this study were selected by Sanger sequencing of patients’ samples. The results obtained using endonuclease restriction, ARMS and HRM were verified with those obtained using Sanger sequencing to determine the specificity of the assays. Sensitivity was measured as the minimal percentage of mutated allele in a sample detected by the assay. The initial portion of mutation was determined using Sanger sequencing.
DNMT3A mutation analysis
Endonuclease restriction analysis identified DNMT3A R882H G>A mutations in 28 out of 230 patients with AML (12.2%) and HRM analysis identified 2 additional R882X G>C mutations (0.9%), which are consistent with the frequency published by Lin et al. [28]. The age of the patients ranged from 24 to 87 years (median, 58 years). Among these patients, 53% had a normal karyotype. None of the patients in the prognostic favourable group had DNMT3A mutations. Of 30 patients, 16 had FLT3 mutations.
Figure 1 provides a representative result of restriction analysis with 5 positive and 2 negative samples. Point mutation at R882H (GCCGC to GCCAC) led to the loss of one recognition site of Fnu4HI, thus creating a larger 289 bp fragment. Because of heterozygosity, the 190 bp wt fragment and the smaller 114 bp fragment are present in every sample. Sensitivity of the assay was analysed using serial dilutions of wt and DNMT3A R882H-mutated DNA (initial mutation ratio in Sanger sequencing was 59%, Figure 2.1). The fragment containing the mutation was explicitly apparent with a mutational content of 0.05%, indicating a very high sensitivity of the assay. In addition mutations in exon 23 of DNMT3A were detected using HRM analysis. Results of HRM analysis were plotted as a difference in the fluorescence of the tested sample versus that of a wt control (normalisation line), referred to as a temperature-shifted difference plot (Figure 3.1). Discrepancies between mutated and wt samples could also be observed in the melting plot profiles. Sample containing R882H mutation showed 2 peaks at 84.5°C and 85.6°C, whereas the wt samples showed only 1 peak at 85.7°C. Compared to the wt allele, R882X allele was slightly shifted to the left, with a melting temperature of 85.6°C (Figure 3.2). Sensitivity of the HRM assay was assessed similar to that of restriction analysis. The assay had high confidence (97%-99%) for the mutated allele up to a mutation ratio of 5.9% (Figure 2.2). Lower mutation ratios could not be assigned as positive and were identified as false negative with a confidence of 92%-98%. Thus our results indicated that HRM analysis had a lower sensitivity than that of endonuclease restriction analysis but had the benefit of identifying different mutations in 1 PCR reaction.
Figure 1.
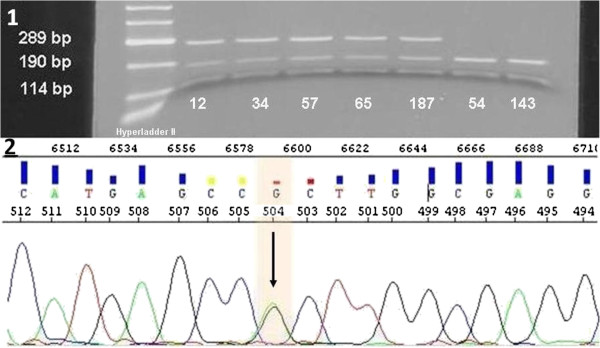
Restriction analysis of DNMT3A R882H mutations. 1) Agarose gel analysis of restricted products of 5 positive (12, 34, 57, 65, 187) and 2 negative (54, 143) patients. Wt samples showed 2 bands at 190 bp and 114 bp. Positive samples showed 3 bands at 289 bp, 190 bp, 114 bp because of the loss of a restriction site of Fnu4HI caused by the mutation. Hyperladder II (Bioline) was used as the marker. 2) Representative sequence analysis of patient 187 showing heterozygote mutation CGC to CAC.
Figure 2.
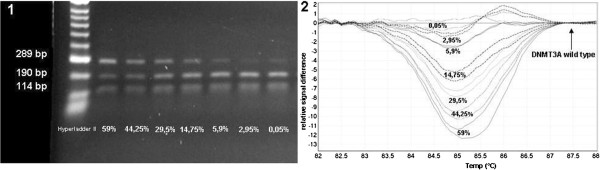
Sensitivity analysis of DNMT3A R882H detection. 1) Endonuclease restriction analysis of serial dilutions of DNMT3A R882H; Undiluted mutation ratio was 59% (estimated by sequencing). Mutated allele wa detected up to a degree of 0.05%. 2) Difference plot for HRM analysis of serial dilutions of DNMT3A R882H: Correct estimation was possible up to a mutation ratio of 5.9%; lower mutation ratios were identified false-negative. Normalisation was performed to the wt allele.
Figure 3.
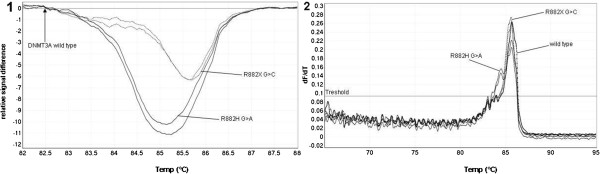
HRM analysis of DNMT3A mutations. 1) Difference plot for HRM analysis of DNMT3A R882H G>A and R882X G>C mutations. Normalisation was performed to the wt allele. R882X showed a right-shifted peak compared to R882H. 2) Melting curve profiles of DNMT3A R882H G>A, R882X G>C and wt allele. Vertical axis corresponds to changes in the fluorescence signal over time (dF/dT). R882H G>A displayed 2 peaks (84.5°C and 85.6°C), while the wt allele had only one peak at 85.7°C. R882X G>C had a left shifted peak at 85.6°C.
IDH2 mutation analysis
The mutational frequency of IDH2 R140Q G>A was 6.69% (16 out of 230 patients with AML), which was similar to the frequency published by Paschka et al. [23] and other studies [29,30]. Most patients with AML with IDH2 mutations were older than 50 years and had de novo AML and a normal karyotype. Of 16 patients, 7 had an NPM1 mutation.
The ARMS analysis allowed differentiation between mutated and wt DNA of IDH2 through specific differences in the amplification properties of the reaction. In the presence of a mutation the PCR reaction generated 3 different fragments with sizes 613 bp (control band), 446 (mutation band) and 233 bp (wt band, Figure 4.1). No 446 bp mutation band was detected in the wt samples and results were confirmed by sequencing (Figure 4.2). In addition some faint unspecific bands of size ≥613 bp were detected. Given that the diagnostic approach was not handicapped, the assay was acceptable for further applications. HRM screening of IDH2 showed no additional mutations in our AML patient group. IDH2 amplification showed a bimodal melting profile with a smaller peak at 79.8°C and a bigger peak at 82.7°C. Differences in mutated and wt allele were visible during melting point analysis, because IDH2 R140Q mutations shifted to lower temperatures than those in wt allele (Figure 5). Sensitivity tests were performed as those described for DNMT3A. For the IDH2 allele, the sensitivities of ARMS and HRM analyses were the same, with a limitation at a mutational ratio of 2.25%. As shown in Figure 6.1 the mutation band at 446 bp was not present in dilutions with 2.25% and 0.45% mutated DNA. Furthermore, HRM analysis showed that dilutions from 45% to 4.5% were clearly positive with a confidence ranging from 77.68% to 98.41%, while the last 2 dilutions were false-negative, with a confidence of 82% to 94.39% (Figure 6.2).
Figure 4.
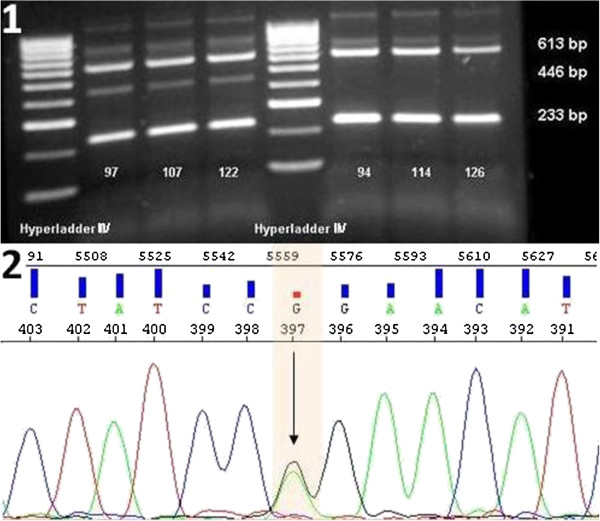
ARMS analysis of IDH2 R140Q mutation. 1) Agarose gel analysis of PCR products of 3 positive (97, 107, 122) and 3 negative (94, 114, 126) patients. All patients showed control (613 bp) and wt (233 bp) bands, while only the positive patients showed a product at 446 bp. Hyperladder II (Bioline) was used as the marker. 2) Representative sequence analysis of patient 97 showing the heterozygote mutation CGG to CAG.
Figure 5.
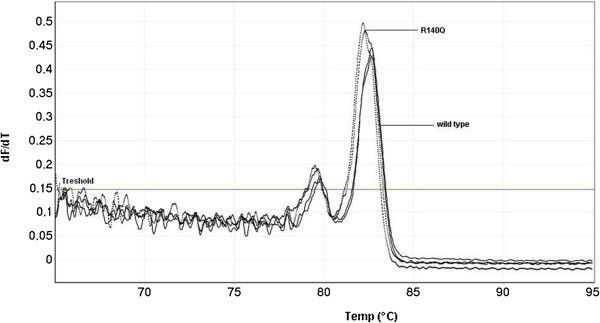
Melting curve profiles of wt allele and IDH2 140Q G>A. Vertical axis corresponds to changes in the fluorescence signal over time (dF/dT). IDH2 analysis showed a bimodal peak; R140Q was shifted to lower temperatures compared to the wt allele.
Figure 6.
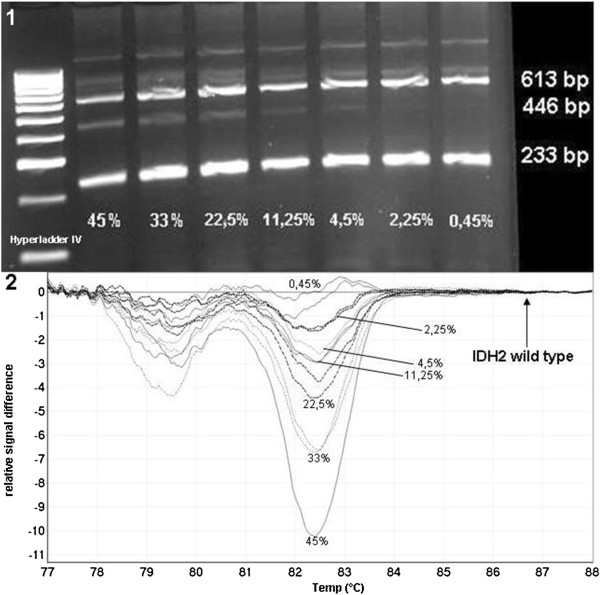
Sensitivity analysis of IDH2 R140Q detection. 1) Serial dilutions of IDH2 R140Q: Undiluted mutation ratio was 45% (estimated by sequencing). Mutated allele was detected up to a degree of 4.5%. 2) Difference plot for HRM analysis of serial dilutions of IDH2 R140Q: Correct estimation was possible up to a mutation ratio of 4.5%; lower mutation ratios were identified false-negative. Normalisation was performed to the wt allele.
IDH1 mutation analysis
An assay to detect specific mutations is not applicable because of the heterogeneity of IDH1 aberrations. Therefore, the HRM assay was evaluated for IDH1, as previously described by Patel et al. [30]. Mutated and wt IDH1 was distinguished through their melting profiles because mutated DNA had a melting point between 80.3°C and 80.5°C while wt IDH1 had a melting point of 81°C (Figure 7.1). However, the distinction between the different mutations of IDH1 was difficult with this analysis as well as with the differentiation plot normalised to the wt control (Figure 7.2). During this study we observed that the temperature-shifted difference plot normalised to R132S C>A control sample was the best to determine different IDH1 mutations (Figure 7.3). Thus, we performed sensitivity tests for G105 C>T and R132C C>T with normalisation to R132S C>A and for R132S C>A with normalisation to G105 C>T (Figure 8). HRM analysis showed sensitivity of 6%-7.8% for all three mutations. Using this method, we determined that 36 out of 230 (15.65%) patients with AML had IDH1 mutations. Of these 19 (8.3%) had G105 C>T, 11 (4.8%) had R132C C>T and 6 (2.6%) had R132S C>A; this frequency is consistent with the data published by Nomdedéu et al. [22,29].
Figure 7.
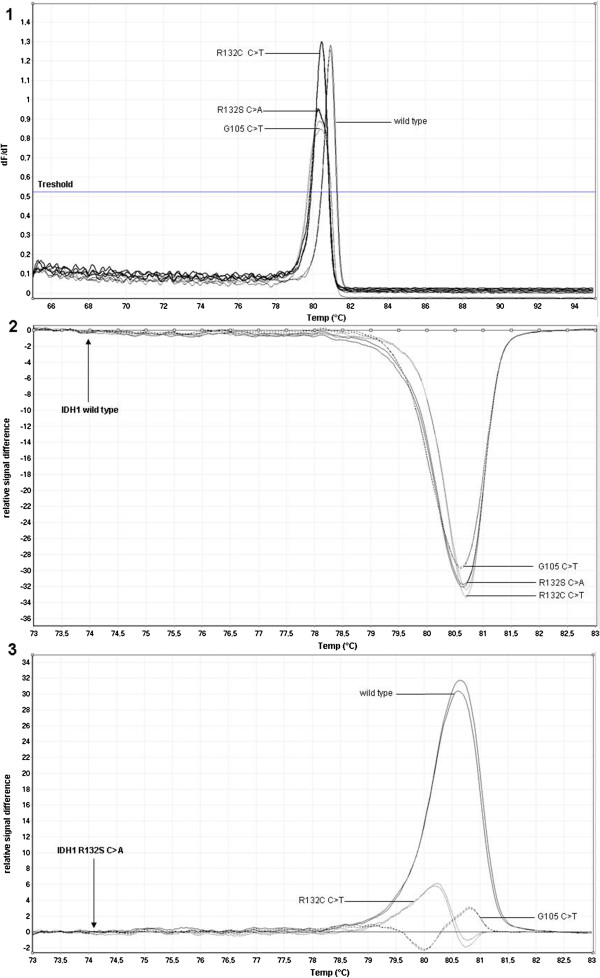
HRM analysis of IDH1 mutations. 1) Melting curve profiles of IDH1 mutated and wt alleles. Vertical axis corresponds to changes in the fluorescence signal over time (dF/dT). Mutated alleles were shifted to lower temperatures, but differentiation between different mutations was not possible. 2) Difference plot for HRM analysis of IDH1 mutations normalised to wt allele, discrimination of different mutations was difficult because of similar graphs. 3) Difference plot for HRM analysis of IDH1 mutations normalised to the R132S C>A allele, determination of different mutations was easier because of clearly separated graphs.
Figure 8.
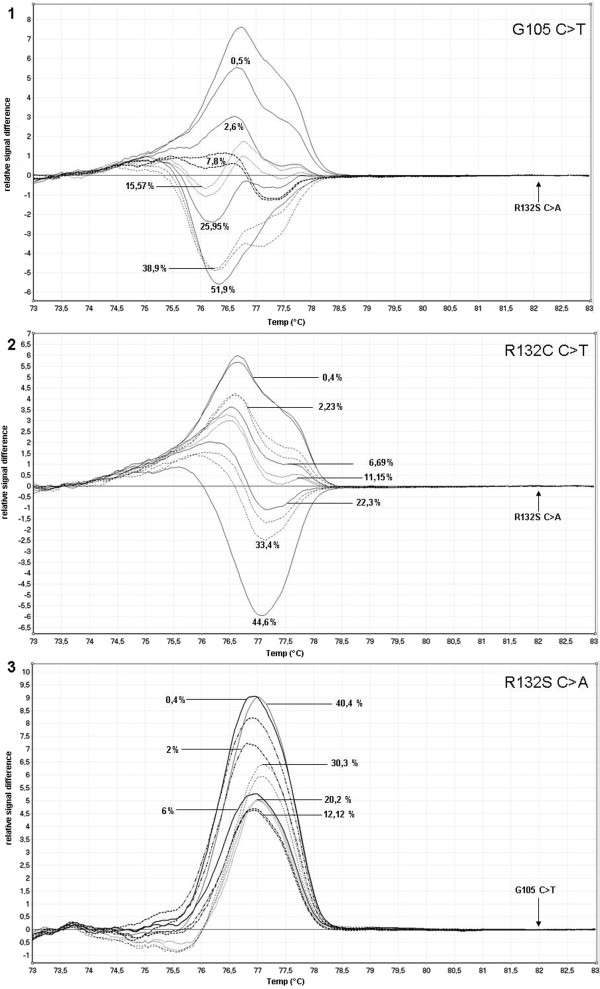
Sensitivity analysis of different IDH1 mutations. 1) Difference plot for HRM analysis of serial dilutions of IDH1 G105 C>T: Undiluted mutation ratio was 51.9% (estimated by sequencing). Correct estimation was possible up to a mutation ratio of 7.8%; lower mutation ratios were identified false-negative. Normalisation was performed to the R132S C>A allele. 2) Difference plot for HRM analysis of serial dilutions of IDH1 R132C C>T: Undiluted mutation ratio was 44.6% (estimated by sequencing). Correct estimation was possible up to a mutation ratio of 6.69%; lower mutation ratios were identified false-negative. Normalisation was performed to the R132S C>A allele. 3) Difference plot for HRM analysis of serial dilutions of IDH1 R132S C>A: Undiluted mutation ratio was 40.4% (estimated by sequencing). Correct estimation was possible up to a mutation ratio of 6%, lower mutation ratios were identified false-negative. Normalisation was performed to the G105 C>T allele.
Combination of different methods is essential to identify DNMT3A and IDH1/2 mutations in routine laboratory analyses
Both the assays designed in this study for the detection of DNMT3A R882H and IDH2 R140Q mutations were completely compliant with Sanger sequencing and had a high specificity. No false-positive results were determined with HRM analysis. Two (0.9%) samples showed variations for DNMT3A but were subsequently determined as wt by endonuclease restriction and sequencing. IDH1 analysis with HRM showed that 6 (2.6%) samples had inaccuracies in melting profiles and hence were determined false negative with this method. Sequencing showed the presence of a R132C C>T mutation in this samples. IDH2 analysis showed no discrepancies with Sanger sequencing.
Compared to Sanger sequencing, HRM analysis represents a timesaving, cost-efficient and more sensitive method to screen mutations in patients with AML at diagnosis. However, an efficient application presumes the presence of specific mutations and wt control samples. Because of the lack of cell lines with DNMT3A, IDH2 and IDH1 mutations, controls have to be established by sequencing different patient samples. Therefore, an effective application of HRM depends on the identification of high amounts of good-quality control samples, availability of a sequencer and HRM competent real-time PCR cycler. In addition, some results obtained with HRM analysis are difficult to interpret because of the variations in the melting curve of 1 mutation and can lead to uncertain conclusions or false-negative results [31]. Because new studies indicate the prognostic significance of IDH1/2 and DNMT3A mutations, which affect the choice of therapy, a steady laboratory diagnosis is essential [10,17,18,21,22,32]. We developed ARMS-PCR to identify IDH2 R140Q mutation and endonuclease restriction analysis to identify DNMT3A R882H mutations; both these methods are rapid and easy to use and interpret. Thus, these methods can be used to verify unclear results obtained using HRM analysis. In addition, these methods provide a possibility to identify the most common mutations in DNMT3A and IDH2 in laboratories that do not have HRM-competent real-time PCR cyclers at their disposal. Secondary endonuclease restriction has higher sensitivity than HRM analysis that allows earlier identification of mutations at relapse during follow-up analysis [33]. For future applications this assay could also be adapted to the quantitative PCR (qPCR) technique. The forward primer can be modified to amplify only the genomic region containing the restriction position that is lost in the mutated state, thus allowing the exclusion of wt and mutated alleles as well as the quantitative assessment of DNMT3A mutation. The main characteristics of all the methods analysed in this study are summarised in Table 1.
Table 1.
Comparative characteristics of all the methods used in this study
| |
DNMT3A
|
IDH2
|
IDH1
|
|||||
|---|---|---|---|---|---|---|---|---|
| Restriction endonuclease | HRM | Sanger sequencing | ARMS | HRM | Sanger sequencing | HRM | Sanger sequencing | |
| Sensitivity*, % |
0.05 |
5.9 |
10 |
4.5 |
4.5 |
10 |
6 to 7.8 |
10 |
| Turnaround time, days |
1 |
1 |
2 to 3 |
1 |
1 |
2 to 3 |
1 |
2 to 3 |
| Technician time, hours |
4 |
3.5 |
10 to 12 |
3 |
3.5 |
10 to 12 |
3.5 |
10 to 12 |
| Cost of diagnosis method, € |
32.13 |
28 |
122 |
44.16 |
28 |
122 |
28 |
122 |
| Interpretation |
Easy |
Medium -difficult |
Medium |
Easy |
Medium -difficult |
Medium |
Medium -difficult |
Medium |
| Identification of different/rare mutations |
No |
Yes |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
| Special equipment | PCR cycler | HRM real time PCR cycler | Sequencer | PCR cycler | HRM real time real time PCR cycler | Sequencer | HRM real time real time PCR cycler | Sequencer |
*Sensitivity was measured as the minimal percentage of mutated allele in a sample detected by the assay.
The measured sensitivities depend on assay conditions and equipment. For example, small amounts of non-specific amplicons and different salt or inhibitory concentrations can influence assay sensitivity [34,35]. Therefore, each laboratory should validate the presented methods with their equipment before application. Both HRM analysis and ARMS-PCR had only low sensitivity, which possibly could lead to false-negative results. Therefore, low mutational ratios could be overlooked and these patients would receive an imprecise laboratory diagnostic report. Potential reduction of amplicon size for both HRM and ARMS analyses could optimise sensitivities [36]. Moreover, adaption of the qualitative endonuclease restriction assay to a quantitative assay could further increase sensitivity and provide objective measurements of mutated cells [37].
In the future, sensitivity limitations for screening DNMT3A and IDH1/2 mutations can be overcome by using allele-specific next-generation sequencing (NGS). This method provides high multiplexing possibilities together with high sensitivity and broad spectrum of detected mutations [38]. However NGS is associated with high costs, high hands-on time and high computational expertise. Because standardisation and validation of NGS can be challenging establishment of this method is an ongoing process in laboratory routine [39]. Conventional PCR-based methods are easy to standardise and validate and therefore could be used when NGS is being implemented in order to provide routine mutational screening of patients with AML.
Possible laboratory workflow for identifying DNMT3A and IDH1/2 mutations
Although Sanger sequencing is considered as the “gold standard” for identifying mutations, this method is time-consuming and cost-intensive in routine laboratory practice, because the diagnostic findings need to be available as fast as possible. Based on the findings of this study, we developed a laboratory workflow for identifying IDH1/2 and DNMT3A mutations in the first diagnosis and relapse without using of sequencing (Figure 9). HRM analysis should be the method of choice for differentiating between wt and all the analysed mutations in primary AML samples. In case of uncertainty results can be verified using the above presented methods. In addition, ARMS and endonuclease restriction provide a possibility to identify the most common IDH2 and DNMT3A mutations when no HRM-compatible real-time PCR cycler is available. Because of the multiplicity of IDH1 mutations, it was not possible to generate a valid method for analysing 1 specific mutation. For this reason HRM analysis is the best alternative to Sanger sequencing. After therapy, follow-up analysis should be chosen depending on the identified mutations at the first diagnosis. Because endonuclease restriction had higher sensitivity for R882H mutations, this method is more suitable for detecting low mutational ratio of known mutations in patients after therapy or relapse and progression of disease. Because of the ease of interpretation ARMS can also be used to identify IDH2 R140Q mutations at relapse or disease progression.
Figure 9.
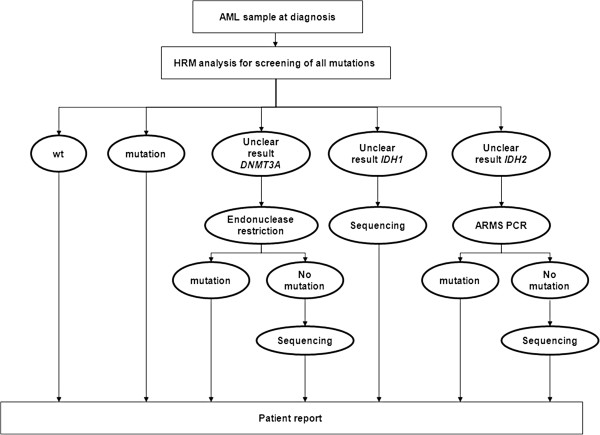
Possible diagnostic workflow to identify DNMT3A, IDH2 and IDH1 mutations in routine laboratory analysis. HRM analysis can be performed in the first diagnosis for all mutations because of high mutational ratios prior to therapy. Unclear results can be verified by endonuclease restriction or ARMS-PCR. Unclear IDH1 results can be checked by sequencing because of the heterogeneity of possible mutations. Effective combination of all the available methods enables more reliable results and a cost-effective and time-saving routine laboratory analysis.
Conclusion
In summary, we generated highly specific, sensitive and rapid methods for identifying the most common mutations in IDH2 (R140Q) and DNMT3A (R882H), which can be used separately or in combination with HRM analysis to provide more reliable diagnostic results. All the developed methods were rapid, specific and easy to use and interpret. PCR-based methods are a useful tool for the routine laboratory identification of relevant prognostic mutations. We propose that early screening of mutations in patients with AML with normal karyotype could facilitate risk stratification and improve treatment opportunities.
Abbreviations
AML: Acute myeloid leukemia; IDH: Isocitrate dehydrogenase; DNMT: DNA methyltransferase; HRM: High resolution melt; PCR: Polymerase chain reaction; α-KG: α-ketoglutarate; 2-HG: 2-hydroxyglutarate; NADP: Nicotinamide adenine dinucleotide phosphate; ARMS: Amplification-refractory mutation system; NPM: Nucleophosmin; FLT: fms-related tyrosine kinase.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
BR carried out design of the study and drafted the manuscript. BO and BIW conceived of the study, and participated in its design and coordination and helped to draft the manuscript. KA and CR carried out the molecular genetic studies. SA and SC participated in sample collection and sequencing. All authors read and approved the final manuscript.
Supplementary Material
Characteristics of patients with AML according to mutation status.
Primers used in this study.
PCR reaction mixtures and conditions.
Contributor Information
Rimma Berenstein, Email: rimma.berenstein@charite.de.
Igor Wolfgang Blau, Email: igor.blau@charite.de.
Asiye Kar, Email: asiye.kar@charite.de.
Ruhiye Cay, Email: ruhiye.cay@charite.de.
Annette Sindram, Email: annette.sindram@laborberlin.com.
Claudia Seide, Email: claudia.seide@laborberlin.com.
Olga Blau, Email: olga.blau@charite.de.
Acknowledgment
This work was supported by the Stefan-Morsch-Stiftung for Leukemia Tumour Patients.
References
- Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88(4):318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, Boyiadzis M. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014. Epub ahead of print, doi:10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed]
- Li KK, Luo LF, Shen Y, Xu J, Chen Z, Chen SJ. DNA methyltransferases in hematologic malignancies. Semin Hematol. 2013;50(1):48–60. doi: 10.1053/j.seminhematol.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O'Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ. et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrozek K, Radmacher MD, Kohlschmidt J, Nicolet D, Whitman SP, Wu YZ, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Moore JO, Caligiuri MA, Larson RA, Bloomfield CD. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30(7):742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, Takada S, Miyazaki Y, Kiyoi H, Ito E, Naoe T, Tomonaga M, Toyota M, Tajima S, Iwama A, Mano H. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Dinardo CD, Patel KP, Garcia-Manero G, Luthra R, Pierce S, Borthakur G, Jabbour E, Kadia T, Pemmaraju N, Konopleva M, Faderl S, Cortes J, Kantarjian HM, Ravandi F. Lack of association of IDH1, IDH2 and DNMT3A mutations with outcome in older patients with acute myeloid leukemia treated with hypomethylating agents. Leuk Lymphoma. 2014. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- Feng JH, Guo XP, Chen YY, Wang ZJ, Cheng YP, Tang YM. Prognostic significance of IDH1 mutations in acute myeloid leukemia: a meta-analysis. Am J Blood Res. 2012;2(4):254–264. [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Patel J, Levine RL. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am. 2011;25(6):1119–1133. doi: 10.1016/j.hoc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27(8):836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen FQ, Sun YH, Zhou SY, Li TY, Chen R. Effects of DNMT1 silencing on malignant phenotype and methylated gene expression in cervical cancer cells. J Exp Clin Cancer Res. 2011;30:98. doi: 10.1186/1756-9966-30-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Chen Y, Wang H, Li Z. DNMT3A mutations and clinical features in Chinese patients with acute myeloid leukemia. Cancer Cell Int. 2013;13(1):1. doi: 10.1186/1475-2867-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova J, Michkova P, Burckova K, Brezinova J, Michalova K, Dohnalova A, Maaloufova JS, Soukup P, Vitek A, Cetkovsky P, Schwarz J. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur J Haematol. 2012;88(2):128–135. doi: 10.1111/j.1600-0609.2011.01716.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, Figueroa ME, Zeilemaker A, Melnick A, Lowenberg B, Valk PJ, Delwel R. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- Schwarz J, Markova J. DNMT3A mutations in AML: a new prognostic factor? Leuk Res. 2013;37(11):1432–1433. doi: 10.1016/j.leukres.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, Yun H, Gohring G, Schlegelberger B, Hoelzer D, Lubbert M, Kanz L, Fiedler W, Kirchner H, Heil G, Krauter J, Ganser A, Heuser M. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- Wang Z, Bao Z, Yan W, You G, Wang Y, Li X, Zhang W. Isocitrate dehydrogenase 1 (IDH1) mutation-specific microRNA signature predicts favorable prognosis in glioblastoma patients with IDH1 wild type. J Exp Clin Cancer Res. 2013;32(1):59. doi: 10.1186/1756-9966-32-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszarska M, Bors A, Feczko A, Meggyesi N, Batai A, Csomor J, Adam E, Kozma A, Orban TI, Lovas N, Sipos A, Karaszi E, Dolgos J, Fekete S, Reichardt J, Lehoczky E, Masszi T, Tordai A, Andrikovics H. Type and location of isocitrate dehydrogenase mutations influence clinical characteristics and disease outcome of acute myeloid leukemia. Leuk Lymphoma. 2013;54(5):1028–1035. doi: 10.3109/10428194.2012.736981. [DOI] [PubMed] [Google Scholar]
- Nomdedeu J, Hoyos M, Carricondo M, Esteve J, Bussaglia E, Estivill C, Ribera JM, Duarte R, Salamero O, Gallardo D, Pedro C, Aventin A, Brunet S, Sierra J. Adverse impact of IDH1 and IDH2 mutations in primary AML: experience of the Spanish CETLAM group. Leuk Res. 2012;36(8):990–997. doi: 10.1016/j.leukres.2012.03.019. [DOI] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, Horst HA, Germing U, Dohner H, Dohner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- Julie Schanz M Friederike Braulke P Katayoon Shirneshan M Kathrin Nachtkamp M Ulrich Germing M Stephan Schmitz M Peter Haas M Michael Lübbert M Müller-Thomas C Katharina G Uwe Platzbecker M Florian Nolte M Wolf-Karsten Hofmann M Detlef Haase M Therapy with demethylating agents significantly improves overall- and AML-free survival in patients with MDS classified as high-risk by IPSS or very high risk by IPSS-R and partial or total monosomy 7-results from a German Multicenter Study Blood 201312221278424009232 [Google Scholar]
- Mani S, Herceg Z. DNA demethylating agents and epigenetic therapy of cancer. Adv Genet. 2010;70:327–340. doi: 10.1016/B978-0-12-380866-0.60012-5. [DOI] [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borovitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of the myeloid neoplasms and leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- Jaatinen T, Laine J. Isolation of mononuclear cells from human cord blood by Ficoll-Paque density gradient. Curr Protoc Stem Cell Biol. 2007;Chapter 2:Unit 2A.1. doi: 10.1002/9780470151808.sc02a01s1. [DOI] [PubMed] [Google Scholar]
- Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6(10):e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KP, Barkoh BA, Chen Z, Ma D, Reddy N, Medeiros LJ, Luthra R. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13(6):678–686. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancik S, Drabek J, Berkovcova J, Xu YZ, Stankova M, Klein J, Kolek V, Skarda J, Tichy T, Grygarkova I, Radzioch D, Hajduch M. A comparison of Direct sequencing, Pyrosequencing, High resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res. 2012;31:79. doi: 10.1186/1756-9966-31-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivarov V, Gueorguieva R, Stoimenov A, Tiu R. DNMT3A mutation is a poor prognosis biomarker in AML: results of a meta-analysis of 4500 AML patients. Leuk Res. 2013;37(11):1445–1450. doi: 10.1016/j.leukres.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Cikota BM, Tukic LJ, Tarabar OT, Magic ZM. Detection of t(14;18), P53 and RAS gene mutations and quantification of residual disease in patients with B-cell non-Hodgkin's lymphoma. J Exp Clin Cancer Res. 2007;26(4):535–542. [PubMed] [Google Scholar]
- Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, Guelly C, Bauernhofer T, Samonigg H, Hoefler G, Dandachi N. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11(2):140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison G, Donald E, McWalter G, Knight L, Fletcher L, Sherwood J, Cantarini M, Orr M, Speake G. A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res. 2010;29:132. doi: 10.1186/1756-9966-29-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Trasler JM, Robaire B. Restriction digestion and real-time PCR (qAMP) Methods Mol Biol. 2009;507:271–280. doi: 10.1007/978-1-59745-522-0_20. [DOI] [PubMed] [Google Scholar]
- Altimari A, de Biase D, De Maglio G, Gruppioni E, Capizzi E, Degiovanni A, D'Errico A, Pession A, Pizzolitto S, Fiorentino M, Tallini G. 454 next generation-sequencing outperforms allele-specific PCR, Sanger sequencing, and pyrosequencing for routine KRAS mutation analysis of formalin-fixed, paraffin-embedded samples. Onco Targets Ther. 2013;6:1057–1064. doi: 10.2147/OTT.S42369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle MA, Fassunke J, Konig K, Grunewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Buttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of patients with AML according to mutation status.
Primers used in this study.
PCR reaction mixtures and conditions.


